Abstract
Cells proliferate by division into similar daughter cells, a process that lies at the heart of cell biology. Extensive research on cell division has led to the identification of the many components and control elements of the molecular machinery underlying cellular division. Here we provide a brief review of prokaryotic and eukaryotic cell division and emphasize how new approaches such as systems and synthetic biology can provide valuable new insight.
Keywords: Cell division, Systems biology, Cellular dynamics, Biological networks, Synthetic biology, Bioengineering, Origin of life
Cell division: one of life’s key processes
“Any system capable of replication and mutation is alive”, wrote Alexander Oparin in 1961 (Popa 2004). Indeed, replication is one of the central features that defines a living object (Trifonov 2011). Through replication, copies of a living organism are generated. An important principle in biology is that cells replicate by division. Unicellular organisms typically replicate by symmetric division while multi-cellular organisms often replicate by an initial cell fusion followed by development of a multicellular individual through many cycles of cell division, starting from that primary cell.
Division poses an array of daunting challenges to the cell. As cell division typically leads to two very similar daughter cells, the primary cell has to grow and synthesize enough biological material for generating two cells. This includes duplicating the genome and segregating the two genomes to both cell halves for an even distribution to the two daughter cells (Fig. 1). This is a nontrivial task as the physical size of the genome is many orders of magnitude larger than that of the cell, necessitating a strong condensation through DNA-binding proteins and DNA supercoiling. For segregation, a number of intriguing mechanisms have been proposed, including entropic segregation (Jun and Mulder 2006), force-driven segregation (Dworkin and Losick 2002), and a Brownian ratchet coupling to protein waves (Di Ventura et al. 2013). Interestingly, in bacteria, the genome orientation remains invariant after segregation (Toro and Shapiro 2010). After segregation of the DNA, bacterial division occurs via the formation of a mid-cell septum and a centripetal invagination of the cell membrane (Fig. 2a). A conserved molecular machinery is responsible for this process, although morphological differences exist among bacterial species.
Fig. 1.
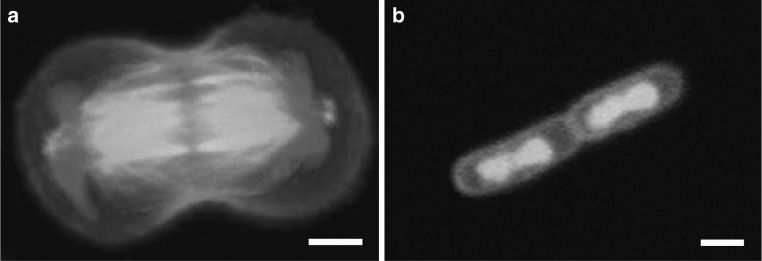
Segregation of the duplicated genomes to both cell halves during cell division. a Chinese hamster ovary cell in anaphase. The genome is labeled in blue while actin and microtubules are seen in red and green, respectively. Scale bar 5 μm. Image courtesy of Ahna Skop, UW-Madison. b E. coli bacterium that is just about to finish cell division. The genome is stained in green; the cell envelope in red. Scale bar 1 μm. Image courtesy of Fabai Wu, TU Delft. (Color figure online)
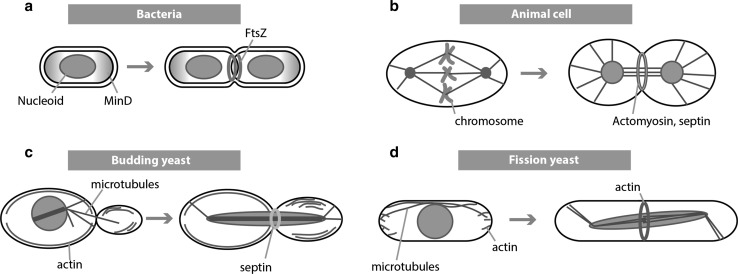
Fig. 2.
Mechanistic and morphological differences between cell division processes in a prokaryotic (E. coli or B. subtilis) and eukaryotic cells: b animal cells, c Budding yeast and d Fission yeast. A microtubule-based apparatus called the mitotic spindle conducts genome segregation during eukaryotic cell division. Bacterial cells (with some exceptions (Ptacin et al. 2010)) do not have mitotic spindle. While in fission and budding yeasts nuclear envelopes remain intact, the envelopes in animal cells break down during mitosis (open mitosis)
In eukaryotic cells, the vast majority of the genetic material is located within the membrane-encapsulated nucleus. Next to doubling the genome, cell division involves the distribution and the organization of the chromosomal mass (in a process called mitosis) and the formation of two new nuclei, as well as the distribution of the cytoplasm among the two daughter cells (Fig. 2b–d). Segregation of the chromosomes is conducted by an apparatus called the mitotic spindle which is composed of a large set of proteins. During mitosis, the chromosomes are tightly controlled to evenly segregate so that the resulting daughter cells have the same amount of chromosomes. In exceptional cases (e.g. in the formation of sperms and oocytes), division occurs without chromosome duplication, which leads to the formation of cells with half the amount of chromosomes of the primary cell (in a process called meiosis).
While the above modes of division are the most common, cell division can also occur asymmetrically, leading to the generation of daughter cells with a molecular composition that is different from the mother (Gonczy 2008). Under physiological conditions, asymmetric cell division involves a symmetric division of the genetic material followed by an asymmetric division of the cytosol. Asymmetric division of the genetic material is typically pathologic and occurs in cancer cells.
While cell division—symmetric or asymmetric—typically yields two daughter cells, multi-daughter cell division also occurs in biology, though with low occurrence. For instance, protozoans of the genus Plasmodium commonly undergo division into three or more cells. In engineered environments, multi-daughter divisions can be induced by confinement (Tse et al. 2012). Such daughter cells often suffer from chromosomal abnormalities, so-called aneuploidy. Multi-polar mitosis and aneuploidy are frequently observed in cancer cell divisions (Kops et al. 2005; Vitale et al. 2010). Here the spindle apparatus adopts a multi-polar configuration, in contrast to a typical bi-polar structure. Multi-polar segregation of genetic material may be associated with multipolar cytokinesis, bipolar cytokinesis or no cytokinesis (Gisselsson et al. 2010).
Dissecting cell division
Since the first observations of cell division early in the 19th century, the process has been studied using a large variety of microscopic, genetic and biochemical approaches, which has led to important advances in our understanding (Table 1). In parallel to these classic approaches, cell division is increasingly being examined with biophysical methods, both theoretically and experimentally (Bassereau et al. 2012). To name just one example, forces acting on spindle poles can be probed by cutting the mitotic spindle in living cells undergoing division (Grill et al. 2001).
Table 1.
Timeline of key discoveries in cell division
| 1882 | Symmetric chromosomal division |
| 1902 | Mitotic spindle |
| 1951 | Chiasmata |
| 1953 | DNA replication |
| 1968 | Dynamic instability |
| 1970 | Cell-cycle control |
| 1970 | Cdc mutants |
| 1971 | Maturation-promoting factor |
| 1973 | Actomyosin system in cytokinesis |
| 1974 | The restriction point |
| 1980 | Yeast centrosome |
| 1982 | Cell-cycle observation |
| 1983 | Cyclin characterization |
| 1987 | Replication origin |
| 1988 | Checkpoint control |
| 1988 | Retinoblastom/E2F |
| 1989 | P53 |
| 1991 | Mitotic checkpoints |
| 1991 | APC and proteolysis |
| 1993 | CDK inhibitors |
| 1994 | SCF and F-box proteins |
| 1994 | Par proteins |
| 1997 | Sister-chromatid cohesion; |
| 1997 | In vitro assembly of microtubule aster |
| 1998 | FtsZ |
| 1999 | Min oscillations |
| 2000 | Division genes in C. elegans |
| 2007 | Self organization of Min surface waves |
| 2008 | Reconstitution of contractile FtsZ rings in liposomes |
| 2010 | Division genes in humans |
| 2011 | In vitro assembly of centromere and kinetochore |
| 2013 | Bottom-up reconstruction of cell cortex |
New approaches to biology, namely systems biology and synthetic biology, have recently emerged that are also increasingly used to study cell division. The systems biology approach relies on the usage of statistical physics, probability theory and kinetics to understand the structural and dynamical complexity of biology at the full system level. Systems biology integrates large data sets to capture how function is generated from its components. Synthetic biology, on the other hand, involves building and engineering biological or biology-inspired components that are subsequently studied inside or outside of the cell. In this way it aims at developing a range of useful applications as well as at understanding how biological complexity emerges from its components. A common approach in synthetic biology is to build in vitro synthetic systems. Synthetic variants of biological systems provide a great opportunity to decode the constraints imposed by the complexity of natural evolved biological systems. The two approaches provide important complementary information about cell division.
Systems biology approach to cell division
Traditionally, biochemistry and molecular biology examine the properties of the biomolecules in bulk. With single-molecule techniques, biophysicists can now study the properties of even single biomolecules in details. To understand the supramolecular functional properties of living organisms, of which replication is an example, one needs to know, however, not only which molecules are involved and how they function, but also which are the many interactions between them (Jeong et al. 2001; Mashaghi et al. 2004). In the systems approach to biology, properties of biomolecular networks are studied with an emphasis on the dynamics of the molecular networks at the system level.
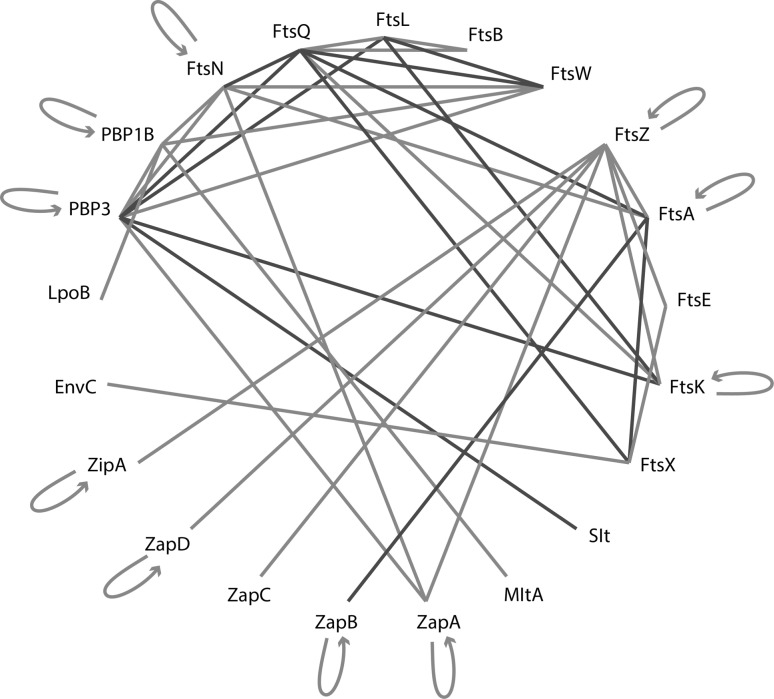
Systems biology of cell division is an emerging research field. We still do not know all of the genes and proteins required for cell division, let alone that we know how all the components interact to give rise to a well-orchestrated cell division. A major step towards unraveling the molecular network underlying cell division came from the Mitocheck project: Using a high-throughput phenotypic screening platform combining gene silencing by RNA interference, time-lapse microscopy and computational image processing, all human genes were screened for their involvement in cell division (Neumann et al. 2010). With the availability of data from such high-throughput studies, the next challenge will be to extract useful system-level information from the data, particularly in the direction of understanding the orchestration of the cell division. Some steps have recently been taken towards understanding the regulatory coordination of cell division events, with, for example, the discovery of positive feedback loops responsible for cell-cycle synchronization (Santos and Ferrell 2008). An integrated analysis of existing biochemical, genetic and protein interaction data has just started to shed light on the hierarchical recruitment of division components (Egan and Vollmer 2013) as well as the structure of protein–protein interaction network that conducts the cell division (Fig. 3).
Fig. 3.
Interaction network of E. coli divisome proteins. Blue lines indicate interactions that were observed both in vitro and in cells. Red lines represents interactions identified only in bacterial two-hybrid screening (in two-hybrid assays (Karimova et al. 2005), protein interactions bring together the binding and activating domains of a transcription factor that turns on a reporter gene). For an extensive review of the raw interaction data that were used to build this interaction graph, see Egan and Vollmer (2013). (Color figure online)
Another important topic in systems biology of cell division is the connection between the cell cycle and cell growth. For some cell types, the cell size increases to a threshold, beyond which division occurs (Leslie 2011). For other cells, however, there is no critical size but rather a critical growth rate (Son et al. 2012) at which cell division occurs. Furthermore, on rare occasions, cells duplicate their genomes, sometimes even multiple times, without dividing. These cells are called polyploids and are commonly seen in plants, the animal kingdom (Sher et al. 2013) and in human cancers (Storchova et al. 2006). With the development of new technologies (such as microfluidics systems with a mass sensor), precise monitoring of cell size has become possible (Son et al. 2012). Considering the ongoing systems biology research on cell growth (Ferrezuelo et al. 2012) and metabolism (Slavov et al. 2011), a better understanding of the interplay between cell growth and division can be expected in the near future. The systems approach can also be used to address epigenetic factors. Recent evidence, for example, indicates a connection between cell cycle dynamics and circadian rhythm (Kowalska et al. 2013). For example a nuclear protein is found to convey circadian gating to the cell cycle (Kowalska et al. 2013). Importantly the circadian clock is known to govern processes such as sleep, functioning of the gastrointestinal, respiratory and cardiovascular systems (Gachon et al. 2004). The discovery of the circadian gating of the cell cycle thus raises questions about connections between irregular sleep, meal times and health.
Synthetic biology approach to cell division
As one of the fundamental processes of life, cell division is of immediate interest to synthetic biologists. There are many interesting questions: What is the mechanism of cell division? Can engineering the components of the divisome help disentangling this? What are the minimal requirements for a cell to divide? Could we even engineer cellular division from the bottom up? And if so, how can we ensure high fidelity and multiple-cycle continuation in engineered cell division?
Efforts have been made to assemble in vitro various of the components that are critically involved in cell division, namely the cell cortex (Vogel et al. 2013), the mitotic spindle (Hannak and Heald 2006; Guse et al. 2011) and FtsZ division rings (Jimenez et al. 2011; Cabre et al. 2013; Osawa and Erickson 2013) (Fig. 4a, b show two examples). Assembly of an entire synthetic divisome (Fig. 4c) is very challenging and still far away, as it does require a thorough understanding of the multiple steps and physical forces involved in cell division as well as a controlled robust assembly of the full molecular system. Yet, various groups are now starting efforts in this direction.
Fig. 4.
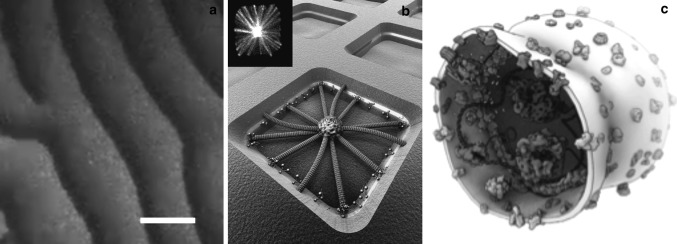
Examples that illustrate how in vitro synthetic biology can help to unravel the mechanism of the cell-division machinery. a Waves of Min proteins (involved in bacterial cell division of E. coli) on a supported lipid bilayer, from Loose et al. (2008). Orange and green color denotes MinE and MinD proteins, respectively. Scale bar is 50 μm. b Artist’s impression of microtubule asters (involved in eukaryotic cell division) that are positioned by cortical motor proteins in microfabricated chambers. Chamber size is 15 μm. Image courtesy Marileen Dogterom. The top left inset shows the experimental result, from Laan et al. (2012). c Artistic sketch that illustrates the future dream of a vesicle with reconstituted components with an engineered minimal divisome that will cause the vesicle to divide. Image courtesy Bert Poolman. (Color figure online)
Concluding remarks
With the rapid progress in systems analysis of cell division as well as bottom-up engineering of this process, there is growing confidence for new insights into cell division, yielding hope that this will lead to better therapeutics and improved treatment of human pathologies. Cell division is impaired in many diseases such as cancer, neurodegenative disorders, cardiovascular and rheumatologic pathologies (Zhivotovsky and Orrenius 2010). Drugs targeting cell division, such as those targeting mitotic entry and the mitotic spindle, have been proposed and tested in animal models of cancer with remarkable success. An even better strategy turned out to be targeting mitotic exit and cytokinesis (Manchado et al. 2012; Domenech and Malumbres 2013). So far, none of these strategies have gained clinical success in humans, but the potential of this approach is clear. Cell division inhibitors are also obvious potential antibacterial drugs (Lock and Harry 2008). A number of molecules have recently been designed and successfully used to selectively target FtsZ, leading to division impairment and antibacterial activity (Ruiz-Avila et al. 2013). It is our hope that systems and synthetic biology of cell division will furthermore lead to new cell-cycle based therapeutic strategies for cancers, antibiotics-resistant bacterial infections and other relevant pathologies.
This special issue on cell division provides scientists with an up-to-date selection of papers from internationally leading laboratories that, through their diversity of backgrounds, help to display the interesting complexity and variety of cell division. We hope that this thematic issue will serve as a platform that will provide insight and stimulate further discussion and follow-up research. Finally, we wish to express our sincere gratitude to the authors of the articles in this special issue.
Acknowledgments
The authors are grateful to Dr. Bertus Beaumont for a critical reading of the article. One if us (C.D.) is supported by the European Research Council ERC Advanced Grant #247072 (NanoForBio).
Contributor Information
Alireza Mashaghi, Email: A.MashaghiTabari@tudelft.nl.
Cees Dekker, Email: C.Dekker@tudelft.nl.
References
- Bassereau P, Phillips R, Schwille P (2012) Focus on the physics of the cell membrane. New J Phys 14:1–4
- Cabre EJ, Sanchez-Gorostiaga A, Carrara P, Ropero N, Casanova M, Palacios P, et al. Bacterial division proteins FtsZ and ZipA induce vesicle shrinkage and cell membrane invagination. J Biol Chem. 2013;288:26625–26634. doi: 10.1074/jbc.M113.491688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ventura B, Knecht B, Andreas H, Godinez WJ, Fritsche M, Rohr K, et al. Chromosome segregation by the Escherichia coli min system. Mol Syst Biol. 2013;9:686. doi: 10.1038/msb.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech E, Malumbres M. Mitosis-targeting therapies: a troubleshooting guide. Curr Opin Pharmacol. 2013;13:519–528. doi: 10.1016/j.coph.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Dworkin J, Losick R. Does RNA polymerase help drive chromosome segregation in bacteria? Proc Natl Acad Sci USA. 2002;99:14089–14094. doi: 10.1073/pnas.182539899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan AJ, Vollmer W. The physiology of bacterial cell division. Ann NY Acad Sci. 2013;1277:8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- Ferrezuelo F, Colomina N, Palmisano A, Gari E, Gallego C, Csikasz-Nagy A et al (2012) The critical size is set at a single-cell level by growth rate to attain homeostasis and adaptation. Nat Commun 3:1–11 [DOI] [PubMed]
- Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004;113:103–112. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- Gisselsson D, Jin Y, Lindgren D, Persson J, Gisselsson L, Hanks S, et al. Generation of trisomies in cancer cells by multipolar mitosis and incomplete cytokinesis. Proc Natl Acad Sci USA. 2010;107:20489–20493. doi: 10.1073/pnas.1006829107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- Grill SW, Gonczy P, Stelzer EHK, Hyman AA. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF (2011) In vitro centromere and kinetochore assembly on defined chromatin templates. Nature 477:354–358 [DOI] [PMC free article] [PubMed]
- Hannak E, Heald R. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat Protoc. 2006;1:2305–2314. doi: 10.1038/nprot.2006.396. [DOI] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Jimenez M, Martos A, Vicente M, Rivas G. Reconstitution and organization of Escherichia coli proto-ring elements (FtsZ and FtsA) inside giant unilamellar vesicles obtained from bacterial inner membranes. J Biol Chem. 2011;286:11236–11241. doi: 10.1074/jbc.M110.194365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc Natl Acad Sci USA. 2006;103:12388–12393. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Dautin N, Ladant D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol. 2005;187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJPL, Weaver BAA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- Kowalska E, Ripperger JA, Hoegger DC, Bruegger P, Buch T, Birchler T, et al. NONO couples the circadian clock to the cell cycle. Proc Natl Acad Sci USA. 2013;110:1592–1599. doi: 10.1073/pnas.1213317110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, Lopez MP, et al. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148:502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie M. How does a cell know its size? Science. 2011;334:1047–1048. doi: 10.1126/science.334.6059.1047. [DOI] [PubMed] [Google Scholar]
- Lock RL, Harry EJ. Cell-division inhibitors: new insights for future antibiotics. Nat Rev Drug Discov. 2008;7:324–338. doi: 10.1038/nrd2510. [DOI] [PubMed] [Google Scholar]
- Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science. 2008;320:789–792. doi: 10.1126/science.1154413. [DOI] [PubMed] [Google Scholar]
- Manchado E, Guillamot M, Malumbres M. Killing cells by targeting mitosis. Cell Death Differ. 2012;19:369–377. doi: 10.1038/cdd.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashaghi AR, Ramezanpour A, Karimipour V. Investigation of a protein complex network. Eur Phys J B. 2004;41:113–121. doi: 10.1140/epjb/e2004-00301-0. [DOI] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Erickson HP. Liposome division by a simple bacterial division machinery. Proc Natl Acad Sci USA. 2013;110:11000–11004. doi: 10.1073/pnas.1222254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa R (2004) In: Between necessity and probability: searching for the definition and origin of life. Springer, Berlin, 252 p
- Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, Comolli LR, et al. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol. 2010;12:791–798. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Avila LB, Huecas S, Artola M, Vergonos A, Ramirez-Aportela E, Cercenado E, et al. Synthetic inhibitors of bacterial cell division targeting the GTP-binding site of FtsZ. ACS Chem Biol. 2013;8:2072–2083. doi: 10.1021/cb400208z. [DOI] [PubMed] [Google Scholar]
- Santos SDM, Ferrell JE. Systems biology—on the cell cycle and its switches. Nature. 2008;454:288–289. doi: 10.1038/454288a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher N, Von Stetina JR, Bell GW, Matsuura S, Ravid K, Orr-Weaver TL. Fundamental differences in endoreplication in mammals and Drosophila revealed by analysis of endocycling and endomitotic cells. Proc Natl Acad Sci USA. 2013;110:9368–9373. doi: 10.1073/pnas.1304889110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N, Macinskas J, Caudy A, Botstein D. Metabolic cycling without cell division cycling in respiring yeast. Proc Natl Acad Sci USA. 2011;108:19090–19095. doi: 10.1073/pnas.1116998108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S, Tzur A, Weng Y, Jorgensen P, Kim J, Kirschner MW, et al. Direct observation of mammalian cell growth and size regulation. Nat Methods. 2012;9:910–912. doi: 10.1038/nmeth.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova Z, Breneman A, Cande J, Dunn J, Burbank K, O’Toole E, et al. Genome-wide genetic analysis of polyploidy in yeast. Nature. 2006;443:541–547. doi: 10.1038/nature05178. [DOI] [PubMed] [Google Scholar]
- Toro E, Shapiro L. Bacterial chromosome organization and segregation. Cold Spring Harb Perspect Biol. 2010;2:a000349. doi: 10.1101/cshperspect.a000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov EN. Vocabulary of definitions of life suggests a definition. J Biomol Struct Dyn. 2011;29:259–266. doi: 10.1080/073911011010524992. [DOI] [PubMed] [Google Scholar]
- Tse HTK, Weaver WM, Di Carlo D (2012) Increased asymmetric and multi-daughter cell division in mechanically confined microenvironments. Plos One 7:1–8 [DOI] [PMC free article] [PubMed]
- Vitale I, Senovilla L, Jemaa M, Michaud M, Galluzzi L, Kepp O, et al. Multipolar mitosis of tetraploid cells: inhibition by p53 and dependency on Mos. EMBO J. 2010;29:1272–1284. doi: 10.1038/emboj.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel SK, Petrasek Z, Heinemann F, Schwille P. Myosin motors fragment and compact membrane-bound actin filaments. Elife. 2013;2:e00116. doi: 10.7554/eLife.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky B, Orrenius S. Cell cycle and cell death in disease: past, present and future. J Intern Med. 2010;268:395–409. doi: 10.1111/j.1365-2796.2010.02282.x. [DOI] [PubMed] [Google Scholar]