Abstract
Solid-state NMR spectroscopy is used to determine the membrane-bound topological structure of a cationic β-hairpin antimicrobial peptide in which the number of Arg residues has been halved. The parent peptide, PG-1, was previously found to form transmembrane β-barrels in anionic membranes where the Arg residues complex with the lipid phosphate groups to cause toroidal pore defects in the membrane. In comparison, the charge-attenuated and less active mutant studied here forms β-sheets that lie on the surface of the zwitterionic membrane and only partially insert into the anionic membrane. The mutant also exhibits much looser contact with the lipid headgroups. These results indicate that transmembrane insertion and tight Arg-phosphate association are two important elements for strong antimicrobial activities of this class of peptides. Comparison with other β-hairpin antimicrobial peptides studied so far further suggests a relative potency scale for the various mechanisms of action for the β-sheet family of antimicrobial peptides. The transmembrane insertion – toroidal pore mechanism is the most potent in disrupting the lipid bilayer, followed by the large-amplitude in-plane motional mechanism. The carpet model, where peptides aggregate on the membrane surface to cause lateral expansion and eventual micellization of the membrane, is a weaker mechanism of action.
Keywords: antimicrobial peptides, depth of insertion, oligomeric structure, arginine, solid-state NMR, β-hairpin
1. Introduction
Antimicrobial peptides are small cationic peptides of the innate immune systems of many animals and plants. They kill a broad spectrum of bacteria, fungi and viruses by destroying the barrier function of the microbial cell membrane (for review, see [1]). Protegin-1 (PG-1) is an antimicrobial peptide found in porcine leukocytes [2] and belongs to the family of disulfide-stabilized β-hairpin antimicrobial peptides [3]. The 18-residue PG-1 contains six Arg residues that are located at the termini and the turn of the hairpin, separated by hydrophobic residues in the middle of each strand (Fig. 1a). Extensive solid-state NMR studies of the membrane-bound structure of PG-1 and its analogs indicate that this amphipathic structure along the hairpin axis is important for the peptide’s antimicrobial activity [4–6]. For example, when the β-hairpin fold is destroyed by replacing the Cys residues by Ala, the antimicrobial activity is significantly attenuated [7] and the peptide becomes random coil both in solution and in association with the lipid membrane [5, 6].
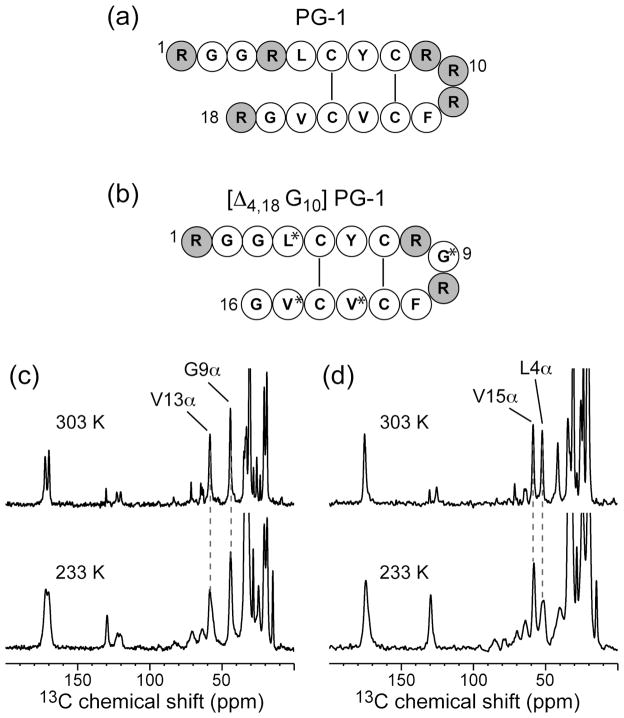
Fig. 1.
Amino acid sequences of wild-type PG-1 (a) and its mutant, [Δ4,18 G10] PG-1 (b). (c) 13C CP-MAS spectra of Gly9 and Val13 of [Δ4,18 G10] PG-1 in POPE/POPG membranes at 303 K (top) and 233 K (bottom). (d) 13C CP-MAS spectra of Leu4 and Val15 of [Δ4,18 G10] PG-1 in POPE/POPG membranes at 303 K (top) and 233 K (bottom).
It is generally accepted and experimental verified that the insertion of charged residues into the hydrophobic part of the lipid bilayer is energetically unfavorable [8–10]. The free energies of insertion of charged amino acid residues (Arg, Lys, Glu, and Asp) into lipid bilayers have been measured by biological assays to be 2.5 – 3.5 kcal/mol [8]. These measurements were carried out on α-helical peptides, which are stabilized by intra-molecular hydrogen bonds between covalently adjacent residues. In addition, polar but neutral residues in multimeric helical membrane proteins have been shown to be stabilized by intermolecular hydrogen bonds [11, 12]. In comparison, β-strand peptides can only be stabilized by inter-strand hydrogen bonds between covalently remote residues, and the energetics of the membrane insertion of charged β-sheet proteins have not been examined so far. In principle, the rules governing the accommodation of charged residues in β-sheet proteins by lipid bilayers may be different from those for α-helical membrane proteins. Recent free energy perturbation calculations and MD simulations suggested that Arg residues remain protonated and charged in the lipid membrane, stabilized by water and membrane deformations [13, 14]. Therefore, elucidating the membrane-bound structure of cationic β-hairpin antimicrobial peptides is not only important for developing more potent antibiotics but also for understanding the fundamental biophysics of the insertion of membrane proteins containing charged residues into lipid bilayers.
While crystal structures of a few β-sheet antimicrobial peptides are available [15, 16], they were determined in the absence of lipids and thus do not directly give information on the mechanism of action and interaction of these peptides with lipid bilayers. Direct studies of the interaction of β-hairpin antimicrobial peptides with lipid bilayers have been reported for a few defensins [17–22], polyphemusins [23] and tachyplesins [24–26], but atomic-level structural information of the peptides in the lipid bilayer remains scarce. Structurally the most comprehensively characterized β-sheet antimicrobial peptide is PG-1. Solid-state NMR distance measurements found that the peptide oligomerizes into β-barrels that insert into the bacteria-mimetic negatively charged lipid membranes in a transmembrane fashion [27, 28]. The positively charged Arg residues in these β-barrels are stabilized in the membrane by complexation with the negatively charged lipid phosphate groups, with distances as short as 4.0 Å between the guanidinium Cζ and the lipid 31P [29]. When the guanidinium groups are dimethylated to reduce the number of possible N-H to O-P hydrogen bonds, both the membrane disorder and the peptide antimicrobial activity were noticeably weakened, and the β-hairpin axis is no longer transmembrane but becomes roughly parallel to the membrane plane [30].
Given the central role of the positively charged residues and its amphipathic distribution to the function of this β-hairpin peptide, it is natural to hypothesize that the number of Arg residues should also impact the antimicrobial activity. Indeed, when three out of six Arg residues were removed without perturbing the amphipathic hairpin structure (Fig. 1b), the antimicrobial activity decreased by an order of magnitude [7]. The membrane disorder induced by this charge-attenuated PG-1 mutant is ~35% lower than wild-type PG-1 based on static 31P NMR spectra of aligned membranes [6]. However, exactly how the lower Arg density changes the topological structure of the peptide in the membrane is still unknown.
In this work, we use solid-state NMR spectroscopy to determine the topological structure, peptide-lipid interaction and dynamics of the PG-1 mutant, [Δ4,18 G10] PG-1, where Arg4 and Arg18 of the wild-type peptide are deleted and Arg10 is converted to Gly (Fig. 1b). We found that this charge-attenuated PG-1 mutant is similarly immobilized in the lipid bilayer as the parent compound, but its distances to the lipid headgroups are longer than for wild-type PG-1. More importantly, the depth of insertion of [Δ4,18 G10] PG-1 now depends on the negative charge density of the membrane surface. In the neutral lipid bilayer the mutant remains on the membrane surface. Even in the completely anionic membrane the mutant is still less inserted than wild-type PG-1. Thus, the lower Arg density of the peptide results in weaker peptide-lipid-headgroup interactions and shallower peptide insertion into the lipid membrane. These structural changes provide a molecular basis for the weaker antimicrobial activity of this mutant, and substantiate the important role of the positively charged residues to the function of this class of β-hairpin antimicrobial peptides.
2. Materials and Methods
2.1. Materials and sample preparation
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylethanolamine (POPE), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol (POPG) were purchased from Avanti Polar Lipids (Alabaster, AL). Uniformly 13C- and 15N-labeled amino acids were purchased from Isotec (Miamisburg, OH) and Cambridge Isotope Laboratory (Andover, MA). [Δ4,18 G10] PG-1 (NH2-RGGLCYCRGR FCVCVG-CONH2) was synthesized using Fmoc chemistry as previously described [31]. The purity of the peptides is greater than 95%, as estimated by comparison of the UV/VIS absorption and gravimetric values of the peptide amount. Two [Δ4,18 G10] PG-1 samples were synthesized: one contains U-13C, 15N-labeled Gly9 and Val13, and the other contains U-13C, 15N-labeled Leu4 and Val15.
POPC/POPG and POPE/POPG (3:1) membranes were prepared by mixing the lipids in chloroform, blowing them dry under N2 gas, then redissolving the mixture in cyclohexane and lyophilizing. The dry lipid powder was dissolved in water and subjected to five cycles of freeze-thawing to form vesicles. An appropriate amount of [Δ4,18 G10] PG-1 to reach a peptide-lipid molar ratio (P/L) of 1:12.5 was then dissolved in water and mixed with the lipid vesicle solution, incubated overnight, then centrifuged at 55,000 rpm for 2.5 hours. The membrane pellet was packed into a 4 mm MAS rotor, giving fully hydrated membrane samples. The final NMR sample in the rotor contained ~30 mg lipids, 5–6 mg of peptide, and ~15 mg of water and has a pH of 6–7. The POPC/POPG mixture has a low phase transition temperature, thus is suitable for 1H spin diffusion experiments that require liquid-crystalline membranes. The POPE/POPG mixture has a higher phase transition temperature, thus is preferred for REDOR experiments where peptide and lipid motions need to be frozen for accurate measurement of intermolecular distances.
It is commonly believed that the peptide concentrations in NMR studies are much higher than in antimicrobial assays. This is incorrect. The peptide concentrations in typical NMR samples are no higher than, and are often lower than, the peptide concentrations in antimicrobial assays. For PG-1, a direct binding study reported that a single P. aeruginosa cell exposed to 2.5 μg/mL PG-1, which is an inhibitory concentration, bound 1.7×106 PG-1 molecules [32]. A typical P. aeruginosa cell contains about 22×106 lipid molecules and 1.4×106 LPS molecules [33]. If all PG-1 molecules bound to plasma membrane lipids, then P/L is ~1:13. If half the PG-1 bound to LPS rather than plasma membrane lipids, then P/L would be ~1:26. Thus, the P/L ratios in the assays are similar to the highest peptide concentrations used in solid-state NMR experiments. Some other studies that reported the number of lipid molecules per cell (~107 cells/lipid) [34] and the number of bacteria colony forming units per mL solution (typically ~106) [35] translate to even higher P/L ratios of 10:1, in excess of the peptide.
2.2 NMR spectroscopy
Solid-state NMR experiments were carried out on a Bruker DSX-400 (9.4 Tesla) spectrometer (Karlsruhe, Germany) with a wide-bore magnet. Two magic-angle spinning (MAS) probes with a 4 mm spinning module and tuned to either 1H/31P/13C or 1H/13C frequencies were used. Low temperatures were reached using a Kinetics Thermal Systems XR air-jet sample cooler (Stone Ridge, NY). Typical 90° pulse lengths were 4 – 5 μs for 13C and 15N, and 1H decoupling fields of 50–80 kHz were used. 13C chemical shifts were referenced externally to the α-Gly 13CO signal at 176.49 ppm on the TMS scale. 31P chemical shifts were referenced externally to the hydroxyapatite 31P signal at 2.73 ppm.
13C-31P distances were measured using a selective rotational-echo double-resonance (REDOR) experiment [36], in which the 13C-13C J couplings in the U-13C, 15N-labeled residues were suppressed by a selective 180° 13C pulse in the middle of the REDOR period. The 13C 180° pulse is Gaussian in shape, centered at the 13C frequency of interest, and synchronized with the rotor period to be either 888 μs or 1333 μs long. This soft pulse recouples the desired 13C-31P dipolar coupling while removing the 13C-13C J-coupling between the 13C spin on resonance and its directly bonded 13C. On the 31P channel, composite 90°180°90° pulses were applied to reduce the flip angle error and increase the distance accuracy [37]. For each REDOR mixing time (tm), a control experiment (S0) with the 31P pulses off and a dephasing experiment (S) with the 31P pulses on were carried out. The normalized intensity, S/S0, as a function of tm gives the 13C-31P dipolar coupling. The experiments were conducted under 4.5 kHz MAS at 230 K for the POPE/POPG membranes. Relatively short 31P 180° pulses of 9 μs were used to achieve complete inversion of the 31P resonance, which has a large chemical shift anisotropy in the rigid limit. The REDOR decay was simulated using an in-house Fortran program. The uncertainties of the best-fit distances are approximately 0.3 Å.
To assess the mobility of [Δ4,18 G10] PG-1, 13C-1H dipolar couplings were measured using the 2D dipolar chemical-shift correlation (DIPSHIFT) experiment [38]. The experiments were carried out under 3.5 kHz MAS at 295 K. The MREV-8 sequence [39], which has a scaling factor of 0.47, was used for 1H homonuclear decoupling. The 1H pulse length in the MREV-8 pulse train was 3.5 μs. The measured DIPSHIFT curves were first symmetrized and then simulated using a Fortran program to obtain the motionally averaged dipolar couplings. The unsymmetrized raw data show final intensities that are about 80% of the initial intensity due to relaxation effects. The uncertainties in the best-fit C-H couplings are ±0.5 kHz. In addition to quantitative C-H dipolar couplings, 1D 13C cross polarization (CP) spectra were measured as a function of temperature to qualitatively assess the dynamics of the membrane-bound peptide.
The depth of insertion of [Δ4,18 G10] PG-1 was investigated using the 2D 13C-detected 1H spin diffusion technique [40]. A 1H T2 filter of 1.2 ms was used to remove the peptide 1H magnetization. Spin diffusion mixing times ranged from 25 ms to 400 ms. Cross-peak intensities were read off from peak heights in the 1D 1H cross sections. The experiments were performed at 295 K under 5.0 kHz MAS. To fit the spin diffusion buildup curve, we used a 1D lattice model where the magnetization (Mi) transfer along the bilayer normal is treated as a discrete process following the equation ΔMi/Δt = −2ΩMi + ΩMi+1 + ΩMi−1. The transfer rate Ω in the equation depends on the diffusion coefficients of the lipid, water, peptide, and the peptide-lipid interface or the peptide-water interface. The single-phase diffusion coefficients have been calibrated on various membrane systems before [27, 40, 41]. The interfacial diffusion coefficients are adjusted to a moderate degree in the simulation to fit the experimental data [5, 27, 40, 41]. The number of discrete steps needed to simulate the measured buildup curve gives the lipid-peptide distance of interest. The uncertainties of the best-fit distances are approximately 2 Å.
3. Results
We investigate the structure of [Δ4,18 G10] PG-1 in the POPE/POPG and POPC/POPG (3:1) membranes, since they mimic the composition of the negatively charged inner membrane of bacteria. Fig. 1(c, d) shows the 13C CP-MAS spectra of (U-13C, 15N-Gly9, Val13)-labeled [Δ4,18 G10] PG-1 and (U-13C, 15N-Leu4, Val15)-labeled [Δ4,18 G10] PG-1 in POPE/POPG membranes below (233 K) and above (303 K) the phase transition temperature. The same 13C chemical shifts are found at the two temperatures, indicating that the secondary structure of the peptide is the same, as expected for the disulfide-linked peptide. Table 1 lists the 303 K Cα and Cβ isotropic chemical shifts of the four labeled residues in the deletion mutant and the isotropic shifts of Leu5 in wild-type PG-1. All residues except Gly9 exhibit chemical shifts characteristic of the β-sheet conformation: the Cα chemical shifts are smaller than the random coil values, while the Cβ chemical shifts are larger [42]. This indicates that [Δ4,18 G10] PG-1 maintains a similarβ-hairpin structure as PG-1 in the lipid membrane. Closer inspection shows that in the liquid-crystalline phase, the Cα carbons of all four residues exhibit relatively narrow peaks with linewidths of ~1.5 ppm, indicating that the peptide is either well ordered and immobilized or undergoes fast motion. At the low temperature, the Cα peaks broaden to 2–3 ppm, except for Leu4 Cα which has a much larger linewidth of 4.3 ppm. The moderate increase in linewidths for the first three residues may be due to stronger 13C-1H dipolar couplings or larger membrane disorder at the low temperature. Thus the additional line broadening of Leu4 Cα must have an additional contribution from conformational disorder.
Table 1.
13C chemical shifts (ppm) of [Δ4,18 G10] PG-1 and PG-1 Leu5 in POPE/POPG membranes at 303 K.
| Sites | Gly9 | Val13 | Leu4 | Val15 | PG-1 Leu5 |
|---|---|---|---|---|---|
| Cα | 43.9 | 58.6 | 52.0 | 58.4 | 51.1 |
| Cβ | - | 33.6 | 41.2 | 34.6 | 42.5 |
The effect of [Δ4,18 G10] PG-1 on the membrane order has been previously examined from 31P NMR lineshapes of oriented POPC/POPG lipids [6]. The 31P spectra showed much less disorder compared to PG-1-containing lipid membranes at the same peptide concentrations. This is reproduced by the 31P spectra of unoriented membranes (not shown), where the isotropic peak intensity is weak compared to PG-1. Thus, the charge-reduced mutant [Δ4,18 G10] PG-1 causes less membrane disorder, consistent with its weaker antimicrobial activity than PG-1.
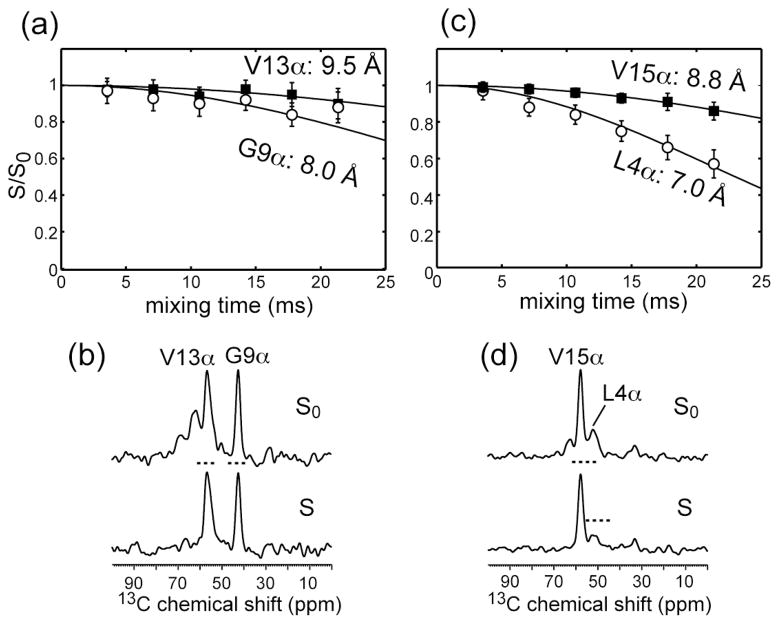
To investigate whether [Δ4,18 G10] PG-1 retains the same distances to the lipid headgroups as PG-1, we measured the 13C-31P distances using REDOR. Fig. 2a, c shows the 13C{31P} REDOR data of several backbone Cα sites of [Δ4,18 G10] PG-1 in the POPE/POPG membrane at 230 K. The low temperature is necessary for suppressing peptide and lipid motions that would average the dipolar couplings of interest. Gly9, Val13 and Val15 Cα show relatively weak REDOR dephasing, with S/S0 values close to 1 up to a mixing time of 25 ms. These S/S0 values translate to long 13C-31P distances of 8.0–9.5 Å. In comparison, Leu4 Cα exhibits stronger dephasing, giving a shorter 13C-31P distance of 7.0 Å. Fig. 2b, d show representative S0 and S spectra of the peptide for a mixing time of 21.3 ms, where Leu4 Cα shows more pronounced dephasing than the other three sites. Overall, the average 13C-31P distance of [Δ4,18 G10] PG-1 backbone (8.3 Å) is longer than the average distance of PG-1 backbone (6.1 Å) to the lipid phosphate groups [29], indicating that the decreased number of Arg residues weakened the peptide-lipid headgroup interaction. In reporting these two-spin simulated 13C-31P distances, we take into account the multi-spin nature of the 31P atoms in the lipid membrane by approximating the vertical distance from the 31P plane to the 13C atom with the two-spin distance. We showed recently [29] that this approximation is valid for two-spin distances of > 7 Å. Thus, all distances reported here should be considered as the vertical distance of 13C to the plane of 31P atoms.
Fig. 2.
13C-31P REDOR data of Gly9, Val13, Leu4 and Val15 Cα of [Δ4,18 G10] PG-1 in POPE/POPG membranes measured at 230 K. (a) 13Cα-31P REDOR data of Gly9 (open circles) and Val13 (filled squares). Best-fit distances are 8.0 Å for Gly9 and 9.5 Å for Val13. (b) A pair of S0 and S spectra of Gly9 and Val13 at the mixing time of 21.3 ms. (c) 13Cα-31P REDOR data of Leu4 (open circles) and Val15 (filled squares). Best-fit distances are 7.0 Å for Leu4 and 8.8 Å for Val15. (d) A pair of S0 and S spectra of Leu4 and Val15 at the mixing time of 21.3 ms. Dashed lines in S spectra indicate the intensities of the corresponding peaks in the S0 spectra.
We next investigated the dynamics of [Δ4,18 G10] PG-1 in the liquid-crystalline membrane. Quantitative information on the amplitude of backbone motion can be obtained from Cα-Hα dipolar couplings, which can be measured using the 2D DIPSHIFT experiment. The ratio of the measured dipolar coupling to the rigid-limit coupling gives the bond order parameter, SCH = δ̄CH/δCH, which is related to the motional amplitude [43]. Fig. 3 shows the 13C-1H DIPSHIFT curves of Gly9, Val13, Leu4 and Val15 in POPC/POPG membranes at 295 K. All four residues show Cα-Hα dipolar couplings of 19.1 – 22.9 kHz, close to the rigid limit of 22.7 kHz. The dephasing of the Gly CH2 group could not be fit to a single coupling but required a distribution of order parameters from 0.83 to 1.0 (Table 2). Thus, overall the backbone of the deletion mutant is mostly immobilized in the liquid-crystalline membrane, with only small-amplitude local segmental motions present. Compared to wild-type PG-1, the strand residues in the mutant, Leu4, Val13 and Val15, have similar order parameters as the strand residues Arg4 and Leu5 in the wild-type peptide [44]. However, the turn residue Gly9 of [Δ4,18 G10] PG-1 is much more rigid than the turn residue Arg11 in PG-1 [44], indicating that the deletion mutant has different peptide-lipid or peptide-peptide interactions at the β-turn position.
Fig. 3.
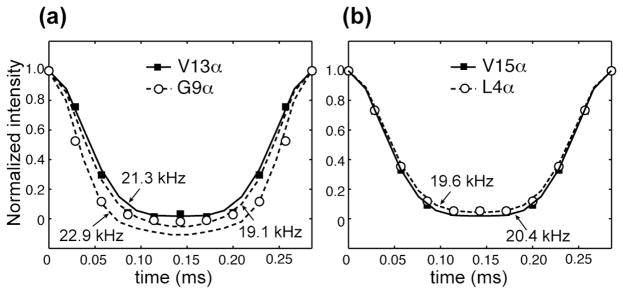
13C-1H DIPSHIFT data of Gly9, Val13, Leu4 and Val15 Cα of [Δ4,18 G10] PG-1 in POPC/POPG membranes measured at 295 K. (a) DIPSHIFT data of Gly9 CH2 (open circles) and Val13 Cα-Hα (filled squares). Best-fit couplings are 19.1–22.9 kHz for Gly9 and 21.3 kHz for Val13. (b) DIPSHIFT data of Leu4 Cα-Hα (open circles) and Val15 Cα-Hα (filled squares). Best-fit couplings are 19.6 kHz for Leu4 and 20.4 kHz for Val15.
Table 2.
C-H order parameters of [Δ4,18 G10] PG-1 residues at 295 K.
| Sites | Gly9 | Val13 | Leu4 | Val15 |
|---|---|---|---|---|
| Cα | 0.83–1.0 | 0.93 | 0.85 | 0.89 |
Since [Δ4,18 G10] PG-1 is mostly immobilized in the lipid membrane, we can exploit the dynamic difference between the rigid peptide and the mobile lipids to determine the depth of insertion of the peptide in the lipid membrane. By applying a 1H T2 filter, we can select the 1H magnetization of the mobile lipids while suppressing that of the rigid peptide, then allow the lipid 1H magnetization to transfer to the peptide through distance-dependent 1H spin diffusion. Fig. 4a shows a representative 2D 13C-detected 1H spin diffusion spectrum of (U- Gly9, U-Val13) [Δ4,18 G10] PG-1 in the POPC/POPG (3 : 1) membrane. The 2D spectrum correlates the lipid or water 1H signals with the peptide 13C signals. The resulting intermolecular cross peaks in the 2D spectra indicate spin-diffusion contact between the lipid moieties and the peptide or between water and the peptide. Mapping the cross-peak intensity as a function of mixing times gives semi-quantitative distances from the peptide to the membrane-surface water or from the peptide to the center of the bilayer. Fig. 4b shows the sum of 1H 1D cross sections at all 13C peaks of the peptide, including both the backbone and sidechain carbons. Peptide cross peaks with the lipid CH2 and CH3 protons are detected at mixing times as short as 25 ms and become prominent by 400 ms. Plotting the cross-peak intensities from the 1D cross sections as a function of the square root of the mixing time [40], we obtained the buildup curves for water and the lipid CH3 as shown in Fig. 4c. For the lipid buildup curve, the intensities were not only corrected for 1H T1 relaxation, but also normalized to the water peak intensity in the 100 ms 2D spectrum, then finally scaled by the intensity ratio between the CH3 and H2O peaks in the direct-excitation 1H 1D spectrum (Fig. 4b top),. The last two steps of correction are necessary for obtaining the correct overall scaling factor for all intensities, which is crucial when the lipid chain - peptide cross peaks do not show a plateau at the longest mixing time used. This plateau-dependent scaling factor affects the initial slope of the buildup curve, which in turn affects distance extraction.
Fig. 4.
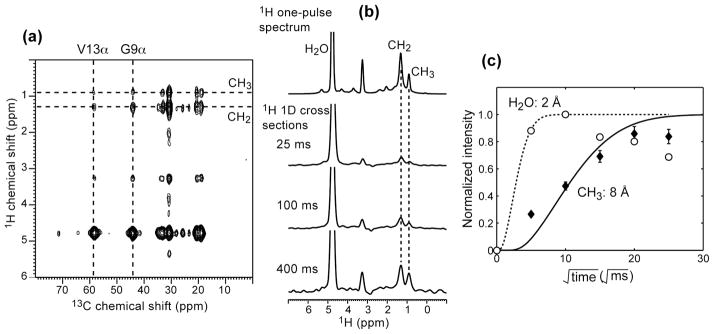
1H spin diffusion data of [Δ4,18 G10] PG-1 in the POPC/POPG (3:1) membrane at 295 K. (a) 2D 13C-detected 1H spin diffusion spectrum of (U- Gly9, U-Val13) [Δ4,18 G10] PG-1 at tm = 400 ms. (b) 1H one-pulse spectrum and the sum of peptide’s 1H cross-sections at various tm. (c) CH3 (filled diamonds) and H2O (open circles) buildup curves. Best-fit distances are 2 Å from H2O to the peptide and 8 Å from CH3 to the peptide.
To extract the peptide-water and peptide-bilayer-center distance, we fitted the experimental buildup curves using diffusion coefficients of 0.03, 0.012 and 0.3 nm2/ms for water, lipid and peptide, respectively [40, 45]. A peptide-lipid interfacial diffusion coefficient (DI) of 0.0012 nm2/ms was used to reproduce the shape of the CH3 buildup curve. This value is on the same order of magnitude as that used for PG-1 previously [5, 27]. Using these diffusion coefficients, we found best-fit distances of 8 Å from the lipid chain end CH3 to the peptide and 2 Å from water to the peptide. The former is 6 Å longer than the distance from CH3 to wild-type PG-1 [27], thus [Δ4,18 G10] PG-1 appears to be only partially inserted into the anionic membrane, not sufficiently deep to reach the center of the bilayer.
The lipid CH3 – peptide buildup curve in Fig. 4c was measured in POPC/POPG (3:1) membranes, and shows a relatively high intensity at 25 ms that is not well reproduced by simulation. We hypothesized that this initial high intensity may result from a small population of the peptide deeply inserted into the bilayer, since the zwitterionic POPC lipids and the anionic POPG lipids could in principle cause different insertion states of the peptide. To test this hypothesis, we measured the depth of insertion of [Δ4,18 G10] PG-1 in a pure POPC membrane and in a pure POPG membrane. The buildup curves of the peptide in these two different lipid membranes are shown in Fig. 5. It can be readily seen that the CH3 buildup curve is much slower for the neutral POPC membrane than for the POPC/POPG membrane, while the CH3 buildup of the negatively charged POPG membrane is faster than for the mixed membrane. The best-fit lipid-chain - peptide distance for the POPC sample is 20 Å with a DI of 0.0005 nm2/ms, whereas the best-fit distance for the POPG membrane is 4 Å with a DI of 0.0012 nm2/ms. Thus, the deletion mutant lies on the surface of the neutral POPC bilayer, far away from the center, but is inserted close to the center of the anionic POPG bilayers. In the mixed POPC/POPG bilayers, the peptide is inserted partially into the acyl chain region. Thus, the depth of insertion of [Δ4,18 G10] PG-1 depends on the negative charge density of the membrane surface. In contrast, the water-peptide cross peaks in the two additional membrane samples show similarly fast intensity buildup as the mixed membrane sample, indicating that in all three cases, part of the peptide is directly exposed to the hydration water.
Fig. 5.
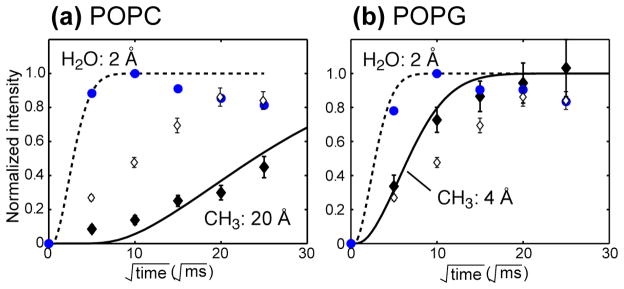
1H spin diffusion data of (U- Gly9, U-Val13) [Δ4,18 G10] PG-1 in POPC (a) and POPG (b) membranes at 295 K. (a) CH3 (filled diamonds) and H2O (blue circles) buildup curves of [Δ4,18 G10] PG-1 in the POPC membrane. Best-fit distance is 20 Å from CH3 to the peptide. (b) CH3 (filled diamonds) and H2O (open circles) buildup curves of [Δ4,18 G10] PG-1 in the POPG membrane. Best-fit distance is 4 Å from CH3 to the peptide. The CH3 buildup curve of the POPC/POPG (3:1) membrane (Fig. 4c) is shown as open diamonds in both (a) and (b) for comparison.
4. Discussion
The goal of this study is to compare the membrane-bound structure of the less positively charged [Δ4,18 G10] PG-1 with the previously determined structure of wild-type PG-1 [27–29, 31, 44, 46, 47], to understand how Arg density affects the structure and membrane interaction of this family of β-hairpin peptides. Static 31P spectra [6] already showed that the charged-attenuated mutant causes much less membrane disorder than wild-type PG-1, consistent with its 10-fold weaker antimicrobial activity [7]. The 13C CP-MAS spectra of [Δ4,18 G10] PG-1 here show that four residues spread throughout the sequence all have similarly narrow linewidths of ~1.5 ppm at ambient temperature. At the same time, their Cα-Hα bond order parameters are close to 1 at ambient temperature, indicating that the mutant is immobilized in the fluid phase of the lipid membrane. Thus, [Δ4,18 G10] PG-1 forms well structured and immobile aggregates in the lipid bilayer, giving rise to narrow lines as well as high rigidity. These ordered aggregates are most likely β-sheets, given the intrinsic β-hairpin fold of the peptide monomer as constrained by the two intramolecular disulfide bonds.
The rigidity of the mutant merits comparison with the wild-type peptide. While PG-1 is also immobile in its two β-strands, its β-turn undergoes large-amplitude motion, as manifested by low order parameters and short relaxation times for the Arg11 backbone [44]. We attributed the mobility of the PG-1 β-turn to its exposure to water on the membrane surface and the lack of intermolecular hydrogen bonds in this region [44]. Conversely, the rigidity of the β-turn of [Δ4,18 G10] PG-1 implies that either the mutant β-turn no longer lies on the membrane surface or it may be involved in intermolecular hydrogen bonding. The first possibility is supported by the relatively long Gly9 13Cα-31P distance of 8.0 Å measured here, which suggests that the β-turn is more deeply inserted into the membrane. The second possibility of intermolecular hydrogen bonding at the turn also cannot be excluded. Overall, the uniformly high order parameters of the mutant supports the formation of extended β-sheets (Fig. 6a) [27].
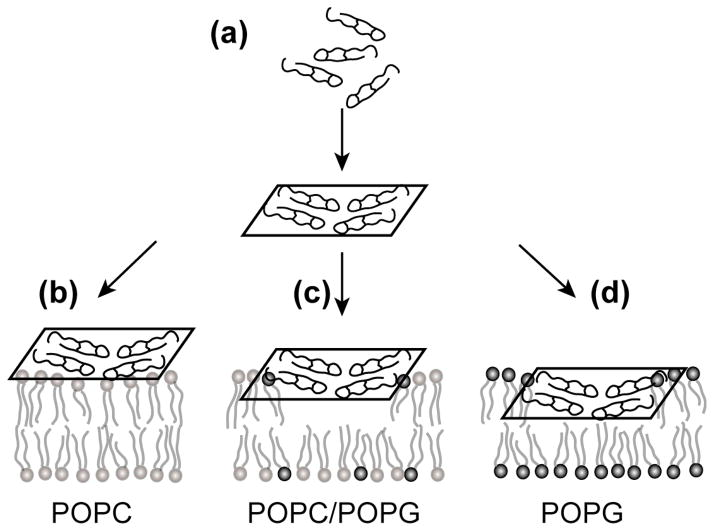
Fig. 6.
Membrane-bound structure of [Δ4,18 G10] PG-1 in three lipid membranes. (a) [Δ4,18 G10] PG-1 first oligomerizes to form β-sheets. (b) The β-sheet lies on the surface of the zwitterionic POPC bilayer. (c) The β-sheet inserts partially into the acyl chain region of the POPC/POPG (3:1) bilayer. (d) The β-sheet inserts nearly to the middle of the POPG bilayer. Grey and black circles denote POPC and POPG headgroups, respectively.
All 13C-31P distances measured for [Δ4,18 G10] PG-1 in the POPE/POPG membrane are longer than those found for wild-type PG-1 [29]. Since the charged nature of the individual Arg residues has not changed, the longer 13C-31P distances can only be attributed to the lower total positive charge of the peptide, which weakens the electrostatic attraction between the peptide and the lipid and strengthens the peptide – peptide association.
The most interesting finding about [Δ4,18 G10] PG-1 is its membrane-dependent depth of insertion. In the neutral POPC membrane, the peptide is at least 20 Å from the membrane center, in other words lying on the bilayer surface. In the 25% negatively charged POPC/POPG membrane, [Δ4,18 G10] PG-1 inserts partially into the bilayer, with a minimum distance of 8 Å from the membrane center. In the 100% negatively charged POPG membrane, the minimum distance of the peptide from the bilayer center decreases to 4 Å. Thus, the depth of insertion of [Δ4,18 G10] PG-1 depends on the negative charge of the membrane surface. This suggests that the lipid bilayer compensates for the lower positive charge of the peptide by requiring a higher negative membrane-surface charge for the peptide to bind.
In principle, the 1H spin diffusion data of the mixed POPC/POPG membrane could be interpreted as [Δ4,18 G10] PG-1 adopting a mixture of surface-bound and transmembrane topologies. The peptide may lie on the surface of the POPC-rich region of the membrane while being fully inserted in the POPG-rich region. However, careful examination of the buildup curve of the mixed membrane shows that it can only be fit by a combination of 30% of the POPC buildup curve and 70% of the POPG buildup curve, which is contrary to the higher amount of POPC in the actual membrane than POPG. Thus, the 1H spin diffusion data of the mixed membrane is best interpreted by a single intermediate depth of insertion of the peptide. The single depth fitting is also consistent with the lack of resolved 31P isotropic or anisotropic chemical shifts between POPC and POPG, suggesting that the membrane is not phase-separated by the peptide.
Compared to the tri-Arg mutant, the hexa-Arg wild-type PG-1 is fully inserted into the mixed anionic membranes [27]. Our previous studies showed that wild-type PG-1 achieves its membrane-disruptive function by forming transmembrane β-barrels [27] in which the Arg residues drag the lipid headgroups into the center of the bilayer, thus causing toroidal pores [27, 29]. In contrast, [Δ4,18 G10] PG-1 no longer inserts fully into the bilayer and no longer associates tightly with the phosphate groups. The change of these two important aspects of PG-1 interaction with the lipid membrane results in weaker antimicrobial activities.
Based on the peptide-headgroup distances, the peptide depth of insertion and the peptide mobility, we propose the following structural model to explain the weaker antimicrobial activity of [Δ4,18 G10] PG-1. Binding to the membrane induces the peptide to form extended β-sheets, since it is energetically more favorable for polar backbone functional groups (C=O and N-H) to be hydrogen-bonded than to be exposed to the hydrophobic part of the lipid bilayer (Fig. 6a). The β-sheets insert into the membrane to a depth that depends on the negative charge density of the bilayer surface. In zwitterionic lipid bilayers the β-sheet lies mostly on the membrane surface (Fig. 6b). With increasing negative charge density of the membrane surface, the β-sheets insert more deeply (Fig. 6c, d), in so doing causes lateral expansion and increased disorder of the membrane. Thus, the antimicrobial activity of the charged-reduced PG-1 mutant is best described by the carpet mechanism [48].
The weakness of the carpet mechanism for [Δ4,18 G10] PG-1 compared to the toroidal pore mechanism for wild-type PG-1 calls for comparison with other PG-1 mutants and similar β-hairpin antimicrobial peptides. For example, in the tachyplesin (TP) family, the most potent peptide, wild-type TP-I, undergoes large-amplitude uniaxial motion on the membrane surface to disrupt the bilayer integrity [26, 49], while the least active TP mutant, TPA4 [24], forms immobile β-sheets, or carpets, on the membrane surface. In the protegrin family, the most potent peptide, wild-type PG-1, uses a β-barrel – toroidal pore mechanism to destroy the membrane integrity [27, 29]. The second most potent peptide, Argmm-PG-1, where the guanidinium group of each Arg is dimethylated, uses large-amplitude motion on the membrane surface to achieve its antimicrobial function [30], similar to TP-I. [Δ4,18 G10] PG-1 has even weaker activity than Argmm-PG-1, and adopts an immobilized carpet mechanism, similar to TPA4. The least active PG-1 mutant studied so far, Ala-PG1, where all four Cys residues are replaced by Ala, is mostly unstructured and does not bind the membrane significantly [5]. Thus, the activities of the protegrin peptides follow the trend of PG-1 > Argmm-PG-1 > [Δ4,18 G10] PG-1 > Ala-PG1, with the corresponding mechanisms of β-barrel-toroidal pore, large-amplitude in-plane motion, carpet, and random-coil poor binding. The TP peptides show decreasing activities from TP-I to TPA4, with the corresponding mechanisms of large-amplitude in-plane motion and carpet, respectively.
While the number of β-hairpin antimicrobial peptides whose structures have been characterized so far is still small [21, 22], the above comparison nevertheless suggests a potency scale for the mechanisms of action of antimicrobial peptides. A transmembrane peptide assembly that induces toroidal pores in the lipid bilayer is the most potent mechanism for disrupting the membrane. In its absence, the next most potent mechanism is a mobile in-plane peptide, which causes severe membrane defects by virtue of its large-amplitude rotation around the bilayer normal. The third, and weaker, mechanism involves massively aggregated peptides in the bilayer plane, and its weakness may be attributed to the requirement of a high amount of peptides to effectively cover the membrane surface. This mechanism – activity correlation may be useful for the future design of novel antimicrobial peptides.
A corollary of the above observations is that β-hairpin antimicrobial peptides do not all insert into lipid membranes with the same topology even at the same peptide concentration. The charge-attenuated PG-1 mutant inserts in a manner that depends on the bilayer surface charge density, and at the physiological charge density the peptide is partially inserted. TP-I and its mutants remain at the membrane-water interface [25, 26] at the same peptide concentration as PG-1. Thus, there are multiple mechanisms of membrane disruption for the same peptide concentrations.
In summary, we have determined the membrane-bound topological structure of the Arg-depleted PG-1 mutant, [Δ4,18 G10] PG-1, using solid-state NMR spectroscopy. We found that the charge-reduced mutant forms immobilized β-sheets that insert into the lipid bilayer to varying depths depending on the negative charge density of the membrane surface. In the 100% anionic bilayer the peptide is incompletely inserted while in the neutral membrane it lies on the membrane surface. The peptide does not form tight complexes with the lipid phosphate groups. These data indicate that the reduced antimicrobial activity of [Δ4,18 G10] PG-1 results from weaker peptide-lipid-headgroup interactions and incomplete insertion of the peptide at the physiological surface charge density of bacterial cell membranes. Therefore, decreasing the number of Arg residues of antimicrobial peptides has a significant effect on the antimicrobial activity. The positive charge density of antimicrobial peptides should thus be tightly controlled in the design of effective new antibiotics.
References
- 1.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Kokryakov VN, Harwig SS, Panyutich EA, Shevchenko AA, Aleshina GM, Shamova OV, Korneva HA, Lehrer RI. Protegrins: leukocyte antimicrobial peptides that combine features of corticostatic defensins and tachyplesins. FEBS Lett. 1993;327:231–236. doi: 10.1016/0014-5793(93)80175-t. [DOI] [PubMed] [Google Scholar]
- 3.Bellm L, Lehrer RI, Ganz T. Protegrins: new antibiotics of mammalian origin. Exp Opin Invest Drugs. 2000;9:1731–1742. doi: 10.1517/13543784.9.8.1731. [DOI] [PubMed] [Google Scholar]
- 4.Hong M. Structure, topology, and dynamics of membrane peptides and proteins from solid-state NMR spectroscopy. J Phys Chem B. 2007;111:10340–10351. doi: 10.1021/jp073652j. [DOI] [PubMed] [Google Scholar]
- 5.Mani R, Waring AJ, Hong M. Conformation, dynamics, and insertion of a non-cysteine-containing protegrin-1 analogue in lipid membranes from solid-state NMR Spectroscopy. Chembiochem. 2007;8:1877–1884. doi: 10.1002/cbic.200700335. [DOI] [PubMed] [Google Scholar]
- 6.Mani R, Waring AJ, Lehrer RI, Hong M. Membrane-disruptive abilities of beta-hairpin antimicrobial peptides correlate with conformation and activity: A P-31 and H-1 NMR study. Biochim Biophys Acta. 2005;1716:11–18. doi: 10.1016/j.bbamem.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Falla TJ, Liu HJ, Hurst MA, Fujii CA, Mosca DA, Embree JR, Loury DJ, Radel PA, Chang CC, Gu L, Fiddes JC. Development of protegrins for the treatment and prevention of oral mucositis: Structure-activity relationships of synthetic protegrin analogues. Biopolymers. 2000;55:88–98. doi: 10.1002/1097-0282(2000)55:1<88::AID-BIP80>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 8.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 9.Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 10.White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 11.Lear JD, Gratkowski H, Adamian L, Liang J, DeGrado WF. Position-dependence of stabilizing polar interactions of asparagine in transmembrane helical bundles. Biochemistry. 2003;42:6400–6407. doi: 10.1021/bi020573j. [DOI] [PubMed] [Google Scholar]
- 12.Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Vorobyov I, Allen TW. Potential of mean force and pKa profile calculation for a lipid membrane-exposed arginine side chain. J Phys Chem B. 2008;112:9574–9587. doi: 10.1021/jp7114912. [DOI] [PubMed] [Google Scholar]
- 14.Yoo J, Cui Q. Does arginine remain protonated in the lipid membrane? Insights from microscopic pKa calculations. Biophys J. 2008;94:L61–L63. doi: 10.1529/biophysj.107.122945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill CP, Yee J, Selsted ME, Eisenberg D. Crystal structure of defensin HNP-3, an amphiphilic dimer: mechanisms of membrane permeabilization. Science. 1991;251:1481–1485. doi: 10.1126/science.2006422. [DOI] [PubMed] [Google Scholar]
- 16.Hoover DM, Chertov O, Lubkowski J. The structure of human beta-defensin-1: new insights into structural properties of beta-defensins. J Biol Chem. 2001;276:39021–39026. doi: 10.1074/jbc.M103830200. [DOI] [PubMed] [Google Scholar]
- 17.Wimley WC, Selsted ME, White SH. Interactions between human defensins and lipid bilayers: evidence for formation of multimeric pores. Protein Sci. 1994;3:1362–1373. doi: 10.1002/pro.5560030902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daly NL, Chen YK, Rosengren KJ, Marx UC, Phillips ML, Waring AJ, Wang W, Lehrer RI, Craik DJ. Retrocyclin-2: Structural analysis of a potent anti-HIV theta-defensin. Biochemistry. 2007;46:9920–9928. doi: 10.1021/bi700720e. [DOI] [PubMed] [Google Scholar]
- 19.Dhople V, Krukemeyer A, Ramamoorthy A. The human beta-defensin-3, an antibacterial peptide with multiple biological functions. Biochim Biophys Acta. 2006;1758:1499–1512. doi: 10.1016/j.bbamem.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Kluver E, Schulz-Maronde S, Scheid S, Meyer B, Forssmann WG, Adermann K. Structure-activity relation of human-defensin 3: Influence of disulfide bonds and cysteine substitution on antimicrobial activity and cytotoxicity. Biochemistry. 2005;44:9804–9816. doi: 10.1021/bi050272k. [DOI] [PubMed] [Google Scholar]
- 21.Buffy JJ, McCormick MJ, Wi S, Waring A, Lehrer RI, Hong M. Solid-State NMR Investigation of the Selective Perturbation of Lipid Bilayers by the Cyclic Antimicrobial Peptide RTD-1. Biochemistry. 2004;43:9800–9812. doi: 10.1021/bi036243w. [DOI] [PubMed] [Google Scholar]
- 22.Tang M, Waring AJ, Lehrer RI, Hong M. Orientation of a beta-hairpin Antimicrobial Peptide in Lipid Bilayers from 2D Dipolar Chemical-Shift Correlation NMR. Biophys J. 2006;90:3616–3624. doi: 10.1529/biophysj.105.062075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powers JP, Tan A, Ramamoorthy A, Hancock RE. Solution structure and interaction of the antimicrobial polyphemusins with lipid membranes. Biochemistry. 2005;44:15504–15513. doi: 10.1021/bi051302m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doherty T, Waring AJ, Hong M. Peptide-lipid interactions of the beta-hairpin antimicrobial peptide tachyplesin and its linear derivatives from solid-state NMR. Biochim Biophys Acta. 2006;1758:1285–1291. doi: 10.1016/j.bbamem.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 25.Doherty T, Waring AJ, Hong M. Membrane-bound conformation and topology of the antimicrobial peptide tachyplesin-I by solid-state NMR. Biochemistry. 2006;45:13323–13330. doi: 10.1021/bi061424u. [DOI] [PubMed] [Google Scholar]
- 26.Doherty T, Waring AJ, Hong M. Dynamic structure of disulfide-removed linear analogs of tachyplesin-I in the lipid bilayer from solid-state NMR. Biochemistry. 2008;47:1105–1116. doi: 10.1021/bi701390t. [DOI] [PubMed] [Google Scholar]
- 27.Mani R, Cady SD, Tang M, Waring AJ, Lehrert RI, Hong M. Membrane-dependent oligomeric structure and pore formation of beta-hairpin antimicrobial peptide in lipid bilayers from solid-state NMR. Proc Natl Acad Sci USA. 2006;103:16242–16247. doi: 10.1073/pnas.0605079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mani R, Tang M, Wu X, Buffy JJ, Waring AJ, Sherman MA, Hong M. Membrane-bound dimer structure of a beta-hairpin antimicrobial peptide from rotational-echo double-resonance solid-state NMR. Biochemistry. 2006;45:8341–8349. doi: 10.1021/bi060305b. [DOI] [PubMed] [Google Scholar]
- 29.Tang M, Waring AJ, Hong M. Phosphate-mediated arginine insertion into lipid membranes and pore formation by a cationic membrane peptide from solid-state NMR. J Am Chem Soc. 2007;129:11438–11446. doi: 10.1021/ja072511s. [DOI] [PubMed] [Google Scholar]
- 30.Tang M, Waring AJ, Lehrer RI, Hong M. Effects of guanidinium-phosphate hydrogen bonding on the membrane-bound structure and activity of an arginine-rich membrane peptide from solid-state NMR spectroscopy. Angewandte Chemie International Edition. 2008;47:3202–3205. doi: 10.1002/anie.200705993. [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi S, Hong T, Waring A, Lehrer RI, Hong M. Solid-state NMR investigations of peptide-lipid interaction and orientation of a beta-sheet antimicrobial peptide, protegrin. Biochemistry. 2002;41:9852–9862. doi: 10.1021/bi0257991. [DOI] [PubMed] [Google Scholar]
- 32.Albrecht MT, Wang W, Shamova O, Lehrer RI, Schiller NL. Binding of protegrin-1 to Pseudomonas aeruginosa and Burkholderia cepacia. Respir Res. 2002;3:18. doi: 10.1186/rr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neihardt FC. Chemical composition of Escherichia coli in E. coli and Salmonella typhimurium, cellular and molecular biology. ASM Press; 1987. [Google Scholar]
- 34.Heefner DL, Claus GW. Change in quantity of lipids and cell size during intracytoplasmic membrane formation in Gluconobacter oxydans. J Bacteriol. 1976;125:1163–1171. doi: 10.1128/jb.125.3.1163-1171.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinberg DA, Hurst MA, Fujii CA, Kung AHC, ho JF, Cheng FC, Loury DJ, Fiddes JC. Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimic Agents and Chemothr. 1997;41:1738–1742. doi: 10.1128/aac.41.8.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaroniec CP, Tounge BA, Rienstra CM, Herzfeld J, Griffin RG. Measurement of C-13-N-15 distances in uniformly C-13 labeled biomolecules: J-decoupled REDOR. J Am Chem Soc. 1999;121:10237–10238. [Google Scholar]
- 37.Sinha N, Schmidt-Rohr K, Hong M. Compensation for pulse imperfections in rotational-echo double-resonance NMR by composite pulses and EXORCYCLE. J Magn Reson. 2004;168:358–365. doi: 10.1016/j.jmr.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 38.Munowitz MG, Griffin RG, Bodenhausen G, Huang TH. Two-Dimensional Rotational Spin-Echo Nuclear Magnetic-Resonance in Solids - Correlation of Chemical-Shift and Dipolar Interactions. J Am Chem Soc. 1981;103:2529–2533. [Google Scholar]
- 39.Rhim WK, Elleman DD, Vaughan RW. Analysis of multiple-pulse NMR in solids. J Chem Phys. 1973;59:3740–3749. [Google Scholar]
- 40.Huster D, Yao XL, Hong M. Membrane protein topology probed by H-1 spin diffusion from lipids using solid-state NMR spectroscopy. J Am Chem Soc. 2002;124:874–883. doi: 10.1021/ja017001r. [DOI] [PubMed] [Google Scholar]
- 41.Luo W, Hong M. Sensitivity-enhanced 1H spin diffusion from lipids to protein for determining membrane protein topology. Solid State Nucl Magn Reson. 2006;29:163–169. doi: 10.1016/j.ssnmr.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Wishart DS, Sykes BD, Richards FM. Relationship between Nuclear-Magnetic-Resonance Chemical-Shift and Protein Secondary Structure. J Mol Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 43.Huster D, Xiao LS, Hong M. Solid-State NMR Investigation of the dynamics of colicin Ia channel-forming domain. Biochemistry. 2001;40:7662–7674. doi: 10.1021/bi0027231. [DOI] [PubMed] [Google Scholar]
- 44.Tang M, Waring AJ, Hong M. Arginine dynamics in a membrane-bound cationic beta-hairpin peptide from solid-state NMR. Chembiochem. 2008;9:1487–1492. doi: 10.1002/cbic.200800005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clauss J, Schmidtrohr K, Spiess HW. Determination of Domain Sizes in Heterogeneous Polymers by Solid-State Nmr. Acta Polymerica. 1993;44:1–17. [Google Scholar]
- 46.Buffy JJ, Hong T, Yamaguchi S, Waring A, Lehrer RI, Hong M. Solid-State NMR Investigation of the Depth of Insertion of Protegin-1 in Lipid Bilayers Using Paramagnetic Mn2+ Biophys J. 2003;85:2363–2373. doi: 10.1016/s0006-3495(03)74660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buffy JJ, Waring AJ, Lehrer RI, Hong M. Immobilization and Aggregation of Antimicrobial Peptide Protegrin in Lipid Bilayers Investigated by Solid-State NMR. Biochemistry. 2003;42:13725–13734. doi: 10.1021/bi035187w. [DOI] [PubMed] [Google Scholar]
- 48.Pouny Y, Rapaport D, Mor A, Nicolas P, Shai Y. Interaction of antimicrobial dermaseptin and its fluorescently labeled analogues with phospholipid membranes. Biochemistry. 1992;31:12416–12423. doi: 10.1021/bi00164a017. [DOI] [PubMed] [Google Scholar]
- 49.Hong M, Doherty T. Orientation determination of membrane-disruptive proteins using powder samples and rotational diffusion: a simple solid-state NMR approach. Chem Phys Lett. 2006;432:296–300. doi: 10.1016/j.cplett.2006.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]