Abstract
The physiological responses to TGF-β stimulation are diverse and vary amongst different cell types and environmental conditions. Even though the principal molecular components of the canonical and the noncanonical TGF-β signaling pathways have been largely identified, the mechanism that underlies the well-established context dependent physiological responses remains a mystery. Understanding how the components of TGF-β signaling function as a system and how this system functions in the context of the global cellular regulatory network requires a more quantitative and systematic approach. Here, we review the recent progress in understanding TGF-β biology using integration of mathematical modeling and quantitative experimental analysis. These studies reveal many interesting dynamics of TGF-β signaling and how cells quantitatively decode variable doses of TGF-β stimulation.
Keywords: TGF-β, Smad, mathematical model, dose response, switch-like response
1. The basics of transforming growth factor-β signaling
TGF-β is the prototypical ligand of the TGF-β superfamily, which signal through receptor serine/threonine kinases. The superfamily is subdivided into two branches: (1) the TGF-β/Activin branch and (2) the Bone Morphogenetic Protein (BMP)/Growth and Differentiation Factor (GDF) branch. Each branch is further divided into subgroups based on sequence similarity [1]. The TGF-β/Activin branch includes TGF-β, Activin, Inhibin, Nodal, and Lefty ligands. The BMP/GDF branch includes BMP, GDF, and Mullerian Inhibitory Substance (MIS) ligands. This review will focus on the quantitative analyses of TGF-β signaling, which is the most studied ligand in terms of basic signal transduction mechanisms. There are also substantial quantitative studies of BMP signaling, especially in the context of morphogen gradient formation and interpretation, which have been covered in several excellent reviews [2-5].
TGF-β is expressed in most cell types and is translated into a proprotein that is proteolytically cleaved into a noncovalently linked mature TGF-β and latency-associated protein (LAP) [6,7]. The active TGF-β ligand is a 25 kDa dimer, covalently linked by a disulfide bond between cysteine residues from each monomeric peptide. When bound to LAP, TGF-β cannot bind to its receptors, resulting in a ligand that lacks bioavailability. Its bioavailability is further restricted by binding to another protein called Latent TGF-β Binding Protein (LTBP). The LAP-TGF-β complex is bound by LTBP during the secretion process [6]. LTBP binds the extracellular matrix and sequesters LAP-TGF-β in vivo [6]. Various proteases cleave LAP and LTBP to liberate the bioactive TGF-β [8]. Bioactive TGF-β can bind various non-receptor cell surface proteins such as decorin, biglycan, and betaglycan, which also serve to regulate its bioavailability, most likely through the enrichment of TGF-β at the plasma membrane [6,7]. Therefore, multiple mechanisms serve to regulate the bioavailability of TGF-β in vivo.
Once bioavailable TGF-β reaches the surface of the target cell, it binds a homodimer of TGF-β type II receptors (TβRII) [1]. The TGF-β-TβRII complex provides a structural interface that facilitates stable complex formation with a homodimer of the TGF-β type I receptor (TβRI) [8]. Therefore, the active ligand-receptor complex is a heterotetrameric complex consisting of a dimer of TGF-β and homodimers of both TβRII and TβRI. Within the active receptor complex, the TβRII, which is a constitutively active kinase, undergoes autophosphorylation, as well as catalyzes transphosphorylation of the TβRI [8]. Transphosphorylation of the TβRI activates kinase activity. In the TGF-β pathway, Smad2 and Smad3 are receptor-regulated effector proteins (R-Smads), which are phosphorylated by the activated TβRI at a C-terminal SSXS motif, resulting in R-Smad nuclear accumulation [8].
TGF-β signaling amplitude and duration can be regulated through the control of receptor trafficking. The ligand bound activated receptor complex is internalized via endoytosis [9]. Internalization of cell surface receptors can occur through either clathrin-mediated or caveolae-mediated endocytosis [10]. Through the clathrin pathway, activated ligand-receptor complexes are brought into early endosomes which are enriched with scaffold proteins such as SARA and Hrs [9]. The proximity of the activated receptor complex and scaffold proteins enhances the phosphorylation of Smad2/3 and their affinity towards Smad4, ultimately activating the nuclear accumulation of Smad4 (also known as the co-Smad) [11]. Therefore, clathrin-mediated endocytosis may promote TGF-β signaling by providing a platform for R-Smad phosphorylation and the formation of active Smad signaling complexes. Ligand-bound receptor complexes in the early endosomes are further sorted to late endosomes, where TGF-β and the receptors are separated. Some of the unbound receptors can be recycled to the plasma membrane, while others are degraded, along with TGF-β, upon fusion with the lysosomes [10]. Since TGF-β is not recycled, internalization of TGF-β by endocytosis is the primary means of removing active TGF-β from the cell surface, and lysosomal degradation is the primary means of termination of TGF-β signaling [12,13].
Activation of TGF-β receptors initiates both Smad-dependent and Smad-independent signaling events [14-16]. Since the majority of the quantitative studies of TGF-β signaling have focused on Smad-dependent events, we will focus our discussion on the dynamics of the canonical pathway (Smad-dependent). In unstimulated cells, Smads constitutively shuttle between the cytoplasm and nucleus. Upon ligand stimulation, the Smads accumulate in the nucleus as the R-Smad/Co-Smad complex formation, which leads to a decrease in their nuclear export rate [17-19]. The Smad complex binds DNA in conjunction with other transcription factors and interacts with the general transcription machinery to regulate the expression of approximately 100-300 target genes [11]. Phosphatase(s) such as PPM1A can deactivate phospho-R-Smads, resulting in the disassembly of the Smad complex and providing a means for negative regulation of TGF-β signaling in the nucleus [18]. Therefore, the intracellular Smad signaling module is a dynamic circuit for ligand sensing.
2. Mathematical modeling of the TGF-β signaling pathway
Conventional cell signaling studies have largely focused on understanding the identity and the functions of the individual parts of each pathway. It is now realized that cell behaviors are not only shaped by the identity of the individual system components, but also by the weighted interactions of components that act together as a system. The systems biology approach using mathematical models has been proven as a powerful tool in studying such complex networks [20]. Mathematical modeling is helpful in predicting emergent cell behaviors and uncovering how the dynamic interactions of signal transducers lead to context dependent cellular responses [21]. Several mathematical models have been established for the canonical TGF-β/Smad signaling [13,17,22-29]. These mathematical models provide quantitative analyses of TGF-β signaling dynamics, leading to a better understanding of the role of feedbacks in regulating TGF-β signaling responses.
The most common way to describe TGF-β signaling dynamics is through a set of deterministic ordinary differential equations (ODEs), assuming the signaling molecules are well-mixed or homogenous in each compartment [20]. The ODEs represent the change of each signaling molecule over time. In order to perform model simulations and predictions, it is necessary to know the values for two types of model parameters: the initial conditions of the pathway that correspond to the concentrations of the signaling molecules at time 0 (before TGF-β stimulation) and the rate constants that characterizes the signaling reactions. The principal molecular components of TGF-β signaling have been identified, yet relatively little is known about the quantitative values of particular components' abundance and rate constants. The lack of experimental data on initial conditions and rate constants is currently one of the bottlenecks for developing high-quality predictive mathematical models for signaling networks.
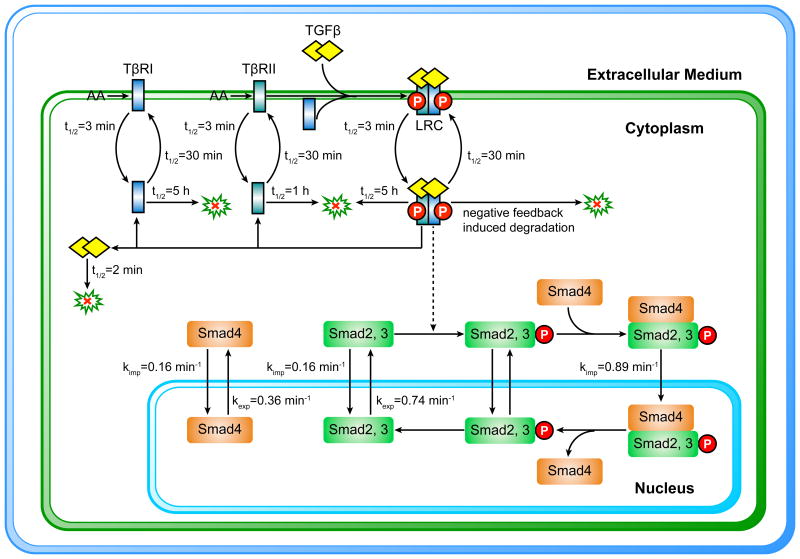
Different approaches have been applied to estimate the model parameters. The initial conditions of the signaling network can be determined by the absolute abundance of the signaling proteins and the volume of the signaling compartment. It is possible to estimate the absolute abundance of a signaling protein when the recombinant protein is available. For example, the absolute abundance of Smad2 protein per cell can be quantified by immunoblotting with a standard curve of recombinant tagged-Smad2 protein [30]. Cell volume is sometimes roughly estimated from cell diameter or it can be measured more accurately by confocal microscopy [31]. On the other hand, direct measurement of the rate constants for different signaling reactions is still experimentally challenging. In order to estimate the unknown model parameters, optimization algorithms are applied to find the most feasible parameters that make model simulations fit the experimental datasets as close as possible [32,33]. During the past few years, some quantitative data has been experimentally measured for the TGF-β signaling pathway, aiding the modeling efforts for this network [10,13,19,25]. Figure 1 summarizes our current knowledge about the model parameters for the canonical TGF-β/Smad signaling network.
Figure 1.
Scheme of the compartmentalized TGF-β model. The kinetic parameter information is approximately estimated according to the experimental data from different cell types. The nuclear export rate constants of Smads are scaled with the ratio of cytoplasmic to nuclear volume size. It is necessary to calibrate these parameter values with quantitative data sets for modeling of TGF-β signaling responses in a specific cell type.
3. Quantitative analysis of signaling responses to different TGF-β stimulations
The cellular responses to TGF-β superfamily ligands depend on the quantity to which the cells are exposed. In development, TGF-β superfamily members form morphogen gradients to determine the fates of cells [2]. Cells read the TGF-β concentration with high precision, as they can distinguish subtle differences in the concentration gradients and orchestrate different cell fates [34]. One of the best examples is the responses of embryonic Xenopus cells to activin, in which five distinct cell responses or fates are observed by varying activin doses [34]. However, the mechanism by which cells are able to accurately decode the concentration of bioavailable TGF-β and elicit a corresponding cellular response remains largely unknown.
3.1 The Smad signaling response correlates with TGF-β molecules per cell rather than the concentration of TGF-β
An early study with mathematical models has shown that cell density affects signaling dynamics in response to the same concentration of ligand [35]. Through modeling analyses of receptor trafficking networks, Zi et al. showed that with the same concentration of ligand stimulation, cells have distinct signaling durations that depend upon cell density, where signaling persists longer when cell density is decreased [35]. Additionally, the model analyses indicate that the dose–response curve of signaling is shifted to the right as the cell density is increased, suggesting that increasing cell density allows for insensitivity to lower doses of ligand. Thus, the key parameter for successful experimental design cannot be “concentration of ligand”, but rather must be “molecules of ligand per cell”, which takes into account the number of cells in the experiment. Further model simulations indicate that signaling responses are regulated by the ratio of ligand to cell surface receptor number [35]. In cell culture experiments, individual cells are likely to express different amounts of receptors at cell surface. Thus, when all cells are exposed to the same amount of ligand, the ratio of ligand to cell surface receptor number in each cell will be different, which might cause heterogeneous signaling responses at single cell level.
To provide additional evidence that cellular responses to ligand occur in terms of “molecules per cell” rather than by the concentration of the ligand, Clarke et al. investigated how cells transduce TGF-β doses into variable kinetic profiles of Smad2 phosphorylation at C-terminal SSXS motif (P-Smad2) by quantitative experimental assays [12]. Clarke et al. measured P-Smad2 levels in a two-level factorial experiment by varying four experimental parameters (TGF-β concentration, cell number at seeding, plate type, and medium volume). When the P-Smad2 data is plotted versus TGF-β concentration, large variations are observed for the same TGF-β concentration among different experimental setups. In contrast, the variation of P-Smad2 levels is significantly reduced if these levels are plotted versus the number of TGF-β molecules per cell. This result implies that the ligand dose expressed as TGF-β molecules per cell is a better predictor of P-Smad2 levels, which is in agreement with early modeling studies about the impact of cell density on signaling response.
Ligand depletion in the TGF-β network provides additional complexity and increases the difficulty of predicting the time dependent signaling responses. For example, the number of TGF-β molecules per cell in the media changes substantially with time because the cells deplete TGF-β from the surrounding medium. It was shown that TβRII defective, but not TβRI defective, cell lines lost their ability to deplete TGF-β from the medium [12]. Thus, TβRII helps to shape the Smad signal amplitude and duration by constitutively depleting extracellular TGF-β. TGF-β depletion most likely occurs through TβRII-mediated endocytosis. However, direct evidence that supports this notion remains to be presented in the literature. In this aspect, TGF-β degradation shares many similarities to EGF or TGF-α [36,37] in that ligand-induced endocytosis does not merely serve as a mechanism for “down-regulation” of signaling, but also provides a mechanism whereby the receptor can continuously track the changes in the secretion of TGF-β by nearby cells. It should be noted that most studies of TGF-β signaling assume that TGF-β concentration in the medium is sufficient to describe the input variable (potency of ligand). Consequently, most modeling studies have assumed a constant level of TGF-β during signaling over time. However, in cell based experiments TGF-β concentration in the medium changes substantially with time, especially for low doses of TGF-β. Therefore, the assumption that TGF-β concentration is constant in medium might be appropriate for high doses of TGF-β, but it is invalid for low doses of TGF-β.
3.2 TGF-β dose-responses are time dependent
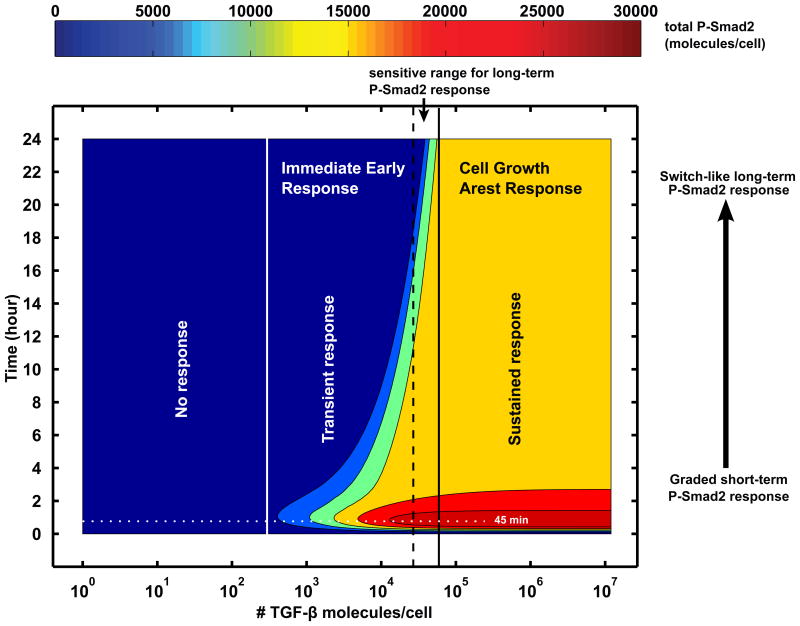
Earlier experimental and modeling analyses showed that Smad signal amplitude is gradually increased with the increments of TGF-β doses [23,24,38]. This leads to an important question about how cells convert continuous TGF-β doses into discrete or binary cellular fate decisions. Since most of modeling studies in the TGF-β field do not account for the TGF-β input variable by assuming constant TGF-β concentration in medium, this omission often results in unreliable predictions of Smad signaling kinetics in response to variable doses of TGF-β stimulation [13]. Recently, we developed an improved mathematical model to describe the dynamics of Smad signaling in response to TGF-β [13]. The model was composed of TGF-β receptor trafficking, Smad phosphorylation and Smad nucleocytoplasmic shuttling. More importantly, it took into account the dynamics of TGF-β depletion. Model parameters were estimated by fitting to several time course datasets, which include variables of TGF-β depletion in the medium, Smad2 nuclear localization and P-Smad2 dynamics. The model was further tested for its ability to predict the P-Smad2 signaling upon pulsation of TGF-β stimulations. With this data-calibrated model, novel predictions were made through model simulations. It was shown that TGF-β signaling responses display different sensitivities to ligand doses at different time scales [13]. In this study, modeling simulations and experimental results show that while short-term P-Smad2 is graded, long-term P-Smad2 response is switch-like to changes in TGF-β doses (Figure 2). In the short-term graded response, P-Smad2 signal gradually increases with the increment of TGF-β dose. In the long-term switch-like response, a small change of TGF-β dose within a certain range results in a large change in P-Smad2 response. Furthermore, a switch-like response is observed for TGF-β induced long-term gene expression and growth inhibitory responses. Additional model perturbation experiments predicted that the long-term switchlike P-Smad2 response is mainly affected by the parameters related to the ligand depletion. This prediction was experimentally confirmed [13].
Figure 2.
Model simulations for the time-course profile of P-Smad2 dose-response in HaCaT cells. Cells have distinct interpretations to TGF-β doses at different time scales. The early P-Smad2 signal (e.g. before 2 hour) displays a graded response to different doses of TGF-β, while the late P-Smad2 response (e.g. at 24 hour) is switch-like. The long-term ultrasensitive signaling response is critical for cellular fate decisions, for example, cell growth arrest.
3.3 The TGF-β pathway is insensitive to high frequency noise
While extensive studies have focused on signaling responses to continuous TGF-β stimulations, little is known about how cells respond to short pulses of TGF-β stimulations. Taking advantage of model simulations, Zi et al. have shown that short-term TGF-β pulse stimulation results in transient P-Smad2, whereas serial pulses result in sustained P-Smad2, similar to that seen with continuous stimulation [13]. To generate a sustained response, the gap between repeated pulse stimulation is ∼30 min. This result suggests that with a strong TGF-β stimulation, the pre-bound receptors are capable of sustaining the signaling response for half of an hour and tiding it to the next stimulus. Incidentally, this optimal gap period is approximately the time to reach maximum Smad2 phosphorylation and nuclear accumulation of Smad2 after TGF-β stimulation [13]. Therefore, there is a time-delay in the TGF-β signaling system, which may be attributed to ligand-bound receptor endocytosis or Smad nucleocytoplasmic shuttling dynamics. Because of this built-in time-delay, the TGF-β signaling system can filter high frequency changes (short time pulsations) in ligand stimulations. Thus, combining mathematical modeling with guided experiments enables new discoveries in systems properties of TGF-β signaling that would have been difficult to reveal using the traditional biochemical approaches alone. The functional significance of pulses of TGF-β has yet to be shown in vivo, but is theoretically occurring within tissues, where the extracellular volume and local secretion of TGF-β is extremely small in magnitude, resulting in a largely noisy extracellular level of TGF-β. Such noise would then be dampened by the delayed TGF-β response that has been observed in cell culture models.
4. Quantitative analysis of transient and sustained signaling responses
The duration of signaling responses can be critical for alerting cell fate decisions in response to growth factor. Previous work with PC12 cells showed that epidermal growth factor (EGF) induces transient activation of extracellular signal-regulated kinase (ERK) and results in cell proliferation, while sustained ERK activation triggered by nerve growth factor (NGF) leads to cell differentiation [39,40]. Although some of these effects could be due to non-ERK dependent responses to the different ligands, these experiments propose an interesting notion, where the duration of ligand stimulation can determine the prevailing cellular response.
In the case of the TGF-β signaling network, the duration of Smad signaling response seems to be context dependent. Experimental studies have shown that keratinocyte epithelial cells have sustained phospho-Smad responses to TGF-β, while some fibroblast and tumor cells display transient Smad activation [41,42]. It was hypothesized that sustained TGF-β signaling may be required for growth inhibition, while transient signaling may cause the resistance to anti-proliferative effects of TGF-β in certain tumor cells [41]. However, the exact mechanism underlying the variation of Smad signal duration in different cell types remains to be elucidated. Here we summarize the time dependent changes in the cellular responses to TGF-β.
4.1 Sequestration of TGF-β receptors by endocytosis
TGF-β signaling is initiated by the binding of TGF-β to TβRI and TβRII. The activation of the ligand-receptor complex is a relatively fast step in the TGF-β signal transduction pathway. An early study from Wrana et al. shows that the phosphorylation of TβRI in the receptor complex peaks at about 2 minutes after TGF-β stimulation [43]. The signal is relayed to the activation of Smad proteins, which arrive at their maximal levels in about 30-60 minutes. The time delay between ligand receptor complex and R-Smad activation may due to intermediate processes, including receptor endocytosis, the recruitment of Smads to receptor complex and Smad activation. After 30-60 minutes, the phosphorylation of Smads correlates with the degree of TGF-β-receptor complex level, which might be due continuous nucleocytoplasmic shuttling of the Smads, but this shuttling fails to explain why there is a prominent delay following receptor activation and prior to Smad phosphorylation [25,44].
Although it has been shown that two main types of endocytosis mediate the internalization of TGF-β receptors, clathrin-dependent and clathrin-independent [9,45], different lines of experimental evidence display discrepancies in the requirement of receptor internalization for Smad phosphorylation. Using potassium depletion, which interferes with clathrin-dependent internalization of receptors, P-Smad2 levels are reduced in Mv1Lu cells [10,46]. On the other hand, potassium depletion or mutant dynamin (K44A) in L17-TβRI cells reveals that receptor trafficking is entirely dispensable for TGF-β signaling to occur [9,47]. Even though there is little debate about whether the TGF-β receptors undergo endocytosis, the precise role of receptor endocytosis in signaling remains controversial [9]. Several lines of evidence support a positive role of endocytosis on R-Smad phosphorylation [10,46,48,49], while there are several reports describing Smad activation immediately at the cell surface without need of receptor endocytosis [47,50,51]. The disagreement among these studies could be attributed to different cell types and experimental systems. Despite the variable effects of receptor endocytosis on Smad phosphorylation, activation of non-Smad signaling pathways by TGF-β appears to require receptor internalization [51,52].
To understand the role of receptor endocytosis on TGF-β signaling, mathematical models were established by focusing on TGF-β receptor endocytosis. Vilar et al. developed a concise model for TGF-β receptor trafficking and the ligand-receptor interactions. This model predicts that the duration of signaling activity is determined by the ratio of the constitutive to the ligand-induced degradation rate of the receptors, termed the “constitutive-to-induced degradation ratio” (CIR). Model analyses suggest that signal activity is transient with a low CIR, while sustained signal response is observed with a high CIR. The model has some assumptions including: (1) TGF-β signaling activity is proportional to the level of ligand-receptor complexes in the internalized endosomes, and (2) Ligand-receptor complexes between type I and type II receptors have the same constitutive degradation rate. In addition, the model lumps the processes including non-clathrin dependent internalization, recycling and the degradation of the receptors into one reaction as “the ligand-induced receptor degradation from plasma membrane”. Thus, CIR defined in Vilar's model is not directly determined by the ratio of the reaction rate constants for constitutive and ligand-induced degradations. Instead, it refers to the balance of the overall effect of the combined processes of the two branches of receptor degradation machinery.
Subsequently, a mathematical model developed by Zi and Klipp includes two major types of TGF-β receptor endocytosis, Smad activation and Smad nuclearcytolasmic shuttling [29]. Similar as Vilar's model, the extended model assumes that Smad activation is proportional to the ligand-receptor complex in early endosomes. The model simulations using different sets of parameter values suggest that Smad activation is regulated by the balance between clathrin dependent endocytosis (kiEE) and caveolar/lipid-raft mediated (clathrin-independent) endocytosis (kiCave). If clathrin-dependent internalization is dominant (kiEE ≫ kiCave), Smad activation becomes a sustained response. On the other hand, if clathrin-independent endocytosis is overwhelming (kiCave ≫ kiEE), Smad activation displays a transient response. Interestingly, the simulation results suggest that changing the balance between the two branches of endocytosis has relatively little effect on the early Smad signal, and has larger effect in reshaping long term Smad activity. This hypothesis might explain the variations among experimental observations about the impact of inhibiting TGF-β receptor endocytosis on TGF-β signaling responses, where the different model systems may have different ratio of clathrin-dependent to clathrin-independent endocytosis. Thus, in a cell line with similar rates of clathrin-dependent and clathrin-independent endocysis, an inhibition of clathrin-dependent will make clathrin-independent endocysis (ligand-induced receptor degradation) dominant and result in a reduction in Smad activity. In other cell types, if the clathrin-dependent endocytosis is overwhelmed by clathrin-independent endocytosis, inhibiting clathrin-dependent internalization will not shift the balance between these two endocytosis branches and should result in little effect on the perturbation of signaling responses. Moreover, the inhibitors of endocytosis used in previous experimental studies may not be very specific and could have some off target effects. In the future, a systems biology approach can be useful to clarify this issue by combining mathematical models with quantitative experimental data of receptors and Smad kinetics.
4.2 Quantitative analysis of the dynamics of Smad nuclear import and export
A prominent feature of TGF-β signaling is the continuous shuttling of Smads in and out the nucleus, in both treated and untreated cells [44,53]. Ligand-induced nuclear accumulation of R-Smad and Co-Smad is relatively slow and peaks at ∼45 min after TGF-β exposure. Ligand-induced Smad shuttling dynamics is considered to be a prevailing mechanism for Smads to continuously monitor receptor activity [44,53]. Different mechanisms have been invoked to account for Smad nucleocytoplasmic shuttling dynamics. Mathematical modeling analysis has been instrumental in differentiating various hypotheses [18]. Clarke et al. [17] was the first to publish a kinetic model of Smad signaling dynamics. The simple kinetic model includes R-Smad phosphorylation/dephosphorylaton, heterodimerization with Smad4, and nucleocytoplasmic shuttling steps. Analyzing the existing data with this model led to several novel hypotheses for Smad nuclear accumulation during TGF-β signaling. Through parameter sensitivity analysis, Clarke et al. posited that (1) the balance between the rates of R-Smad phosphorylation in the cytoplasm and phospho-R-Smad dephosphorylation in the nucleus determines Smad nuclear accumulation. (2) Smad homo- or hetero-oligomerization could protect the phospho-R-Smads from rapid dephosphorylation and therefore promote Smad nuclear accumulation. (3) Nuclear retention factors alone are insufficient for induction of Smad nuclear accumulation. Schmierer et al. investigated the relationship between receptor activation and Smad shuttling dynamics using a combined mathematical modeling and systematic experiment approach [25]. Two competing models were developed based on different assumptions of the Smad import mechanism. The retention/enhanced complex import (RECI) model assumes Smad complexes to be imported at a faster rate than the monomeric species, while the retention only (RO) model sets identical import rates for both Smad complexes and monomeric species but only allows nuclear retention of complexes. The two competing models were simultaneously fit to four sets of kinetic empirical data. The result of comparative analysis shows that RECl is clearly a more accurate model. Moreover, the RECl model also shows excellent agreement with fluorescence recovery after photobleaching (FRAP) data of GFP-Smad2 that were not used to construct the model. The FRAP experiment was used to infer the shuttling of GFP-Smad2 between nucleus and cytoplasmic compartments. Therefore, quantitative analysis of Smad shuttling led to the notion that the Smad complex must have a faster nuclear import rate and that the phosphatase(s) that deactivate R-Smads likely resides in the nucleus. A novel insight that comes out of the modeling analysis is that time-delayed coupling between receptor activity and Smad nuclear accumulation could function as a low-pass filter to dampen the noise in receptor activity [25,54].
4.3 Negative feedbacks in TGF-β signaling
Negative feedback in a signal transduction cascade is one of the major mechanisms for desensitization of sustained ligand stimulation and generation of transient, sometimes oscillating signaling outputs [55]. Even though transient or oscillatory responses associated with sustained TGF-β exposure are not ubiquitous, potential negative feedback motifs have been identified. The most well characterized example is Smad7, a TGF-β-inducible early response gene [56,57]. Smad7 antagonizes TGF-β signaling through multiple mechanisms, both in the cytosol and the nucleus. Since Smad7 can bind the TGF-β receptors, but lacks the SXSS motif commonly found in R-Smads, it has been proposed that Smad7 is likely to be a pseudo substrate and a competitive inhibitor of R-Smads [56,57]. Owing to its ability to form stable complexes with the TGF-β receptors, PP1 phosphatase [58], or the ubiquitination E3 ligase Smurf1 and Smurf2 [59], Smad7 serves as an adapter to mediate inactivation of TGF-β receptors through dephosphorylation or ubiquitin-proteasomal degradation. Besides targeting receptors for inactivation, Smad7 also appears to be involved in repressing TGF-β-dependent transcription through either the disruption of R-Smad/Co-Smad/DNA complexes or the recruitment of histone deacetylases [60]. Regardless of the exact biochemical mechanisms of Smad7, the induction of Smad7 by TGF-β and its function as an antagonist for TGF-β signaling are very compelling. A key question is, “what is the strength of this feedback and to what degree does it impact TGF-β signal transduction.” Melke at al. analyzed a more comprehensive model of TGF-β/Smad signaling in endothelial cells that included canonical Smad signaling. They found that negative feedback through Smad7 was important for terminating signaling and for conferring global robustness to the TGF-β pathway [24].
TMEPAI is another antagonist of TGF-β signaling and is transcriptionally induced by ligand exposure [61]. TMEPAI possesses both transmembrane and Smad interacting domain (SIM). The inhibition of TGF-β signaling can be attributed to sequestration of both unphosphorylation and phosphorylated R-Smads from interacting with the receptors or Smad4 [61]. While inhibition of TGF-β signaling by Smad7 and TMEPAI predominantly occurs in the early step of signaling propagation, the negative feedback loop that involves SnoN appears to target the downstream signaling cascade at the chromatin level. SnoN and its relative Ski are both transcriptional co-repressors of the Smad signaling complex [62,63]. In the early phase of TGF-β stimulation, SnoN is destabilized by association with activated R-Smads and ubiquitin E3 ligases such as Arkadia [64-67]. Degradation of SnoN/Ski unleashes the full activity of the Smad complex, resulting in the transcriptional activation of target genes. SnoN is one of the TGF-β inducible genes and the elevated SnoN restrains the activity of Smad complexes [64]. Therefore, there are at least three negative feedback loops associated with TGF-β signaling. All of these are initiated by transcriptional induction of the antagonist, although spatial and temporal variations exist among them.
4.4 Are there oscillations in TGF-β signaling responses?
Oscillations of signaling responses have been observed in some pathways with negative feedbacks, for example, NF-κB, p53 and Erk systems [37,68,69]. Modeling analyses have shown that biochemical oscillations can occur if four general requirements are satisfied: negative feedback, time delay, nonlinearity of the reaction kinetics, and tightly controlled timescales of opposing chemical reactions [70]. Since different negative feedbacks have been proposed for regulating TGF-β signaling [71], theoretically, it is possible to generate oscillating responses in the TGF-β network. Recent modeling analyses by extensive sampling of parameter space show that oscillating Smad signaling can appear by fine-tuning only a few parameters related to negative feedback (e.g. Smad7) [22,28]. However, it is still an open question about whether there are oscillations in TGF-β network because no oscillations of TGF-β signaling have yet been observed in cells. One reason could be that the negative feedback regulations of TGF-β network may not be strong enough at the endogenous level. Moreover, the time-delay between negative feedback and Smad activation might not be coupled in a proper time scale at in vivo conditions. Last but not least, cell signaling often results in heterogeneous responses within single cells. Averaging signaling dynamics at cell population levels can mask dynamic signaling mechanisms within individual cells [72]. Oscillation responses may appear upon investigation at the single cell level in a variety of cell lines and culture conditions.
4.5 Modeling cell context specific TGF-β signaling
It has been long recognized that the cellular responses triggered by TGF-β are often cell type specific and stimulation specific [73]. For example, TGF-β is a potent inhibitor of normal epithelial cell proliferation but acts to stimulate fibroblast growth [73]. Even in the same cell type, TGF-β can produce opposite proliferation effects depending on the presence of other growth factors. In the presence of PDGF, TGF-β stimulates growth of Myc-1 cells, and in the presence of EGF, TGF-β inhibits growth of the exact same cells [73]. The exact molecular mechanisms underlying these contradictory cellular responses remain largely elusive. One possible explanation to account for these observations is that the pathways activated by TGF-β vary among different cell types and are restrained by crosstalk with other signaling pathways. Even though TGF-β signals through Smad2 and Smad3 in most cell types through ALK-5, in endothelial and hepatic stellate cells TGF-β induces phosphorylation of both Smad1/5 and Smad2 via ALK-1 and ALK-5, respectively [24,38,74]. Based on this evidence, Melke et al. developed a mathematical model for the TGF-β pathway in endothelial cells, taking parallel activation of ALK1 and ALK5 into consideration. Their model recapitulates the kinetics of the experimental data and correctly predicts the behavior in experiments where the system is perturbed [24]. This study highlights a need to develop mathematical modeling tailored to a specific biological context in order to understand the multifunctional nature of TGF-β signaling.
5. Outlook and concluding remarks
TGF-β signaling is spatiotemporally regulated in at least three compartments (extracellular, cytosol and nucleus). Quantitative analysis of the TGF-β signaling pathway is still very much in its infancy. At present, modeling efforts have been focused on the canonical TGF-β signaling cascade. As TGF-β signals through both canonical and noncanonical pathways, it is imperative to develop mathematical models that comprise both pathways in order to accurately predict overall TGF-β signaling. Since the noncanonical pathway is often cell type dependent and operates in the context of other signaling networks, it will be a major challenge to develop such models. Another challenge is to understand the quantitative coupling between Smad signaling kinetics and gene expression profiles. Most of the mathematical models developed so far assume that the dynamics of Smad2 and Smad3 phosphorylation is similar and indistinguishable. However, the biological functions of Smad2 and Smad3 are clearly different, based on mouse knockout studies and DNA microarray analysis [75,76]. The abundance of Smad2 and Smad3 varies significantly in different cell types and the ratio of Smad2 and Smad3 influences the resulting cellular responses to TGF-β stimulation [17,77]. Future modeling efforts need to consider how to model the R-Smads separately. Finally, given the emerging role of TGF-β type III receptor in modulating TGF-β signaling [78], it would be interesting to quantitatively assess their contributions to the signaling dynamics. As more quantitative TGF-β signaling data becomes readily accessible, we anticipate that the innovative systems biology approach to study TGF-β/Smad signaling will fundamentally advance our knowledge in this major signaling network.
Acknowledgments
This work was supported by the Excellence Initiative of the German Federal and State Governments (EXC 294, Z.Z.). D.A.C. was supported by a predoctoral training grant from NIGMS (T32GM08759). We thank Edward Kee for critical reading of the manuscript. X.L. is supported by grants from the National Institutes of Health R01GM083172 and R01CA107098.
Abbreviations
- TGF-β
Transforming Growth Factor-β
- TβRI
TGF-β type I receptors
- TβRII
TGF-β type II receptors
- BMP
Bone Morphogenetic Protein
- GDF
Growth and Differentiation Factor
- ODEs
Ordinary Differential Equations
- P-Smad2
Smad2 phosphorylation at C-terminal SSXS motif
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–91. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 2.Rogers KW, Schier AF. Morphogen gradients: from generation to interpretation. Annu Rev Cell Dev Biol. 2011;27:377–407. doi: 10.1146/annurev-cellbio-092910-154148. [DOI] [PubMed] [Google Scholar]
- 3.Nahmad M, Lander AD. Spatiotemporal mechanisms of morphogen gradient interpretation. Curr Opin Genet Dev. 2011;21:726–31. doi: 10.1016/j.gde.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wartlick O, Mumcu P, Julicher F, Gonzalez-Gaitan M. Understanding morphogenetic growth control -- lessons from flies. Nat Rev Mol Cell Biol. 2011;12:594–604. doi: 10.1038/nrm3169. [DOI] [PubMed] [Google Scholar]
- 5.Barkai N, Shilo BZ. Robust generation and decoding of morphogen gradients. Cold Spring Harb Perspect Biol. 2009;1:a001990. doi: 10.1101/cshperspect.a001990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. J Cell Sci. 2003;116:217–24. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- 7.Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor beta in human disease. N Engl J Med. 2000;342:1350–8. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 8.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 9.Chen YG. Endocytic regulation of TGF-beta signaling. Cell research. 2009;19:58–70. doi: 10.1038/cr.2008.315. [DOI] [PubMed] [Google Scholar]
- 10.Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL. Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat Cell Biol. 2003;5:410–21. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- 11.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 12.Clarke DC, Brown ML, Erickson RA, Shi Y, Liu X. Transforming growth factor beta depletion is the primary determinant of Smad signaling kinetics. Mol Cell Biol. 2009;29:2443–55. doi: 10.1128/MCB.01443-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zi Z, Feng Z, Chapnick DA, Dahl M, Deng D, Klipp E, Moustakas A, Liu X. Quantitative analysis of transient and sustained transforming growth factor-beta signaling dynamics. Molecular systems biology. 2011;7:492. doi: 10.1038/msb.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–84. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 15.Moustakas A, Heldin CH. Non-Smad TGF-beta signals. J Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 16.Mu Y, Gudey SK, Landstrom M. Non-Smad signaling pathways. Cell Tissue Res. 2011 doi: 10.1007/s00441-011-1201-y. [DOI] [PubMed] [Google Scholar]
- 17.Clarke DC, Betterton MB, Liu X. Systems theory of Smad signalling. Systems Biology (Stevenage) 2006;153:412–424. doi: 10.1049/ip-syb:20050055. [DOI] [PubMed] [Google Scholar]
- 18.Clarke DC, Liu X. Decoding the quantitative nature of TGF-beta/Smad signaling. Trends Cell Biol. 2008;18:430–42. doi: 10.1016/j.tcb.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmierer B, Hill CS. Kinetic analysis of Smad nucleocytoplasmic shuttling reveals a mechanism for transforming growth factor beta-dependent nuclear accumulation of Smads. Mol Cell Biol. 2005;25:9845–58. doi: 10.1128/MCB.25.22.9845-9858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK. Physicochemical modelling of cell signalling pathways. Nat Cell Biol. 2006;8:1195–203. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- 21.Arkin AP, Schaffer DV. Network news: innovations in 21st century systems biology. Cell. 2011;144:844–9. doi: 10.1016/j.cell.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Celliere G, Fengos G, Herve M, Iber D. plasticity of TGF-beta signaling. BMC Systems Biology. 2011;5:184. doi: 10.1186/1752-0509-5-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung SW, Miles FL, Sikes RA, Cooper CR, Farach-Carson MC, Ogunnaike BA. Quantitative modeling and analysis of the transforming growth factor beta signaling pathway. Biophysical Journal. 2009;96:1733–50. doi: 10.1016/j.bpj.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melke P, Jonsson H, Pardali E, ten Dijke P, Peterson C. A rate equation approach to elucidate the kinetics and robustness of the TGF-beta pathway. Biophysical Journal. 2006;91:4368–80. doi: 10.1529/biophysj.105.080408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmierer B, Tournier AL, Bates PA, Hill CS. Mathematical modeling identifies Smad nucleocytoplasmic shuttling as a dynamic signal-interpreting system. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6608–13. doi: 10.1073/pnas.0710134105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vilar JM, Jansen R, Sander C. Signal processing in the TGF-beta superfamily ligand-receptor network. PLoS Comput Biol. 2006;2:e3. doi: 10.1371/journal.pcbi.0020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vilar JM, Saiz L. Trafficking coordinate description of intracellular transport control of signaling networks. Biophysical Journal. 2011;101:2315–23. doi: 10.1016/j.bpj.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wegner K, et al. Dynamics and feedback loops in the transforming growth factor beta signaling pathway. Biophysical chemistry. 2012;162:22–34. doi: 10.1016/j.bpc.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Zi Z, Klipp E. Constraint-based modeling and kinetic analysis of the Smad dependent TGF-beta signaling pathway. PLoS One. 2007;2:e936. doi: 10.1371/journal.pone.0000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clarke DC, Liu X. Measuring the absolute abundance of the Smad transcription factors using quantitative immunoblotting. Methods Mol Biol. 2010;647:357–76. doi: 10.1007/978-1-60761-738-9_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Errington RJ, White NS. Measuring dynamic cell volume in situ by confocal microscopy. Methods in molecular biology. 1999;122:315–40. doi: 10.1385/1-59259-722-x:315. [DOI] [PubMed] [Google Scholar]
- 32.Moles CG, Mendes P, Banga JR. Parameter estimation in biochemical pathways: a comparison of global optimization methods. Genome research. 2003;13:2467–74. doi: 10.1101/gr.1262503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zi Z. SBML-PET-MPI: a parallel parameter estimation tool for Systems Biology Markup Language based models. Bioinformatics. 2011;27:1028–9. doi: 10.1093/bioinformatics/btr038. [DOI] [PubMed] [Google Scholar]
- 34.Green JB, New HV, Smith JC. Responses of embryonic Xenopus cells to activin and FGF are separated by multiple dose thresholds and correspond to distinct axes of the mesoderm. Cell. 1992;71:731–9. doi: 10.1016/0092-8674(92)90550-v. [DOI] [PubMed] [Google Scholar]
- 35.Zi Z, Klipp E. Cellular signaling is potentially regulated by cell density in receptor trafficking networks. FEBS Lett. 2007;581:4589–95. doi: 10.1016/j.febslet.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 36.Reddy CC, Wells A, Lauffenburger DA. Receptor-mediated effects on ligand availability influence relative mitogenic potencies of epidermal growth factor and transforming growth factor alpha. J Cell Physiol. 1996;166:512–22. doi: 10.1002/(SICI)1097-4652(199603)166:3<512::AID-JCP6>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 37.Shankaran H, Ippolito DL, Chrisler WB, Resat H, Bollinger N, Opresko LK, Wiley HS. Rapid and sustained nuclear-cytoplasmic ERK oscillations induced by epidermal growth factor. Molecular systems biology. 2009;5:332. doi: 10.1038/msb.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goumans MJ, Valdimarsdottir G, Itoh S, Rosendahl A, Sideras P, ten Dijke P. Balancing the activation state of the endothelium via two distinct TGF-beta type I receptors. The EMBO journal. 2002;21:1743–53. doi: 10.1093/emboj/21.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–85. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 40.Fowler T, Sen R, Roy AL. Regulation of primary response genes. Molecular Cell. 2011;44:348–60. doi: 10.1016/j.molcel.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicolas FJ, Hill CS. Attenuation of the TGF-beta-Smad signaling pathway in pancreatic tumor cells confers resistance to TGF-beta-induced growth arrest. Oncogene. 2003;22:3698–711. doi: 10.1038/sj.onc.1206420. [DOI] [PubMed] [Google Scholar]
- 42.Ahn JY, Kim MH, Lim MJ, Park S, Lee SL, Yun YS, Song JY. The inhibitory effect of ginsan on TGF-beta mediated fibrotic process. Journal of cellular physiology. 2011;226:1241–7. doi: 10.1002/jcp.22452. [DOI] [PubMed] [Google Scholar]
- 43.Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370:341–7. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 44.Inman GJ, Nicolas FJ, Hill CS. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-beta receptor activity. Molecular Cell. 2002;10:283–94. doi: 10.1016/s1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- 45.Le Roy C, Wrana JL. Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nature reviews. Molecular cell biology. 2005;6:112–26. doi: 10.1038/nrm1571. [DOI] [PubMed] [Google Scholar]
- 46.Penheiter SG, Mitchell H, Garamszegi N, Edens M, Dore JJ, Jr, Leof EB. Internalization-dependent and -independent requirements for transforming growth factor beta receptor signaling via the Smad pathway. Mol Cell Biol. 2002;22:4750–9. doi: 10.1128/MCB.22.13.4750-4759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu Z, Murray JT, Luo W, Li H, Wu X, Xu H, Backer JM, Chen YG. Transforming growth factor beta activates Smad2 in the absence of receptor endocytosis. The Journal of biological chemistry. 2002;277:29363–8. doi: 10.1074/jbc.M203495200. [DOI] [PubMed] [Google Scholar]
- 48.Runyan CE, Schnaper HW, Poncelet AC. The role of internalization in transforming growth factor beta1-induced Smad2 association with Smad anchor for receptor activation (SARA) and Smad2-dependent signaling in human mesangial cells. The Journal of biological chemistry. 2005;280:8300–8. doi: 10.1074/jbc.M407939200. [DOI] [PubMed] [Google Scholar]
- 49.Hayes S, Chawla A, Corvera S. TGF beta receptor internalization into EEA1-enriched early endosomes: role in signaling to Smad2. J Cell Biol. 2002;158:1239–49. doi: 10.1083/jcb.200204088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Y, Scolavino S, Funderburk SF, Ficociello LF, Zhang X, Klibanski A. Receptor internalization-independent activation of Smad2 in activin signaling. Mol Endocrinol. 2004;18:1818–26. doi: 10.1210/me.2004-0079. [DOI] [PubMed] [Google Scholar]
- 51.Meyer C, et al. Distinct role of endocytosis for Smad and non-Smad TGF-beta signaling regulation in hepatocytes. Journal of hepatology. 2011;55:369–78. doi: 10.1016/j.jhep.2010.11.027. [DOI] [PubMed] [Google Scholar]
- 52.Zuo W, Chen YG. Specific activation of mitogen-activated protein kinase by transforming growth factor-beta receptors in lipid rafts is required for epithelial cell plasticity. Mol Biol Cell. 2009;20:1020–9. doi: 10.1091/mbc.E08-09-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hill CS. Nucleocytoplasmic shuttling of Smad proteins. Cell research. 2009;19:36–46. doi: 10.1038/cr.2008.325. [DOI] [PubMed] [Google Scholar]
- 54.Shankaran H, Wiley HS. Smad signaling dynamics: insights from a parsimonious model. Sci Signal. 2008;1:pe41. doi: 10.1126/scisignal.136pe41. [DOI] [PubMed] [Google Scholar]
- 55.Brandman O, Meyer T. Feedback loops shape cellular signals in space and time. Science. 2008;322:390–5. doi: 10.1126/science.1160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakao A, et al. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631–5. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 57.Hayashi H, et al. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell. 1997;89:1165–73. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 58.Shi W, Sun C, He B, Xiong W, Shi X, Yao D, Cao X. GADD34-PP1c recruited by Smad7 dephosphorylates TGFbeta type I receptor. J Cell Biol. 2004;164:291–300. doi: 10.1083/jcb.200307151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Molecular Cell. 2000;6:1365–75. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 60.Simonsson M, Heldin CH, Ericsson J, Gronroos E. The balance between acetylation and deacetylation controls Smad7 stability. The Journal of biological chemistry. 2005;280:21797–803. doi: 10.1074/jbc.M503134200. [DOI] [PubMed] [Google Scholar]
- 61.Watanabe Y, et al. TMEPAI, a transmembrane TGF-beta-inducible protein, sequesters Smad proteins from active participation in TGF-beta signaling. Molecular Cell. 2010;37:123–34. doi: 10.1016/j.molcel.2009.10.028. [DOI] [PubMed] [Google Scholar]
- 62.Liu X, Sun Y, Weinberg RA, Lodish HF. Ski/Sno and TGF-beta signaling. Cytokine Growth Factor Rev. 2001;12:1–8. doi: 10.1016/s1359-6101(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 63.Luo K. Ski and SnoN: negative regulators of TGF-beta signaling. Curr Opin Genet Dev. 2004;14:65–70. doi: 10.1016/j.gde.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 64.Stroschein SL, Wang W, Zhou S, Zhou Q, Luo K. Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science. 1999;286:771–4. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- 65.Sun Y, Liu X, Ng-Eaton E, Lodish HF, Weinberg RA. SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor beta signaling. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:12442–7. doi: 10.1073/pnas.96.22.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Levy L, Howell M, Das D, Harkin S, Episkopou V, Hill CS. Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol Cell Biol. 2007;27:6068–83. doi: 10.1128/MCB.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nagano Y, et al. Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-beta signaling. The Journal of biological chemistry. 2007;282:20492–501. doi: 10.1074/jbc.M701294200. [DOI] [PubMed] [Google Scholar]
- 68.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–5. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 69.Geva-Zatorsky N, et al. Oscillations and variability in the p53 system. Molecular systems biology. 2006;2:2006 0033. doi: 10.1038/msb4100068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Novak B, Tyson JJ. Design principles of biochemical oscillators. Nature reviews. Molecular cell biology. 2008;9:981–91. doi: 10.1038/nrm2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Itoh S, ten Dijke P. Negative regulation of TGF-beta receptor/Smad signal transduction. Current opinion in cell biology. 2007;19:176–84. doi: 10.1016/j.ceb.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 72.Spiller DG, Wood CD, Rand DA, White MR. Measurement of single-cell dynamics. Nature. 2010;465:736–45. doi: 10.1038/nature09232. [DOI] [PubMed] [Google Scholar]
- 73.Sporn MB. The early history of TGF-beta, and a brief glimpse of its future. Cytokine Growth Factor Rev. 2006;17:3–7. doi: 10.1016/j.cytogfr.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 74.Wiercinska E, et al. Id1 is a critical mediator in TGF-beta-induced transdifferentiation of rat hepatic stellate cells. Hepatology. 2006;43:1032–41. doi: 10.1002/hep.21135. [DOI] [PubMed] [Google Scholar]
- 75.Weinstein M, Yang X, Deng C. Functions of mammalian Smad genes as revealed by targeted gene disruption in mice. Cytokine Growth Factor Rev. 2000;11:49–58. doi: 10.1016/s1359-6101(99)00028-3. [DOI] [PubMed] [Google Scholar]
- 76.Piek E, et al. Functional characterization of transforming growth factor beta signaling in Smad2- and Smad3-deficient fibroblasts. The Journal of biological chemistry. 2001;276:19945–53. doi: 10.1074/jbc.M102382200. [DOI] [PubMed] [Google Scholar]
- 77.Kim SG, Kim HA, Jong HS, Park JH, Kim NK, Hong SH, Kim TY, Bang YJ. The endogenous ratio of Smad2 and Smad3 influences the cytostatic function of Smad3. Mol Biol Cell. 2005;16:4672–83. doi: 10.1091/mbc.E05-01-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gatza CE, Oh SY, Blobe GC. Roles for the type III TGF-beta receptor in human cancer. Cell Signal. 2010;22:1163–74. doi: 10.1016/j.cellsig.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]