Abstract
The membrane-bound conformation of a cell-penetrating peptide, penetratin, is investigated using solid-state NMR spectroscopy. The 13C chemical shifts of 13C, 15N-labeled residues in the peptide indicate a reversible conformational change from β-sheet at low temperature to coil-like at high temperature. This conformational change occurs for all residues examined between positions 3 and 13, at peptide/lipid molar ratios of 1:15 and 1:30, in membranes with 25–50% anionic lipids, and in both saturated DMPC/DMPG (1,2-dimyristoyl-sn-glycero-3-phosphatidylchloline/1,2-dimyristoyl-sn-glycero-3-phosphatidylglycerol) membranes and unsaturated POPC/POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol) membranes. Thus, it is an intrinsic property of penetratin. The coil state of the peptide has C– H order parameters of 0.23–0.52 for Cα and Cβ sites, indicating that the peptide backbone is unstructured. Moreover, chemical shift anisotropy lineshapes are uniaxially averaged, suggesting that the peptide backbone undergoes uniaxial rotation around the bilayer normal. These observations suggest that the dynamic state of penetratin at high temperature is a structured turn instead of an isotropic random coil. The thermodynamic parameters of this sheet–turn transition are extracted and compared to other membrane peptides reported to exhibit conformational changes. We suggest that the function of this turn conformation may be to reduce hydrophobic interactions with the lipid chains and facilitate penetratin translocation across the bilayer without causing permanent membrane damage.
Keywords: cell-penetrating peptide, solid-state NMR, conformational change, lipid membrane, sheet, coil transition
Introduction
Cell-penetrating peptides (CPPs) are small cationic peptides that are able to enter cells carrying macro-molecular cargos such as DNA and proteins without leaking the cell content or causing permanent damage to the cell membrane.1–3 The promise of these peptides as drug-delivery compounds has made them the objects of many studies to understand their mechanism of cell entry. Earlier investigations using fluorescence microscopy and flow cytometry found that the internalization was independent of both temperature and the D,L-configuration of the amino acids,4,5 suggesting that these peptides directly translocate across the plasma membrane rather than entering by endocytosis. More recent analysis of HIV Tat(48–60) and (Arg)9, two commonly studied CPPs, found that the cell fixation procedure used in the microscopy experiments caused artifactual peptide uptake into cells and that the actual uptake in living cells was temperature dependent,6 suggesting the involvement of the energy-dependent transport mechanism, endocytosis. However, reduced cell internalization at low temperatures does not rule out direct membrane permeation as another route of entry, since this mechanism is also less efficient at low temperature in the gel-phase membrane, as shown by experiments comparing cell lines with different membrane fluidities.7 Furthermore, even peptides internalized via the endocytotic pathway, as detected by biological readout assays,8 must still cross the endosomal membrane in order to enter the cytosol and must cross additional membranes if they are internalized in intracellular compartments. A large number of biophysical studies of CPPs in model membranes have shown the strong affinity of CPPs to lipid membranes.9 This raises the fundamental question of how these peptides, whose amino acid sequences contain 40–100% of the cationic residues Arg and Lys, translocate across the hydrophobic part of the lipid membrane against large free-energy barriers.10
CPPs share their cationic sequences with antimicrobial peptides, which are also Arg- or Lys-rich peptides but which cause permanent damages to the cell membranes of microbial organisms to achieve their cell-killing function. Antimicrobial peptides generally have distinct amphipathic structures, which are considered essential to their membrane-disruptive function.11,12 In comparison, the role of conformation to the translocation of CPPs remains ambiguous. It has been suggested that no specific conformation is required for translocation, since some CPPs such as the HIV Tat peptide show no distinct secondary structures13 while other CPPs such as penetratin are structurally plastic depending on the environment.9 Molecular dynamics simulations of Tat(47–58) showed a random coil that crosses the membrane by electrostatic interactions between the Arg side chains and the lipid phosphate groups.14 For penetratin, there is general consensus that it is a random coil in aqueous solution and becomes significantly α-helical in SDS micelles.15–17 However, in lipid vesicles, both α-helical and β-sheet conformations have been reported depending on the anionic lipid content and the peptide/lipid molar ratio (P/L).16,18–20 Since most conformational studies on membrane-bound CPPs used circular dichroism (CD), which is complicated by light scattering of lipid vesicles and spectral deconvolution uncertainties, the CD structural information has limited accuracy. Thus, the conformation of CPPs in lipid membranes remains to be elucidated to understand the potential role of conformation in membrane translocation and to gain insight into the general mechanism of cationic protein interaction with the lipid membrane.
In this work, we use solid-state NMR spectroscopy to investigate the conformation and dynamics of penetratin in lipid membranes. Penetratin is a 16-residue peptide corresponding to the third helix (residues 43–58) of the Antennapedia homeodomain of Drosophila. It is the first discovered CPP4,5 and one of the most extensively characterized so far. We studied the penetratin conformation in hydrated multi-lamellar liposomes, which have no large intravesicular aqueous compartments as in real cells. Thus, the peptide is mostly partitioned in the lipid membrane instead of water. This sample condition allows us to capture the structure adopted by the peptide when it interacts transiently with the membranes of real cells. We show chemical shift and dynamic data that indicate a clear temperature-induced reversible sheet↔coil conformational change of the membrane-bound penetratin. The nature of this “coil-like” conformation and its potential relevance to membrane translocation are discussed.
Results
Temperature-induced penetratin conformational change in the membrane
To determine the conformation of penetratin in the lipid membrane, we measured the 13C chemical shifts and assigned them by 2D 13C–13C correlation spectra. The main membrane composition in this study is DMPC/DMPG (1,2-dimyristoyl-sn-glycero-3-phosphatidylchloline/1,2-dimyristoyl-sn-glycero-3-phosphatidylglycerol; 8:7). The high anionic lipid content of ∼50% is used since it was reported to be necessary for the peptide to bind to both leaflets of the bilayer.21 Figure 1 shows the aliphatic region of the 2D spectra of (I3, N9)-labeled penetratin in DMPC/DMPG (8:7) bilayers. The spectra were obtained with a short mixing time of 20 ms, thus showing mainly intraresidue cross peaks that are relevant for chemical shift assignment. The spectra were measured at two temperatures, 243 K, which corresponds to the gel phase of the membrane, and 310 K, which corresponds to the liquid-crystalline (LC) phase of the membrane. All expected intra-residue cross peaks are observed, with the C′,Cα, and Cβ chemical shifts reflecting the peptide conformation. Interestingly, both I3 and N9 α/β cross peaks exhibit frequency changes between the two temperatures. At the higher temperature, the Cα chemical shifts increased by 2.8 ppm while the Cβ chemical shifts decreased by 2.2 and 3.5 ppm compared to the low temperature values. The Cα and Cβ isotropic shifts depend on the (φ, ψ) torsion angles in a counterdirectional fashion: the α-helical conformation has larger Cα chemical shifts and smaller Cβ chemical shifts compared to the random coil, while the β-sheet conformation has negative Cα and positive Cβ secondary shifts.22,23 The observed chemical shift changes of I3 and N9 thus indicate a change from a β-sheet-like conformation at low temperature to a helix or coil-like conformation at high temperature.
Fig. 1.

2D 13C–13C correlation spectra of (I3, N9)-labeled penetratin in DMPC/DMPG (8:7) membranes at P/L = 1:15. Black: 243 K; red: 310 K. Note the frequency changes of the α/β cross peaks, which denote conformational change. The spectra were measured under 6 kHz MAS with a 20-ms DARR mixing time.
To determine whether the appearance of the second conformation is gradual or sudden, we measured the 13C cross-polarization (CP)-magic-angle spinning (MAS) spectra of the peptide as a function of temperature. Figure 2 shows the 283–310 K 13C spectra of (I3, N9)-labeled penetratin in the DMPC/DMPG (8:7) membrane. It can be seen that the second conformation grows gradually: as the temperature decreases, the random-coil peaks decrease in intensity while the β-sheet peaks increase in intensity. Around 303 K, both conformations coexist. Below 283 K, only β-sheet peaks are present and the spectra no longer change. For each site, only two chemical shifts are observed, without intermediate values between them. Table S1 lists the intensity fractions of the two sets of peaks for I3 and N9 Cα sites at the temperatures examined.
Fig. 2.

13C CP-MAS spectra of (I3, N9)-labeled penetratin in DMPC/DMPG (8:7) membranes (P/L = 1:15) as a function of temperature. Broken and continuous lines guidethe eye for β-sheet and random-coil peaks, respectively.
To assess if the conformational changes occur in multiple residues in the peptide, we measured the 13C chemical shifts of three other sites, I5, Q8, and K13. Figure 3 shows the 2D spectra at high and low temperatures for two samples containing the labeled sites. It can be seen that the Cα/Cβ chemical shift changes are present for all three residues, indicating that the sheet–turn conformational change occurs for a large fraction of the peptide. Figure 3c shows a long mixing-time 2D spectrum at 310 K, where cross peaks between the lipid head group Cα and the peptide K13 signals are detected, indicating that the high-temperature state of the peptide is in intimate contact with the lipids. The variable-temperature 1D 13C spectra of (I5, Q8, K13)-labeled penetratin in DMPC/DMPG (8:7) bilayers (Fig. 4) further illustrate the temperature-induced chemical shift changes.
Fig. 3.

2D 13C–13C correlation spectra of labeled penetratin in DMPC/DMPG membranes (8:7) at P/L = 1:15. (a) (I5, Q8, K13)-labeled penetratin at 249 K with 50 ms PDSD mixing (black) and at 310 K with 100 ms PDSD mixing (red). (b) K13-labeled penetratin at 273 K with 50 ms DARR mixing (black) and at 310 K with 30 ms DARR mixing (red). (c) 2D PDSD spectrum of K13-labeled penetratin at 310 K with 100 ms mixing. Note the lipid/peptide cross peaks.
Fig. 4.

Variable-temperature 1D 13C CP-MAS spectra of (I5, Q8, K13)-labeled penetratin in DMPC/DMPG (8:7) membranes at P/L = 1:15. Broken and continuous lines guide the eye for sheet and coil peaks, respectively.
Table S2 tabulates the high- and low-temperature 13C chemical shifts of all measured residues in the DMPC/DMPG membrane. The differences of these chemical shifts from the random-coil values give information on the secondary structure of the peptides. Using the random-coil values of Zhang et al.,24 we plotted the secondary chemical shifts in Fig. 5. The low-temperature secondary shifts are negative for Cα and CO but positive for Cβ, which clearly indicate a β-sheet conformation. In comparison, the high-temperature secondary shifts are near the random-coil values and much less than the α-helical secondary shifts. Thus, while the peptide adopts an α-helical conformation in its parent protein, as an independent molecule bound to the LC lipid bilayer, it adopts neither a β-sheet nor an α-helical conformation but is more like a random coil.
Fig. 5.

Experimental Cα, CO, and Cβ secondary shifts of all labeled residues in penetratin in DMPC/DMPG (8:7) bilayers at P/L = 1:15. (a) 243 K. (b) 310 K. Yellow and red lines in (a) and (b) are database secondary chemical shifts for β-sheet and α-helical conformations, respectively.24
Generality of the sheet ↔ coil transition of penetratin
We next examine the generality of this conformational change of penetratin in lipid membranes. Does it occur only at high peptide concentrations or also at low P/L ratios? Is it unique to the nearly 50% anionic DMPC/DMPG membrane or is it present also at other membrane compositions? To address these questions, we prepared a low-concentration sample, with P/L= 1:30, in the DMPC/DMPG (8:7) membrane, a high-concentration (P/L= 1:15) sample in 25% anionic DMPC/DMPG (3:1) membrane, and a high-concentration (P/L = 1:15) sample in the un-saturated POPC/POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol; 8:7) membrane. Figure 6 shows the variable-temperature 13C CPMAS spectra of (I3, N9)-labeled penetratin in DMPC/DMPG (8:7) bilayers at the concentration of P/L = 1:30. The same chemical shift changes are detected, but the relative intensities of the two components at each temperature differ from those of the 1:15 sample (Table S1). Figure 7 shows the 13C spectra of the peptide in the 25% anionic DMPC/DMPG (3:1) membrane. This sample allows us to assess whether the conformational change is mainly induced by the anionic lipid. Again, the same sheet ↔ coil chemical shift changes are observed between low and high temperatures. Thus, the conformational change is not a result of the peptide preferentially partitioning into anionic lipid domains; it also occurs in zwitterionic membranes.
Fig. 6.

Variable-temperature 13C CP-MAS spectra of (I3, N9)-labeled penetratin in DMPC/DMPG (8:7) membranes at P/L = 1:30. Broken and continuous lines guide the eye for the sheet and coil peaks, respectively.
Fig. 7.

Variable-temperature 13C CP-MAS spectra of (I3, N9)-labeled penetratin in DMPC/DMPG (3:1) membranes at P/L = 1:15. Broken and continuous lines guide the eye for the sheet and coil peaks, respectively.
In the unsaturated POPC/POPG (8:7) membrane, we found the same chemical shift changes indicative of a sheet ↔ coil conformational change (Fig. 8). However, the spectral linewidths of the POPC/POPG-bound peptide are much broader than those of the DMPC/DMPG samples near and below the phase transition temperature. This suggests that penetratin exhibits more pronounced intermediate-timescale motion in the POPC/POPG membrane than in the DMPC/DMPG membrane, possibly due to the higher dynamic disorder of the unsaturated chains of POPC and POPG lipids.
Fig. 8.

Variable-temperature 13C CP-MAS spectra of (I3, N9)-labeled penetratin in POPC/POPG (8:7) membranes at P/L = 1:15. Broken and continuous lines guide the eye for the sheet and coil peaks, respectively.
Nature of the coil-like conformation in the membrane from penetratin dynamics
It has been proposed that random coils, by definition without any fixed structure due to stochastic isotropic motion of the protein segments, are not compatible with the lipid membrane due to the energetically unfavorable exposure of non-hydrogen-bonded backbone polar functional groups to the hydrophobic lipid chains.25 Peptides that are unstructured in solution often adopt helical or sheet structures upon association with the lipid membrane.26 Thus, it is puzzling that penetratin exhibits random-coil chemical shifts in the LC lipid membranes.
To better understand the nature of the penetratin structure at high temperature, we measured the C–H dipolar couplings of the peptide. A true random coil should exhibit vanishing dipolar couplings, corresponding to C–H order parameters of 0. In contrast, a β-strand conformation is usually only stable in the membrane if interstrand hydrogen bonds are present.27,28 Such a multistrand β-sheet is usually immobilized and should have order parameters near 1.29 Figure 9 shows the C–H DIPSHIFT curves of two backbone Cα sites, Q8 and K13, in penetratin in DMPC/DMPG bilayers (P/L = 1:15). The coil peaks give clearly less dipolar dephasing, or weaker dipolar couplings, than the corresponding β-sheet peaks. The Cα–Hα and Cβ–Hβ order parameters of the β-sheet signals range from 0.84 to 0.94, while the order parameters of the coil signals range from 0.23 to 0.52 (Table 1). Thus, the backbone of the coil peptide indeed undergoes large-amplitude motion in the lipid membrane. However, since the Cα and Cβ order parameters are much higher than 0, the motion is not isotropic, and the peptide retains significant residual anisotropy.
Fig. 9.
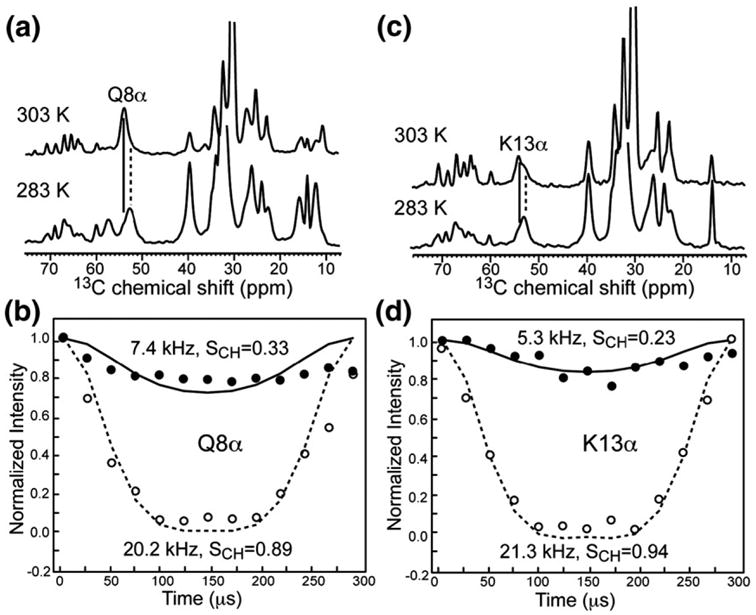
C–H dipolar couplings of coil and sheet Cα peaks of Q8 and K13 at 303 and 283 K, respectively. The lipid membrane is DMPC/DMPG (8:7) and P/L = 1:15. (a) 13C dimensions of the 2D DIPSHIFT spectra showing the Q8α chemical shift change. (b) C–H dipolar de-phasing of Q8α at 303 K (filled symbols, continuous line) and 283 K (open symbols, broken line). (c) 13C dimensions of the 2D DIPSHIFT spectra showing the K13α chemical shift change. (d) C–H dipolar dephasing of K13α at 303 K (filled symbols, continuous line) and 283 K (open symbols, broken line). Best-fit couplings and the corresponding order parameters are given in (b) and (d).
Table 1. C–H order parameters of penetratin in DMPC/DMPG membranes at 303 and 283 K.
| Residue | Site | 303 K | 283 K |
|---|---|---|---|
| I3 | Cα | 0.94s | 0.94s |
| Cβ | 0.33c,o, 0.66s,o | 0.84s,o | |
| Cδ | 0.19 | 0.23 | |
| Cε | 0.14 | 0.23 | |
| I5 | Cα | — | 0.94s |
| Cγ | 0.33 | 0.52 | |
| Cδ | 0.14 | 0.33 | |
| Cε | 0.14 | 0.28 | |
| Q8 | Cα | 0.33c | 0.89s |
| N9 | Cα | 0.52c | 0.84s |
| Cβ | 0.33c,o, 0.66s,o | 0.84s,o | |
| K13 | Cα | 0.23c | 0.94s |
| Cε | 0.33 | 0.37 |
Superscripts c and s denote coil and sheet conformations, respectively. Superscript o indicates partial overlap with another peptide peak. Side-chain groups other than Cβ have no conformational dependence.
Table 1 also shows the side-chain order parameters for all resolved sites. Interestingly, while the ends of the long Ile side chains exhibit the expected small order parameters of 0.14–0.19 at 303 K, the end of the long Lys side chain Cε exhibits a much higher-order parameter of 0.33 at 303 K. This suggests that the cationic Lys amino group may form transient associations with the lipid phosphate groups, similar to what has been observed for Arg residues in an antimicrobial peptide.30,31
For protein backbones, bond order parameters may be smaller than 1 for two different types of motion. They may reflect internal segmental motion, which is the case, for example, for a random coil, or they may reflect overall motion of a rigid molecule. If rigid-body motion is axially symmetric around an external axis, which is typically the bilayer normal for membrane-bound proteins, then the different order parameters simply reflect the different orientations of the individual bond vectors with respect to the bilayer normal. Distinguishing segmental motion from rigid-body motion is a complex task requiring the determination of the order tensor for each segment of the molecule.32,33 Determining each order tensor, which is a traceless and symmetric second-rank tensor, requires five independent NMR couplings.34 In the limiting case of rigid-body uniaxial rotation, the situation is much simpler: all segments have the same order tensor, with a diagonalized unique principal value of 1. Further, the uniaxiality of the motion imposes an axially symmetric lineshape to all NMR interactions, described by an asymmetry parameter η of 0.
One can assess which motional model is more likely for penetratin backbone in the LC membrane by determining the asymmetry parameter of motion. If the only motion giving rise to the reduced dipolar couplings is uncorrelated segmental motion, then the motionally averaged NMR spectra should, in general, exhibit η̄ different from 0. We can measure the asymmetry of the lineshape through chemical shift anisotropy (CSA) spectra, since the static-limit CSAs of Cα and Cβ sites of proteins are generally nonuniaxial. We used the ROCSA (recoupling of CSA) technique35 to recouple the 13C CSA while suppressing the 13C–13C dipolar couplings. Figure 10 shows the 2D ROCSA spectrum of (I3, N9)-labeled penetratin in DMPC/DMPG bilayers at 303 K, along with the CSA cross sections of the N9α coil peak and the mixed I3β and N9β coil peak. It can be seen that the CSA lineshapes are close to uniaxial and are clearly narrower than the static-limit CSA. The N9α line-shape is well fit by a motionally averaged anisotropy parameter δ of 9.9 ppm and η̄ = 0. This is very different from the static-limit δ of 14.4 ppm and η = 0.90. The static-limit CSA principal values were taken from ab initio calculated shielding values36 for the β-sheet conformation. For the mixed I3β and N9β peak, the predicted rigid-limit β-sheet CSA (δ, η) is (21.1 ppm, 0.82) for Asn Cβ and (11.8 ppm, 0.82) for Ile Cβ. Their average gives the broken line pattern in Fig. 10c, which is clearly broader than the measured CSA pattern. The experimental pattern is well fit with (δ, η̄) = (9.1 ppm, 0). For both sites, the measured CSA lineshapes not only are much narrower than the static-limit patterns but also are axially symmetric within experimental uncertainty,37 indicating that the peptide undergoes fast uniaxial rotation around the bilayer normal. This backbone uniaxial motion suggests that the coil chemical shifts may result from a rigid turn conformation that rotates around the membrane normal rather than an isotropic random coil. The chemical shifts of various β- and γ-turns are intermediate between helix and sheet chemical shifts and are not well distinguished from random-coil chemical shifts in protein databases. Since the coil chemical shifts are observed for residues from I3 to K13, the proposed turn structure involves a large segment of the peptide in the fluid membrane.
Fig. 10.

2D ROCSA spectra of (I3, N9)-labeled penetratin in DMPC/DMPG (8:7) membranes at 303 K and P/L=1:15. (a) 2D spectrum. (b) N9α cross section at 52.2 ppm. (c) I3β and N9β cross section at 36.5 ppm. In the 1D panels (b and c), the thick line indicates the experimental data, the thin line denotes the best fit, and the broken line represents the calculated static-limit CSA lineshape based on ab initio calculations for the β-sheet conformation of these residues.36
Thermodynamics of the sheet ↔ turn transition
The temperature-induced reversible sheet↔turn transition of penetratin is an equilibrium reaction. Assuming a simple two-state model, sheet↔turn+ ΔH, we can estimate the enthalpy and entropy of the conformational change using the van't Hoff equation:
| (1) |
The equilibrium constant Keq can be obtained from the intensity ratio between the turn and sheet peaks at each temperature. These Keq values are listed in Table 2 for I3α and N9α in the DMPC/DMPG (8:7) membrane at both high and low peptide concentrations. Figure 11a and b plot Keq for the two sites at both concentrations as a function of temperature. All four curves plateau below 283 K, where the β-sheet conformation dominates. The lower-concentration curves are shifted up from the high-concentration curves, indicating increased populations of the turn conformation. This is expected since the β-sheet conformation requires intermolecular association and is, thus, favored by high peptide concentrations. The turn conformation does not yet exhibit a plateau at the highest temperature measured (310 K) that is still deemed safe for the samples. Figure 11a and b show that the conformational change occurs broadly around the DMPC/DMPG phase transition temperature of 296 K.
Table 2.
Temperature-dependent equilibrium constant Keq = [turn]/[sheet] for the sheet ↔ turn transition of I3α and N9α of penetratin in DMPC/DMPG membranes (8:7) at P/L = 1:15 and 1:30
| T (K) | P/L = 1:15 | P/L = 1:30 | ||
|---|---|---|---|---|
|
|
|
|||
| I3α | N9α | I3α | N9α | |
| 243 | 0.33 | 0.58 | 0.68 | 1.00 |
| 263 | 0.34 | 0.56 | 0.29 | 0.96 |
| 283 | 0.26 | 0.52 | 0.54 | 1.25 |
| 288 | 0.37 | 1.78 | — | — |
| 293 | 0.49 | 2.68 | 2.21 | 4.08 |
| 298 | 1.00 | 2.62 | 7.29 | 32.8 |
| 303 | 1.00 | 2.76 | 4.21 | 4.76 |
| 305 | 1.45 | 5.85 | — | — |
| 310 | 3.20 | 7.13 | 18.0 | 15.5 |
Fig. 11.

Thermodynamics of the sheet ↔ turn conformational change. The turn/sheet intensity ratios, Keq, are plotted as a function of temperature. (a) I3α. (b) N9α. (c) ln Keq versus 1000/T for I3α at P/L = 1:15. The slope of the linear fit gives the ΔH and the intercept gives the ΔS of the conformational change.
Figure 11c shows a representative plot of ln Keq versus 1000/T for I3α at P/L = 1:15. For the temperature regime where the transition occurs, the data fit well to a straight line with a negative slope. This linear regime covers 263–310 K for I3α at P/L = 1:30 and 283–310 K for the other three conditions. As indicated by Eq. (1), the slope of the line gives the enthalpy ΔH of the conformation change, while the intercept gives the entropy change, ΔS. The negative slope indicates a positive enthalpy, or an endothermic sheet↔ turn reaction, consistent with the fact that high temperature induces the turn conformation. The entropy change is also positive, consistent with the higher mobility of the turn state. Table 3 lists the enthalpy and entropy changes for the conformational transition. The ΔH and ΔS values are similar between the two residues, suggesting that the conformational change is global rather than local. If all 16 residues are involved in the conformational change, then ΔH per residue is +0.89 kcal/mol, while ΔS per residue is 3.1 cal/(mol K).
Table 3. Enthalpy and entropy changes of the sheet ↔ turn transition of penetratin in DMPC/DMPG (8:7) membranes.
| Residue, P/L | ln Keq versus 1000/T | ΔH (kcal/mol) | ΔS (kcal/mol K) | |
|---|---|---|---|---|
|
| ||||
| Slope | Intercept | |||
| I3α, 1:15 | −7.52 | 25.1 | 14.9 | 0.050 |
| N9α, 1:15 | −7.07 | 24.8 | 14.0 | 0.049 |
| I3α, 1:30 | −6.79 | 24.1 | 13.5 | 0.048 |
| N9α, 1:30 | −7.20 | 25.8 | 14.3 | 0.051 |
| Mean | — | — | 14.2 | 0.050 |
The thermodynamic values are for 1 mol of peptide.
Discussion
Plasticity of penetratin conformation
The conformation of penetratin has been studied extensively and has been found to be highly plastic, sensitive to the nature of the solvent (buffer, organic solution, micelles, bicelles, or lipid vesicles), peptide concentration, and membrane composition. Most studies employed CD spectroscopy. In water, the peptide is clearly a random coil. In helix-promoting solvents such as trifluoroethanol,15,38 the peptide becomes partly α-helical. In SDS micelles, the helical content is even higher (∼50%) but the random-coil fraction remains significant from both CD and 1H NMR data.15–17,39 In comparison, in lipid bilayers, CD spectra show that penetratin has significant helicity only in largely zwitterionic membranes at low peptide concentrations.18–41 In zwitterionic membranes at high peptide concentrations, the CD spectra indicate a high percentage of random coil.16,42 In anionic membranes with high peptide concentrations, penetratin conformation shifts to β-structures, including β-strands as well as β-turns.42
Since CD spectroscopy of membrane-bound peptides is complicated by light scattering of lipid vesicles and uncertainties in spectral deconvolution, the conformation information is only approximate. In comparison, NMR gives site-specific and accurate information on the peptide conformation in lipid membranes. So far, the only NMR study of penetratin in lipid membranes used DMPC/DMPG (10:1) bicelles with a low anionic lipid content and a low peptide concentration (P/L = 1:130). Under these conditions, 1H chemical shifts and 1H–1H nuclear Overhauser effects indicate that the nine central residues of the peptide are α-helical but with somewhat irregular geometries,18 reminiscent of the random coils observed under other conditions.
Coil or turn conformation at high temperature?
The current solid-state NMR chemical-shift-based conformational constraints are obtained for penetratin bound to 50% and 25% anionic lipid membranes and at P/L ratios of 1:15 and 1:30. We found that in the gel-phase membrane, the peptide is clearly a β-sheet from residue 3 to 13, but in the fluid phase, penetratin changes to a coil-like structure with significant residual anisotropy and uniaxial mobility. The latter two observations suggest that the high-temperature conformation may be a structured turn rather than an unstructured coil. The (φ, ψ) torsion angles and three-dimensional fold of this turn-rich conformation are not yet determined, since most existing solid-state NMR techniques for quantitative structure determination require immobilized molecules, which would require freezing penetratin, which would destroy the very structure of interest.
These solid-state NMR results are in qualitative agreement with CD studies done on membranes with similar compositions. CD spectra measured in POPC/POPG (70:30) vesicles at high peptide concentrations of P/L = 1:17 to 1:33 yielded estimates of 40–60% β-structure, 20–25% random coil, and 20–30% helix.42 The β-structure included not only antiparallel and parallel β-strands but also β-turns; thus, the result is consistent with the NMR data. The remaining α-helical content estimated from the CD spectra may be attributed to the different vesicle sizes used. The CD experiments were carried out on sonicated small unilamellar vesicles while the solid-state NMR data were measured on large unilamellar vesicles. Since penetratin has large helicity in curved SDS micelles, high curvature of the bilayer should also promote α-helix formation.
We found that the sheet ↔ turn conformational change of penetratin occurs under a range of peptide concentrations, anionic lipid content, and lipid chain saturation. Thus, it appears to be an intrinsic property of the peptide in lipid bilayers. It is possible that short segments of β-strand residues, not included among the five labeled residues here, connect the turns. Turn-rich proteins with very short β-strand segments have been observed before. For example, the elastin-mimetic (VPGVG)n polypep-tides consist of repeated β-turns, called β-spirals, centered at Pro and Gly residues in each repeat.43 Turn-rich conformations have also been found in a number of cationic antimicrobial peptides such as indolicidin and tritrpticin.44–46 Solution NMR structures indicate that these turn-rich conformations lead to an amphipathic organization of the peptides, which facilitate their membrane-disruptive activity. For penetratin, whose translocation function leaves the membrane intact, the turn conformation may disfavor an amphipathic three-dimensional fold.
It is important to note that the high-temperature turn state of penetratin is embedded in the membrane rather than floating in the aqueous phase, in contrast to other peptides that undergo coil ↔ helix or coil ↔ sheet changes as a result of membrane binding. Evidence for the strong membrane association includes cross peaks between the lipid Cα and the coil peptide (Fig. 3c) and paramagnetic relaxation enhancement data that show that the turn conformation is well shielded from the Mn2+ ions on the membrane surface.47
Energetics of penetratin sheet ↔ turn transition and comparison with other peptides
Coil↔sheet conformational changes have been reported for several membrane peptides before26,48,49 and are usually triggered by the partitioning of the peptides from aqueous solution to the lipid membrane.25 The thermodynamics of this bilayer-induced secondary structure formation has been studied using CD and isothermal titration calorimetry,50 which yielded quite variable ΔH and ΔS values. For example, the hexapeptide AcWL5 is a monomeric random coil in solution, aggregates to β-sheets upon membrane binding, and unfolds back to a coil upon heating. The per-residue enthalpy change, ΔHresidue, of the sheet → coil unfolding was found to be +1.3 kcal/mol.48 The antimicrobial peptide (KIGAKI)3 is random coil in solution and becomes a nearly perfect β-sheet upon membrane binding. The ΔHresidue of the β-sheet formation was measured to be −0.23 kcal/mol,26 which corresponds to +0.23 kcal/mol for the reverse sheet→coil transition. The significant ΔH variation among different β-sheet formers has been noted before26 and contrasts with the relatively uniform helix ↔ coil enthalpy change of +0.7 to +1.1 kcal/mol. For penetratin, assuming that the sheet↔turn transition occurs for all residues of the peptide, ΔHresidue is +0.89 kcal/mol, which is within the range reported in the literature. However, since penetratin is much closer to (KIGAKI)3 than AcWL5 in terms of sequence length and the cationic nature, ΔH for penetratin might be expected to be much closer to the value of +0.23 kcal/mol for (KIGAKI)3.Experimental conditions such as peptide concentration and membrane composition may explain the different ΔH values.
The entropy change per residue, ΔSresidue, of the sheet ↔ turn transition from the intercept of the linear fit is + 3.1 cal/mol K. Combined with ΔHresidue, it means that the free-energy change, ΔGresidue = ΔHresidue − TΔSresidue, is only slightly negative, −0.05 kcal/mol, at 298 K, which is consistent with the coexistence of turn and sheet peaks at this temperature. Increasing the temperature to 310 K makes ΔGresidue slightly more negative, −0.07 kcal/mol; thus, the sheet ↔ turn transition is an entropically driven process. Overall, the small free-energy reduction of penetratin's sheet ↔ turn transition contrasts with the previously reported ΔGresidue of about −0.5 kcal/mol for bilayer-induced secondary structure formation.25 This lends further support to our conclusion that the conformational change of penetratin occurs within the membrane, rather than involving a change from the membrane to the aqueous phase.
What is the significance of this turn conformation to the translocation function of penetratin? The structural polymorphism of penetratin has been suggested to be important for cell internalization by facilitating the peptide's passage through solvents with very different dielectric constants.51 We propose a more specific hypothesis for the existence of the turn structure in the LC membrane after initial peptide binding. Since the turn structure has a small surface-to-volume ratio and lacks amphipathic organization, it may facilitate the peptide's translocation across the membrane by minimizing hydrophobic interactions with the lipid chains (Fig. 12). Such hydrophobic interactions are implicated in the stabilization of amphipathic antimicrobial peptides in the membrane, leading to pores or other permanent damage to the membrane.27,52 In comparison, penetratin's function requires it to have only transient association with the lipid bilayer, without leaking the cell content, and a compact turn conformation may be better suited than amphipathic β-sheet or α-helical structures for rapid crossing through and eventual dissociation from the membrane. This model suggests that peptide conformation may, in fact, be important in cell internalization. It does not, however, explain the driving force for passage through the membrane. That driving force has been suggested to be the voltage potential across the cell membrane created by differential intracellular and extracellular ion concentrations.53
Fig. 12.

Proposed model of membrane translocation by penetratin. A compact turn-rich conformation reduces hydrophobic interactions with the lipid chains and enables the peptide to cross the lipid bilayer without causing long-lasting pores.
Materials and Methods
Membrane sample preparation
All lipids, including d54-DMPC, DMPG, POPC, and POPG, were purchased from Avanti Polar Lipids (Alabaster, AL) and used without further purification. Penetratin (RQIKIWFQNR RMKWKK) was synthesized using Fmoc solid-phase protocols (PrimmBiotech, Cambridge, MA) and purified by HPLC to >95% purity. U-13C, 15N-labeled residues were incorporated at positions I3, I5, Q8, N9, and K13 in three different samples.
Hydrated membrane samples were prepared by an aqueous-phase mixing protocol. First, a zwitterionic lipid and an anionic lipid were mixed in chloroform at the desired molar ratio and dried under N2 gas. The lipid film was dissolved in cyclohexane and lyophilized overnight. The mixed lipid powder was redissolved in deionized water and vortexed thoroughly and then freeze–thawed eight times. The lipid vesicle solution was then extruded successively through 1.0-, 0.4-, and 0.1-μm polycarbonate filters to obtain large unilamellar vesicles of ∼100 nm diameter. The vesicle solution was added to an appropriate amount of peptide-containing aqueous solution. The mixture was incubated overnight and then centrifuged at 55,000 rpm for 3 h above the lipid phase transition temperature to obtain a pellet. The pellet was center-packed into a 4-mm MAS rotor with Teflon spacer at the bottom to give a hydrated membrane sample for NMR experiments. This study used three membrane compositions—DMPC/DMPG (8:7), POPC/POPG (8:7), and DMPC/DMPG (3:1)—and two P/L ratios—1:15 and 1:30 —to examine the environmental dependence of penetratin conformation. Chain-perdeuterated d54-DMPC was used in the membrane mixtures to reduce the intensities of the lipid background signals.
NMR experiments
Solid-state NMR experiments were carried out on a Bruker AVANCE-600 (14.1 T) spectrometer and a DSX-400 (9.4 T) spectrometer (Karlsruhe, Germany) using MAS probes. Low temperatures were achieved with a Bruker BCU-Xtreme unit on the AVANCE-600 spectrometer and a Kinetics Thermal Systems XR air-jet sample cooler (Stone Ridge, NY) on the DSX-400 spectrometer. Typical 90° pulse lengths were 5.0 μs for 13C and 3.5–4.0 μs for 1H. The α-glycine C′ resonance of 176.49 ppm on the tetramethylsilane scale was used as the external reference for the 13C chemical shifts. 1H–13C CP experiments were carried out with a contact time of 500 μs and a 1H decoupling field of 62 kHz. Variable-temperature 13C spectra were acquired after stabilizing the sample for at least 25 min at each temperature.
2D 1H-driven 13C spin diffusion experiments were carried out without 1H decoupling (PDSD) or with 1H decoupling at the spinning frequency (DARR) during the mixing time. Mixing times of 20–100 ms and spinning speeds of 5.0 and 6.0 kHz were used. 13C–1H dipolar couplings were measured using the 2D dipolar chemical shift correlation (DIPSHIFT) experiment at a spinning speed of 3401 Hz. For 1H homonuclear decoupling, the MREV-8 sequence54 was used. The t1 dipolar data were fit using a home-written Fortran program. The fit values were divided by the scaling factor, 0.47, of the MREV-8 sequence to obtain the true C–H dipolar couplings. The ratio of the true coupling with the rigid-limit one-bond C–H dipolar coupling, 22.7 kHz, gives the bond order parameter, SCH. For CH2 groups, the three-spin system gives rise to an η = 1 dipolar pattern with principal values of (−δ, 0, δ), where δ is the coupling strength. For four-spin CH3 systems, the 1:3:3:1 multiplet pattern was simulated using a 3:1 mixture of dipolar couplings δ and 3δ.
13C CSA was measured using the 2D ROCSA experiment,35 where the CSA interaction is recoupled by a sequence and the 13C–13C homonuclear dipolar coupling is mostly suppressed. The 13C recoupling spin-lock field strength is 4.283 times the spinning speed. The experiment has a CSA scaling factor of 0.272. The experiments were carried out at 310 K under 8.0 kHz MAS.
Supplementary Material
Acknowledgments
This work is funded by National Institutes of Health grant GM-066976.
Abbreviations used
- CPP
cell-penetrating peptide
- P/L
peptide/lipid molar ratio
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphatidylchloline
- DMPG
1,2-dimyristoyl-sn-glycero-3-phosphatidylglycerol
- LC
liquid crystalline
- CP
cross-polarization
- MAS
magic-angle spinning
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol
- CSA
chemical shift anisotropy
- ROCSA
recoupling of CSA
Footnotes
Supplementary Data: Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j. jmb.2008.06.007
References
- 1.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 2.Torchilin VP, Rammohan R, Weissig V, Levchenko TS. TAT peptide on the surface of liposomes affords their efficient intracellular delivery even at low temperature and in the presence of metabolic inhibitors. Proc Natl Acad Sci USA. 2001;98:8786–8791. doi: 10.1073/pnas.151247498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gratton JP, Yu J, Griffith JW, Babbitt RW, Scotland RS, Hickey R, et al. Cell-permeable peptides improve cellular uptake and therapeutic gene delivery of replication-deficient viruses in cells and in vivo. Nat Med. 2003;9:357–362. doi: 10.1038/nm835. [DOI] [PubMed] [Google Scholar]
- 4.Derossi D, Joliot AH, Chassaing G, Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- 5.Derossi D, Calvet S, Trembleau A, Brunissen A, Chassaing G, Prochiantz A. Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J Biol Chem. 1996;271:18188–18193. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 6.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, et al. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J Biol Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 7.Letoha T, Gaál S, Somlai C, Czajlik A, Perczel A, Penke B. Membrane translocation of penetratin and its derivatives in different cell lines. J Mol Recognit. 2003;16:272–279. doi: 10.1002/jmr.637. [DOI] [PubMed] [Google Scholar]
- 8.El Andaloussi S, Guterstam P, Langel U. Assessing the delivery efficacy and internalization route of cell-penetrating peptides. Nat Protoc. 2007;2:2043–2047. doi: 10.1038/nprot.2007.302. [DOI] [PubMed] [Google Scholar]
- 9.Fischer R, Fotin-Mleczek M, Hufnagel H, Brock R. Break on through to the other side—biophysics and cell biology shed light on cell-penetrating peptides. ChemBioChem. 2005;6:2126–2142. doi: 10.1002/cbic.200500044. [DOI] [PubMed] [Google Scholar]
- 10.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 11.Epand RM, Vogel HJ. Diversity of antimicrobial peptides and their mechanisms of action. Biochim Biophys Acta. 1999;1462:11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 12.Powers JP, Hancock RE. The relationship between peptide structure and antibacterial activity. Peptides. 2003;24:1681–1691. doi: 10.1016/j.peptides.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 13.Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, Sugiura Y. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J Biol Chem. 2001;276:5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- 14.Herce HD, Garcia AE. Molecular dynamics simulations suggest a mechanism for trans-location of the HIV-1 TAT peptide across lipid membranes. Proc Natl Acad Sci USA. 2007;104:20805–20810. doi: 10.1073/pnas.0706574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berlose JP, Convert O, Derossi D, Brunissen A, Chassaing G. Conformational and associative behaviours of the third helix of antennapedia home-odomain in membrane-mimetic environments. Eur J Biochem. 1996;242:372–386. doi: 10.1111/j.1432-1033.1996.0372r.x. [DOI] [PubMed] [Google Scholar]
- 16.Magzoub M, Kilk K, Eriksson LE, Langel U, Graslund A. Interaction and structure induction of cell-penetrating peptides in the presence of phospholipid vesicles. Biochim Biophys Acta. 2001;1512:77–89. doi: 10.1016/s0005-2736(01)00304-2. [DOI] [PubMed] [Google Scholar]
- 17.Drin G, Demene H, Temsamani J, Brasseur R. Translocation of the pAntp peptide and its amphipathic analogue AP-2AL. Biochemistry. 2001;40:1824–1834. doi: 10.1021/bi002019k. [DOI] [PubMed] [Google Scholar]
- 18.Lindberg M, Biverstahl H, Graslund A, Maler L. Structure and positioning comparison of two variants of penetratin in two different membrane mimicking systems by NMR. Eur J Biochem. 2003;270:3055–3063. doi: 10.1046/j.1432-1033.2003.03685.x. [DOI] [PubMed] [Google Scholar]
- 19.Persson D, Thoren PE, Herner M, Lincoln P, Norden B. Application of a novel analysis to measure the binding of the membrane-translocating peptide penetratin to negatively charged liposomes. Biochemistry. 2003;42:421–429. doi: 10.1021/bi026453t. [DOI] [PubMed] [Google Scholar]
- 20.Clayton AH, Atcliffe BW, Howlett GJ, Sawyer WH. Conformation and orientation of penetratin in phospholipid membranes. J Pept Sci. 2006;12:233–238. doi: 10.1002/psc.715. [DOI] [PubMed] [Google Scholar]
- 21.Binder H, Lindblom G. Charge-dependent translocation of the Trojan peptide penetratin across lipid membranes. Biophys J. 2003;85:982–995. doi: 10.1016/S0006-3495(03)74537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spera S, Bax A. Empirical correlation between protein backbone conformation and Ca and Cb 13C NMR chemical shifts. J Am Chem Soc. 1991;113:5490–5492. [Google Scholar]
- 23.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Neal S, Wishart DS. RefDB: a database of uniformly referenced protein chemical shifts. J Biomol NMR. 2003;25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- 25.White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 26.Meier M, Seelig J. Thermodynamics of the coil ↔ beta-sheet transition in a membrane environment. J Mol Biol. 2007;369:277–289. doi: 10.1016/j.jmb.2007.02.082. [DOI] [PubMed] [Google Scholar]
- 27.Mani R, Cady SD, Tang M, Waring AJ, Lehrer RI, Hong M. Membrane-dependent oligomeric structure and pore formation of a b-hairpin antimicrobial peptide in lipid bilayers from solid-state NMR. Proc Natl Acad Sci USA. 2006;103:16242–16247. doi: 10.1073/pnas.0605079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mani R, Tang M, Wu X, Buffy JJ, Waring AJ, Sherman MA, Hong M. Membrane-bound dimer structure of a b-hairpin antimicrobial peptide from rotational-echo double-resonance solid-state NMR. Biochemistry. 2006;45:8341–8349. doi: 10.1021/bi060305b. [DOI] [PubMed] [Google Scholar]
- 29.Buffy JJ, Waring AJ, Lehrer RI, Hong M. Immobilization and aggregation of antimicrobial peptide protegrin in lipid bilayers investigated by solid-state NMR. Biochemistry. 2003;42:13725–13734. doi: 10.1021/bi035187w. [DOI] [PubMed] [Google Scholar]
- 30.Tang M, Waring AJ, Hong M. Phosphate-mediated arginine insertion into lipid membranes and pore formation by a cationic membrane peptide from solid-state NMR. J Am Chem Soc. 2007;129:11438–11446. doi: 10.1021/ja072511s. [DOI] [PubMed] [Google Scholar]
- 31.Tang M, Waring AJ, Lehrer RI, Hong M. Effects of guanidinium-phosphate hydrogen bonding on the membrane-bound structure and activity of an arginine-rich membrane peptide from solid-state NMR. Angew Chem, Int Ed Engl. 2008;47:3202–3205. doi: 10.1002/anie.200705993. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature. 2007;450:1263–1267. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Q, Sun X, Watt ED, Al-Hashimi HM. Resolving the motional modes that code for RNA adaptation. Science. 2006;311:653–656. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt-Rohr K, Hong M. Information on bond orientation in lipids and liquid crystals from segmental order parameters. J Phys Chem. 1996;100:3861–3866. [Google Scholar]
- 35.Chan JCC, Tycko R. Recoupling of chemical shift anisotropies in solid-state NMR under high-speed magic-angle spinning and in uniformly 13C-labeled systems. J Chem Phys. 2003;118:8378–8389. [Google Scholar]
- 36.Sun H, Sanders LK, Oldfield E. Carbon-13C NMR shielding in the twenty amino acids: comparisons with experimental results in proteins. J Am Chem Soc. 2002;124:5486–5495. doi: 10.1021/ja011863a. [DOI] [PubMed] [Google Scholar]
- 37.Wylie BJ, Franks WT, Rienstra CM. Determinations of 15N chemical shift anisotropy magnitudes in a uniformly 15N,13C-labeled micro-crystalline protein by three-dimensional magic-angle spinning nuclear magnetic resonance spectroscopy. J Phys Chem B. 2006;110:10926–10936. doi: 10.1021/jp060507h. [DOI] [PubMed] [Google Scholar]
- 38.Czajlik A, Meskó E, Penke B, Perczel A. Investigation of penetratin peptides. Part 1 The environment dependent conformational properties of penetratin and two of its derivatives. J Pept Sci. 2002;8:151–171. doi: 10.1002/psc.380. [DOI] [PubMed] [Google Scholar]
- 39.Lindberg M, Gräslund A. The position of the cell penetrating peptide penetratin in SDS micelles determined by NMR. FEBS Lett. 2001;497:39–44. doi: 10.1016/s0014-5793(01)02433-4. [DOI] [PubMed] [Google Scholar]
- 40.Magzoub M, Eriksson LE, Graslund A. Comparison of the interaction, positioning, structure induction and membrane perturbation of cell-penetrating peptides and non-translocating variants with phospholipid vesicles. Biophys Chem. 2003;103:271–288. doi: 10.1016/s0301-4622(02)00321-6. [DOI] [PubMed] [Google Scholar]
- 41.Thoren PE, Persson D, Karlsson M, Norden B. The antennapedia peptide penetratin translocates across lipid bilayers—the first direct observation. FEBS Lett. 2000;482:265–268. doi: 10.1016/s0014-5793(00)02072-x. [DOI] [PubMed] [Google Scholar]
- 42.Magzoub M, Eriksson LE, Gräslund A. Conformational states of the cell-penetrating peptide penetratin when interacting with phospholipid vesicles: effects of surface charge and peptide concentration. Biochim Biophys Acta. 2002;1563:53–63. doi: 10.1016/s0005-2736(02)00373-5. [DOI] [PubMed] [Google Scholar]
- 43.Yao XL, Hong M. Structural distribution in an elastin-mimetic peptide (VPGVG)3 investigated by solid-state NMR. J Am Chem Soc. 2004;126:4199–4210. doi: 10.1021/ja036686n. [DOI] [PubMed] [Google Scholar]
- 44.Ladokhin AS, Selsted ME, White SH. CD spectra of indolicidin antimicrobial peptides suggest turns, not polyproline helix. Biochemistry. 1999;38:12313–12319. doi: 10.1021/bi9907936. [DOI] [PubMed] [Google Scholar]
- 45.Schibli DJ, Hwang PM, Vogel HJ. Structure of the antimicrobial peptide tritrpticin bound to micelles: a distinct membrane-bound peptide fold. Biochemistry. 1999;38:16749–16755. doi: 10.1021/bi990701c. [DOI] [PubMed] [Google Scholar]
- 46.Chan DI, Prenner EJ, Vogel HJ. Tryptophan- and arginine-rich antimicrobial peptides: structures and mechanisms of action. Biochim Biophys Acta. 2006;1758:1184–1202. doi: 10.1016/j.bbamem.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Su Y, Mani R, Hong M. Asymmetric insertion of membrane proteins in lipid bilayers by solid-state NMR paramagnetic relaxation enhancement: a cell-penetrating peptide example. J Am Chem Soc. 2008;130:8856–8864. doi: 10.1021/ja802383t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wimley WC, Hristova K, Ladokhin AS, Silverstro L, Axelsen PH, White SH. Folding of b-sheet membrane proteins: a hydrophobic hexapeptide model. J Mol Biol. 1998;277:1091–1110. doi: 10.1006/jmbi.1998.1640. [DOI] [PubMed] [Google Scholar]
- 49.Terzi E, Hölzemann G, Seelig J. Self-association of beta-amyloid peptide (1–40) in solution and binding to lipid membranes. J Mol Biol. 1995;252:633–642. doi: 10.1006/jmbi.1995.0525. [DOI] [PubMed] [Google Scholar]
- 50.Ladokhin AS, White SH. Folding of amphipathic alpha-helices on membranes: energetics of helix formation by melittin. J Mol Biol. 1999;285:1363–1369. doi: 10.1006/jmbi.1998.2346. [DOI] [PubMed] [Google Scholar]
- 51.Deshayes S, Decaffmeyer M, Brasseur R, Thomas A. Structural polymorphism of two CPP: an important parameter of activity. Biochim Biophys Acta. 2008;1778:1197–1205. doi: 10.1016/j.bbamem.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 52.Hong M. Structure, topology, and dynamics of membrane peptides and proteins from solid-state NMR spectroscopy. J Phys Chem B. 2007;111:10340–10351. doi: 10.1021/jp073652j. [DOI] [PubMed] [Google Scholar]
- 53.Rothbard JB, Jessop TC, Lewis RS, Murray BA, Wender PA. Role of membrane potential and hydrogen bonding in the mechanism of translocation of guanidinium-rich peptides into cells. J Am Chem Soc. 2004;126:9506–9507. doi: 10.1021/ja0482536. [DOI] [PubMed] [Google Scholar]
- 54.Rhim WK, Elleman DD, Vaughan RW. Analysis of multiple-pulse NMR in solids. J Chem Phys. 1973;59:3740–3749. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


