Abstract
Poultry intestinal material, sewage and poultry processing drainage water were screened for virulent Clostridium perfringens bacteriophages. Viruses isolated from broiler chicken offal washes (O) and poultry feces (F), designated ΦCP39O and ΦCP26F, respectively, produced clear plaques on host strains. Both bacteriophages had isometric heads of 57 nm in diameter with 100-nm non-contractile tails characteristic of members of the family Siphoviridae in the order Caudovirales. The double-strand DNA genome of bacteriophage ΦCP39O was 38,753 base pairs (bp), while the ΦCP26F genome was 39,188 bp, with an average GC content of 30.3%. Both viral genomes contained 62 potential open reading frames (ORFs) predicted to be encoded on one strand. Among the ORFs, 29 predicted proteins had no known similarity while others encoded putative bacteriophage capsid components such as a pre-neck/appendage, tail, tape measure and portal proteins. Other genes encoded a predicted DNA primase, single-strand DNA-binding protein, terminase, thymidylate synthase and a transcription factor. Potential lytic enzymes such as a fibronectin-binding autolysin, an amidase/hydrolase and a holin were encoded in the viral genomes. Several ORFs encoded proteins that gave BLASTP matches with proteins from Clostridium spp. and other Gram-positive bacterial and bacteriophage genomes as well as unknown putative Collinsella aerofaciens proteins. Proteomics analysis of the purified viruses resulted in the identification of the putative pre-neck/appendage protein and a minor structural protein encoded by large open reading frames. Variants of the portal protein were identified, and several mycobacteriophage gp6-like protein variants were detected in large amounts relative to other virion proteins. The predicted amino acid sequences of the pre-neck/appendage proteins had major differences in the central portion of the protein between the two phage gene products. Based on phylogenetic analysis of the large terminase protein, these phages are predicted to be pac-type, using a head-full DNA packaging strategy.
Introduction
Clostridium perfringens is a Gram-positive, spore-forming, anaerobic bacterium that is commonly present in the intestines of humans and animals. C. perfringens strains are classified into five types (A, B, C, D, or E) based on the toxin they produce [69, 75]. Spores of the pathogen can persist in soil, feces or the environment, and the bacterium causes many severe infections of animals and humans. The bacterium can cause food poisoning, gas gangrene (clostridial myonecrosis), enteritis necroticans, and non-foodborne gastrointestinal infections in humans and is also a veterinary pathogen, causing enteric diseases in both domestic and wild animals [69, 75]. This organism is considered the cause of necrotic enteritis in chickens, and although this does not necessarily present a threat of human illness, it could potentially become a far greater problem for the poultry industry if antibiotics are withdrawn from animal feeds, as is the case in the European Union [81].
Although there are now sequences of C. perfringens genomes available [60, 73], there is a paucity of data for C. perfringens bacteriophages. Temperate and virulent phages are associated with the bacterium, for which there is no genomic sequence [12, 19, 37, 40, 50, 51, 62, 65, 76, 79, 82], and a phage-typing system was developed for the organism [86]. Zimmer et al. [88] isolated two temperate phages by UV irradiation (Φ3626 and Φ8533) of lysogenic C. perfringens cultures. The genome of phage Φ3626 was determined to be 33.5 kb, containing 50 potential open reading frames (ORF) with 3′-protruding cohesive ends. Only 19 of these gene products could be assigned to potential biological functions based on bioinformatics analysis. Several of these potentially influence cell spore formation due to the presence of phage genes in the bacterial genome. Subsequently, these investigators also identified a phage-specific enzyme identified as a murein hydrolase. The lysin had lytic activity against 48 test cultures of C. Perfringens but was not active against members of other clostridial species or bacteria belonging to other genera [89].
There has been a resurgent interest in bacteriophage biology and the use of phages or their gene products as antibacterial agents [28, 45, 57]. The potential application of bacteriophages and/or their lytic enzymes has been of considerable interest for human and veterinary medicine as well as the bioindustry worldwide due to antibiotic resistance of human bacterial pathogens [47, 81]. The importance of phages to bacterial evolution [2, 15], the role of phage- or prophage-encoded virulence factors that contribute to bacterial infectious diseases [11] and the contribution of phages to horizontal gene transfer [15] cannot be overstated. Additionally, their contribution to microbial ecology [64] and agricultural production [16, 80] is also extremely important. Bacteriophages play important roles in related clostridial species that include toxin gene transfer and toxin production, and they may even affect physiological functions of the host bacterium such as sporulation [13, 63]. Although the role of phage in the pathogenesis of C. botulinum [25, 26] and, more recently, C. difficile [32, 33, 36] has been documented, this is not the case for C. perfringens.
Clostridium perfringens plays important roles in human food-borne disease and causes diseases among poultry or other animals [69] that are of concern for human health [81]. We are identifying new antimicrobial agents, such as putative lytic enzymes from the genomes of bacteriophages, that are active against C. perfringens. Consequently, bacterial viruses capable of lysing strains of C. perfringens were initially characterized by negative staining and transmission electron microscopy. Two clear-plaque bacteriophage isolates designated ΦCP39O and ΦCP26F were chosen for genomic and proteomic analysis.
Materials and methods
Bacterial hosts, bacteriophage isolation and propagation
Clostridium perfringens isolates 26 and 39, utilized as hosts for propagation of bacteriophages, were cultured in brain heart infusion (BHI) broth or on agar (Remel, Lenexa, KS) and characterized by 16S rRNA-DNA sequence analysis as described previously [23, 74, 85]. Offal washes (O) and feces (F) obtained at a local chicken-processing facility were clarified by low-speed centrifugation (5,000×g for 20 min at 5°C), followed by filtration of the supernatant, first through cheesecloth, and then through 0.45-μm bottle filters (Corning Inc., Corning, NY). Bacterial viruses producing clear plaques on C. perfringens were identified by spot-testing and titration on strains [76, 88] susceptible to the isolated phages. Several clostridial species including C. absonum, C. acetobutylicum, C. beijerinckii, C. novyi, C. rubrum, C. sordellii, C. sporogenes, C. tetani, and C. tetanomorphum [74, 85] were spot-tested for lytic activity [76, 88]. The C. perfringens-specific bacteriophages ΦCP39O and ΦCP26F were propagated by plating with low-melt agar using the C. perfringens isolates 39 and 26 cultured at 37°C in the Anaero Pack™(Mitsubishi Gas Chemical Co., Japan) system with AnaeroGen (OXOID Ltd., Basingstoke, England) sachets [76, 88]. The bacteriophages were subjected to three rounds of plaque purification and consistently suspended in TMGS (10 mM Tris, pH 8, 10 mM Mg++, 0.55% NaCl, 0.1% gelatin) at an average titer of 2 × 108 pfu/ml. Subsequently, bacteriophages ΦCP39O and ΦCP26F were propagated utilizing a plate lysis method [38] under anaerobic conditions for virus purification and DNA extraction.
Purification of bacteriophages, genomic DNA purification and electron microscopy
After plate lysis [38] in anaerobic chambers, bacteriophage genomic DNA was purified using the Qiagen Lambda Phage DNA isolation protocol (Qiagen, Valencia, CA, USA). Additionally, bacteriophages were purified from plate lysates by centrifugation at 2,000×g for 20 min to remove bacterial debris and low-melt agarose. The clarified supernatant was centrifuged at 103,800×g for 90 min, followed by suspension of the phage pellet in 1 ml TBS (20 mM Tris and 500 mM NaCl at pH 7.5). The bacteriophage suspensions were layered over a 15–55% sucrose-TB gradient and centrifuged at 103,800×g in a Beckman JS 24.38 rotor for 90 min [58] and further purified through 20–50% continuous potassium tartrate density gradients [31, 55, 58] by centrifugtion at 105,000×g in a Beckman JS 24.15 rotor for 90 min. The bacteriophage bands were drawn from the gradient, diluted in TBS and concentrated by centrifugation at 105,000×g in a Beckman JS 24.15 rotor for 90 min, followed by suspension in TBS. Bacteriophages were stained with phosphotungstate (270, pH 7.2) and examined in a Philips EM 300 electron microscope. Magnification was controlled by means of T4 phage tails [3]. The bacteriophage pellets were also subjected to proteinase K (20 μg/ml) digestion in the presence of 0.1% sarcosyl and 0.2 M EDTA, followed by phenol–chloroform extraction and ethanol precipitation to obtain genomic DNA [68].
Molecular cloning, sequencing, annotation of genomic DNA and phylogenetic analysis
After purification of bacteriophage genomic DNA, the nucleic acid was subjected to spectrophotometer readings at 260/280 nm and restriction enzyme digestion, followed by agarose gel electrophoresis [67]. Sequencing of the bacteriophage genomes was completed by MWG Biotech, (High Point, NC, USA), by the J. Craig Venter Institute [29], and pyrosequencing [53] was done at the National Microbiology Laboratory (Winnipeg, MB). For Sanger sequencing, phage DNA was sheared using a nebulizer, blunt-end repaired and dephosphorylated [68]. DNA fragments of the desired size (1 to 4 kb) were ligated into pSmart (Lucigen™) for propagation in E. coli after transformation. Clones were sequenced such that approximately 14-fold redundancy was obtained for the genome including primer walking to fill gaps [29]. To check accuracy, phage DNA was cleaved with the restriction enzymes HindIII, EcoRI, EcoRV, AluI and ClaI (New England Biolabs, Ipswich, MA), treated with Taq polymerase [44] and cloned [56] into the TOPO TA vector (Invitrogen, Carlsbad, CA). Additionally, end-repair and G-tailing were used for cloning restriction enzyme fragments into pSmart vectors (Lucigen, Middleton, WI) for nucleotide sequencing [77; Applied Biosystems Inc., Foster City, CA].
Nucleotide sequence assembly, editing, analysis, prediction of amino acid sequences and alignments were conducted using the Celera Assembler [59], MacVector 7.2™ (Accelrys, San Diego, CA) and DNASTAR™ (Madison, WI) software. Open reading frames (ORFs) were predicted using GeneMark.hmm for prokaryotes [http://opal.biology.gatech.edu/GeneMark; 49] and ORF Finder [http://ncbi.nlm.nih.gov/gorf/gorf.html] software. The predicted protein amino acid sequences were searched using BLAST [6] and PSI-BLAST or BLASTP [7, 70] as well as the conserved domain database [52] algorithms. The large terminase protein was utilized to predict the mechanism of DNA packaging and the structure of the genome ends [21, 29]. We inferred the tree with a J. Craig Venter Institute (JCVI) in-house wrapper script around the PAUP* program [84]. The wrapper converted a FASTA-format alignment created by MUSCLE [24] into the NEXUS format expected by PAUP*. In addition, as trees bootstrapped by PAUP lose evolutionarily meaningful branch lengths (see http://paup.csit.fsu.edu/paupfaq/faq.html), retaining only the bootstrap values themselves, the wrapper fed the PAUP-generated tree along with the alignment into TREE-PUZZLE [71] to infer the maximum-likelihood branch length values for the tree.
Preparation of purified virions and two-dimensional (2D) gel electrophoresis
Bacteriophage protein purification was done by adding four volumes of acetone at −20°C to the centrifuged phage pellet for at least 1 h. After centrifugation at 16,000×g for 10 min at 4°C, the pellet was washed three times in cold acetone/water (4:1), followed by centrifugation, and dried under vacuum [22]. To ensure reproducible gel electrophoresis, purified virion protein fractions were digested with N-glycosidase F (PNGase F) to cleave any potential oligosaccharides prior to extraction with acetone as per the manufacturer’s instructions (New England Biolabs, Ipswich, MA) for 1 h at 37°C. The water–acetone pellets were further purified following the procedure for “2-D gel clean-up” to remove interfering substances such as salts, detergents, lipids and phenolics (Amersham Biosciences, Piscataway, NJ).
Proteomics analysis was carried out using 2-D gel electrophoresis [22, 61]. Protein samples were suspended in electrophoresis buffer (5 M urea, 2 M thiourea, 2% CHAPS, 2% SB3-10, 0.2% 3/10 ampholyte with 40 mM Tris, pH 7.4), mixed by vortexing and centrifuged at 16,000×g at 22°C for 10 min. Supernatant proteins suspended in electrophoresis buffer were separated by isoelectric focusing (pH 3–10) in the first dimension, followed by SDS–PAGE and staining with Coomassie blue [22, 61]. Spot detection and pattern matching were done qualitatively on a Bio-Rad VersaDoc 4000 imager, analyzing gels from each isolate in triplicate with PDQuest version 7.3.0 (Bio-Rad, Hercules, CA). Protein spots were cut using a BioRad EXQuest spot cutter and digested using Genomic Solutions Proprep [30].
Identification of purified bacteriophage proteins by mass spectrometry
Tryptic peptide molecular masses obtained by MALDI-TOF-TOF MS [4, 42] were utilized to identify proteins by searching the protein sequence Mascot database at the National Center for Biotechnology Information, PIR and Swiss-Prot with ProFound (Proteomics, New York, NY) and the predicted amino acid sequences from the bacteriophage genomes. Specifically, samples were prepared and spotted onto a MALDI (matrix-assisted laser desorption/ionization) target using ZipTipu-C18 from Millipore (Billerica, MA). Samples were aspirated and dispensed with ZipTipu-C18 and eluted with conditioning solution (70% acetonitrile, 0.2% formic acid) containing 5 mg/ml MALDI matrix (α-cyano-4-hydroxycinnamic acid), and 0.5 μl was spotted onto the MALDI target.
Samples were analyzed using a model 4700 Proteomics Analyzer with TOF/TOF Optics (Applied Biosystems, Foster City, CA). MALDI-MS data were collected in the m/z range of 700 to 4,000 using trypsin autolysis products with m/z ratios of 842.51 and 2211.10 as internal standards. Data were analyzed using GPS Explorer Software (Applied Biosystems). The data were acquired in reflector mode from a mass range of 700–4,000 Da, and 1,250 laser shots were averaged for each mass spectrum. Each sample was internally calibrated if both the 842.51 and 2211.10 ions from trypsin autolysis were present. The eight most intense ions from the MS analysis that were not on the exclusion list were subjected to MS/MS. For MS/MS analysis, the mass range was 70 to precursor ion with a precursor window of −1 to 3 Da, with an average 5,000 laser shots for each spectrum.
The peptide data were extracted from the Oracle database, and a peak list was created by GPS Explorer software (Applied Biosystems Inc.,) from the raw data generated from the ABI 4700. This peak list was based on signal-to-noise filtering and an exclusion list and included de-isotoping. The resulting file was then searched by Mascot (Matrix Science, Boston, MA). A tolerance of 20 ppm was used if the sample was internally calibrated, and 200 ppm tolerance was used if the default calibration was applied. Protein identification was validated by the following criteria:>20 ppm mass accuracy for all MS ions, and all ions, which were not modified, had to be accounted for in at least two MS/MS spectra. Database search parameters included one missed cleavage, oxidation of methionines and carbamidomethylation of cysteines.
Results and discussion
Isolation of bacteriophages that are virulent for Clostridium perfringens
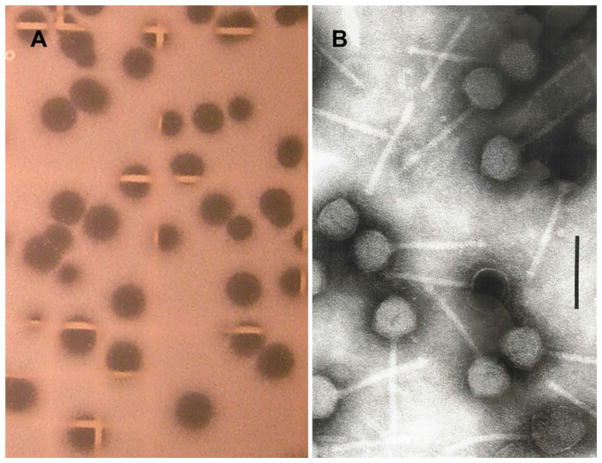
Bacteriophages ΦCP39O and ΦCP26F, which are capable of lysing C. perfringens isolates 39 and 26, were identified by limit dilution, followed by spot-testing and titration on the host with 0.2-μm-filtered chicken offal wash (O) or feces (F). Both the of viruses produced clear plaques after limit dilution plating of the virus on their respective hosts (Fig. 1a), and both viruses were restricted in range to their specific hosts. Also, only clear plaques were detected upon repeated propagation, with no indication of lysogeny. Rather than CsCl density gradient centrifugation for purification, the bacteriophages were purified by potassium tartrate gradient centrifugation [31], which produced pure phage preparations with morphologically intact particles containing nucleic acid. Both bacteriophages had equivalent morphologies that were characteristic of members of the family Siphoviridae, order Caudovirales [1, 2], and had isometric heads of 57 nm between apices of rigid 100-by-7 nm non-contractile tails with terminal knobs (Fig. 1b). ΦCP39O and ΦCP26F differed in their dimensions from other known C. perfringens phages, most of which were members of the family Podoviridae that had short non-contractile tails with a collar and collar fibers [12, 62, 76]. The bacteriophages resemble some C. perfringens siphoviruses [51, 88] but differ from others by their considerably shorter tails [37, 40].
Fig. 1.
Bacteriophage ΦCP39O and ΦCP26F plaques and electron microscopy of the purified virions. a Plaques produced on the host strain Clostridium perfringens strain 39. b Morphology of bacteriophage ΦCP39O following purification by density gradient purification. The bar represents 100 nm
Nucleotide sequences of bacteriophage genomes and predicted open reading frames
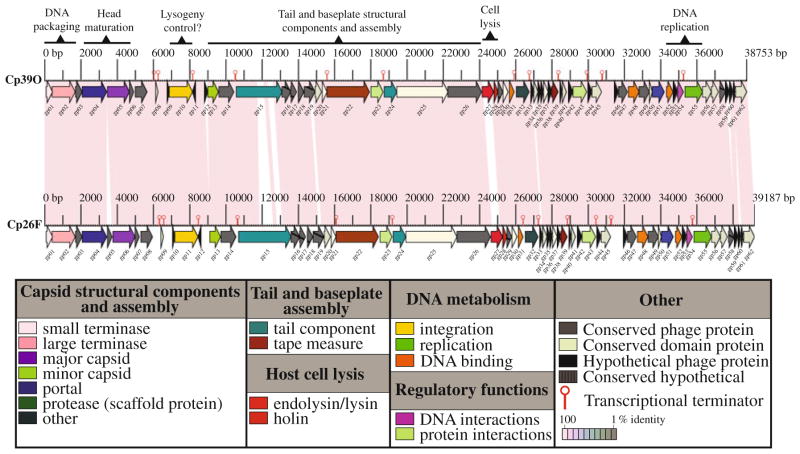
The double-strand (ds) DNA genome of bacteriophage ΦCP39O was 38,753 base pairs (bp) long with a 30.4% GC content. It contained 62 predicted open reading frames (ORFs). The ds DNA genome of bacteriophage ΦCP26F was 39,188 bp with a 30.3% GC content and also contained 62 predicted ORFs. The genomes of ΦCP39O and ΦCP26F shared 96.6% identity, and the genome maps were arranged as a linear model (Fig. 2) encoding proteins predicted or similar to putative proteins found in the ORFs of other bacteriophages or bacterial genomes (Tables S1 and S2, as online resources 1 and 2, respectively). However, 29 ORFs encode putative proteins that had no known similarity to others in the databases. All but one ORF were predicted to be transcribed from one strand, and two genes, CDS05 of ΦCP26F (Fig. 2; Table S2) and CDS12 of ΦCP39O (Fig. 2; Table S1), were present and absent, respectively, in the bacteriophage genomes. The genomes of ΦCP39O and ΦCP26F were similar in size to that of the previously reported C. perfringens Φ3626 [88], but smaller than those of bacteriophages isolated from C. difficile [32, 33, 36, 54]. Although the genome organization of these bacteriophages had a modular arrangement similar to that of many phages [20], the overall sequence similarity was comparatively low, being only 46% with the ΦSM101 genome [NC_008265; 60] and less than 20% similar to the only other published C. perfringens phage genome, Φ3626 [NC_003524; 88]. Also, no putative genes encoding clostridial toxin were identified in the genomes of ΦCP39O and ΦCP26F.
Fig. 2.
Physical map of the bacteriophage ΦCP39O and ΦCP26F genomes. The linear map is based on nucleotide sequences of the phage genomes and predicted open reading frames. Forward arrows represent directional transcription from what would be considered the plus strand, while the single reverse arrow represents possible transcription on the opposite strand. Colored boxes indicate functional gene groups, while transcriptional terminators are indicated by hairpin-type structures
The genomes of both bacteriophages contained genes (Tables S1 and S2) encoding several predicted proteins involved in DNA packaging and morphogenesis, including a putative small terminase (CDS01) and a predicted terminase large subunit (CDS02) that was homologous to a protein detected in the C. thermocellum genome [YP_001038869]. The putative large terminase identified in the ΦCP39O and ΦCP26F genomes was most similar to those of Staphylococcus spp. phages [41]. A gene encoding the putative bacteriophage portal protein (CDS04) for DNA packaging and injection was also present in clostridial genomes [10, 72] other than C. perfringens. Reportedly, the two most conserved tailed-bacteriophage proteins are those involved in DNA packaging, the large terminase and portal proteins [20]. In a BLAST search of ΦCP39O and ΦCP26F, the predicted portal proteins (from CDS04) were most closely related to a protein found in Clostridium difficile bacteriophages ΦCD119 [35] and ΦC2 [33], Bacillus subtilis phage ΦSPP1 [5] and staphylococcal phages [41].
Putative minor head structural proteins from CDS05 of ΦCP39O and CDS06 of ΦCP26F were similar to Bacillus subtilis phage ΦSPP1gp7, which is required for viral head morphogenesis [8]. A Staphylococcus phage ΦROSA ORF19-like protein [41] was also encoded in the genomes (CDS07 of ΦCP39O; CDS08 of ΦCP26F). The putative scaffold protein (CDS13), which is presumably involved in conformational changes to ensure efficiency and fidelity of assembly [27], and a pre-neck/appendage-like protein (CDS15) were most similar to putative proteins encoded in the genomes of C. novyi [10] and C. tetani [14] as well as to those reported in Staphylococcus phages [41]. A conserved hypothetical protein (CDS14) of ΦCP39O and ΦCP26F that was also predicted in C. perfringens genomes [ZP_02864210] was most similar to the gp6 protein of several mycobacteriophages [NP_817344; 66] and another phage protein that has been reported to be the capsid protein of a freshwater cyanophage [YP_001285797; 46]. The putative bacteriophage tail protein (CDS24) encoded in the ΦCP39O and ΦCP26F genomes was similar to that reported for the C. perfringens Φ3626 gp14 [88]. The ΦCP39O and ΦCP26F tail proteins were also similar to the C. difficile ΦCD119 ORF17 product [35] and the C. difficile ΦCD27 gp16 [54]. The predicted tape-measure proteins (CDS22) of ΦCP39O and ΦCP26F were similar to the ORF13 gene product of C. perfringens Φ3626, which was also reported to be a tape-measure protein [88].
Several ORFs encoded proteins that are potentially involved in nucleic acid replication or regulation (Tables S1 and S2). One of the predicted phage anti-repressor/transcription factor proteins encoded by CDS23 was similar to a predicted ORF product in C. difficle [72]. Another potential phage anti-repressor protein gene (CDS32) was located adjacent to a predicted DNA-binding protein gene (CDS31) that was also reported in Streptococcus phage ΦSfi21 [48] and Staphylococcus spp. phage Φ71 [41]. A putative phage anti-repressor of ΦCP39O and ΦCP26F (CDS43) was similar to a predicted C. difficile phage ΦC2 anti-repressor protein [33]. Also present in the ΦCP39O and ΦCP26F genomes were a single-strand DNA-binding protein gene (CDS52) that was also identified in the C. perfringens genome [60] and a recombination protein gene (CDS48) that is distantly related to the bacteriophage Φ42e DNA-binding protein gene [41]. A putative flavin-dependent thymidylate synthase gene that has been reported in the C. perfringens genome [60] was also encoded in the ΦCP39O genome (CDS51). The gene product of CDS54 was a predicted XRE-family-like transcriptional regulator protein similar to elements identified in Streptococcus thermophilus with possible integrative and conjugative functions [17], and its gene was located adjacent to an ORF (CDS55) encoding a potential DNA primase Zn-finger domain.
Two ORFs located contiguously on the ΦCP39O and ΦCP26F genomes (Tables S1 and S2) encoded a predicted N-acetylmuramoyl-L-alanine amidase/hydrolase (CDS27) and the putative phage holin (CDS28). These proteins are required for host-cell lysis and release of phage progeny during late infection [9, 47]. The putative amidase of phage ΦCP39O was predicted to be a 213-amino-acid protein, while the putative amidase of phage ΦCP26F was a 212-residue protein presumably involved in cleaving the amide bond between N-acetylmuramoyl and L-amino acids in bacterial cell walls or cell envelope biogenesis of the outer membrane [78]. The predicted holin protein gene encoded a 75-aminoacid molecule that had sequence similarity to hypothetical proteins found in C. perfringens [YP_695712; NP_561927], Listeria phage ΦB054 [YP_001468736] and Streptococcus bacteriophages [NP_049941; NP_056698]. The predicted phage ΦCP39O and ΦCP26F holin proteins do not have the M-L-M motif of class I or II holins, so it is most likely a class III holin with a single transmembrane domain [87], since the protein was predicted to primarily be hydrophobic with low surface probability in the central region of the molecule, while the N- and C-terminal ends were predicted to be hydrophilic.
Proteomics analysis of purified bacteriophage ΦCP39O
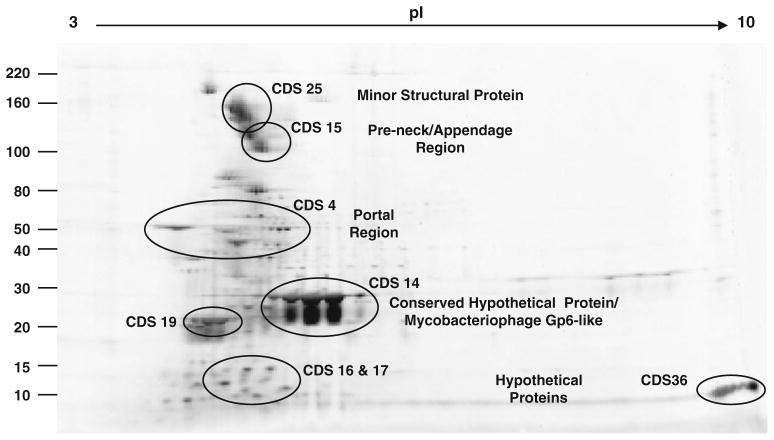
Following 2D-gel electrophoresis of the purified virion (Fig. 3), the proteins were analyzed by mass spectrometry (Table S3 as online resource 3). Four principal virion protein regions were identified, including a portal protein (CDS04), a conserved hypothetical clostridial protein (CDS14) that was determined to be mycobacteriophage gp6-like, a pre-neck appendage protein (CDS15) and a minor structural protein (CDS25). The predicted portal protein region corresponding to CDS04 was identified as a protein of approximately 50 kDa (Fig. 3). The predicted size of the conserved hypothetical mycobacteriophage gp6-like protein from CDS14 was approximately 30 kDa (Table S1), and this is consistent with its relative mobility in the gel (Fig. 3). This protein is also homologous to the pfam11651 coat protein of gene product 5 from bacteriophage P22, which is involved in the formation of procapsid shells [43]. There are 415 molecules of the bacteriophage P22 coat protein arranged in an icosahedral morphology [43], and the CDS14 gene product occurred in the greatest abundance. Consequently, this protein is probably the major capsid protein of the bacteriophage. The proteins included several variants with differences in isoelectric point, possibly due to posttranslational modification [34], since they were predicted to be highly myristilated and phosphorylated at many residues.
Fig. 3.
Two-dimensional gel electrophoresis of purified bacteriophage ΦCP39O virion proteins. After velocity and density gradient centrifugation, proteins of the purified virions were subjected to isoelectric focusing in the first dimension at pI 4–10, followed by vertical SDS–PAGE (8–12%) in the second dimension. The genome sequences from which the proteins were encoded (CDS) are also noted in the figure. Results of the mass spectrometry analyses can be found in supplemental Table S3
A predicted minor structural protein with an approximate size of greater that 110 kDa was identified. This minor structural protein, which is a product of CDS25 (Table S1), shares approximately 30% sequence similarity to ORF15 of C. perfringens phage Φ3626, which encodes a putative endopeptidase [88]. Interestingly, the minor structural protein also had similarity in the C-terminal portion of the molecule to a putative choline-binding protein with potential hydrolase activity found in the genome of C. perfringens [NP_562049]. These bacterial enzymes have an N-terminal catalytic module and a C-terminal choline-binding module (CBM) that attaches them to the bacterial surface [39].
The pre-neck/appendage protein, a product of CDS15 (Table S1), was identified at an approximate relative mobility of 100 kDa (Fig. 3). The pre-neck/appendage proteins of ΦCP39O and ΦCP26F had predicted amino acid sequences with an overall identity of 79% between them. However, the proteins shared 100% sequence identity from residues one through 170 and in the C-terminal portion of the protein beyond residue 828 of the alignment. The major sequence differences occurred in the central portion of the protein (Fig. S1 as online resource 4). The pre-neck/appendage-like protein is most similar to predicted sequences encoded by C. novyi [YP_878235] and C. tetani [NP_782154] genomes, with a region from residues 79 through 237 that has similarity to the bacterial TolA proteins with a myosin-like domain involved in the translocation of group A colicins [67]. The TolA protein is important to filamentous phages and colicins during the process of import into E. coli [83], and consequently, the CDS15 gene product could also potentially be involved in entry of phage ΦCP39O and ΦCP26F into their hosts.
Several proteins with no identifiable homologous function were identified as part of the virion or co-purified with the virus. These corresponded to products of CDS16 and CDS17 with an approximate relative mobility of 10–15 kDa and the gene product of CDS36, with an approximate relative mobility of less than 10 kDa and an extremely high pI of >9 (Fig. 3; Table S1). The proteins encoded by CDS16 and CDS17 had low identity (30%) to proteins predicted in the Collinsella aerofaciens genome [ZP_01771283; ZP_01771282]. Another hypothetical protein with no known homologous function was identified, with a relative mobility of approximately 20 kDa, corresponding to a protein of CDS19 (Fig. 3; Table S1).
Phylogenetic analysis of the predicted terminase protein
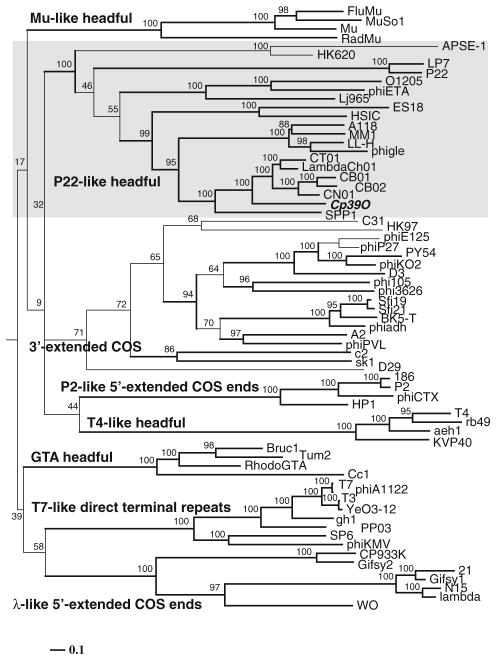
Following BLAST analysis of the predicted phage proteins, the large terminase was determined to be encoded by CDS02 of the ΦCP39O and ΦCP26F phage genomes (Tables S1 and S2). Specifically, the predicted 50-kDa protein was most closely related to terminase proteins encoded by putative prophage sequences identified in the genomes of various clostridial species such as C. thermocellum [YP_001038869], C. botulinum [YP_001254847; YP_001254222] and less closely related to C. kluyveri [YP_001396654] or C. difficile [ZP_01804380]. The terminase was not closely related to the currently published clostridial phage terminase proteins, but it was closer in sequence similarity to the prophage ΦLambd-aCh01 terminase large subunit reported from Carboxydo-thermus hydrogenoformans [YP_360500]. The most closely related bacteriophage terminase protein was from Staphylococcus spp. phage Φ27 [YP_240074] reported by Kwan et al. [41]. Phylogenetic analysis of the available bacteriophage terminase proteins (Fig. 4; Table S4 as online resource 5) demonstrated that the ΦCP39O and ΦCP26F phage termin-ases were most closely related to ΦCh01, ΦCB01, ΦCB02, ΦCN01 and bacteriophage ΦSPP1 (Fig. 4). This indicates that our clostridial phages are potentially pac-type bacteriophages, which use a headfull genomic DNA packaging strategy [21].
Fig. 4.
Phylogenetic analysis of amino acid sequences of the large terminase of bacteriophage ΦCP39O compared with other bacteriophage terminase proteins. Accession numbers for the large terminase proteins can be found in the online supplemental information, Table S4
The phage ΦCP39O and ΦCP26F portal proteins were most similar in sequence to the bacteriophage ΦSPP1 portal protein, which, along with the terminase, functions to package its genomic DNA by a head-full packaging mechanism [18]. Also, the principal virion protein (CDS04) is similar to a coat protein of ΦP22 that undergoes head-full DNA packaging [43]. This correlates well with the phylogenetic prediction for phages ΦCP39O and ΦCP26F being pac-type bacteriophages.
Supplementary Material
Acknowledgments
The authors thank Mary Ard at the University of Georgia, College of Veterinary Medicine, for preliminary electron microscopy. The investigations were supported by the Agricultural Research Service, USDA CRIS project #6612-32000-046 (BSS, GRS, MS) and a non-funded CRADA #58-6612-7-175 with DEF at JCVI. The proteomics portion of the project was supported by NIH Grant Number P20 RR-016464 from the INBRE Program of the National Center for Research Resources (RW and KS). Sequencing was also supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (AK). The genome sequences were submitted to GenBank with accession numbers EU588980 for the ΦCP39O genome and GQ443085 for the ΦCP26F genome.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00705-010-0812-z) contains supplementary material, which is available to authorized users.
Contributor Information
Bruce S. Seal, Email: bruce.seal@ars.usda.gov, Poultry Microbiology Safety Research Unit, Richard B. Russell Agricultural Research Center, Agricultural Research Service, USDA, 950 College Station Road, Athens, GA 30605, USA
Derrick E. Fouts, J. Craig Venter Institute, 9712 Medical Center Drive, Rockville, MD 20850, USA
Mustafa Simmons, Poultry Microbiology Safety Research Unit, Richard B. Russell Agricultural Research Center, Agricultural Research Service, USDA, 950 College Station Road, Athens, GA 30605, USA.
Johnna K. Garrish, Poultry Microbiology Safety Research Unit, Richard B. Russell Agricultural Research Center, Agricultural Research Service, USDA, 950 College Station Road, Athens, GA 30605, USA
Robin L. Kuntz, Poultry Microbiology Safety Research Unit, Richard B. Russell Agricultural Research Center, Agricultural Research Service, USDA, 950 College Station Road, Athens, GA 30605, USA
Rebekah Woolsey, Nevada Proteomics Center, University of Nevada, Reno/MS200, 1664 North Virginia Street, Reno, NV 89557, USA.
Kathleen M. Schegg, Nevada Proteomics Center, University of Nevada, Reno/MS200, 1664 North Virginia Street, Reno, NV 89557, USA
Andrew M. Kropinski, Laboratory for Foodborne Diseases, Public Health Agency of Canada, Guelph, ON N1G 3W4, Canada. Molecular and Cellular Biology, University of Guelph, Guelph, ON N1G 3W4, Canada
Hans-W. Ackermann, Felix d’Herelle Reference Center for Bacterial Viruses, Department of Medical Biology, Faculty of Medicine, Laval University, Laval, QC G1K 7P4, Canada
Gregory R. Siragusa, Poultry Microbiology Safety Research Unit, Richard B. Russell Agricultural Research Center, Agricultural Research Service, USDA, 950 College Station Road, Athens, GA 30605, USA
References
- 1.Ackermann H-W. Tailed bacteriophages: the order Caudovirales. Adv Virus Res. 1998;51:135–201. doi: 10.1016/S0065-3527(08)60785-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackermann H-W. Bacteriophage observations and evolution. Res Microbiol. 2003;154:245–251. doi: 10.1016/S0923-2508(03)00067-6. [DOI] [PubMed] [Google Scholar]
- 3.Ackermann H-W. Basic phage electron microscopy. Methods Mol Biol. 2008;501:113–126. doi: 10.1007/978-1-60327-164-6_12. [DOI] [PubMed] [Google Scholar]
- 4.Aebersold RH, Leavitt J, Saavedra RA, Hood LE, Kent SB. Internal amino acid sequence analysis of proteins separated by one-or two-dimensional gel electrophoresis after in situ protease digestion on nitrocellulose. Proc Natl Acad Sci USA. 1987;84:6970–6974. doi: 10.1073/pnas.84.20.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alonso JC, Lüder G, Stiege AC, Chai S, Weise F, Trautner TA. The complete nucleotide sequence and functional organization of Bacillus subtilis bacteriophage SPP1. Gene. 1997;204:201–212. doi: 10.1016/s0378-1119(97)00547-7. [DOI] [PubMed] [Google Scholar]
- 6.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 7.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker B, De la Fuente N, Gassel M, Günther D, Tavares P, Lurz R, Trautner TA, Alonso JC. Head morphogenesis genes of the Bacillus subtilis bacteriophage SPP1. J Mol Biol. 1997;268:822–839. doi: 10.1006/jmbi.1997.0997. [DOI] [PubMed] [Google Scholar]
- 9.Bernhardt TG, Wang IN, Struck DK, Young R. Breaking free: “protein antibiotics” and phage lysis. Res Microbiol. 2002;153:493–501. doi: 10.1016/s0923-2508(02)01330-x. [DOI] [PubMed] [Google Scholar]
- 10.Bettegowda C, Huang X, Lin J, Cheong I, Kohli M, Szabo SA, Zhang X, Diaz LA, Jr, Velculescu VE, Parmigiani G, Kinzler KW, Vogelstein B, Zhou S. The genome and transcriptomes of the anti-tumor agent Clostridium novyi-NT. Nat Biotechnol. 2006;24:1573–1580. doi: 10.1038/nbt1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyd EF, Brüssow H. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 2002;10:521–529. doi: 10.1016/s0966-842x(02)02459-9. [DOI] [PubMed] [Google Scholar]
- 12.Bradley DE, Hoeniger JF. Structural changes in cells of Clostridium perfringens infected with a short-tailed bacteriophage. Can J Microbiol. 1971;17:397–402. doi: 10.1139/m71-066. [DOI] [PubMed] [Google Scholar]
- 13.Brüggemann H. Genomics of clostridial pathogens: implication of extrachromosomal elements in pathogenicity. Curr Opin Microbiol. 2005;8:601–605. doi: 10.1016/j.mib.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Brüggemann H, Baumer S, Fricke WF, Wiezer A, Liesegang H, Decker I, Herzberg C, Martinez-Arias R, Merkl R, Henne A, Gottschalk G. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc Natl Acad Sci USA. 2003;100:1316–1321. doi: 10.1073/pnas.0335853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brüssow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brussow H, Desiere F. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol Microbiol. 2001;39:213–222. doi: 10.1046/j.1365-2958.2001.02228.x. [DOI] [PubMed] [Google Scholar]
- 17.Burrus V, Pavlovic G, Decaris B, Guedon G. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid. 2002;48:77–97. doi: 10.1016/s0147-619x(02)00102-6. [DOI] [PubMed] [Google Scholar]
- 18.Camacho AG, Gual A, Lurz R, Tavares P, JAlonso JC. Bacillus subtilis bacteriophage SPP1 DNA packaging motor requires terminase and portal proteins. J Biol Chem. 2003;278:23251–23259. doi: 10.1074/jbc.M301805200. [DOI] [PubMed] [Google Scholar]
- 19.Conard B, Cole ST. Lysogenic phages of Clostridium perfringens: mapping of the chromosomal attachment sites. FEMS Microbiol Lett. 1990;54:323–326. doi: 10.1111/j.1574-6968.1990.tb04020.x. [DOI] [PubMed] [Google Scholar]
- 20.Casjens SR. Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol. 2005;8:451–458. doi: 10.1016/j.mib.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Casjens SR, Gilcrease EB. Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol Biol. 2009;502:91–111. doi: 10.1007/978-1-60327-565-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Champion KM, Nishihara JC, Joly JC, Arnott D. Similarity of the Escherichia coli proteome upon completion of different bio-pharmaceutical fermentation processes. Proteomics. 2001;1:1133–1148. doi: 10.1002/1615-9861(200109)1:9<1133::AID-PROT1133>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 23.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JA. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 24.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eklund MW, Poysky FT, Reed SM, Smith CA. Bacteriophage and the toxigenicity of Clostridium botulinum type C. Science. 1971;172:480–482. doi: 10.1126/science.172.3982.480. [DOI] [PubMed] [Google Scholar]
- 26.Eklund MW, Poysky FT, Reed SM. Bacteriophage and the toxigenicity of Clostridium botulinum type D. Nat New Biol. 1972;235:16–17. doi: 10.1038/newbio235016a0. [DOI] [PubMed] [Google Scholar]
- 27.Fane BA, Prevelige PE., Jr Mechanism of scaffolding-assisted viral assembly. Adv Protein Chem. 2003;64:259–299. doi: 10.1016/s0065-3233(03)01007-6. [DOI] [PubMed] [Google Scholar]
- 28.Fischetti VA. Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol. 2005;13:491–496. doi: 10.1016/j.tim.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Fouts DE, Rasko DA, Cer RZ, Jiang L, Fedorova NB, Shvartsbeyn A, Vamathevan JJ, Tallon L, Althoff R, Arbogast TS, Fadrosh DW, Read TD, Gill SR. Sequencing Bacillus anthracis typing phages Gamma and Cherry reveals a common ancestry. J Bacteriol. 2006;188:3402–3408. doi: 10.1128/JB.188.9.3402-3408.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finhout E, Lee K. Comparison of automated in-gel digest methods for femtomole level samples. Electrophoresis. 2003;24:3508–3516. doi: 10.1002/elps.200305615. [DOI] [PubMed] [Google Scholar]
- 31.Garwes DJ, Pike BV, Wyld SG, Pocock DH, Gourlay RN. Characterization of Mycoplasmatales virus-laidlawii 3. J Gen Virol. 1975;29:11–24. doi: 10.1099/0022-1317-29-1-11. [DOI] [PubMed] [Google Scholar]
- 32.Goh S, Riley TV, Chang BJ. Isolation and characterization of temperate bacteriophages of Clostridium difficile. Appl Environ Microbiol. 2005;71:1079–1083. doi: 10.1128/AEM.71.2.1079-1083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goh S, Ong PF, Song KP, Riley TV, Chang BJ. The complete genome sequence of Clostridium difficile phage ΦC2 and comparisons to ΦCD119 and inducible prophages of CD630. Microbiology. 2007;153:676–685. doi: 10.1099/mic.0.2006/002436-0. [DOI] [PubMed] [Google Scholar]
- 34.Gorg A, Weiss W, Dunn M. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 2004;4:3665–3685. doi: 10.1002/pmic.200401031. [DOI] [PubMed] [Google Scholar]
- 35.Govind R, Fralick JA, Rolfe RD. Genomic organization and molecular characterization of Clostridium difficile bacteriophage ΦCD119. J Bacteriol. 2006;188:2568–2577. doi: 10.1128/JB.188.7.2568-2577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Govind R, Vediyappan G, Rolfe RD, Dupuy B, Fralick JA. Bacteriophage-mediated toxin gene regulation in Clostridium difficile. J Virol. 2009;83:12037–12045. doi: 10.1128/JVI.01256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grant RB, Riemann HP. Temperate phages of Clostridium perfringens type C1. Can J Microbiol. 1976;22:603–610. doi: 10.1139/m76-090. [DOI] [PubMed] [Google Scholar]
- 38.Helms C, Graham MY, Dutchik JE, Olson MV. A new method for purifying lambda DNA from phage lysates. DNA. 1985;4:39–49. doi: 10.1089/dna.1985.4.39. [DOI] [PubMed] [Google Scholar]
- 39.Hermoso JA, Monterroso B, Albert A, Galán B, Ahrazem O, García P, Martínez-Ripoll M, García JL, Menéndez M. Structural basis for selective recognition of pneumococcal cell wall by modular endolysin from phage Cp-1. Structure. 2003;11:1239–1249. doi: 10.1016/j.str.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Hirano S, Yonekura Y. The structure of Clostridium per-fringens bacteriophages. Acta Med Univ Kagoshima. 1967;9:41–56. [Google Scholar]
- 41.Kwan T, Liu J, DuBow M, Gros P, Pelletier J. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci USA. 2005;102:5174–5179. doi: 10.1073/pnas.0501140102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lahm HW, Langen H. Mass spectrometry: a tool for the identification of proteins separated by gels. Electrophoresis. 2000;21:2105–2114. doi: 10.1002/1522-2683(20000601)21:11<2105::AID-ELPS2105>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 43.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 44.Lewis JG, Chang G-J, Lanciotti RS, Trent DW. Direct sequencing of large flavivirus PCR products for analysis of genome variation and molecular epidemiological investigations. J Virol Meth. 1992;38:11–24. doi: 10.1016/0166-0934(92)90165-a. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Dehbi M, Moeck G, Arhin F, Bauda P, Bergeron D, Callejo M, Ferretti V, Ha N, Kwan T, McCarty J, Srikumar R, Williams D, Wu JJ, Gros P, Pelletier J, DuBow M. Antimicrobial drug discovery through bacteriophage genomics. Nat Biotechnol. 2004;22:185–191. doi: 10.1038/nbt932. [DOI] [PubMed] [Google Scholar]
- 46.Liu X, Kong S, Shi M, Fu L, Gao Y, An C. Genomic analysis of freshwater cyanophage Pf-WMP3 Infecting cyano-bacterium Phormidium foveolarum: the conserved elements for a phage. Microb Ecol. 2008;56:671–680. doi: 10.1007/s00248-008-9386-7. [DOI] [PubMed] [Google Scholar]
- 47.Loessner MJ. Bacteriophage endolysins—current state of research and applications. Curr Opin Microbiol. 2005;8:480–487. doi: 10.1016/j.mib.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 48.Lucchini S, Desiere F, Brüssow H. The structural gene module in Streptococcus thermophilus bacteriophage ΦSfi11 shows a hierarchy of relatedness to Siphoviridae from a wide range of bacterial hosts. Virology. 1998;246:63–73. doi: 10.1006/viro.1998.9190. [DOI] [PubMed] [Google Scholar]
- 49.Lukashin AV, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahony DE, Easterbrook KB. Intracellular development of a bacteriophage of Clostridium perfringens. Can J Microbiol. 1970;16:983–988. doi: 10.1139/m70-167. [DOI] [PubMed] [Google Scholar]
- 51.Mahony DE, Kalz GG. A temperate bacteriophage of Clostridium perfringens. Can J Microbiol. 1968;14:1085–1093. doi: 10.1139/m68-183. [DOI] [PubMed] [Google Scholar]
- 52.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Margulies M, Egholm M, Altman WE, Attiya S, JBader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayer MJ, Narbad A, Gasson MJ. Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J Bacteriol. 2008;190:6734–6740. doi: 10.1128/JB.00686-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCrea JF, Epstein RS, Barry WH. Use of potassium tartrate for equilibrium density-gradient centrifugation of animal viruses. Nature. 1961;189:220–221. doi: 10.1038/189220a0. [DOI] [PubMed] [Google Scholar]
- 56.Mead DA, Pey NK, Herrnstadt C, Marcil RA, Smith LM. A universal method for the direct cloning of PCR amplified nucleic acid. Biotechnology. 1991;9:657–662. doi: 10.1038/nbt0791-657. [DOI] [PubMed] [Google Scholar]
- 57.Merril CR, Scholl D, Adhya SL. The prospect for bacteriophage therapy in Western medicine. Nat Rev Drug Discov. 2003;2:489–497. doi: 10.1038/nrd1111. [DOI] [PubMed] [Google Scholar]
- 58.Misra V, Blumenthal RM, Babiuk LA. Proteins specified by bovine herpesvirus 1 (infectious bovine rhinotracheitis virus) J Virol. 1981;40:367–378. doi: 10.1128/jvi.40.2.367-378.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, Anson EL, Bolanos RA, Chou HH, Jordan CM, Halpern AL, Lonardi S, Beasley EM, Brandon RC, Chen L, Dunn PJ, Lai Z, Liang Y, Nusskern DR, Zhan M, Zhang Q, Zheng X, Rubin GM, Adams MD, Venter JC. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 60.Myers GS, Rasko DA, Cheung JK, Ravel J, Seshadri R, DeBoy RT, Ren Q, Varga J, Awad MM, Brinkac LM, Daugherty SC, Haft DH, Dodson RJ, Madupu R, Nelson WC, Rosovitz MJ, Sullivan SA, Khouri H, Dimitrov GI, Watkins KL, Mulligan SJ, Benton J, Radune D, Fisher DJ, Atkins HS, Hiscox T, Jost BH, Billington SJ, Songer JG, McClane BA, Titball RW, Rood JI, Melville SB, Paulsen IT. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 2006;16:1031–1040. doi: 10.1101/gr.5238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 62.Paquette G, Fredette V. Properties of four temperate bacteriophages active on Clostridium perfringens type A. Rev Can Biol. 1977;36:205–215. [PubMed] [Google Scholar]
- 63.Paredes CJ, Alsaker KV, Papoutsakis ET. A comparative genomic view of clostridial sporulation and physiology. Nat Rev Microbiol. 2005;3:969–978. doi: 10.1038/nrmicro1288. [DOI] [PubMed] [Google Scholar]
- 64.Paul JH, Sullivan MB, Segall AM, Rohwer F. Marine phage genomics. Comp Biochem Physiol B Biochem Mol Biol. 2002;133:463–476. doi: 10.1016/s1096-4959(02)00168-9. [DOI] [PubMed] [Google Scholar]
- 65.Payment P, Franco E. Clostridium perfringens and somatic coliphages as indicators of the efficiency of drinking water treatment for viruses and protozoan cysts. Appl Environ Microbiol. 1993;59:2418–2424. doi: 10.1128/aem.59.8.2418-2424.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker W, Kumar V, Kandasamy J, Keenan L, Bardarov S, Kriakov J, Lawrence JG, Jacobs WR, Jr, Hendrix RW, Hatfull GF. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–182. doi: 10.1016/s0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 67.Pommier S, Gavioli M, Cascales E, Lloubès R. Tol-dependent macromolecule import through the Escherichia coli cell envelope requires the presence of an exposed TolA binding motif. J Bacteriol. 2005;187:7526–7534. doi: 10.1128/JB.187.21.7526-7534.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sambrook J, Maniatis T, Fritsch EF. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 69.Sawires YS, Songer JG. Clostridium perfringens: insight into virulence evolution and population structure. Anaerobe. 2006;12:23–43. doi: 10.1016/j.anaerobe.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 70.Schäffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmidt HA, von Haeseler A. Maximum-likelihood analysis using TREE-PUZZLE. Curr Protoc Bioinformatics. 2007;Chapter 6(Unit 6.6) doi: 10.1002/0471250953.bi0606s17. [DOI] [PubMed] [Google Scholar]
- 72.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. The multidrug-resistant human pathogen Clos-tridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 73.Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc Natl Acad Sci USA. 2002;99:996–1001. doi: 10.1073/pnas.022493799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Siragusa GR, Danyluk MD, Hiett KL, Wise MG, Craven SE. Molecular subtyping of poultry-associated type A Clostridium perfringens isolates by repetitive-element PCR. J Clin Microbiol. 2006;44:1065–1073. doi: 10.1128/JCM.44.3.1065-1073.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smedley JG, 3rd, Fisher DJ, Sayeed S, Chakrabarti G, McClane BA. The enteric toxins of Clostridium perfringens. Rev Physiol Biochem Pharmacol. 2004;152:183–204. doi: 10.1007/s10254-004-0036-2. [DOI] [PubMed] [Google Scholar]
- 76.Smith HW. The bacteriophages of Clostridium perfringens. J Gen Microbiol. 1959;21:622–630. doi: 10.1099/00221287-21-3-622. [DOI] [PubMed] [Google Scholar]
- 77.Smith LM, Sanders JZ, Kaiser RJ, Hughs P, Dodd C, Connell CR, Heins C, Kent SBH, Hood LE. Fluorescent detection in automated DNA sequence analysis. Nature. 1986;321:673–681. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 78.Smith TJ, Blackman SA, Foster SJ. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology. 2000;146:249–262. doi: 10.1099/00221287-146-2-249. [DOI] [PubMed] [Google Scholar]
- 79.Stewart AW, Johnson MG. Increased numbers of heat-resistant spores produced by two strains of Clostridium perfringens bearing temperate phage s9. J Gen Microbiol. 1977;103:45–50. doi: 10.1099/00221287-103-1-45. [DOI] [PubMed] [Google Scholar]
- 80.Sturino JM, Klaenhammer TR. Bacteriophage defense systems and strategies for lactic acid bacteria. Adv Appl Microbiol. 2004;56:331–378. doi: 10.1016/S0065-2164(04)56011-2. [DOI] [PubMed] [Google Scholar]
- 81.Van Immerseel F, De Buck J, Pasmans F, Huyghebaert G, Haesebrouck F, Ducatelle R. Clostridium perfringens in poultry: an emerging threat for animal and public health. Avian Pathol. 2004;33:537–549. doi: 10.1080/03079450400013162. [DOI] [PubMed] [Google Scholar]
- 82.Vieu JF, Guélin A, Dauguet C. Morphology of the Welchia perfringens bacteriophage 80. Ann Inst Pasteur (Paris) 1965;109:157–160. [PubMed] [Google Scholar]
- 83.Webster R. The tol gene products and the import of macromolecules into E. coli. Mol Microbiol. 1991;5:1005–1011. doi: 10.1111/j.1365-2958.1991.tb01873.x. [DOI] [PubMed] [Google Scholar]
- 84.Wilgenbusch JC, Swofford D. Inferring evolutionary trees with PAUP*. Curr Protoc Bioinform. 2003;Chapter 6(Unit 6.4) doi: 10.1002/0471250953.bi0604s00. [DOI] [PubMed] [Google Scholar]
- 85.Wise MG, Siragusa GR. Quantitative detection of Clostridium perfringens in the broiler fowl gastrointestinal tract by real-time PCR. Appl Environ Microbiol. 2005;71:3911–3916. doi: 10.1128/AEM.71.7.3911-3916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yan WK. Use of host modified bacteriophages in development of a phage typing scheme for Clostridium perfringens. Med Lab Sci. 1989;46:186–193. [PubMed] [Google Scholar]
- 87.Young R. Bacteriophage holins: deadly diversity. J Mol Microbiol Biotechnol. 2002;4:21–36. [PubMed] [Google Scholar]
- 88.Zimmer M, Scherer S, Loessner MJ. Genomic analysis is Clostridium perfringes bacteriophage Φ3626, which integrates into guaA and possibly affects sporulation. J Bacteriol. 2002;184:4359–4368. doi: 10.1128/JB.184.16.4359-4368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zimmer M, Vukov N, Scherer S, Loessner MJ. The murein hydrolase of the bacteriophage Φ3626 dual lysis system is active against all tested Clostridium perfringes strains. Appl Environ Microbiol. 2002;68:5311–5317. doi: 10.1128/AEM.68.11.5311-5317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.