Abstract
Interaction of DNA methylation and sequence variants that are methylation quantitative trait loci (mQTLs) may influence susceptibility to diseases such as alcohol dependence (AD). We used genome-wide genotype data from 268 African Americans (AAs: 129 AD cases and 139 controls) and 143 European Americans (EAs: 129 AD cases and 14 controls) to identify mQTLs that were associated with promoter CpGs in 82 AD risk genes. 282 significant mQTL-CpG pairs (9.9×10-100≤Pnominal≤7.7×10-8) in AAs and 313 significant mQTL-CpG pairs (2.7×10-53≤Pnominal≤9.9×10-8) in EAs were identified [i.e., mQTL-CpG associations survived multiple-testing correction, q-values (false discovery rate) ≤ 0.05]. The most significant mQTL was rs1800759, which was strongly associated with CpG cg12011299 in both AAs (Pnominal=9.9×10-100; q=6.7×10-91) and EAs (Pnominal=2.7×10-53; q=1.4×10-44). Rs1800759 (previously known to be associated to AD) and CpG cg12011299 (distance: 37 bp) are both located in alcohol dehydrogenase (ADH) 4 gene (ADH4) promoter region. In general, the strength of association between mQTLs and CpGs was inversely correlated with the distance between them. Association was also influenced by race and AD. Additionally, 48.3% of the mQTLs identified in AAs and 65.6% of the mQTLs identified in EAs were predicted to be expression QTLs. Three mQTLs (rs2173201, rs4147542, and rs4147541 in ADH1B-AHD1C gene cluster region) found in AAs were previously identified by our genome-wide association studies as being significantly associated with AD in AAs. Thus, DNA methylation, which can be influenced by sequence variants and is implicated in gene expression regulation, appears to at least partially underlie the association of genetic variation with AD.
Keywords: methylation quantitative trait loci (mQTL), promoter CpGs, alcohol dependence risk genes, genome-wide association, functional annotation, alcohol dehydrogenase (ADH)
Introduction
DNA methylation usually occurs at CpG sites in somatic cells, and is non-randomly distributed across the genome. It is one of the major epigenetic mechanisms controlling gene expression. An inverse relationship is often observed between promoter DNA methylation levels and gene expression levels (Abdolmaleky et al. 2006; Baer et al. 2012; Egger et al. 2004; Etcheverry et al. 2010; Tekpli et al. 2013; Woloszynska-Read et al. 2007), although findings are inconsistent (Bell et al. 2011; Fraser et al. 2012; Lam et al. 2012). Altered DNA methylation in the promoter regions of genes may influence gene transcription and thus affect phenotypic variation or disease susceptibility.
To understand the mechanism by which DNA methylation regulates gene transcription, it is critical to understand the various biological influences on DNA methylation. Demographic factors, such as sex (Liu et al. 2010), age (Huang et al. 2013), and ethnicity (Nielsen et al. 2010; Zhang et al. 2011), covary with certain patterns of DNA methylation. Variation in DNA methylation has been linked to diverse environmental exposures, including pre- and post-natal nutrition (Lan et al. 2013; Soubry et al. 2013; Supic et al. 2013), early-life stress (Tyrka et al. 2012; Yang et al. 2013; Zhang et al. 2013b), psychosocial stress (de Rooij et al. 2012; Unternaehrer et al. 2012), and inflammation (Lam et al. 2012). Genetic variation can also impact DNA methylation patterns. DNA sequence variants that are associated with DNA methylation patterns are referred to as methylation quantitative trait loci (mQTLs). mQTLs are widely dispersed throughout the genome across different tissues (Bell et al. 2011; Gibbs et al. 2010; Kerkel et al. 2008; Schalkwyk et al. 2010; Zhang et al. 2010). The study by Quon et al.(Quon et al. 2013) demonstrated that methylation patterns of 3-4% of CpG sites in the genome were heritable. Given the close relationship between sequence variants and DNA methylation patterns, the heritability of DNA methylation is potentially mediated by genetic variation.
DNA methylation is important for normal development and is associated with key processes such as genomic imprinting and X-chromosome inactivation. Significant associations between aberrant DNA methylation and susceptibility to numerous diseases have been reported. For example, we compared the DNA methylation levels of 384 CpGs in the promoter regions of 82 candidate genes of subjects with alcohol dependence (AD) and healthy controls, and identified several promoter CpGs that were differentially methylated in AD subjects (Zhang et al. 2013a). Nevertheless, it is unknown whether DNA methylation alterations in AD subjects are due to DNA sequence variants (potentially enriched in the genome of AD subjects) or chronic alcohol consumption (an environmental factor). Alcohol consumption can alter DNA methylation status (Chen et al. 2013). Alternatively or additionally, considering the widespread presence of mQTLs in the genome, aberrant DNA methylation patterns may exist in individuals at risk for AD due to sequence variants (or mQTLs) that can moderate gene transcription and thus confer vulnerability to AD.
The present study aimed to identify mQTLs that regulate the methylation patterns of CpGs in the promoter regions of 82 candidate genes potentially involved in AD-relevant biological pathways. Selection of genes for study based on a priori hypothesis (i.e., there is a potential role of these genes in AD development) should be more appropriate than random selection of genes in the genome, as suggested by previous studies (Zhao et al. 2013) and with the availability of well-established alcohol-related gene database (Guo et al. 2009). In this study, we examined the association between genotypes of genome-wide single nucleotide polymorphisms (SNPs) and methylation levels (as quantitative traits) of 384 CpGs located in the promoter regions of 82 AD candidate genes (Supplementary Table S1), resulting in the identification of both cis- and trans-mQTLs for the 384 promoter CpGs.
Materials and methods
Human subjects
We included 411 subjects [268 African Americans (AAs) and 143 European Americans (EAs)] in this study. They were recruited for DNA methylation (Zhang et al. 2013a) and genome-wide association (Gelernter et al. 2014) studies of AD. The subjects were interviewed using an electronic version of the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA) (Pierucci-Lagha et al. 2005), and life-time diagnoses for AD were made according to the criteria of the Diagnostic and Statistical Manual of Mental Disorders, 4th edition (American Psychiatric Association 1994). Control subjects were not affected with alcohol or drug abuse or dependence. Both AD and control subjects were free of major psychotic disorders such as schizophrenia and bipolar disorders. The ancestry proportions of the 411 subjects were estimated with the program STRUCTURE (Pritchard et al. 2000) using 36 short tandem repeat markers and five SNPs (Xie et al. 2009). Among 268 AAs, 129 (48.1%) were AD cases and 139 (51.9%) were healthy controls, 97 (36.2%) were males, and the average AA ancestry proportion was 0.94 ± 0.08. Among 143 EAs, 129 (90.2%) were AD cases and 14 (9.8%) were healthy controls, 73 subjects (51.5%) were males, and the average EA ancestry proportion was 0.97 ± 0.07. No age difference was observed between AAs (39 ± 11) and EAs (41 ± 13) (P > 0.05).
DNA methylation data
Methylation data for 384 CpGs in the promoter regions [from 2,000 bp upstream to 1,000 bp downstream of the transcription start sites (TSS)] of 82 candidate genes (Supplementary Table S1) for the 411 subjects were generated using Illumina's GoldenGate DNA methylation assays (Zhang et al. 2013a). The 82 candidate genes are involved in several brain neurotransmission systems (dopaminergic, opioidergic, serotonergic, GABAergic/glutamatergic, cholinergic, and cannabinoidergic), alcohol metabolism, DNA methylation, or signal transduction related to alcohol reward and reinforcement (Hodgkinson et al. 2008; Zhang et al. 2013a). One to 12 CpGs in each of the 82 candidate genes were included in the custom methylation profiling panel. The background normalization algorithm minimized background variation within the array by using built-in negative control probe signals. Raw DNA methylation data were further processed with the R package sva (Johnson et al. 2007) to remove batch effects. The processed DNA methylation data were used for the mQTL (or SNP genotype-CpG methylation association) analysis.
Genome-wide SNP genotype data
The 411 subjects were among 5,697 subjects (3,318 AAs and 2,379 EAs) included in our recent AD genome-wide association study (GWAS) (Gelernter et al. 2014). All subjects were genotyped on the Illumina HumanOmni1-Quad v1.0 microarray containing 988,306 autosomal SNPs. Genotypes of SNPs were called using the GenomeStudio software V2011.1 and genotyping module V1.8.4 (Illumina, San Diego, CA, USA). The extent of identity-by-descent (IBD) sharing among the 411 subjects was estimated by PLINK (Purcell et al. 2007), and all subjects passed the sample quality examination. SNPs were excluded if the: 1) minor allele frequency (MAF) was ≤ 0.05; 2) missing rate per SNP was > 10%; or 3) P values of Hardy-Weinberg disequilibrium tests were ≤ 0.001. After data cleaning and quality control, 784,514 SNPs for 268 AAs and 699,437 SNPs for 143 EAs remained for the mQTL analysis.
Statistical and bioinformatics analyses
All statistical analyses were implemented using the open-source program R 3.0.1 (http://www.r-project.org/). To identify mQTLs for the 384 CpGs in the promoter regions of 82 candidate genes, we analyzed the association of genotypes (in an additive model) of genome-wide SNPs and methylation levels of each of the 384 promoter CpGs using PLINK (Purcell et al. 2007) in 268 AAs and 143 EAs. Multiple linear regression analysis was used to predict the effect of SNPs on the methylation of CpGs, with adjustment for sex, age, ancestry proportion, and AD status. Nominal P values (Pnominal) were obtained for > 260 million SNP-CpG pairs [in 268 AAs: 784,514 (SNPs) × 384 (CpGs) pairs; in 143 EAs: 699,437 (SNPs) × 384 (CpGs) pairs]. To study the influence of AD on mQTL-CpG associations, multiple linear regression analysis was used to validate significant mQTL-CpG pairs (identified in AAs or EAs) in AD cases and healthy controls separately, with adjustment for sex, age, and ancestry proportion. All regression models were summarized in Supplementary Table S2. Qvalue (Storey 2002) was used to account for multiple testing by controlling the false discovery rate (FDR) at 0.05. mQTL (or SNP)-CpG pairs were considered to be significant if the q value ≤ 0.05. Based on the distance between mQTLs and CpGs, mQTLs were further classified as cis-mQTLs (located within 1 Mb from a specific CpG site) or trans-mQTLs (located beyond 1Mb from a specific CpG site or on different chromosomes). Regional association results from genome-wide association scans were plotted using LocusZoom (Pruim et al. 2010).
To explore the function of mQTLs (or SNPs), the web-based functional annotation tool SNPnexus (Dayem Ullah et al. 2012) was queried to determine whether the identified mQTLs were located in coding or non-coding regions, transcription factor binding sites (TFBS), microRNA binding sites, or vertebrate conserved regions, and whether they were previously known to be associated with phenotypes or diseases. We also examined whether the identified mQTLs were among AD-associated SNPs obtained in our recent GWAS (Gelernter et al. 2014). The identified mQTLs were further analyzed to see whether they were expression quantitative trait loci (eQTLs) by querying the gene expression datasets (generated from the lymphoblastoid cell lines derived from peripheral blood lymphocytes of HapMap YRI and CEU subjects) in the GTEx eQTL database (GTExConsortium 2013) using the web server SCAN (Nicolae et al. 2010). The influence of the distance between SNPs and CpGs, race, and AD on the strength of the mQTL (or SNP)-CpG association was also investigated.
Results
Significant mQTL-CpG pairs identified in African Americans (AAs)
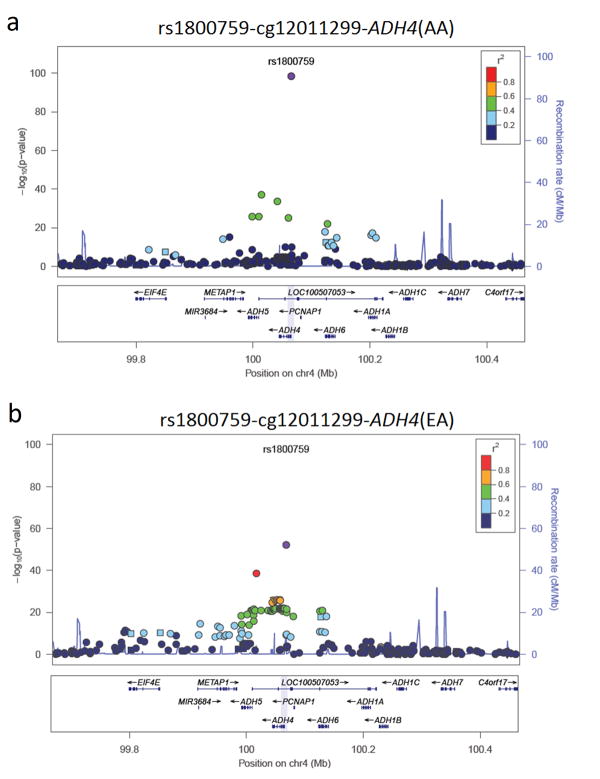
In AAs, we identified 282 significant mQTL-CpG pairs with P values survived multiple-testing correction when false discovery rate (FDR) q value was controlled at the 0.05 level (9.9×10-100 ≤ Pnominal ≤ 7.7×10-8 or 6.7×10-91 ≤ q ≤ 0.05) (Supplementary Table S3a). These 282 mQTL-CpG pairs (including 202 cis-mQTL-CpG and 80 trans-mQTL-CpG pairs) included 238 unique SNPs (mapped to 99 genes, including 20 of the 82 candidate genes) and 65 unique promoter CpGs (mapped to 44 of the 82 candidate genes). The top 10 cis-mQTL-CpG and top 10 trans-mQTL-CpG pairs identified in AAs are listed in Table 1. The most significant cis-mQTL was rs1800759, which was strongly associated with CpG cg12011299 (Pnominal = 9.9×10-100; q = 6.7×10-91). Both rs1800759 and CpG cg12011299 are located in the promoter region of the alcohol dehydrogenase 4 gene (ADH4), 37 bp apart. A regional plot by LocusZoom displays the association of SNPs (400 kb up or downstream of rs1800759) and cg12011299 as well as the linkage disequilibrium (LD) between rs1800759 and the surrounding SNPs in AAs (Fig. 1a). Six additional cis-mQTLs (rs1154415, rs2851301, rs2602836, rs3828541, rs13148577, and rs4147547) located in the ADH5-ADH4-ADH6 gene cluster region (about 128 kb) were also strongly associated with cg12011299 (1.7×10-37 ≤ Pnominal ≤ 8.0×10-23 or 6.7×10-30 ≤ q ≤ 2.2×10-15) (Table 1). The two top trans-mQTLs identified in AAs were rs3097793 and rs173567. Both of them are located in the intergenic region between CCDC69 (coding for coiled-coil domain containing 69) and GM2A (coding for GM2 ganglioside activator), and strongly associated with CpG cg14150516 in the promoter region of CREB1 (coding for cAMP responsive element binding protein 1) (rs3097793: Pnominal = 3.1×10-43, q = 6.5×10-35; rs173567: Pnominal = 3.8×10-32, q = 8.5×10-25) (Table 1).
Table 1. Top 10 cis- and top 10 trans-mQTLs identified in 268 African Americans (AAs).
| SNPs | CpGs | SNP-CpG linear regression analysis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||
| SNPs | Chr.a | Coordinatesa | Genes | SNP Type | CpGs | Chr.a | Coordinatesa | Genes | Allele | Betab | Pnominalc | qd |
| Top 10 cis-mQTLs in AAs: | ||||||||||||
| rs1800759 | 4 | 100065509 | ADH4 | Promoter | cg12011299 | 4 | 100065546 | ADH4 | C | 0.12 | 9.9E-100 | 6.7E-91 |
| rs8028779 | 15 | 27034731 | GABRB3 | promoter | cg07763397 | 15 | 27020865 | GABRB3 | G | 0.09 | 4.6E-38 | 4.5E-30 |
| rs2602836 | 4 | 100014805 | ADH5 | promoter | cg12011299 | 4 | 100065546 | ADH4 | G | 0.08 | 1.7E-37 | 6.7E-30 |
| rs3828541 | 4 | 100042262 | ADH4 | Downstream | cg12011299 | 4 | 100065546 | ADH4 | G | 0.08 | 2.6E-34 | 1.8E-26 |
| rs4486851 | 15 | 27022876 | GABRB3 | promoter | cg07763397 | 15 | 27020865 | GABRB3 | A | 0.07 | 9.5E-30 | 3.8E-22 |
| rs732821 | 13 | 47472879 | HTR2A | promoter | cg06020661 | 13 | 47472138 | HTR2A | A | 0.08 | 8.7E-30 | 5.7E-22 |
| rs1154415 | 4 | 99999033 | ADH5 | Intron | cg12011299 | 4 | 100065546 | ADH4 | G | 0.12 | 1.2E-26 | 4.3E-19 |
| rs2851301 | 4 | 100010127 | ADH5 | promoter | cg12011299 | 4 | 100065546 | ADH4 | A | 0.12 | 1.2E-26 | 4.3E-19 |
| rs13148577 | 4 | 100060836 | ADH4 | Intron | cg12011299 | 4 | 100065546 | ADH4 | A | 0.11 | 7.6E-26 | 2.6E-18 |
| rs4147547 | 4 | 100127311 | ADH6 | Intron | cg12011299 | 4 | 100065546 | ADH4 | A | 0.11 | 8.0E-23 | 2.2E-15 |
| Top 10 trans-mQTLs in AAs: | ||||||||||||
| rs3097793 | 5 | 150617370 | CCDC69 - GM2A | intergenic | cg14150516 | 2 | 208394784 | CREB1 | A | 0.04 | 3.1E-43 | 6.5E-35 |
| rs173567 | 5 | 150618829 | CCDC69 - GM2A | intergenic | cg14150516 | 2 | 208394784 | CREB1 | G | 0.04 | 3.8E-32 | 8.5E-25 |
| rs628825 | 12 | 110436233 | ATXN2 | Intron | cg10959984 | 12 | 112204380 | ALDH2 | G | 0.00 | 3.2E-22 | 4.3E-15 |
| rs11552148 | 16 | 48385957 | LONP2 | 3′ UTR | cg25195998 | 11 | 113345607 | DRD2 | G | -0.01 | 9.9E-22 | 1.8E-14 |
| rs8055764 | 16 | 48384370 | LONP2 | Intron | cg25195998 | 11 | 113345607 | DRD2 | A | -0.01 | 6.6E-21 | 7.1E-14 |
| rs11866529 | 16 | 48247523 | ABCC11 | Intron | cg25195998 | 11 | 113345607 | DRD2 | C | -0.01 | 5.5E-21 | 1.1E-13 |
| rs8054314 | 16 | 48394330 | SIAH1 | Downstream | cg25195998 | 11 | 113345607 | DRD2 | A | -0.01 | 7.3E-21 | 1.1E-13 |
| rs8190 | 16 | 48394626 | SIAH1 | 3′ UTR | cg25195998 | 11 | 113345607 | DRD2 | A | -0.01 | 7.3E-21 | 1.1E-13 |
| rs153462 | 5 | 150605070 | CCDC69 | promoter | cg14150516 | 2 | 208394784 | CREB1 | G | 0.03 | 7.6E-20 | 5.9E-13 |
| rs9926192 | 16 | 48306394 | LONP2 | Intron | cg25195998 | 11 | 113345607 | DRD2 | A | -0.01 | 4.2E-19 | 4.2E-12 |
Chromosomes and coordinates of SNPs and CpGs in GRCh37/hg19.
Regression coefficients by multiple linear regression analysis with adjustment for sex, age, ancestry proportion, and the status of alcohol dependence (AD).
Observed P values by multiple linear regression analysis with adjustment for sex, age, ancestry proportion, and the status of alcohol dependence (AD).
Adjusted P values by R package qvalue, controlling the False Discovery Rate (FDR) at 0.05.
Fig. 1.
Regional plot of associations between SNPs around the top mQTL rs1800759 and CpG cg12011299.
a. Regional plot of associations between SNPs around the top mQTL rs1800759 and CpG cg12011299 in African Americans (AAs). b. Regional plot of associations between SNPs around the top mQTL rs1800759 and CpG cg12011299 in European Americans (EAs). The graph was made by LocusZoom using the 1000 Genomes data (hg19). The left Y-axis indicates -log10(P values) of mQTL-CpG associations, the right Y-axis indicates recombination rates, and the X-axis shows the physical coordinates of genes and SNPs on chromosome 4. The color of dots indicates the strength of correlation between rs1800759 and nearby SNPs. Gene annotation is displayed at the bottom of the figure, and the light blue shadow refers to the location of cg12011299.
Significant mQTL-CpG pairs identified in European Americans (EAs)
In EAs, we identified 313 significant mQTL-CpG pairs with P values that survived correction for multiple testing (2.7×10-53 ≤ Pnominal ≤ 9.9×10-8 or 1.4×10-44 ≤ q ≤ 0.05) (Supplementary Table S3b). These 313 mQTL-CpG pairs (including 223 cis-mQTL-CpG and 90 trans-mQTL-CpG pairs) included 305 unique SNPs (mapped to 73 genes, including 10 of the 82 candidate genes) and 44 unique promoter CpGs (mapped to 30 of the 82 candidate genes). The top 10 cis-mQTL-CpG and top 10 trans-mQTL-CpG pairs identified in EAs are listed in Table 2. As in AAs, the most significant cis-mQTL identified in EAs was rs1800759, which was also strongly associated with CpG cg12011299 (Pnominal = 2.7×10-53; q = 1.4×10-44). A regional plot by LocusZoom shows the association of SNPs (400 kb upstream or downstream of rs1800759) and cg12011299 as well as the LD between rs1800759 and the surrounding SNPs in EAs (Fig. 1b). Eight additional cis-mQTLs (rs2602836, rs1126670, rs1126671, rs6837685, rs1126673, rs4699710, rs6532798, and rs10017466) located in ADH4 or ADH5 were also strongly associated with cg12011299 (1.3×10-39 ≤ Pnominal ≤ 3.0×10-26 or 3.4×10-31 ≤ q ≤ 4.8×10-19) (Table 2). The two top trans-mQTLs identified in EAs were the same as those identified in AAs, i.e., rs3097793 and rs173567 (two intergenic SNPs between CCDC69 and GM2A). Both of these were strongly associated with CpG cg14150516 in the promoter region of CREB1 (rs3097793: Pnominal = 1.2×10-33, q = 2.0×10-25; rs173567: Pnominal = 4.5×10-33, q = 5.8×10-25) (Table 2).
Table 2. Top 10 cis- and top 10 trans-mQTLs identified in 143 European Americans (EAs).
| SNPs | CpGs | SNP-CpG linear regression analysis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||
| SNPs | Chr.a | Coordinatesa | Genes | SNP Type | CpGs | Chr.a | Coordinatesa | Genes | Allele | Betab | Pnominalc | qd |
| Top 10 cis-mQTLs in EAs: | ||||||||||||
| rs1800759 | 4 | 100065509 | ADH4 | Promoter | cg12011299 | 4 | 100065546 | ADH4 | A | -0.15 | 2.7E-53 | 1.4E-44 |
| rs2602836 | 4 | 100014805 | ADH5 | Promoter | cg12011299 | 4 | 100065546 | ADH4 | A | -0.14 | 1.3E-39 | 3.4E-31 |
| rs732821 | 13 | 47472879 | HTR2A | Promoter | cg06020661 | 13 | 47472138 | HTR2A | G | -0.07 | 2.9E-27 | 2.2E-19 |
| rs1126670 | 4 | 100052733 | ADH4 | Exon | cg12011299 | 4 | 100065546 | ADH4 | C | -0.13 | 1.7E-26 | 2.7E-19 |
| rs1126671 | 4 | 100048414 | ADH4 | Exon | cg12011299 | 4 | 100065546 | ADH4 | A | -0.13 | 1.7E-26 | 2.7E-19 |
| rs6837685 | 4 | 100049145 | ADH4 | Intron | cg12011299 | 4 | 100065546 | ADH4 | G | -0.13 | 1.7E-26 | 2.7E-19 |
| rs1126673 | 4 | 100045616 | ADH4 | Exon | cg12011299 | 4 | 100065546 | ADH4 | G | -0.13 | 1.9E-26 | 3.3E-19 |
| rs4699710 | 4 | 100051819 | ADH4 | Intron | cg12011299 | 4 | 100065546 | ADH4 | G | -0.13 | 1.9E-26 | 3.3E-19 |
| rs6532798 | 4 | 100054827 | ADH4 | Intron | cg12011299 | 4 | 100065546 | ADH4 | G | -0.13 | 1.9E-26 | 3.3E-19 |
| rs10017466 | 4 | 100055800 | ADH4 | Intron | cg12011299 | 4 | 100065546 | ADH4 | G | -0.13 | 3.0E-26 | 4.8E-19 |
| Top 10 trans-mQTLs in EAs: | ||||||||||||
| rs3097793 | 5 | 150617370 | CCDC69 - GM2A | intergenic | cg14150516 | 2 | 208394784 | CREB1 | A | 0.04 | 1.2E-33 | 2.0E-25 |
| rs173567 | 5 | 150618829 | CCDC69 - GM2A | intergenic | cg14150516 | 2 | 208394784 | CREB1 | G | 0.04 | 4.5E-33 | 5.8E-25 |
| rs6536733 | 4 | 141069646 | MAML3 | Intron | cg23463131 | 16 | 10276410 | GRIN2A | G | 0.02 | 1.7E-29 | 1.5E-21 |
| rs17005603 | 4 | 141083754 | MAML3 | Promoter | cg23463131 | 16 | 10276410 | GRIN2A | G | 0.02 | 8.2E-26 | 3.5E-18 |
| rs153462 | 5 | 150605070 | CCDC69 | Promoter | cg14150516 | 2 | 208394784 | CREB1 | G | 0.03 | 6.5E-18 | 3.7E-11 |
| rs7917960 | 10 | 113155506 | ADRA2A - GPAM | intergenic | cg15408490 | 12 | 112204591 | ALDH2 | A | -0.05 | 8.7E-17 | 8.2E-10 |
| rs9420082 | 10 | 113166617 | ADRA2A - GPAM | intergenic | cg15408490 | 12 | 112204591 | ALDH2 | G | -0.05 | 2.7E-16 | 1.8E-09 |
| rs10885154 | 10 | 113151980 | ADRA2A - GPAM | intergenic | cg15408490 | 12 | 112204591 | ALDH2 | A | -0.05 | 2.6E-15 | 2.0E-08 |
| rs4372376 | 10 | 113231124 | ADRA2A - GPAM | intergenic | cg15408490 | 12 | 112204591 | ALDH2 | A | -0.05 | 1.5E-14 | 9.8E-08 |
| rs7896658 | 10 | 113176902 | ADRA2A - GPAM | intergenic | cg15408490 | 12 | 112204591 | ALDH2 | G | -0.05 | 2.0E-14 | 1.3E-07 |
Chromosomes and coordinates of SNPs and CpGs in GRCh37/hg19.
Regression coefficients by multiple linear regression analysis with adjustment for sex, age, ancestry proportion, and the status of alcohol dependence (AD).
Observed P values by multiple linear regression analysis with adjustment for sex, age, ancestry proportion, and the status of alcohol dependence (AD).
Adjusted P values by R package qvalue, controlling the False Discovery Rate (FDR) at 0.05.
Influence of race on mQTL-CpG association
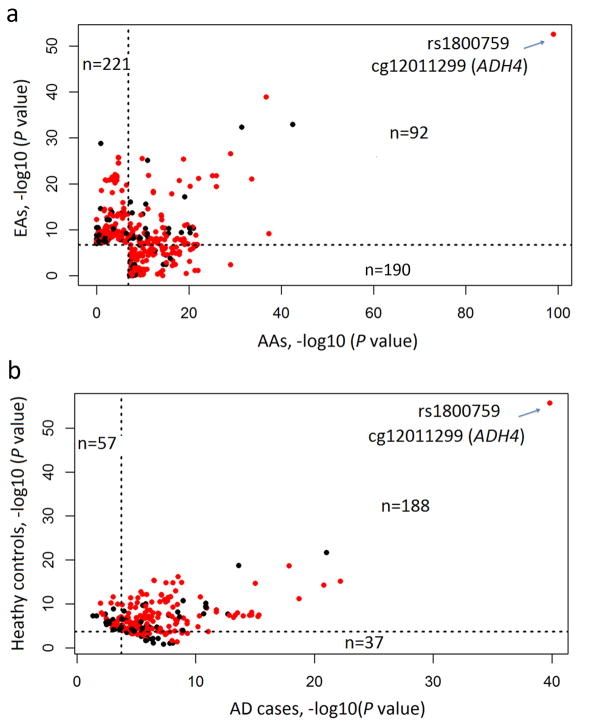
We searched for significant mQTL-CpG pairs (q ≤ 0.05) that were common to both AAs and EAs. As shown in Fig. 2a, 92 significant mQTL-CpG pairs (i.e., 32.6% of the 282 significant mQTL-CpG pairs identified in AAs or 29.4% of the 313 significant mQTL-CpG pairs identified in EAs) were shared between AAs and EAs. These 92 common mQTL-CpG pairs (including 57 cis-mQTL-CpG and 35 trans-mQTL-CpG pairs) were comprised of 89 unique SNPs (i.e., 37.4% of the 238 unique SNPs involved in the 282 mQTL-CpG pairs in AAs or 29.2% of the 305 unique SNPs involved in the 313 mQTL-CpG pairs in EAs) and 16 unique CpGs. However, 190 (or 67.4%) of the 282 significant mQTL-CpG pairs (including 145 cis-mQTL-CpG and 45 trans-mQTL-CpG pairs) identified in AAs were specific for AAs, and 221 (or 70.6%) of the 313 significant mQTL-CpG pairs (including 166 cis-mQTL-CpG and 55 trans-mQTL-CpG pairs) identified in EAs were specific for EAs. The most significant cis-mQTL-CpG pair, consisting of rs1800759 and CpG cg12011299 (both located in the promoter region of ADH4), was identified in both populations.
Fig. 2.
The effect of race and AD on mQTL-CpG association.
a. The effect of race on mQTL-CpG association in African Americans (AAs) and European Americans (EAs). The X axis represents the P values [converted to sign(linear regression beta value)×(-log10P value)] of 282 (92 + 190) significant mQTL-CpG pairs in AAs (q ≤ 0.05), and the Y axis represents the P values [converted to sign(linear regression beta value)×(-log10P value)] of 313 (92 + 221) significant mQTL-CpG pairs in EAs (q ≤ 0.05). The dashed line indicates the threshold of significance (q ≤0.05) (vertical, for AAs; horizontal, for EAs). The color of the dots indicates the type of mQTLs (red, cis-mQTLs; black, trans-mQTLs). b. The effect of alcohol dependence (AD) on mQTL-CpG association in African Americans (AAs). The X axis represents the P values [converted to sign(linear regression beta value)×(-log10P value)] of 225 (188 + 37) significant mQTL-CpG pairs identified in AD subjects (q ≤ 0.05), and the Y axis represents the P values [converted to sign(linear regression beta value)×(-log10P value)] of 245 (188 + 57) significant mQTL-CpG pairs identified in control subjects (q ≤ 0.05). The dashed line indicates the threshold of significance (0.05/282 = 1.8×10-4) (vertical, for AD subjects; horizontal, for control subjects).
Influence of AD on mQTL-CpG association
Because chronic drinking or other features associated with AD may influence CpG methylation status and thus alter the association of mQTLs (or SNPs) and CpGs, we further examined the validity of the 282 significant mQTL-CpG pairs (identified in all AA subjects) in 129 AA cases and 139 AA controls separately. After correcting for multiple testing (i.e., the P value for significance was set at 0.05/282 = 1.8×10-4), 188 (or 66.7%) of the 282 mQTL-CpG pairs were shared between AD and control subjects (Fig. 2b). These 188 common mQTL-CpG pairs (including 144 cis-mQTL-CpG and 44 trans-mQTL-CpG pairs) included 154 unique SNPs (64.7% of the 238 unique SNPs involved in the 282 mQTL-CpG pairs in AAs) and 36 unique CpGs. However, 37 (or 13.1%) of the 282 mQTL-CpG pairs (including 22 cis-mQTL-CpG and 15 trans-mQTL-CpG pairs) were observed only in AD subjects, and 57 (or 20.2%) of the 282 mQTL-CpG pairs (including 36 cis-mQTL-CpG and 21 trans-mQTL-CpG pairs) only in control subjects. The influence of AD on the mQTL-CpG association was not analyzed in EAs because the size of the EA control sample was small (n = 14).
Association of identified mQTLs and AD
We next evaluated whether there was overlap between the mQTLs identified as associated with CpGs and SNPs associated with AD in our recent GWAS (Gelernter et al. 2014). Of the 238 mQTLs identified in AAs, 21 (or 8.8%) were associated with AD in AAs at the level of overall meta P ≤ 0.05. Permutation tests showed that the 21 mQTLs were over-represented by AD-associated SNPs identified in our recent GWAS (Gelernter et al. 2014) (Ppermutaion = 0.041, 1 million times). The P values of three mQTLs identified in AAs reached genome-wide significance [rs2173201 (an intergenic SNP between ADH1B and ADH1C): overall meta P = 2.4 ×10-9; rs4147542 (in ADH1C intron 2): overall meta P = 9.1 ×10-9; and rs4147541 (in AHD1C promoter region): overall meta Pmeta = 1.1 ×10-8] (Supplementary Table S4a). Of the 305 mQTLs identified in EAs, 58 (19.0%) were associated with AD at the level of overall meta P ≤ 0.05. Permutation tests showed that the 58 mQTLs were over-represented by AD-associated SNPs identified in our recent GWAS (Gelernter et al. 2014) (Ppermutaion<1.0 ×10-6, 1 million iterations); however, none of the 58 mQTLs was associated with AD at the genome-wide level. The most significant mQTL associated with AD in EAs was rs1800759 (Pmeta = 0.008), which is located in the promoter region of ADH4 (Supplementary Table S4b).
Prediction of the function of identified mQTLs
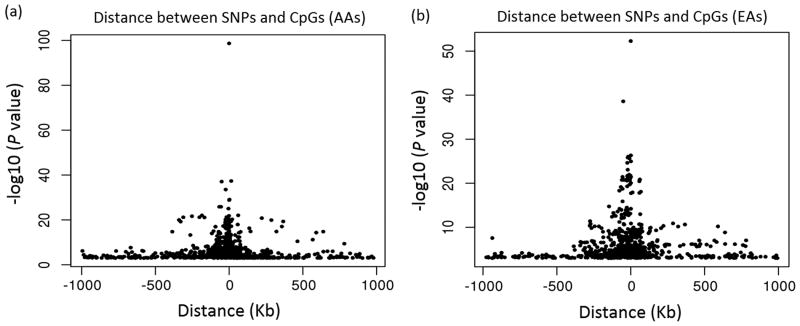
First, the correlation of cis-mQTL-CpG physical distance and cis-mQTL-CpG association strength was examined. As shown in Fig. 3, the distance between cis-mQTLs and CpGs was highly correlated with the magnitude of association between cis-mQTLs and CpGs, i.e., the shorter the distance from cis-mQTLs to CpGs, the stronger the association between cis-mQTLs and CpGs. This result was obtained in both AAs (Fig. 3a) and EAs (Fig. 3b). The distance between rs1800759 and CpG cg12011299 (both in the promoter region of ADH4), which formed the most significant SNP-CpG pair in both AAs and EA, was only 37 bp. Second, the function of the identified mQTLs was predicted using the online server SNPnexus (Dayem Ullah et al. 2012). The numbers of cis- and trans-mQTLs that were predicted to impact protein function, regulate gene transcription, or influence any disease phenotypes (including alcohol dependence or abuse) are summarized in Table 3. Third, the functional role of the identified mQTLs in regulating gene expression was predicted by querying the SCAN eQTL database. As shown in Table 3, 76 (46.9%) of the 202 cis-mQTLs and 39 (51.3%) of the 80 trans-mQTLs identified in AAs as well as 151 (70.2%) cis-mQTLs of the 223 cis-mQTLs and 49 (54.4%) of the 90 trans-mQTLs identified in EAs were predicted to be expression quantitative trait loci (eQTLs).
Fig. 3.
The correlation of SNP-CpG physical distance (bp) and SNP-CpG association strength in (a) African Americans (AA) and (b) European Americans (EA). X-axis, the distance (bp) between SNPs and CpGs; Y-axis, the strength of SNP-CpG association [P values were converted to -log10(P value)]. SNP-CpG pairs (represented as black dots) with P nominal ≤ 0.001 are shown in the figure. The most significant mQTL (or SNP)-CpG association was derived from rs1800759 and cg12011299 (both located in the promoter region of ADH4, with a distance between them of only 37 bp), and this association was observed in both AAs and EAs.
Table 3. Prediction of the function of mQTLs by querying the SNPnexus and SCAN databases.
| African Americans (AAs) | European Americans (EAs) | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Cis-mQTLs | Trans-mQTLs | All | Cis-mQTLs | Trans-mQTLs | All | |
| No. of mQTL-CpG pairs (q ≤ 0.05) | 202 | 80 | 282 | 223 | 90 | 313 |
| No. of unique mQTLs | 162 | 76 | 238 | 215 | 90 | 305 |
| No. of unique CpGs | 33 | 36 | 69 | 18 | 29 | 47 |
| No. of mQTLs in coding regions | 3e | 2f | 5 | 5g | 2h | 7 |
| Protein function prediction by Sift | 3 tolerated | 2 tolerated | 4 tolerated 1 damaging(rs1126671) | 2 tolerated | ||
| No. of mQTLs in conserved TFBSa | 2 | 1 | 3 | 4 | 0 | 4 |
| No. of mQTLs in CpG Islands | 8 | 1 | 9 | 10 | 0 | 10 |
| No. of mQTLs in conserved regions by PHASTb | 11 | 5 | 16 | 12 | 6 | 18 |
| No. of mQTLs in conserved regions by GERP++c | 17 | 8 | 25 | 18 | 8 | 26 |
| No. of mQTLs associated with any disease phenotypes | 118 | 26 | 144 | 150 | 31 | 181 |
| No. of mQTLs associated with alcohol dependence/abuse | 15 | 0 | 15 | 50 | 0 | 50 |
| No. of eQTLsd | 76 (46.9%) | 39 (51.3%) | 115 (48.3%) | 151 (70.2%) | 49 (54.4%) | 200 (65.6%) |
Number of mQTLs located in conserved transcription factor binding sites (TFBS).
Number of mQTLs located in conserved genomic regions identified by the Vertebrate Alignment and Conservation (PHAST) method.
Number of mQTLs located in conserved genomic regions identified by the Genomic Evolutionary Rate Profiling (GERP++) method.
Number of mQTLs that were also expression quantitative trait loci (eQTLs) by querying the SCAN eQTL database.
Three coding cis-mQTLs (rs2246614 in CDHR5, rs1131665 in IRF7, and rs695871 in ATXN2) identified in AAs.
Two coding trans-mQTLs (rs16945988 and rs11863236, both located in ABCC11) identified in AAs.
Five coding cis-mQTLs (rs1126671 and rs1126673 in ADH4, rs2246614 in CDHR5, rs1131665 in IRF7, and rs3752630 in TMEM116) identified in EAs.
Two coding trans-mQTLs (rs16945988 and rs11863236, both located in ABCC11) identified in EAs.
Discussion
The present study demonstrated that mQTL-CpG distance, race, and AD status could influence the association of mQTLs and CpGs. We also found that 48.3% of the mQTLs identified in AAs and 65.6% of the mQTLs identified in EAs were predicted to be eQTLs. These results help in interpreting our AD GWAS findings, providing a mechanism for the AD risk contribution of certain noncoding variants, i.e., their effect on disease susceptibility may be through altering promoter DNA methylation levels.
Our findings suggest that sequence variants (particularly non-coding variants) may influence AD susceptibility via DNA methylation. As summarized in Table 3, only a small number of the identified mQTLs lead to amino acid changes or potentially participate in gene expression regulation by altering transcription factor binding sites or conserved genomic sequences. A majority of the identified mQTLs may affect AD risk by influencing DNA methylation patterns. Of the 12 coding mQTLs or non-synonymous SNPs (five identified in AAs and seven identified in EAs), only rs1126671 (located in ADH4 exon 7 and resulting in the substitution of valine to isoleucine) identified in EAs was predicted to damage protein function. This SNP marker was also strongly associated with CpG cg12011299, which was located in the promoter region of ADH4 (Table 2). Given the significant association of rs1126671 and AD (Edenberg et al. 2006; Luo et al. 2005), this variant may have more than one functional mechanism, i.e., altering ADH4 protein function and regulating ADH4 expression by impacting promoter CpG methylation levels. About half of the mQTLs identified in AAs and more than half of the mQTLs identified in EAs were predicted to be eQTLs (Table 3). This implies that some mQTLs may mediate the interaction of SNP genotype, DNA methylation, and gene expression. Additionally, by querying the Genetic Association Database (GAD) through the web server SNPnexus, we found that 15 (9.2%) of the 162 cis-mQTLs identified in AAs and 50 (23.2%) of the 215 cis-mQTLs identified in EAs were associated with alcohol dependence or abuse. Three cis-mQTLs (rs2173201, rs4147542 and rs4147541 located in the ADH1B-ADH1C gene region) identified in AAs were all significantly associated with CpG cg25997474 in the promoter region of ADH1C (Supplementary Table S3a), and they were among the SNPs showing genome-wide association with AD in AAs in our recent GWAS (Gelernter et al. 2014). However, none of the 76 trans-mQTLs identified in AAs and the 90-trans-mQTLs identified in EAs were reported to be associated with alcohol dependence or abuse in published studies. Therefore, cis-mQTLs that influence the methylation levels of proximal CpGs in the promoter regions of genes are more likely to increase the risk of diseases such as AD. In addition, we noted that three of the top 10 cis-mQTLs in AAs (Table 1) and four of the top 10 cis-mQTLs in EAs (Table 2) were intronic SNPs. The significant correlation of these intronic SNPs and promoter CpGs may be due to the tight LD between these intronic SNPs and promoter SNPs that are proximal to promoter CpG sites. As showed in Figure 1, SNP markers in the ADH4-ADH5 gene cluster formed a tight LD block.
The distance between cis-mQTLs and CpGs was inversely correlated with the strength of association between cis-mQTLs and CpGs: a shorter distance between cis-mQTLs and CpGs predicted a stronger effect of genetic variation on CpG methylation (Fig. 3). A similar observation was reported in a recent study that performed DNA methylome analysis using Illumina's 450K Methylation array (Moen et al. 2013). Both studies showed that a majority of the significantly associated SNP-CpG pairs were comprised of SNPs and CpGs with distances less than 500 kb. However, unlike the previous study, the present study focused mainly on the effect of genetic polymorphisms on methylation patterns of promoter CpGs in candidate risk genes for AD. As shown in Fig. 3, the most significant mQTL (or SNP)-CpG association was derived from rs1800759 and cg12011299 (both located in the promoter region of ADH4, with a distance between them of only 37 bp), and this association was observed in both AAs and EAs. The present mQTL study provided further evidence that rs1800759 may strongly influence the methylation status of the nearby promoter CpG cg12011299, thus leading to altered transcription of ADH4, a gene encoding for an enzyme involved in alcohol metabolism.
Additionally, the present study demonstrated that the strength of mQTL-CpG association could be affected by non-genetic factors such as race and AD status: about 70% of the identified mQTL-CpG pairs were either AA or EA-specific (Fig. 2a). Previous studies also showed that mQTLs were population-specific (Fraser et al. 2012; Lam et al. 2012; Moen et al. 2013). Our study provides additional evidence that population-specific mQTLs may be due to the combined effect of population differences in DNA methylation and allele frequencies. An additional source for the observed population-specific mQTLs may originate from sex differences of the two populations (i.e., 36.2% of AAs were males vs. 51.5% EAs were males), although sex was adjusted by the multiple linear model. Furthermore, we analyzed the effect of AD on association between mQTLs and CpGs in AAs. Two-thirds of the mQTL-CpG pairs identified in AAs were common to both AD case and control subjects. In other words, a majority of the associations between mQTL and CpGs were not affected by AD. This may be because AD exerts only a minor or moderate effect on DNA methylation, as demonstrated in published studies (Philibert et al. 2012; Zhang et al. 2013a).
There are several limitations to this study. First, we examined the influence of sequence variants (i.e., SNPs) on methylation patterns of only a small number of promoter CpGs in 82 candidate risk genes for AD. For a fuller understanding of the relations among SNP genotypes, DNA methylation, and gene expression, future studies should be performed on a genome-wide scale. Second, the present study analyzed the impact of only two non-genetic factors (i.e., race and AD) on the association of SNPs (or mQTLs) and CpGs. However, numerous studies have demonstrated that environmental factor such as early-life stress (Murgatroyd et al. 2009; Zhang et al. 2013b) and demographic factors such as age (Boks et al. 2009; Numata et al. 2012) and sex (Boks et al. 2009; Xu et al. 2014) can alter DNA methylation levels. Non-genetic factors such as early-life stress (not included in the present study) should be considered when performing mQTL studies. Third, the present study did not consider the linkage disequilibrium (LD) among SNPs in mQTL-CpG association analysis, yielding evidence of association of multiple tightly linked SNP markers with the same promoter CpG. Experimental studies are needed to specify the functional SNP that influences the methylation status of a specific CpG site. Lastly, the present study examined mQTLs in only peripheral blood. Because DNA methylation can be cell-, tissue-, organ-, individual- or population-specific, and there is a dynamic regulation of DNA methylation during development, caution is warranted when interpreting the mQTL-CpG association analysis results.
In conclusion, we found a significant association between certain DNA sequence variants (or mQTLs) and methylation patterns of promoter CpGs in a number of AD candidate genes. The strength of mQTL-CpG association was determined by factors such as mQTL-CpG distance, race, and AD. In addition, a large proportion of the identified mQTLs were predicted to be eQTLs. These findings suggest that some mQTLs may regulate gene expression and thus underlie the association of genetic variation with AD.
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health grants K99/R00 DA022891, P50 AA12870, RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, R01 AA017535, and the VA Connecticut and Philadelphia VA MIRECCs.
Footnotes
Conflict of Interest: Although unrelated to this research, Dr. Kranzler has been a consultant or advisory board member to the following pharmaceutical companies: Alkermes, Lilly, Lundbeck, Pfizer, and Roche. He is also a member of the American Society of Clinical Psychopharmacology's Alcohol Clinical Trials Initiative, which is supported by Lilly, Lundbeck, Abbott, and Pfizer. Other authors declare that they have no competing interests.
References
- Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, Smith CL, Shafa R, Aeali B, Carnevale J, Pan H, Papageorgis P, Ponte JF, Sivaraman V, Tsuang MT, Thiagalingam S. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15:3132–45. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4th. Washington, DC: 1994. [Google Scholar]
- Baer C, Claus R, Frenzel LP, Zucknick M, Park YJ, Gu L, Weichenhan D, Fischer M, Pallasch CP, Herpel E, Rehli M, Byrd JC, Wendtner CM, Plass C. Extensive promoter DNA hypermethylation and hypomethylation is associated with aberrant microRNA expression in chronic lymphocytic leukemia. Cancer Res. 2012;72:3775–85. doi: 10.1158/0008-5472.CAN-12-0803. [DOI] [PubMed] [Google Scholar]
- Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Gilad Y, Pritchard JK. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boks MP, Derks EM, Weisenberger DJ, Strengman E, Janson E, Sommer IE, Kahn RS, Ophoff RA. The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS One. 2009;4:e6767. doi: 10.1371/journal.pone.0006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ozturk NC, Zhou FC. DNA methylation program in developing hippocampus and its alteration by alcohol. PLoS One. 2013;8:e60503. doi: 10.1371/journal.pone.0060503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayem Ullah AZ, Lemoine NR, Chelala C. SNPnexus: a web server for functional annotation of novel and publicly known genetic variants (2012 update) Nucleic Acids Res. 2012;40:W65–70. doi: 10.1093/nar/gks364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij SR, Costello PM, Veenendaal MV, Lillycrop KA, Gluckman PD, Hanson MA, Painter RC, Roseboom TJ. Associations between DNA methylation of a glucocorticoid receptor promoter and acute stress responses in a large healthy adult population are largely explained by lifestyle and educational differences. Psychoneuroendocrinology. 2012;37:782–8. doi: 10.1016/j.psyneuen.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Porjesz B, Rice J, Schuckit M, Tischfield J, Begleiter H, Foroud T. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet. 2006;15:1539–49. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–63. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Etcheverry A, Aubry M, de Tayrac M, Vauleon E, Boniface R, Guenot F, Saikali S, Hamlat A, Riffaud L, Menei P, Quillien V, Mosser J. DNA methylation in glioblastoma: impact on gene expression and clinical outcome. BMC Genomics. 2010;11:701. doi: 10.1186/1471-2164-11-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Lam LL, Neumann SM, Kobor MS. Population-specificity of human DNA methylation. Genome Biol. 2012;13:R8. doi: 10.1186/gb-2012-13-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D, Wodarz N, Zill P, Zhao H, Farrer LA. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry. 2014;19:41–9. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, Arepalli S, Dillman A, Rafferty IP, Troncoso J, Johnson R, Zielke HR, Ferrucci L, Longo DL, Cookson MR, Singleton AB. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTExConsortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo AY, Webb BT, Miles MF, Zimmerman MP, Kendler KS, Zhao Z. ERGR: An ethanol-related gene resource. Nucleic Acids Res. 2009;37:D840–5. doi: 10.1093/nar/gkn816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson CA, Yuan Q, Xu K, Shen PH, Heinz E, Lobos EA, Binder EB, Cubells J, Ehlers CL, Gelernter J, Mann J, Riley B, Roy A, Tabakoff B, Todd RD, Zhou Z, Goldman D. Addictions biology: haplotype-based analysis for 130 candidate genes on a single array. Alcohol Alcohol. 2008;43:505–15. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Chen Z, Huang X. Age-adjusted nonparametric detection of differential DNA methylation with case-control designs. BMC Bioinformatics. 2013;14:86. doi: 10.1186/1471-2105-14-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, Morris M, Haghighi F, Tycko B. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–8. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- Lam LL, Emberly E, Fraser HB, Neumann SM, Chen E, Miller GE, Kobor MS. Factors underlying variable DNA methylation in a human community cohort. Proc Natl Acad Sci U S A. 2012;109(Suppl 2):17253–60. doi: 10.1073/pnas.1121249109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X, Cretney EC, Kropp J, Khateeb K, Berg MA, Penagaricano F, Magness R, Radunz AE, Khatib H. Maternal Diet during Pregnancy Induces Gene Expression and DNA Methylation Changes in Fetal Tissues in Sheep. Front Genet. 2013;4:49. doi: 10.3389/fgene.2013.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Morgan M, Hutchison K, Calhoun VD. A study of the influence of sex on genome wide methylation. PLoS One. 2010;5:e10028. doi: 10.1371/journal.pone.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Yang BZ, Lappalainen J, Gelernter J. ADH4 gene variation is associated with alcohol and drug dependence: results from family controlled and population-structured association studies. Pharmacogenet Genomics. 2005;15:755–68. doi: 10.1097/01.fpc.0000180141.77036.dc. [DOI] [PubMed] [Google Scholar]
- Moen EL, Zhang X, Mu W, Delaney SM, Wing C, McQuade J, Myers J, Godley LA, Dolan ME, Zhang W. Genome-wide variation of cytosine modifications between European and African populations and the implications for complex traits. Genetics. 2013;194:987–96. doi: 10.1534/genetics.113.151381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OF, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–66. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen DA, Hamon S, Yuferov V, Jackson C, Ho A, Ott J, Kreek MJ. Ethnic diversity of DNA methylation in the OPRM1 promoter region in lymphocytes of heroin addicts. Hum Genet. 2010;127:639–49. doi: 10.1007/s00439-010-0807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, Colantuoni C, Weinberger DR, Kleinman JE, Lipska BK. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet. 2012;90:260–72. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Plume JM, Gibbons FX, Brody GH, Beach SR. The impact of recent alcohol use on genome wide DNA methylation signatures. Front Genet. 2012;3:54. doi: 10.3389/fgene.2012.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, Farrer L, Kranzler HR. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug Alcohol Depend. 2005;80:303–12. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quon G, Lippert C, Heckerman D, Listgarten J. Patterns of methylation heritability in a genome-wide analysis of four brain regions. Nucleic Acids Res. 2013;41:2095–104. doi: 10.1093/nar/gks1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalkwyk LC, Meaburn EL, Smith R, Dempster EL, Jeffries AR, Davies MN, Plomin R, Mill J. Allelic skewing of DNA methylation is widespread across the genome. Am J Hum Genet. 2010;86:196–212. doi: 10.1016/j.ajhg.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubry A, Schildkraut JM, Murtha A, Wang F, Huang Z, Bernal A, Kurtzberg J, Jirtle RL, Murphy SK, Hoyo C. Paternal obesity is associated with IGF2 hypomethylation in newborns: results from a Newborn Epigenetics Study (NEST) cohort. BMC Med. 2013;11:29. doi: 10.1186/1741-7015-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2002;64:479–498. [Google Scholar]
- Supic G, Jagodic M, Magic Z. Epigenetics: a new link between nutrition and cancer. Nutr Cancer. 2013;65:781–92. doi: 10.1080/01635581.2013.805794. [DOI] [PubMed] [Google Scholar]
- Tekpli X, Landvik NE, Anmarkud KH, Skaug V, Haugen A, Zienolddiny S. DNA methylation at promoter regions of interleukin 1B, interleukin 6, and interleukin 8 in non-small cell lung cancer. Cancer Immunol Immunother. 2013;62:337–45. doi: 10.1007/s00262-012-1340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka AR, Price LH, Marsit C, Walters OC, Carpenter LL. Childhood adversity and epigenetic modulation of the leukocyte glucocorticoid receptor: preliminary findings in healthy adults. PLoS One. 2012;7:e30148. doi: 10.1371/journal.pone.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unternaehrer E, Luers P, Mill J, Dempster E, Meyer AH, Staehli S, Lieb R, Hellhammer DH, Meinlschmidt G. Dynamic changes in DNA methylation of stress-associated genes (OXTR, BDNF ) after acute psychosocial stress. Transl Psychiatry. 2012;2:e150. doi: 10.1038/tp.2012.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloszynska-Read A, James SR, Link PA, Yu J, Odunsi K, Karpf AR. DNA methylation-dependent regulation of BORIS/CTCFL expression in ovarian cancer. Cancer Immun. 2007;7:21. [PMC free article] [PubMed] [Google Scholar]
- Xie P, Kranzler HR, Poling J, Stein MB, Anton RF, Brady K, Weiss RD, Farrer L, Gelernter J. Interactive effect of stressful life events and the serotonin transporter 5-HTTLPR genotype on posttraumatic stress disorder diagnosis in 2 independent populations. Arch Gen Psychiatry. 2009;66:1201–9. doi: 10.1001/archgenpsychiatry.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wang F, Liu Y, Yu Y, Gelernter J, Zhang H. Sex-biased methylome and transcriptome in human prefrontal cortex. Hum Mol Genet. 2014;23:1260–70. doi: 10.1093/hmg/ddt516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BZ, Zhang H, Ge W, Weder N, Douglas-Palumberi H, Perepletchikova F, Gelernter J, Kaufman J. Child abuse and epigenetic mechanisms of disease risk. Am J Prev Med. 2013;44:101–7. doi: 10.1016/j.amepre.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Cheng L, Badner JA, Chen C, Chen Q, Luo W, Craig DW, Redman M, Gershon ES, Liu C. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 2010;86:411–9. doi: 10.1016/j.ajhg.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, Gonzalez K, Vishwanatha JK, Santella RM, Morabia A. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6:623–9. doi: 10.4161/epi.6.5.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Herman AI, Kranzler HR, Anton RF, Zhao H, Zheng W, Gelernter J. Array-based profiling of DNA methylation changes associated with alcohol dependence. Alcohol Clin Exp Res. 2013a;37(Suppl 1):E108–15. doi: 10.1111/j.1530-0277.2012.01928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang F, Kranzler HR, Zhao H, Gelernter J. Profiling of childhood adversity-associated DNA methylation changes in alcoholic patients and healthy controls. PLoS One. 2013b;8:e65648. doi: 10.1371/journal.pone.0065648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Webb BT, Jia P, Bigdeli TB, Maher BS, van den Oord E, Bergen SE, Amdur RL, O'Neill FA, Walsh D, Thiselton DL, Chen X, Pato CN, International Schizophrenia C, Riley BP, Kendler KS, Fanous AH. Association study of 167 candidate genes for schizophrenia selected by a multi-domain evidence-based prioritization algorithm and neurodevelopmental hypothesis. PLoS One. 2013;8:e67776. doi: 10.1371/journal.pone.0067776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.