Abstract
We previously reported the mechanisms involved in the formation of Mallory-Denk bodies (MDBs) in mice fed DDC. To further provide clinical evidence as to how ubiquitin-like protein (Ubls) modification, gene transcript expression in Ufmylation and FATylation were investigated in human archived formalin-fixed, paraffin-embedded (FFPE) liver biopsies and frozen liver sections from DDC re-fed mice were used. Real-time PCR analysis showed that all Ufmylation molecules (Ufm1, Uba5, Ufc1, Ufl1 and UfSPs) were significantly down regulated, both in DDC re-fed mice livers and patients’ livers where MDBs had formed, indicating that gene transcript changes were limited to MDB-forming livers where the protein quality control system was down regulated. FAT10 and subunits of the immunoproteasome (LMP2 and LMP7) were both up regulated as previously shown. An approximate 176- and 5-fold up regulation (respectively) of FAT10 were observed in the DDC re-fed mice liver and in the livers of human alcoholic hepatitis with MDBs present, implying that there was an important role played by this gene. The FAT10-specific E1 and E2 enzymes Uba6 and USE1, however, were found to be down regulated both in patients’ livers and in the liver of DDC re-fed mice. Interestedly, the down regulation of mRNA levels was proportionate to MDB abundance in the liver tissues. Our results show the first systematic demonstration of transcript regulation of Ufmylation and FATylation in the liver of patients who form MDBs, where protein quality control is down regulated. This was also shown in livers of DDC re-fed mice where MDBs had formed.
Keywords: Ubiquitin-like (Ubl) modifiers, Mallory-Denk bodies (MDBs), Ufm1, FAT10, transcript regulation
Introduction
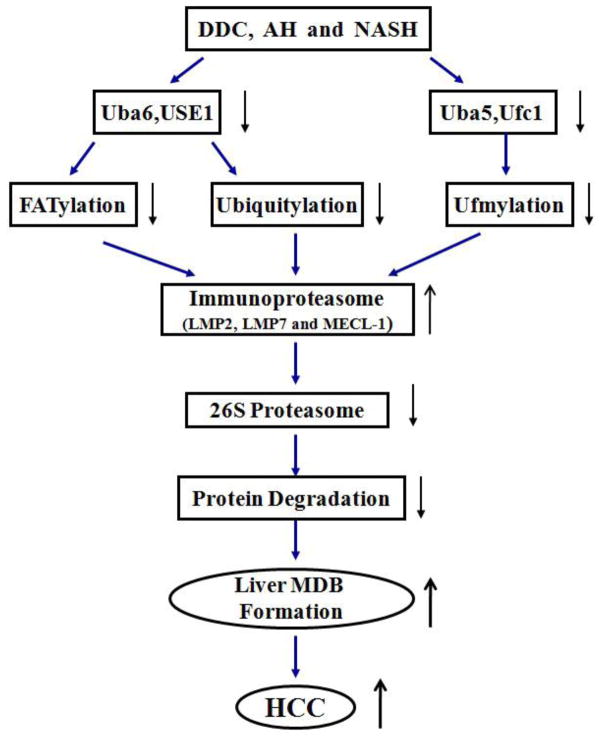
The ubiquitin-like (Ubl) modifiers conjugation pathway plays an essential role in protein degradation, protein quality control, DNA replication, signal transduction, cell cycle control and immune response (Hershko & Ciechanover, 1998; Kerscher et al., 2006; Mukhopadhyay & Riezman, 2007; Cajee et al., 2012; Merbl et al., 2013). It is important to determine what the levels are of the ubiquitylation enzymes involved in cell cycle regulation, because changes in the activity of these enzymes can lead to tumorigenesis (Rape, 2014). Ubls and Ubiquitin (Ub) share a common fold in their molecules and their transfer is carried out by conjugating with substrates through an elaborate enzymatic reaction consisting of E1, E2 and E3. This is done using a multi-step process involving several sequential steps in an ATP-dependent manner (Hershko, 2005; Kerscher et al., 2006). The ubiquitylated substrates are then recognized by highly conserved ubiquitin receptors such as on the 26S proteasome for proteasomal degradation (Fu et al., 2010). Dysregulation of ubiquitylation, however, is implicated in the etiology of various human diseases (Kerscher et al., 2006). The activity of the 26S proteasome is down regulated in the liver of DDC fed mice, leading to the accumulation of undigested proteins and Mallory-Denk body (MDB) formation (Bardag-Gorce et al., 2010). In the DDC fed mouse model where liver cells proliferate, MDBs form and later, after DDC withdrawal, hepatocellular carcinomas (HCCs) develop (Oliva et al., 2008). MDBs contain K18 and 8, ubiquitin and p62 (Zatloukal et al., 2007), which are prevalent in alcoholic hepatitis (AH), cirrhosis, non-alcoholic steatohepatitis (NASH) and in some HCCs. There is a clinical link between MDB formation in human chronic liver diseases as well as in HCC formation (Nakanuma & Ohta, 1985). Although three new mechanisms of MDB formation have recently been revealed (French et al., 2010), the mechanisms involved in the formation of MDB aggresomes is still not fully understood.
At present, nine distinct classes of Ubls are found (Cajee et al., 2012), which are involved in apoptosis, autophagy and signaling pathways. Among the Ubls, the recently identified Ufm1 (ubiquitin fold modifier 1), is activated by a specific E1-like enzyme Uba5 and is converted to an E2-like enzyme Ufc1. It binds to the only identified substrate protein C20orf116 by an E3 ligase named Ufl1. The protein conjugate is then cleaved by the specific cysteine proteases UfSP1 and UfSP2. This constitutes the reversibility of the Ufm1-conjugating system (Ufmylation) (Tatsumi et al., 2011). Interestingly, the Ufm1 conjugation was found to be abundant in the liver of Ufm1-transgenic mice (Tatsumi et al., 2009), pointing to a possible link between this novel Ubl system and liver function.
Another member of Ubls family is FAT10 known as diubiquitin, which shares a moderate sequence similarity to ubiquitin. FAT10 modification is mediated through the Uba6 (E1) and USE1 (E2) enzymes, which are specific to both FAT10 and ubiquitin (Chiu et al., 2007). FAT10 expression is induced by interferon (IFN)-γ and tumor necrosis factor α (TNFα) (Lukasiak et al., 2008; Oliva et al., 2010). Increased FAT10 gene expression and protein levels have been observed in various cancers (Raasi et al., 2001; Lee et al., 2003; Lim et al., 2006; Qing et al., 2011), and interference of FAT10 could suppress cell proliferation by inhibiting the cell cycle S-phase entry and inducing apoptosis (Chen et al., 2014). Despite these reports, the biological significance of the FAT10 system in liver tumor development remains unclear and warrants further research. More recently, it has been found that only FAT10 and Ufm1 exhibited a higher frequency of preferential targets when analyzing the Ubl modification of mitotic control, showing that the functions of FAT10 and Ufm1 system are largely insulated from the activity of other Ubls (Merbl et al., 2013).
To provide key clinical evidence as to whether chronic liver diseases are relevant to the Ufm1 and FAT10 conjugation system, the first systematic study of gene transcript expression of Ufmylation and FATylation was performed on livers of DDC re-fed mice and patients’ liver biopsies. The significant down regulation of Ufm1 and FAT10 conjugation components were observed both in livers of DDC re-fed mice and human liver biopsies from patients with AH, cirrhosis and NASH. Interestingly, the down regulation of mRNA was relevant to MDB abundance in these liver tissues (data not shown).
Materials and Methods
Clinical specimens
Human archived formalin-fixed paraffin-embedded (FFPE) liver biopsies from patients who had alcoholic hepatitis (AH), cirrhosis, and non-alcoholic steatohepatitis (NASH) were obtained from Harbor UCLA hospital archives. In all the cases liver forming MDBs were presented except in the normal control livers. A summary of the detailed clinicopathological information for chronic liver diseases in patients is shown in Table I.
Table I.
Clinical pathological features of human liver biopsies
| Clinical pathological features | Numbers of cases | MDBs |
|---|---|---|
| Alcoholic hepatitis (AH) | N=4 | Yes |
| Cirrhosis | N=4 | Yes |
| Non-alcoholic steatohepatitis (NASH) | N=3 | Yes |
| Normal liver control | N=3 | No |
Mouse liver
Diethyl 1, 4-dehydro-2, 4, 6-trimethyl-3, 5-pyridine-dicarboxylate (DDC) was used as a model to induce the formation of Mallory-Denk bodies (MDBs) in mice. One-month-old C3H male mice were fed 0.1% DDC added to the control diet and a second group were fed control diet for 10 weeks (Li et al., 2008). The mice were then withdrawn from the drug for 1 month and then re-fed DDC for 7 days as was previously done (Oliva et al., 2009). Three mice were used in each of two groups as followed: 1) control, 2) DDC. DDC was re-fed for 7 days. Mice livers were placed in isopentane and were then fast frozen with liquid nitrogen and stored at −80°C. The livers used had been used in a prior study (Oliva et al., 2009). All mice were treated in a humane manner as approved by the Animal Care Committee at Harbor UCLA Laboratory Bio Medical Research Institute according to the Guidelines of the National Academy of Science.
Tissue sectioning
Mice liver frozen sections were performed as standard protocol. Briefly, archived mice liver frozen sections were cut (5 μm thickness) at −20°C and immediately transferred to a micro slide box kept on dry ice and stored at −80°C. These slides were placed under the hood to dry for 1–2 hour and subsequently stored in a micro slide box at −80°C. A new blade was used for each frozen sample.
RNA isolation
For RNA isolation of FFPE tissue sections, paraffin was first removed from tissue samples when human liver biopsies were assayed. The paraffin-embedded tissues sections were mounted on a glass slide and dried at 60°C for 30 minutes. The slides were then submerged in xylene at room temperature for 1 hour changing the xylene once after 30 minutes. The samples were hydrated by washing progressively for 2 minutes in 100%, 70%, 50% ethanol, and then pure RNase-free water before air-drying the samples on the slides for approximately 10 minutes. RNA isolation was processed using the Pinpoint™ Slide RNA isolation System II (ZYMO) by adding Pinpoint™ Solution directly to a small region of the tissue section to allow the solution to dry completely at the room temperature. The embedded tissue was then removed from the slide using a sterile blade or scalpel to scrape tissues from the slides followed by transferring the tissues to a micro-centrifuge tube for subsequent proteinase K digestion. The RNA was extracted and purified according to the manufacturer’s protocol (ZYMO). DNA-free RNA can be obtained with subsequent DNase I treatment following the manufacturer’s recommended protocol (ZYMO). The process described above was also followed for mice frozen liver tissues, except for the deparaffinization step. The quality and yield of the resulting total RNAs were assessed with an absorbance reading at 260 nm (A260) using a spectrophotometer (Thermo) by loading 1 μl of the extracted RNA.
Quantitative Real-time PCR analysis
Synthesis of first-strand cDNAs was performed with the above mentioned total RNA (250ng), and random hexamer primers using SuperSript III First-Strand Synthesis SuperMix (Invitrogen) following the instruction. Real-time PCR was performed using the Fast SYBR Green Master Mix on a StepOnePlus™ Real-time PCR System (Applied Biosystems) with a primer concentration of 200 nM. Primer sequences and the related gene Accession Number are listed in Table II. Reaction conditions consisted of 95°C for 20 sec, followed by 40 cycles of 95°C for 3 sec, 60 °C for 30 sec. Single PCR product was confirmed with the heat dissociation protocol at the end of the PCR cycles. Human α-tubulin and mice β-actin were used as controls to normalize the starting quantity of RNA. Quantitative values were obtained from the threshold PCR cycle number (CT) at which point the increase in signal associated with an exponential growth for PCR product starts to be detected. The target mRNA abundance in each sample was normalized to its endogenous control level and the relative mRNA expression levels were analyzed using the ΔΔCT method. Reaction of each sample was performed in triplicate.
Table II.
Sequences of the forward and reverse real-time PCR primers of Ufm1 and FAT10 conjugation system components
| Gene name (Species) | Accession Number | Sequences of primer |
|---|---|---|
| Ufm1 (Mouse) | NM_026435 | Forward Primer:: 5′-GTA AGC AAC CCC TAA TTA AA-3′ Reverse Primer: 5′-GCA GTT AAA TTT TGA AAC CA-3′ |
| Ufc1 (Mouse) | NM_025388 | Forward Primer: 5′-GTC ACC TCT GGC TGT TAC AA-3′ Reverse Primer: 5′-ACT TGG AGA AGG GAT ACA AA-3′ |
| Ufl1 (Mouse) | NM_026194 | Forward Primer: 5′-GAA CAC TGA AGA AGC AAG CA-3′ Reverse Primer: 5′-AAA CTG GGC TAA TGA CAG AA-3′ |
| UfSP1 (Mouse) | NM_027356 | Forward Primer: 5′-AAC CGT TGT GAA CTA CAA GC-3′ Reverse Primer: 5′-TGG CTG TGT TTA GTC TCA GT-3′ |
| UfSP2 (Mouse) | NM_138668 | Forward Primer: 5′-GTT TGT CAA CCA AGG TTC AG-3′ Reverse Primer: 5′-TCC ATG CGA CTC CTA GTA TT-3′ |
| Uba5 (Mouse) | NM_025692 | Forward Primer: 5′-TCT GGA TGA AAT TGA AGC AG-3′ Reverse Primer: 5′-GGA TAA CCA ATG TTT CCT GA-3′ |
| FAT10 (Mouse) | NM_023137 | Forward Primer: 5′-ACC TCT GTG ATC CCT AAG AA -3′ Reverse Primer: 5′-TTG TCA GAA AGA GCA AAC TG -3′ |
| Uba6 (Mouse) | NM_172712 | Forward Primer: 5′-GAA TCC TCA AAC ATG AAG CA -3′ Reverse Primer: 5′-AAT GAC AAA ATG GTT GTG GA -3′ |
| USE1 (Mouse) | NM_025917 | Forward Primer: 5′-AGG ACA ATC AGA CCC TGT CA-3′ Reverse Primer: 5′-GTG TGC AGG CTT TCT TGT TGT TT-3′ |
| LMP2 (Mouse) | NM_013585 | Forward Primer: 5′-CAA GTA CCG TGA GGA CTT GT-3′ Reverse Primer: 5′-CCA TAA ATG TAG GAG CTT CC-3′ |
| LMP7 (Mouse) | XM_006537283 | Forward Primer: 5′-AAG AAG GGA CCA GGA CTT TA-3′ Reverse Primer: 5′-CTC TTC AGG ACT GAG GTC CT-3′ |
| MECL-1 (Mouse) | NM_013640 | Forward Primer: 5′-TAC ATG TAT GTG CCT GCA GT-3′ Reverse Primer: 5′-CTT TCG TCT TTG CTG CTA GT-3′ |
| Ufm1 (Human) | NM_001256799 | Forward Primer: 5′-ATG TTG AGT CTT AGT TAA GC -3′ Reverse Primer: 5′-TGG CAC AGG ACA ACA TAA AT -3′ |
| Ufc1 (Human) | NM_016406 | Forward Primer: 5′-AAC ACA AAG AGA AAT GCA AC -3′ Reverse Primer: 5′-CAA CTA GAG GTG GAG GGT GT -3′ |
| Uba5 (Human) | NM_024818 | Forward Primer: 5′-CAG AAA GTC TCA ACT ACC AA -3′ Reverse Primer: 5′-AGA CTC ACC AAG GCA ACA TA -3′ |
| Uba6 (Human) | NM_018227 | Forward Primer: 5′-CTC TGG TGA TAT TCA TGG AA -3′ Reverse Primer: 5′-GTG GAA AGA TTT AGG TCA CG-3′ |
| FAT10 (Human) | NM_006398 | Forward Primer: 5′-TCA GAA AGG GCA ACT TAC TC -3′ Reverse Primer: 5′-TTG GGA AAT CAT CAG AAG AT -3′ |
| USE1 (Human) | NM_018467 | Forward Primer: 5′-CAG CTG AGC TAG ACC TCG TC -3′ Reverse Primer: 5′-CTC CGT CTT CAG TTT CTC CA -3′ |
Immunohistochemical staining
Formalin fixed, paraffin embedded tissue slides were double stained for UFM 1 (Abcam Inc., Cambridge MA) and Ubiquitin (Millipore, Temecula, CA). A second set of slides were stained for FAT10 (Enzo Life Sciences, Farmingdale, NY) and Ubiquitin (Millipore, Temecula, CA). UFM 1 and FAT10 were detected using the second antibody donkey anti rabbit Alexa Fluor 488 (Jackson Immuno Research Laboratories Inc. West Grove, PA). Ubiquitin was detected using the second antibody donkey anti mouse Alexa Fluor 594 (Jackson Labs. West Grove, PA). All slides were stained with the nuclear stain DAPI (Molecular Probes, Eugene, OR). The slides were examined with a Nikon 400 fluorescent microscope.
Statistical analysis
All data were presented as the mean ±S.E.M and were representative of at least two-independent experiments done in triplicate. Statistical differences were calculated using SigmaStat software, which unpaired, two-tailed student’s t-test is used to compare two groups’ variance and One Way ANOVA is used to analyze the variance of multiple groups. In general, a p value less than 0.05 denoted statistical significance.
Results
The Ufm1 and FAT10 conjugation system is transcriptionally down regulated in the livers of DDC re-fed mice
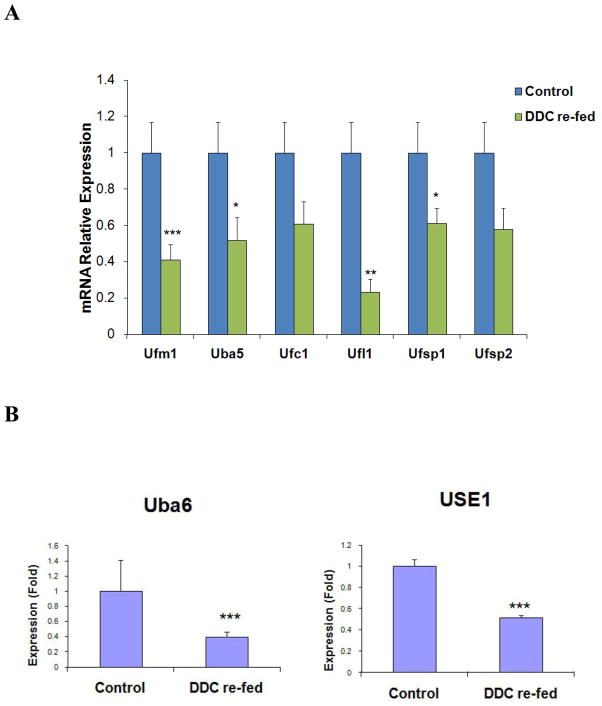
DDC was used to induce MDB formation in mice. Hepatocyte isolation was then performed from mice fed the control diet and mice re-fed DDC for 7 days as described earlier (Oliva et al., 2009). The mRNA expression of Ufmylation components in the livers of DDC re-fed mice was examined using real-time PCR analysis. A 40% transcriptional down regulation of all Ufmylation components tested was observed in the livers of DDC re-fed mice (Fig. 1A). Among them, approximately 80% and 60% (respectively, p<0.01) mRNA down regulation levels of Ufc1 and Ufm1 mRNAs was observed when compare to other mRNAs. This implies that there is an important role played by these two genes during this process.
Figure 1.
Decrease of mice Ufmylation and FATylation components in the livers of DDC re-fed mice. Gene expression levels of different Ufmylation (A) and FATylation (B) components are shown. Data represent mean values ±S.E.M. Statistical significance was determined using the t-test with SigmaStat software. * p<0.05, **p<0.01, and ***p<0.001 by t-test.
To confirm whether the FAT10 conjugation is changed when MDB formation develops, Uba6 (E1) and USE1 (E2) enzymes, which are relevant to FATylation, were further analyzed by real-time PCR in the livers of DDC re-fed mice. As expected, the down regulation expression of Uba6 (60%, p<0.001) and USE1 (50%, p<0.001) were also observed by real-time PCR analysis (Fig. 1B), similar to the mRNA expression of the Ufmylation mRNAs. These results suggest that both Ufm1 and the FAT10 conjugation system are transcriptionally down regulated in the livers of DDC re-fed mice.
Up regulation of FAT10 and the immunoproteasome in the livers of DDC re-fed mice
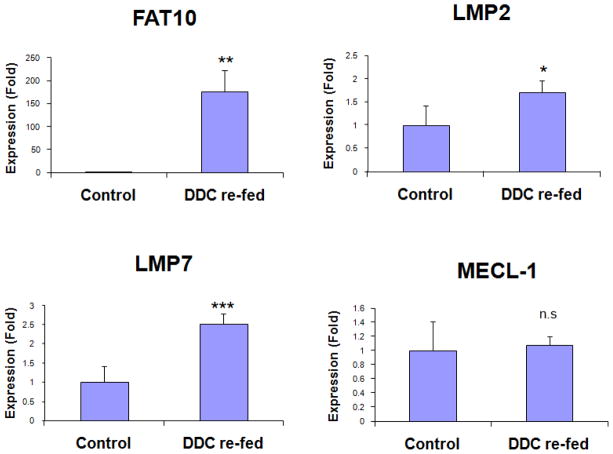
As previously demonstrated, when DDC is re-fed, the activity of the 26S proteasome is decreased by the shift of the expression of the proteasome catalytic subunits from the 26S proteasome proteins to that of the immunoproteasome (Bardag-Gorce et al., 2010). This leads to the accumulation of undigested proteins and MDB formation in FAT10 over expressing hepatocytes (French et al., 2010). Because of this, the catalytic subunits (LMP2, LMP7 and MECL-1) of the immunoproteasome and FAT10 were analyzed by real-time PCR. Similar to previous observations, the mRNA levels of LMP2, LMP7, and FAT10 were all up regulated to various degrees (Figure 2). Among these, FAT10 mRNA was induced by up to a 176-fold expression in liver by DDC re-feeding, suggesting that the shift had occurred in FAT10 positive hepatocytes. The MECL-1 mRNA, however, was only slightly increased in this study (Figure 2). These results clearly indicated that DDC re-feeding caused the up regulation of the immunoproteasome and FAT10 in hepatocytes.
Figure 2.
Induction of FAT10 and subunits of immunoproteasome (LMP2, LMP7 and MECL-1) in the livers of DDC re-fed mice. Quantification of mRNA was carried out by SYBR real-time PCR assays. n.s, not significant, * p<0.05, **p<0.01, and ***p<0.001 by t-test with SigmaStat software.
Ufm1 and FAT10 conjugation are involved in MDB formation, which progresses to HCC in the livers of patients and mice fed DDC
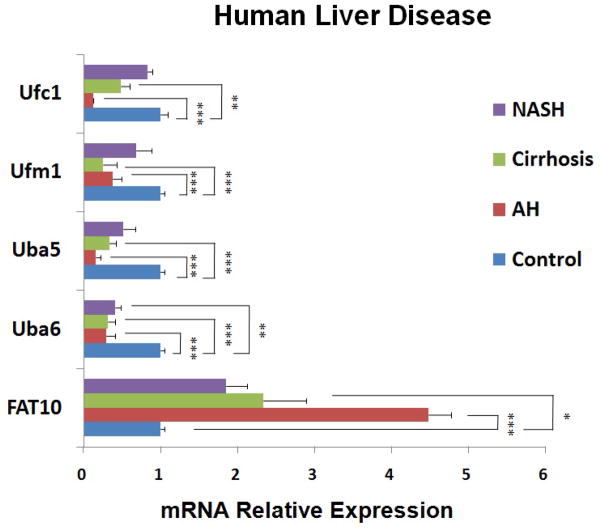
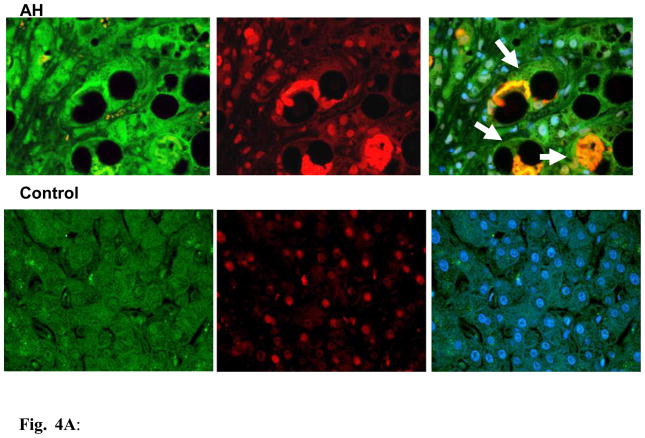
To confirm whether the transcriptional down regulation expression of Ufm1 and FAT10 conjugation system is involved in human chronic liver diseases, in which MDB formation progresses to tumor formation, the mRNA levels of certain Ufm1 and FAT10 system components in liver biopsies, including normal liver tissue, AH, cirrhosis and NASH were measured. As expected, the down regulation of mRNA expression of human Ufm1, Ufc1, Uba5 and Uba6 was also observed in these patients’ livers by real-time PCR analysis (Figure 3), similar to the observation from the livers of DDC re-fed mice. Interestingly, we found that the transcriptional down regulation levels of these genes were relevant to MDB abundance (data not shown). More mRNA down regulation levels were observed in AH and cirrhosis biopsies which had more MDBs (Figure 3). The mRNA expression levels of FAT10, however, were significantly up regulated in these biopsies (Figure 3), suggesting that the FAT10 expression increase during liver MDB formation, is similar to the mRNA expression in the livers of DDC re-fed mice. Immunohistochemical staining showed that MDB containing balloon hepatocytes in AH liver biopsies had reduced intensity of staining of the cytoplasm for UFM1 (Fig. 4B) and increased staining intensity for FAT10 (Fig. 4A).
Figure 3.
Transcriptional expression of human Ufmylation and FATylation in patient livers from archived biopsies with AH, cirrhosis, NASH and normal liver tissue. Data represent mean values ±S.E.M. * p<0.05, **p<0.01, and ***p<0.001 by One Way ANOVA with SigmaStat software.
Figure 4.
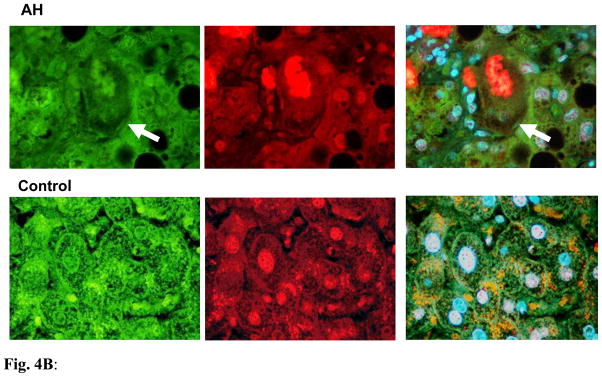
Fig. 4A: The liver sections from AH patients and controls were double stained with antibodies to FAT10 (green), Ubiquitin (red) and DAPI (tricolor). The AH liver sections stained with greater intensity for FAT10 compared to the controls, especially the balloon cells that had formed MDBs (arrows). FAT10 and Ubiquitin co-localized in the MDBs. x520
Fig. 4B: The liver section from AH patients and controls were double stained with antibodies to UFM1 (green), Ubiquitin (red) and DAPI (tricolor). AH liver sections stained with less intensity for UFM1 compared to controls, especially the balloon cells that had formed MDBs (arrows). UFM1 and Ubiquitin did not co-localize in the MDBs. ×780
Discussion
Recent evidence showed that MDB formation in FAT10 positive hepatocytes is the result of a drug-induced shift from the 26S proteasome to the immunoproteasome form (Bardag-Gorce et al., 2010; French et al., 2010). The Ufm1 and FAT10 conjugation system was recently identified as an Ubl system, which shares common features with other Ubl systems. However, the information about their biological significance is still partly unknown. In this study, the transcriptional down regulation of the Ufm1 and FAT10 conjugation system in the livers of DDC re-fed mice and patients’ livers was demonstrated, providing key evidence for possible involvement of the Ufm1 and FAT10 conjugation system in MDB formation and tumor formation. To our knowledge, this is the first clinical evidence of direct involvement of the Ufm1 and FAT10 conjugation system with liver MDB formation. Therefore, a direct link between the gene transcriptional down regulation of Ufmylation and FATylation with liver MDB formation has been established. This may provide a better understanding of the biological function of these two novel Ubl systems in alcoholic hepatitis.
Although it was reported that the Ufm1 system complex was abundant in the liver of Ufm1-transgenic mice (Tatsumi et al., 2009), the actual evidence for the Ufm1 system involved in liver is still elusive. In the present study, taking advantage of the livers of DDC re-fed mice, we first demonstrated that the Ufm1 system components were transcriptionally down regulated by real-time PCR analysis.
An interesting observation is that the mRNA level of Ufl1 was much lower in liver from DDC re-fed mice. Ufl1 is a novel E3 ligase for the Ufm1 system mainly localized in the endoplasmic reticulum (Tatsumi et al., 2009). Recent studies showed that Ufm1 targets are enriched with trans-membrane transporters, ion channels, and cytokine receptors (Merbl et al., 2013). Ufm1-specific proteases, UfSP1 and UfSP2, can cleave both the C terminus of pro-Ufm1 and several Ufm1 conjugates, although the apparent efficiency of these two reactions seems different (Kang et al., 2007). In our experiments, the mRNA down regulation level of UfSP2 was slightly lower than that of UfSP1. These observations are similar to previous reports (Kang et al., 2007), but differ from the observation that UfSP1 was more efficient in de-conjugating Ufm1 from the substrates than UfSP2 was (Tatsumi et al., 2009). The possible causes may be the difference in the models used.
Another interesting observation is an approximate 60% and 40% down regulation level of Uba6 and USE1, which are specific to both FAT10 and ubiquitin. The E1 enzyme Uba6 of the FAT10 conjugation can activate both ubiquitin and FAT10, while FAT10 binds to Uba6 with higher affinity than ubiquitin (Gavin et al., 2012). Also the E2 enzyme USE1 in charge of FAT10 conjugation is bi-specific for ubiquitin and FAT10, and USE1 is the main but not the only E2 enzyme for FAT10 (Schmidtke et al., 2013). However, until now no putative E3 ligases and de-conjugating enzymes of the FAT10 cascade have been discovered. Although we observed the significant transcriptional down regulation of Uba6 and USE1 in MDB formation in hepatocytes, it is still uncertain as to how the Uba6 and USE1 determine which substrates are to be ubiquitylated and which are to be FATylated for protein degradation in MDB-forming hepatocytes.
Linking specific Ubls to their substrates is an essential first step in understanding specificity and selectivity of Ubl modifications and identifying the pathways in which they operate. The ubiquitylated protein substrates can be targeted to the 26S proteasome for degradation, leading to accumulation of altered and ubiquitylated proteins (Fu et al., 2010). In DDC re-fed mice, the activity of the 26S proteasome is decreased by the shift from the 26S proteasome to the immunoproteasome and is limited to FAT10 over-expressing liver cells (Bardag-Gorce et al., 2004; Bardag-Gorce et al., 2010). Because of this, the transcription expression of FAT10 and catalytic subunits of the immunoproteasome (LMP2, LMP7 and MECL-1) were investigated by real-time PCR analysis in the livers of DDC re-fed mice. The prominent up regulation expression of mRNA levels of these genes was observed (Figure 2). A 176-fold induction of FAT10 was determined, indicating that a switch from the 26S proteasome to immunoproteasome had occurred in the FAT10 positive hepatocytes and boosted FAT10 production. These results clearly indicated that DDC re-feeding caused a switch from the 26S proteasome to immunoproteasome, which resulted in the accumulation of undigested proteins in the MDB forming FAT10 positive liver cells, similar to our previous observation.
To provide further clinical evidence of the involvement of FATylation and Ufmylation in chronic liver disease development, human archived formalin-fixed paraffin-embedded (FFPE) liver biopsies from patients who had alcoholic hepatitis (AH), cirrhosis and NASH were examined by real-time PCR. In all cases the presence of liver forming MDBs was the focus of the present examination. It was discovered that Uba5, Uba6, Ufc1 and Ufm1, which were involved in Ufmylation and FATylation were also transcriptionally down regulated to various degrees in AH, cirrhosis and NASH biopsies. Immunohistochemistry analysis also illustrated the decrease in Ufm1 in AH hepatocytes compared to the control (Fig. 4B). Interestingly, the mRNA down regulation levels of these components were much lower in AH and cirrhosis biopsies, which have more MDBs. This implies a close relationship of these gene transcript changes with MDB-forming cells as a signal for proteasome degradation. The FAT10 mRNA levels were significantly up regulated in these biopsies, with a high approximate 4.3-fold induction in AH biopsies, indicating an increase in FAT10 production in livers with MDB formation. Fig. 4A illustrated the increase in FAT10 in AH hepatocytes compared to the control livers. These findings provide important clinical evidence of the involvement of the Ufm1 and FAT10 conjugation pathway of protein quality control in liver pathology. Based on this finding in DDC re-fed mice livers or clinical liver diseases, transcription down regulation of FATylation and Ufymlation which results in liver MDB formation is shown in Figure 5.
Figure 5.
Proposed model of transcription expression of Ufm1 and FAT10 conjugation system in mice and human liver MDB formation. DDC re-fed mice, alcoholic hepatitis (AH) and non-alcoholic steatohepatitis (NASH) transcriptional down regulation of FATylation and Ufmylation are shown.
Why is Ufm1 and FAT10 conjugation transcriptionally down regulated and what are the biological functions or mechanism of these two Ubls in liver MDB formation? A possible answer to this question is the up regulation of FAT10 and LMP2, which are both strongly and synergistically induced at the mRNA and protein levels under the stimulation of pro-inflammatory cytokines, interferon (IFN)-γ and tumor necrosis factor (TNF)-α (Raasi et al., 1999). The fact that pro-inflammatory cytokines are responsible for MDB formation in FAT10 positive hepatocytes has been demonstrated in the DDC induced mouse model (French et al., 2010; Oliva et al., 2010). It needs to be determined as to which inflammatory pathways, among so many are involved in the transcription regulation of Ufmylation and FATylation in the formation of MDBs in the liver.
The complexity of transcriptional regulation of the Ufm1 and FAT10 conjugation system is reflected in this study. For example, the mRNA levels of different components of induction appear to vary in response to DDC or in the chronic liver disease biopsies. Therefore the isolation and characterization of the promoters of individual Ufm1 and FAT10 system components will be crucial in understanding their response to stress in liver MDB formation. Because FAT10 is the only ubiquitin-like modifier which directly targets its substrate for degradation by the 26S proteasome, the complexity of its mRNA induction was shown by IFN-γ and TNF-α stimulation at different times (Raasi et al., 1999; Aichem et al., 2012). It is possible that the mRNA levels of the FAT10 system varies with the time of DDC re-feeding or the different developmental stages of liver disease. Finally, because the mRNA down regulation levels of FATylation and Ufmylation were in proportion to MDB abundance, the accuracy of MDB counts needs to be determined in these tissues to determine the relationship of gene transcription with MDB numbers.
In summary, provided here for the first time is evidence of transcription down regulation of the Ufm1 and FAT10 conjugation system in liver MDB formation. The marked transcription inhibition of Ufmylation and FATylation components was observed and the down regulation of mRNA levels was proportionate to MDB abundance in the liver tissues. These results provide further insight into the understanding of human liver disease development, providing important clinical data concerning the Ufm1 and FAT10 conjugation system in liver pathogenesis.
Acknowledgments
This work was supported by a grant from NIH (AAU01021898-02) and P50-11999, Morphology Core. Some results were presented in a Poster Abstract (No. LB493) in Experimental Biology April 2014, San Diego.
Abbreviations
- AH
alcoholic hepatitis
- DDC
diethyl 1, 4-dehydro-2, 4, 6-trimethyl-3, 5-pyridine-dicarboxylate
- FATylation
FAT10-conjugating system
- FFPE
formalin-fixed, paraffin-embedded
- HCC
hepatocellular carcinoma
- MDB
Mallory-Denk body
- NASH
non-alcoholic steatohepatitis
- Ubl
Ubiquitin-like
- Ufm1
ubiquitin fold modifier 1
- Ufmylation
Ufm1-conjugating system
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aichem A, Kalveram B, Spinnenhirn V, Kluge K, Catone N, Johansen T, Groettrup M. The proteomic analysis of endogenous FAT10 substrates identifies p62/SQSTM1 as a substrate of FAT10ylation. J Cell Sci. 2012;125:4576–4585. doi: 10.1242/jcs.107789. [DOI] [PubMed] [Google Scholar]
- Bardag-Gorce F, Oliva J, Li J, French BA, French SW. SAMe prevents the induction of the immunoproteasome and preserves the 26S proteasome in the DDC-induced MDB mouse model. Exp Mol Pathol. 2010;88:353–362. doi: 10.1016/j.yexmp.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardag-Gorce F, Vu J, Nan L, Riley N, Li J, French SW. Proteasome inhibition induces cytokeratin accumulation in vivo. Exp Mol Pathol. 2004;76:83–89. doi: 10.1016/j.yexmp.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Cajee UF, Hull R, Ntwasa M. Modification by ubiquitin-like proteins: significance in apoptosis and autophagy pathways. Int J Mol Sci. 2012;13:11804–11831. doi: 10.3390/ijms130911804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yang L, Chen H, Yuan T, Liu M, Chen P. Recombinant adenovirus encoding FAT10 small interfering RNA inhibits HCC growth in vitro and in vivo. Exp Mol Pathol. 2014;96:207–211. doi: 10.1016/j.yexmp.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Chiu YH, Sun Q, Chen ZJ. E1-L2 activates both ubiquitin and FAT10. Mol Cell. 2007;27:1014–1023. doi: 10.1016/j.molcel.2007.08.020. [DOI] [PubMed] [Google Scholar]
- French SW, Bardag-Gorce F, Li J, French BA, Oliva J. Mallory-Denk body pathogenesis revisited. World J Hepatol. 2010;2:295–301. doi: 10.4254/wjh.v2.i8.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Lin YL, Fatimababy AS. Proteasomal recognition of ubiquitylated substrates. Trends Plant Sci. 2010;15:375–386. doi: 10.1016/j.tplants.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Gavin JM, Chen JJ, Liao H, Rollins N, Yang X, Xu Q, Ma J, Loke HK, Lingaraj T, Brownell JE, Mallender WD, Gould AE, Amidon BS, Dick LR. Mechanistic studies on activation of ubiquitin and di-ubiquitin-like protein, FAT10, by ubiquitin-like modifier activating enzyme 6, Uba6. J Biol Chem. 2012;287:15512–15522. doi: 10.1074/jbc.M111.336198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 2005;12:1191–1197. doi: 10.1038/sj.cdd.4401702. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Kang SH, Kim GR, Seong M, Baek SH, Seol JH, Bang OS, Ovaa H, Tatsumi K, Komatsu M, Tanaka K, Chung CH. Two novel ubiquitin-fold modifier 1 (Ufm1)-specific proteases, UfSP1 and UfSP2. J Biol Chem. 2007;282:5256–5262. doi: 10.1074/jbc.M610590200. [DOI] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Lee CG, Ren J, Cheong IS, Ban KH, Ooi LL, Yong Tan S, Kan A, Nuchprayoon I, Jin R, Lee KH, Choti M, Lee LA. Expression of the FAT10 gene is highly upregulated in hepatocellular carcinoma and other gastrointestinal and gynecological cancers. Oncogene. 2003;22:2592–2603. doi: 10.1038/sj.onc.1206337. [DOI] [PubMed] [Google Scholar]
- Li J, Bardag-Gorce F, Dedes J, French BA, Amidi F, Oliva J, French SW. S-adenosylmethionine prevents Mallory Denk body formation in drug-primed mice by inhibiting the epigenetic memory. Hepatology. 2008;47:613–624. doi: 10.1002/hep.22029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CB, Zhang D, Lee CG. FAT10, a gene up-regulated in various cancers, is cell-cycle regulated. Cell Div. 2006;1:20. doi: 10.1186/1747-1028-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasiak S, Schiller C, Oehlschlaeger P, Schmidtke G, Krause P, Legler DF, Autschbach F, Schirmacher P, Breuhahn K, Groettrup M. Proinflammatory cytokines cause FAT10 upregulation in cancers of liver and colon. Oncogene. 2008;27:6068–6074. doi: 10.1038/onc.2008.201. [DOI] [PubMed] [Google Scholar]
- Merbl Y, Refour P, Patel H, Springer M, Kirschner MW. Profiling of ubiquitin-like modifications reveals features of mitotic control. Cell. 2013;152:1160–1172. doi: 10.1016/j.cell.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Nakanuma Y, Ohta G. Is mallory body formation a preneoplastic change? A study of 181 cases of liver bearing hepatocellular carcinoma and 82 cases of cirrhosis. Cancer. 1985;55:2400–2404. doi: 10.1002/1097-0142(19850515)55:10<2400::aid-cncr2820551017>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Oliva J, Bardag-Gorce F, French BA, Li J, McPhaul L, Amidi F, Dedes J, Habibi A, Nguyen S, French SW. Fat10 is an epigenetic marker for liver preneoplasia in a drug-primed mouse model of tumorigenesis. Exp Mol Pathol. 2008;84:102–112. doi: 10.1016/j.yexmp.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva J, Bardag-Gorce F, Li J, French BA, Nguyen SK, Lu SC, French SW. Betaine prevents Mallory-Denk body formation in drug-primed mice by epigenetic mechanisms. Exp Mol Pathol. 2009;86:77–86. doi: 10.1016/j.yexmp.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva J, Bardag-Gorce F, Lin A, French BA, French SW. The role of cytokines in UbD promoter regulation and Mallory-Denk body-like aggresomes. Exp Mol Pathol. 2010;89:1–8. doi: 10.1016/j.yexmp.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing X, French BA, Oliva J, French SW. Increased expression of FAT10 in colon benign, premalignant and malignant epithelial neoplasms. Exp Mol Pathol. 2011;90:51–54. doi: 10.1016/j.yexmp.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raasi S, Schmidtke G, de Giuli R, Groettrup M. A ubiquitin-like protein which is synergistically inducible by interferon-gamma and tumor necrosis factor-alpha. Eur J Immunol. 1999;29:4030–4036. doi: 10.1002/(SICI)1521-4141(199912)29:12<4030::AID-IMMU4030>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Raasi S, Schmidtke G, Groettrup M. The ubiquitin-like protein FAT10 forms covalent conjugates and induces apoptosis. J Biol Chem. 2001;276:35334–35343. doi: 10.1074/jbc.M105139200. [DOI] [PubMed] [Google Scholar]
- Rape M. Tumor suppression by mitotic quality control (472.1) The FASEB Journal. 2014;28(1 Supplement):472.1. [Google Scholar]
- Schmidtke G, Aichem A, Groettrup M. FAT10ylation as a signal for proteasomal degradation. Biochim Biophys Acta. 2013;1843:97–102. doi: 10.1016/j.bbamcr.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Tatsumi K, Sou YS, Tada N, Nakamura E, Iemura S, Natsume T, Kang SH, Chung CH, Kasahara M, Kominami E, Yamamoto M, Tanaka K, Komatsu M. A novel type of E3 ligase for the Ufm1 conjugation system. J Biol Chem. 2009;285:5417–5427. doi: 10.1074/jbc.M109.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi K, Yamamoto-Mukai H, Shimizu R, Waguri S, Sou YS, Sakamoto A, Taya C, Shitara H, Hara T, Chung CH, Tanaka K, Yamamoto M, Komatsu M. The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat Commun. 2011;2:181. doi: 10.1038/ncomms1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatloukal K, French SW, Stumptner C, Strnad P, Harada M, Toivola DM, Cadrin M, Omary MB. From Mallory to Mallory-Denk bodies: what, how and why? Exp Cell Res. 2007;313:2033–2049. doi: 10.1016/j.yexcr.2007.04.024. [DOI] [PubMed] [Google Scholar]