Abstract
Stem Cell Antigen-1 (Sca1 or Ly6A/E) is a cell surface marker that is widely expressed in mesenchymal stem cells, including adipose-derived stem cells (ASCs). We hypothesized that the fat depot-specific gene signature of Sca1high ASCs may play the major role in defining adipose tissue function and extracellular matrix (ECM) remodeling in a depot-specific manner. Herein we aimed to characterize the unique gene signature and ECM remodeling of Sca1high ASCs isolated from subcutaneous (inguinal) and visceral (epididymal) adipose tissues. Sca1high ASCs are found in the adventitia and perivascular area of adipose tissues. Sca1high ASCs purified with magnetic-activated cell sorting (MACS) demonstrate dendrite or round shape with the higher expression of cytokines and chemokines (e.g., Il6, Cxcl1) and the lower expression of a glucose transporter (Glut1). Subcutaneous and visceral fat-derived Sca1high ASCs particularly differ in the gene expressions of adhesion and ECM molecules. While the expression of the major membrane-type collagenase (MMP14) is comparable between the groups, the expressions of secreted collagenases (MMP8 and MMP13) are higher in visceral Sca1high ASCs than subcutaneous ASCs. Consistently, slow but focal MMP-dependent collagenolysis was observed with subcutaneous Sca1high ASCs, whereas rapid and bulk collagenolysis was observed with visceral Sca1high ASCs in MMP-dependent and –independent manners. These results suggest that the fat depot-specific gene signatures of ASCs may contribute to the distinct patterns of ECM remodeling and adipose function in different fat depots.
Keywords: Adipocyte, ECM, Collagen, MMP, Adipose-derived stem cell, RNA-seq, Obesity
1. Introduction
Adipose-derived stem cells (ASCs) are multi-potent mesenchymal stem cells that can differentiate into the cells of multiple lineages (Gimble et al., 2007; Gonzalez-Cruz and Darling, 2013; Mizuno et al., 2012). Among a cohort of cell surface markers of stem cells, stem cell antigen 1 (Sca1, also known as Ly6A/E) was originally identified as the cell surface protein that allows the identification of hematopoietic stem cells (Spangrude et al., 1988; van de Rijn et al., 1989). Sca1 (Ly6A/E), however, is expressed not only in hematopoietic stem cells but also in mesenchymal stem cells (Morikawa et al., 2009; Welm et al., 2002). Mesenchymal stem cells of adipose tissues, ASCs, are preferentially found in perivascular area and contribute to adipose tissue development (Tang et al., 2008).
Tissue fibrosis plays a key role in the progression of chronic and metabolic diseases (Chun, 2012). Adipose tissue fibrosis has been recently highlighted as the potential etiology of adipose tissue dysfunction found in obesity (Chun, 2012; Divoux et al., 2010; Sun et al., 2013). Among extracellular matrix (ECM) proteins, the collagen family members constitute the major part of fibrillar ECM network in vivo (Ricard-Blum, 2011). Collagens are actively synthesized by fibroblast-like mesenchymal cells during organ development and tissue regeneration (Duffield et al., 2013). The degree of tissue collagen deposition is determined by the balance between the synthesis and the degradation of collagens (Ricard-Blum, 2011). The major enzymes that degrade fibrillar collagens belong to the matrix metalloproteinase (MMP) family (Kessenbrock et al., 2010). Among them, MMP14 (MT1-MMP) plays the dominant role in regulating physiological, pericellular collagenolysis and adipose ECM remodeling (Chun et al., 2006; Chun et al., 2010), whereas secreted collagenases, i.e., MMP1, -8, and -13, may contribute to pro-inflammatory, bulk collagenolysis (Chavey et al., 2003; Sabeh et al., 2009).
Adipose tissue development requires the pericellular collagen turnover mediated by a membrane-type MMP, MT1-MMP, which is also known as MMP14 (Chun et al., 2006). In adults, adipose tissues change their size and function in adaptation to nutritional challenges. Underscoring the role played by MMP family members in the regulation of adipose tissue size and function, the genetic polymorphisms of MMPs are associated with body mass index (BMI) and diabetes traits (Chun et al., 2010; Nho et al., 2008; Traurig et al., 2006). The genetic program that governs ECM remodeling may play the major role in adipose tissue development and expansion (Chun, 2012).
The role of the tissue mesenchymal stem cells in the regulation of adipose ECM remodeling and function has not been well characterized to this date. To understand the cellular function of ASCs in adipose ECM remodeling, we aimed to determine the gene signature and ECM remodeling of ASCs that express higher levels of Sca1 cell surface marker (Sca1high ASCs). RNA-seq and qPCR analyses unraveled the distinct gene signature of Sca1high ASCs. The transcriptome profiles of Sca1high ASCs also suggested the presence of fat depot-specific gene signature that is coupled with depot-specific pattern of ECM remodeling. While subcutaneous Sca1high cells exerted slow and focused collagenolysis mediated by membrane-bound collagenases (MMP14-MMP2 axis), visceral Sca1high cells displayed rapid, bulk collagenolysis, which is partly mediated by secreted collagenases, i.e., MMP8 or MMP13. (Sabeh et al., 2009; Sato-Kusubata et al., 2011) and a MMP-independent mechanism.
2. Results
2.1. Identification of Sca1high mesenchymal stem cells in adipose tissues
Sca1 (Ly6A/E)-positive cells of dendrite or round shape were found in proximity to vasculatures and in space between adipocytes (Figure 1A). We isolated vascular-stromal cells (VSCs) by digesting inguinal and epididymal white adipose tissues (iWAT and eWAT) with collagenase treatment and examined the cell-surface expression of Sca1 as well as F4/80 (Emr1), a cell surface marker of macrophages (Lumeng et al., 2008). The populations of Sca1highF4/80low cells constitute approximately 20% and 33% of total VSCs in iWAT and eWAT, respectively (Figure 1B). For the functional assessment of Sca1highF4/80low mesenchymal cells found in each fat depot, we used magnetic-activated cell sorting (MACS) to purify adherent Sca1high and Sca1low cells. From iWAT and eWAT, the populations of cells positively selected based on Sca1 expression were approximately 13% and 8% of total VSCs, respectively (Figure 1C). The Ly6a (Sca1) mRNA expression in Sca1high cells was 3- to 5-times higher than in Sca1low cells (Figure 1D). Consistent with the gene expression analysis, Sca1high cells displayed markedly higher Sca1 protein expression on the plasma membranes relative to Sca1low cells (Figure 1E). Emr1 (F4/80) mRNA expression in these adherent cells was very low relative to the level found in undigested adipose tissues (~5%) suggesting that the adherent VSCs isolated from young male mice did not contain a significant number of monocytes/macrophages (data not shown).
Figure 1.
Identification and isolation of Sca1high ASCs from iWAT and eWAT. (A) Immunofluorescent detection of Sca1high cells in inguinal and epididymal white adipose tissues (iWAT and eWAT) isolated from 6-week-old C57/BL6 male mice. Sca1 (red), type IV collagen (green), and nucleus (blue). Scale = 100 μm. Representative figures, N=3. (B) Flow cytometry analysis of Sca1(+)F4/80(−) cells in iWAT- and eWAT-derived VSCs. Representative data, n ≥ 3. (C) The isolation of Sca1high ASCs from iWAT and eWAT using MACS, shown as mean ± SD. N=4. (D) The mRNA expression of Sca1 in the Sca1high ASCs isolated from iWAT and eWAT using MACS. N=4. Mean ± SEM. * P < 0.05 (E) The expression of SCA1 protein on the plasma membrane of Sca1high ASCs isolated from iWAT (red). Nucleus (blue); scale = 10 μm.
2.2. The adipogenic potentials of Sca1high and Sca1low VSCs
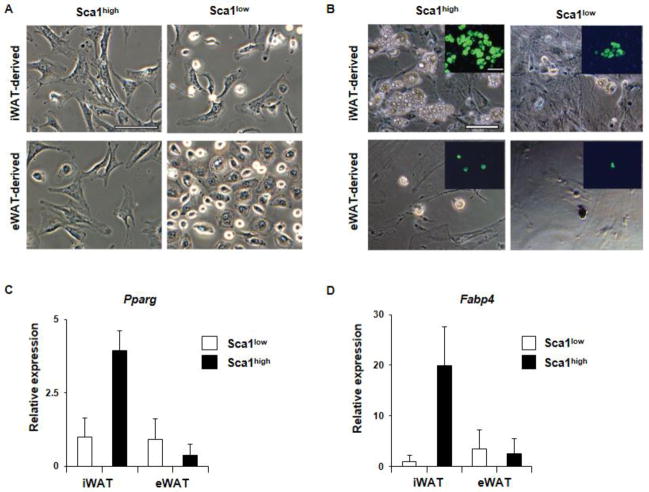
We examined the adipogenic potentials of Sca1high and Sca1low cells. Sca1high cells were fibroblast-like cells displaying well-stretched cell shape (Figure 2A). The difference in cell shape between Sca1high and Sca1low cells was most evident in eWAT-derived VSCs. The eWAT-derived Sca1low cells displayed epithelial cell-like morphology in contrast to the fibroblast-like adherent cell shape of Sca1high cells (Figure 2A). Upon the hormonal induction of adipogenesis, we observed significant lipid accumulation in iWAT-derived Sca1high cells but only to a lesser degree in Sca1low cells (Figure 2B). The lipid accumulation in eWAT-dervied Sca1high cells was markedly suppressed in comparison to iWAT-derived Sca1high cells (Figure 2B). Consistent with the morphological observation, the expression of the dominant adipogenic transcription factor, peroxisome proliferator-activated receptor (Pparg), or the adipocyte-specific lipid carrier protein, fatty acid-binding protein (Fabp4), was robustly induced during adipogenesis of iWAT-derived Sca1high cells but not observed with other types of cells (Figure 2C and D).
Figure 2.
Adipogenic potential of Sca1high and Sca1low ASCs isolated from iWAT and eWAT. (A) The morphology of iWAT- and eWAT-derived Sca1high ASCs and Sca1low cells shown under phase microscopy. Scale = 100 μm. (B) Adipocyte differentiation (7 days after adding adipogenic mix) shown under phase microscopy. The insets show the lipid droplets (green) detected in adipocytes. Scale = 100 μm. (C) Pparg mRNA expression in iWAT- and eWAT-derived ASCs before and after adipogenesis. N=4. (D) Fabp4 expression before and after adipogenesis. N=4. Shown as means ± SEM.
2.3. Genome-wide expression profiles of Sca1high and Sca1low VSCs
To define the set of genes of which expressions co-segregate with Sca1 expression, we used a whole genome RNA sequencing (RNA-seq) analysis coupled with RT-qPCR. Among 40,920 genes to which the sequenced reads were aligned, 13,917 genes (34%) were defined as “expressed” with their expression levels higher than 1.0 Fragments Per Kilobase of exon per Million fragments mapped (FPKM). The average gene expression levels in Sca1low and Sca1high VSCs were 237.0 and 246.0 FPKM, respectively, with the correlation coefficient of 0.999. Among expressed genes, we found 617 genes (4.4% of expressed genes) were differentially expressed between Sca1high and Sca1low VSCs (q-value <0.05) (Storey and Tibshirani, 2003) (Supplemental Table 1). Gene Ontology (GO)-Cellular Component (CC) analyses suggested that the genes differentially expressed between Sca1high and Sca1low cells were found mostly in extracellular region (GO:0005576) and plasma membrane (GO:0005886) (Figure 3A and Supplemental Table 2). GO-Molecular Function (MF) analysis then pointed to the gene enrichment in growth factor activity (GO: 0008083, Supplemental Table 3). Among them, the gene expressions of chemokine (C-X-C motif) ligand 1 (CXCL1), interleukin 6 (IL6), glial cell line derived neurotrophic factor (GDNF), chemokine (C-X-C motif) ligand 12 (CXCL12), interleukin (IL11), inhibin beta-A (INHBA), epiregulin (EREG), growth differentiation factor 10 (GDF10), c-fos induced growth factor (FIGF), and nerve growth factor (NGF) were higher in Sca1high ASCs than in Sca1low cells (Supplemental table 1). Some of these results were validated with respective RT-qPCR; for example, we observed the robust expressions of Il6 and Cxcl1 in Sca1high ASCs (Figure 3B), which may represent the pro-inflammatory characteristics of mesenchymal stem cells reported previously (Hoogduijn et al., 2013). By contrast, we observed a set of genes encoding the proteins located on the plasma membrane was significantly down-regulated in Sca1high cells, e.g., the dominant glucose transporter expressed in VSCs, SLC2A1 (GLUT1) (Figure 3C). These results suggest that Sca1high ASCs may demonstrate a distinct pattern in the gene expression of the molecules found in extracellular region and on the plasma membrane.
Figure 3.
Gene expression of Sca1high ASCs. (A) The heatmap for the genes differentially expressed in “extracellular region” (GO: 0005576) between Sca1high and Sca1low VSCs in iWAT and eWAT. (B) The expression of chemokines and cytokines highly expressed by Sca1high ASCs. Cxcl1 and IL6 expression in iWAT- and eWAT-derived Sca1low versus Sca1high ASCs. N=3. (C) The Glut1 expression is reduced in iWAT- and eWAT-derived Sca1high ASCs compared to Sca1low cells. N=3. * P < 0.05
2.4. Fat depot-specific transcriptome profile of Sca1high ASCs
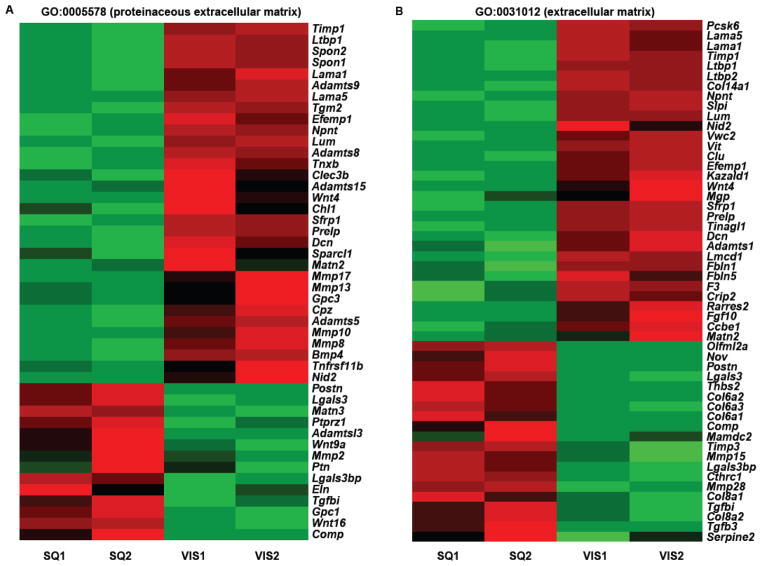
To investigate the fat depot-specific gene expression of Sca1high ASCs, we compared the gene signature of Sca1high cells between iWAT and eWAT. Sequenced reads were aligned to 40,920 genes, and >1.0 FPKM was used as the cutoff to define “expressed” genes. Among the listed genes, 13,857 genes (33.8% of total) were found as “expressed” (FPKM>1.0). The correlation coefficient was 0.994. Among the expressed genes, 1,178 genes (8.5% of expressed genes) were differentially expressed (Supplemental Table 4). When these differentially regulated genes were interrogated with GO analyses, GO-CC pointed to the genes encoding the proteins located in extracellular region (GO: 0005576), extracellular matrix (GO: 0031012), extracellular space (GO: 0005615), proteinaceous extracellular matrix (GO: 0005578), plasma membrane (GO: 0005886), basement membrane (GO: 0005604), anchored to membrane (GO: 0031225), and cell surface (GO: 0009986) (Supplemental Table 5 and Figure 4). Among the extracellular matrix genes showing significant differences in their gene expressions (GO:0031012), GO-Molecular Functions (MF) pointed to the enrichment of the genes for diverse biological functions, e.g., polysaccharide binding, pattern binding, carbohydrate binding, glycosaminoglycan binding, heparin binding, and growth factor activity (Supplemental Table 6). These results suggest that the differential expression of the genes encoding proteins found in the extracellular space or on the plasma membrane of Sca1high ASCs may define the depot-specific characteristics of adipose tissue function. Among fibrillar ECM proteins, RNA-seq suggested that type VI collagen, of which role in adipose tissue has been well characterized (Khan et al., 2009), as well as type VIII collagen genes was expressed more in iWAT-derived Sca1high ASCs (SQ) than eWAT-derived Sca1high ASCs (Vis) (Table 1). Of note, the gene expression of elastin, a determinant of tissue elasticity, is higher in SQ. The gene expression of glycoproteins, such as Lumican, Decorin, and Glypican-3 are expressed more in Vis, whereas Glypican-1 expressed more in SQ. Most of the genes encoding basement membrane proteins (laminin-5 and -1, nidogen-2) and proteins that anchor ECM proteins to the basement membrane of cells (prolargin, fibulin-1 and -5, tenascin-X, transforming growth factor beta-induced protein ig-h3, cartilage oligomeric matrix protein) are expressed more in Vis ASCs. Among proteinases, the family members of a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS), i.e., Adamts-1, -5, -8, -9, and -15, which may cleave aggrecan (Verma and Dalal, 2011) or versican (Dancevic et al., 2013) and act as angiogenesis inhibitors (Vázquez et al., 1999), are expressed more in Vis than SQ ASCs (Table 1). Latent transforming growth factor beta-binding proteins are expressed more in Vis ASCs along with BMP4, which plays a key role in adipogenesis (Macotela et al., 2012) and brown fat-like phenotype (Qian et al., 2013). The WNT family members, which are the major regulators of adipogenesis and adipocyte function (Cristancho and Lazar, 2011; Ross et al., 2000), are expressed differently between Vis and SQ ASCs. Wnt9a and -16 (Kandyba et al., 2013) are expressed more in SQ-ASCs, whereas pro-adipogenic Wnt4 (Nishizuka et al., 2008) is expressed more in Vis-ASCs (Table 1). Moreover, ECM modifiers, such as tissue transgutaminase (TGM2), and the proteins that regulate adhesion and coagulation pathways, e.g., tissue factor, are expressed more in Vis ASCs (Table 1).
Figure 4.
Differential expression of extracellular matrix-related and membrane-associated proteins in iWAT- and eWAT-derived Sca1high ASCs. The results of RNA-seq performed with RNAs isolated from iWAT Sca1high ASCs (SQ1, SQ2) and eWAT Sca1high ASCs (Vis1, Vis2) are shown as heatmaps of the genes belonging to GO:0005578, proteinaceous extracellular matrix (A) and GO:0031012, extracellular matrix (B). N=2 each, FPKM >1.0, adjusted p value (q value) <0.05.
Table 1.
Fat depot-specific gene expression in Sca1high ASCs
| Gene name | Protein name | Fat-depot | Fold-increase |
|---|---|---|---|
| Collagens and other structural ECMs | |||
| Col14a1 | Collagen alpha-1(XIV) chain | Vis | 3.9 |
| Eln | Elastin | SQ | 1.6 |
| Col6a1 | Collagen alpha-1(VI) chain | SQ | 3.6 |
| Col6a2 | Collagen alpha-2(VI) chain | SQ | 4.4 |
| Col6a3 | Collagen alpha-3(VI) chain | SQ | 4.3 |
| Col8a1 | Collagen alpha-1(VIII) chain | SQ | 1.9 |
| Col8a2 | Collagen alpha-2(VIII) chain | SQ | 1.8 |
| Proteoglycans | |||
| Lum | Lumican | Vis | 4.9 |
| Dcn | Decorin | Vis | 1.8 |
| Gpc3 | Glypican-3 | Vis | 2.2 |
| Gpc1 | Glypican-1 | SQ | 1.9 |
| Basement membrane and anchoring proteins | |||
| Lama5 | Laminin subunit alpha-5 | Vis | 2.5 |
| Lama1 | Laminin subunit alpha-1 | Vis | 2.9 |
| Nid2 | Nidogen-2 | Vis | 12.9 |
| Prelp | Prolargin | Vis | 2.0 |
| Fbln1 | Fibulin-1 | Vis | 1.9 |
| Fbln5 | Fibulin-5 | Vis | 2.2 |
| Tnxb | Tenascin-X | Vis | 3.3 |
| Tgfbi | Transforming growth factor-beta-induced protein ig-h3 | SQ | 1.9 |
| Comp | Cartilage oligomeric matrix protein | SQ | 38 |
| Proteinases and proteinase inhibitors | |||
| Pcsk6 | Proprotein convertase subtilisin/kexin type 6 | Vis | 3.3 |
| Timp1 | Metalloproteinase inhibitor 1 | Vis | 3.2 |
| Slpi | Antileukoproteinase | Vis | 5.9 |
| Kazald1 | Kazal-type serine protease inhibitor domain-containing protein 1 | Vis | 6.8 |
| Adamts1 | A disintegrin and metalloproteinase with thrombospondin motifs 1 | Vis | 1.9 |
| Adamts5 | A disintegrin and metalloproteinase with thrombospondin motifs 5 | Vis | 4.7 |
| Adamts8 | A disintegrin and metalloproteinase with thrombospondin motifs 8 | Vis | 7.6 |
| Adamts9 | A disintegrin and metalloproteinase with thrombospondin motifs 9 | Vis | 3.2 |
| Adamts15 | A disintegrin and metalloproteinase with thrombospondin motifs 15 | Vis | 2.7 |
| Mmp8 | Neutrophil collagenase | Vis | 25.6 |
| Mmp10 | Stromelysin-2 | Vis | 5.8 |
| Mmp13 | Collagenase 3 | Vis | 1.8 |
| Mmp17 | Matrix metalloproteinase-17 | Vis | 1.6 |
| Cpz | Carboxypeptidase Z | Vis | 4.2 |
| Mmp2 | 72 kDa type IV collagenase | SQ | 1.9 |
| Timp3 | Metalloproteinase inhibitor 3 | SQ | 2.0 |
| Mmp15 | Matrix metalloproteinase-15 | SQ | 2.6 |
| Mmp28 | Matrix metalloproteinase-28 | SQ | 2.1 |
| Serpine2 | Glia-derived nexin | SQ | 2.0 |
| TGF-beta-related | |||
| Ltbp1 | Latent-transforming growth factor beta-binding protein 1 | Vis | 3.2 |
| Ltbp2 | Latent-transforming growth factor beta-binding protein 2 | Vis | 4.0 |
| Bmp4 | Bone morphogenetic protein 4 | Vis | 10.2 |
| Tgfb3 | Transforming growth factor beta-3 | SQ | 1.7 |
| WNT activity | |||
| Wnt4 | Protein Wnt-4 | Vis | 6.6 |
| Sfrp1 | Secreted frizzled-related protein 1 | Vis | 2.2 |
| Wnt9a | Protein Wnt-9a | SQ | 2.2 |
| Wnt16 | Protein Wnt-16 | SQ | 15.4 |
| ECM modifier, cell adhesion, coagulation, and other functions | |||
| Npnt | Nephronectin | Vis | 4.3 |
| Vwc2 | Brorin | Vis | 5.8 |
| Vit | Vitrin | Vis | 5.9 |
| Clu | Clusterin | Vis | 5.4 |
| Efemp1 | EGF-containing fibulin-like extracellular matrix protein 1 | Vis | 4.5 |
| Mgp | Matrix Gla protein | Vis | 3.6 |
| Tinagl1 | Tubulointerstitial nephritis antigen-like | Vis | 2.3 |
| Lmcd1 | LIM and cysteine-rich domains protein 1 | Vis | 2.2 |
| F3 | Tissue factor | Vis | 1.7 |
| Crip2 | Cysteine-rich protein 2 | Vis | 1.6 |
| Rarres2 | Retinoic acid receptor responder protein 2 | Vis | 2.4 |
| Fgf10 | Fibroblast growth factor 10 | Vis | 2.6 |
| Ccbe1 | Collagen and calcium-binding EGF domain-containing protein 1 | Vis | 2.9 |
| Matn2 | Matrilin-2 | Vis | 1.6 |
| Spon2 | Spondin-2 | Vis | 3.1 |
| Spon1 | Spondin-1 | Vis | 2.9 |
| Chl1 | Neural cell adhesion molecule L1-like protein | Vis | 3.7 |
| Tnfrsf11b | Tumor necrosis factor receptor superfamily member 11B | Vis | 5.0 |
| Tgm2 | Protein-glutamine gamma-glutamyltransferase 2 | Vis | 4.7 |
| Clec3b | Tetranectin | Vis | 3.5 |
| Sparcl1 | SPARC-like protein 1 | Vis | 1.7 |
| Postn | Periostin | SQ | 3.6 |
| Lgals3 | Galectin-3 | SQ | 2.8 |
| Matn3 | Matrilin-3 | SQ | 2.9 |
| Ptprz1 | Receptor-type tyrosine-protein phosphatase zeta | SQ | 4.9 |
| Adamtsl3 | ADAMTS-like protein 3 | SQ | 3.6 |
| Ptn | Pleiotrophin | SQ | 2.8 |
| Lgals3bp | Galectin-3-binding protein | SQ | 1.8 |
| Olfml2a | Olfactomedin-like 2A | SQ | 8.9 |
| Nov | Protein NOV homolog | SQ | 8.0 |
| Thbs2 | Thrombospondin-2 | SQ | 5.1 |
| Mamdc2 | MAM domain-containing protein 2 | SQ | 4.0 |
| Cthrc1 | Collagen triple helix repeat-containing protein 1 | SQ | 2.0 |
2.5. The ECM Remodeling by Sca1high ASCs in adipose tissues
We were particularly interested in a set of genes that degrade collagen fibers in the interfaces between cells and extracellular environments. Based on the RNA-seq data that suggested the enrichment of genes encoding extracellular and cell surface proteins, we further examined the ECM remodeling of Sca1high ASCs isolated from iWAT and eWAT. When iWAT-derived Sca1high ASCs were cultured atop fluorescent-labeled rat type I collagen fibers, type I collagen degradation was observed distinctively underneath individual cells beginning in 48 hours (Figure 5A, upper left, arrows). The subjacent collagenolysis of iWAT-derived Sca1high ASCs was mediated entirely by MMPs as evidenced by the complete blockade of the collagenolysis in the presence of GM6001, a synthetic MMP inhibitor (Figure 5A, +GM6001). By contrast, eWAT-derived Sca1high ASCs exerted rapid, bulk collagenolysis demonstrating the large areas of collagen displacement underneath the groups of cells (Figure 5A, lower left, arrowheads). While GM6001 reduced the areas of collagen turnover by approximately 50%, eWAT-derived Sca1high cells continued to display the significant amount of type I collagen displacement (Figure 5A, +GM6001, lower right), suggesting the presence of an MMP-independent mechanism in the collagen remodeling by eWAT-derived Sca1high ASCs.
Figure 5.
Collagen degradation and MMP gene expression of Sca1high ASCs. (A) iWAT- and eWAT-derived Sca1high ASCs were cultured atop fluorescent-labeled type I collagen fibers (red) and the areas of degradation were observed as the loss of fluorescence (black). Arrows point to the focal collagenolysis mediated by iWAT-derived Sca1high ASCs. Higher magnification is shown in the inset. The arrowheads point to the rapid, bulk collagenolysis mediated by eWAT-derived SCA1high ASCs. GM6001 completely blocked the focal collagenolysis of iWAT-derived Sca1high ASCs, but only partially the bulk collagenolysis of eWAT-derived SCA1high ASCs. (B) The expression of the major membrane-type collagenase, MMP14. The gene expressions of MMP2 (C) and secreted collagenases, MMP8 (D) and MMP13 (E), are shown. Each gene expression is normalized for 36B4 mRNA levels. Means ± SEM, N = 3 for each condition.* P<0.05, **P<0.005
Mmp14 that encodes the dominant pericellular collagenase in mammalian cells is expressed in both iWAT and eWAT-derived Sca1high cells (Figure 5B). The expression of Mmp14 is 33% more in iWAT-derived Sca1high cells than eWAT-derived Sca1high cells. Mmp2 encoding the most abundant gelatinase/type IV collagenase is expressed 67% more in iWAT-derived Sca1high cells than in eWAT-derived Sca1high cells (iWAT 2.2 ± 0.4 vs. eWAT 1.3 ± 0.2, means ± SEM). By contrast, the expression of neutrophil collagenase (MMP8) in eWAT-derived Sca1high cells was increased 120-fold relative to in iWAT-derived Sca1high cells (p = 0.001, Figure 5D). Likewise, the expression of fibroblast collagenase (MMP13) was twice as high in eWAT-derived Sca1high ASCs as in iWAT-derived Sca1high ASCs albeit to a level statistically less significant (p = 0.08, Figure 5E).
2.6. TIMPs and collagen family members
Consistent with the higher expressions of soluble collagenases in eWAT-derived Sca1high ASCs, the endogenous MMP inhibitor, TIMP1, which effectively blocks secreted MMPs (MMP8 and MMP13) but not membrane-type MMPs (MMP14 and MMP15) was expressed three-times as high in eWAT-derived Sca1high ASCs as in iWAT-derived cells (Supplemental Figure 1A). TIMP2, which is an effective inhibitor against both secreted and membrane-type MMPs, was expressed almost equally in eWAT- and iWAT-derived Sca1high cells (Supplemental Figure 1B). TIMP3 expression was 44% less in eWAT-derived Sca1high cells than in iWAT-derived Sca1high cells (p = 0.11, Supplemental Figure 1C). Among collagen family members, the expressions of type I and IV collagens (Col1a1 and Col4a1) were not significantly different between Sca1high and Sca1low cells and also between different fat depots (Supplemental Figure 2 A and C). The expression of fibrogenic type III collagen (Col3a1), however, was higher in eWAT-derived Sca1high cells than in iWAT-derived Sca1high cells (3-fold increase). On the other hand, the expressions of adipose-specific collagen genes, such as Col5a3 (Huang et al., 2011) and Col6a1 (Khan et al., 2009), were higher in iWAT-derived Sca1high cells than in eWAT-derived Sca1high cells (6- and 2-fold increases, respectively) (Supplement Figure 2 D, E).
2.7. In vivo collagen degradation and gene signature
Based on these results, we examined the type I collagen degradation in vivo. Inguinal and epididymal adipose tissues were recovered from 6-week-old male mice and stained for Sca1 and MMP-dependent collagen degradation products. In iWAT, MMP-dependent collagen cleavage was observed in association with Sca1high ASCs (Figure 6). The collagen degradation products were concentrated on the plasma membrane of Sca1high ASCs (Figure 6, left). By contrast, collagen degradation products were observed more diffusely in eWAT and not necessarily concentrated at the plasma membrane of Sca1high ASCs (Figure 6, right). MMP-dependent degradation products were also found in the cytoplasm of Sca1-negative cells, suggesting the possible engulfment of degraded collagens by Sca1-negative cells.
Figure 6.

In vivo collagenolysis mediated by Sca1high ASCs in iWAT and eWAT. (A) The MMP-dependent collagenolysis in iWAT and eWAT was detected with an antibody reactive with MMP-dependent collagen cleavage products (green). Sca1 (red) and nucleus (green) are shown. Focal collagenolysis was increased at Sca1-positive plasma membrane in iWAT, whereas cell-associated collagen cleavage products observed diffusely in eWAT. Scale = 10 μm.
3. Discussion
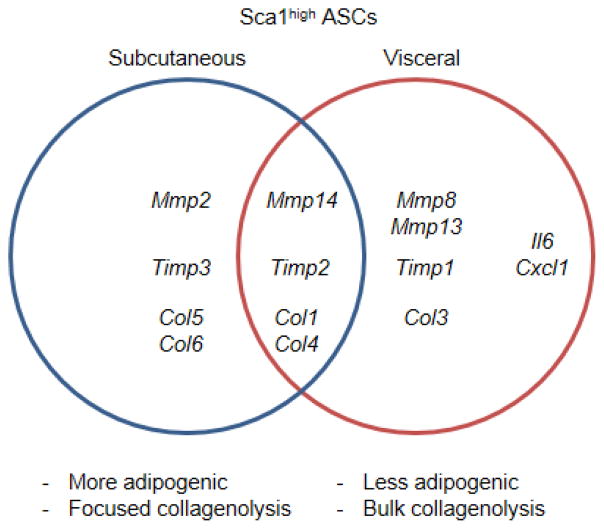
ASCs not only possess a therapeutic potential in regenerative medicine (Gimble et al., 2007; Mizuno et al., 2012), but also provide critical insights into the disease processes underlying chronic and metabolic diseases. In this paper, we have demonstrated that subcutaneous and visceral Sca1high ASCs display depot-specific gene signatures in a manner coupled with different patterns of ECM remodeling. Fat depot-dependent differences in the transcriptome and ECM remodeling of ASCs may contribute to the diverse adipose tissue function observed during development and obesity progression (Figure 7).
Figure 7.
The fat depot-specific expression of collagenolytic MMPs, MMP inhibitors, and collagens in Sca1high ASCs.
A recent study that monitored adipogenesis in vivo with adipocyte lineage tracing suggests that subcutaneous adipose tissues develop earlier than visceral adipose tissues in mice and only visceral adipose tissues actively engage adipogenesis during obesity progression (Wang et al., 2013). In vitro, however, Sca1high ASCs isolated from visceral, epididymal adipose tissues (eWAT) were not able to differentiate into adipocytes as effectively as subcutaneous, iWAT-derived Sca1high ASCs in our study. Similar findings were obtained by others (Han et al., 2011; Macotela et al., 2012). Macotela et al., however, were able to rescue the adipogenic potential of eWAT-derived ASCs by adding BMP4. Han et al., could partially rescue the adipogenic potential of eWAT-derived VSCs by culturing the eWAT-derived VSCs in three-dimensional (3-D) Matrigel (Han et al., 2011). These findings suggest that iWAT-derived ASCs can initiate adipogenesis in vivo and in vitro when appropriate hormonal, nutritional, or spatial cues is provided. On the other hand, iWAT-derived ASCs were able to effectively differentiate into adipocytes in vitro with a simple adipogenic mix despite the reduced adipogenic potential noted in vivo (Wang et al., 2013). The discrepancy found in fat depot-specific adipogenesis between in vitro and in vivo settings is intriguing. Adipocyte progenitor cells or mesenchymal stem cells in subcutaneous adipose tissues of adult mice may remain quiescent until they are challenged with tissue injury (Schmidt and Horsley, 2013) or cold exposure (Wang et al., 2013). The lobules of adipocytes are enwrapped by a dense network of collagen fibers, particularly in subcutaneous adipose tissues (Chun et al., 2006). Type I collagen fibers are suppressive to adipogenic histone modifications, which precede adipocyte gene expression (Sato-Kusubata et al., 2011). Therefore, the collagen-rich environment may restrain the full adipogenic potential of ASCs in subcutaneous adipose tissues in vivo. Once freed from the collagenous confinement, these iWAT-derived ASCs may become fully capable of differentiating into adipocytes in vitro. On the other hand, eWAT-derived ASCs displayed a very limited adipogenic potential in vitro. While the limitation appears to be alleviated by BMP4 (Macotela et al., 2012) or 3-D Matrigel (Han et al., 2011), it is likely that the adipogenesis in visceral adipose tissues in vivo is regulated more by hormonal and nutritional cues (Wang et al., 2013) and tissue microenvironment in vivo as well as angiogenesis (Han et al., 2011; Hausman and Richardson, 2004; Rupnick et al., 2002). Despite those findings from rodents, the fat depot-specific adipogenic potential as well as temporospatial regulation of adipose tissue development may differ across species; therefore, caution should be exercised when extrapolating these findings from rodents to humans (Ailhaud, 1992; Hauner and Entenmann, 1991; Loncar, 1991).
Sca1high ASCs are found in the vicinity of blood vessels as well as between adipocytes. Characterizing the proliferation, migration, and differentiation of these ASCs during development and obesity progression may help us better understand the intricate dynamics of adipocytes and interspersing ASCs in the regulation of adipose tissue function. RNA-seq data suggest that some cytokines or chemokines, such as IL-6 and CXCL1, are expressed more in Sca1high ASCs than in Sca1low cells. The pro-inflammatory or immunomodulatory characteristics of mesenchymal stem cells have been also reported by others (English, 2013; Hoogduijn et al., 2013). The expressions of these cytokines or chemokines are decreased during adipocyte differentiation (unpublished data). As such, the expression of these stem cell-derived cytokines in adipose tissues may represent the number of adipogenic stem cells within adipose tissues, at least in physiological conditions. Consistent with the notion, inverse correlations between inflammatory markers and body mass index have been noted in non-obese children (Zabaleta et al., 2013). IL6 knockout mice develop obesity and diabetes (Wallenius et al., 2002). The expression of IL6 and other chemokines from ASCs may serve as a negative feedback mechanism that suppresses adipogenesis and inhibits the hypertrophic change of neighboring adipocytes (van Hall et al., 2003). In the chronic phase of obesity, M1-like macrophage infiltration dominates in visceral adipose tissues (Olefsky and Glass, 2010); however, in physiological stages or in early stages of obesity, cytokines and chemokines expressed by Sca1high ASCs within adipose tissues may play a collective role in regulating adipose tissue size and function.
Diverse functional molecules are expressed differently between eWAT (Vis)- and iWAT (SQ)-derived ASCs. The gene expressions of basement membrane proteins (laminins, nidogen, fibulins, and tenascin), ADAMTS family members (ADAMTS1, -5, -8, -9, -15), latent TGF-beta binding proteins (LTBP1 and -2), WNT receptor, SFRP1, and ECM modifiers, cell adhesion, and prothromobotic molecules, such as Matrix Gla protein (MGP), tissue transglutaminase (TGM2), and tissue factor (TF) are expressed more in Vis ASCs. The expression of these genes may significantly modify the adhesive and proliferative function of Vis ASCs within epididymal adipose tissues. Pro-adipogenic WNT4 (Nishizuka et al., 2008) and BMP4 (Macotela et al., 2012; Qian et al., 2013) are also expressed more in Vis ASCs. While our assessment using conventional adipogenesis induction did not reveal the adipogenic potential of Vis ASCs, the expressions of these pro-adipogenic genes may contribute to adipogenesis in vivo. In order to define the biological and metabolic roles played by these sets of genes expressed in a fat depot-specific manner, we may need to further identify their genotype-phenotype links in animal models and define the diverse molecular and biological interactomes among them.
Sca1 (Ly6a) is highly expressed in pro-adipogenic mesenchymal stem cells (Staszkiewicz et al., 2008); however, the gene targeting of Sca1 in mice did not cause a significant change in adipose tissue size (Staszkiewicz et al., 2012). These findings suggest that Sca1 serves as a useful cell surface marker to identify pro-adipogenic ASCs; however, Sca1 itself may not play a substantial biological role in the regulation of adipose tissue size and function. Despite the fact that Sca1 may merely function as one of stem cell markers, in both subcutaneous and visceral adipose tissues of mice, we found a cohort of ECM and ECM modifier genes co-segregate with Sca1 expression. As such, Sca1 is a useful cell-surface marker to purify adipose-derived stem cells in mice in order to assess their biological roles in adipose tissue remodeling and function. Our approach of using RNA-seq for the functional characterization of Sca1high ASCs uncovered some of unexpected characteristics of Sca1high ASCs in comparison to Sca1low adipose-derived VSCs. Among four facilitated glucose transporters (Slc2a1, Slc2a2, Slc2a3, Slc2a4; also known as Glut1~4), Glut1 was the dominant glucose transporter expressed in adipose VSCs. The level of Glut3 expression was 1/10 of Glut1 expression (data not shown). Glut2 and Glut4 were not expressed (<1.0 FPKM). Notably, the expression Glut1 was 50% less in Sca1high ASCs than in Sca1low VSCs. It is intriguing to speculate that the glucose metabolism of Sca1high ASCs might significantly differ from that of Sca1low cells in adipose tissues, suggesting that lower glucose uptake or glycolysis may define some of the stem cell-like characteristics of Sca1high cells. Supporting the potentially altered balance between glycolysis and mitochondrial activities in mesenchymal stem cells, Sca1high ASCs displayed two-fold increase in the expression of mitochondrial NPDH dehydrogenases (mt-Nd1m mt-Nd2) (Supplemental table 1). Reduced Glut1 expression may render Sca1high ASCs more sensitive to hypoxia and mitochondrial dysfunction (Heilig et al., 2003). As nutritional stress, such as high-fat diet, induces mitochondrial dysfunction as well as hypoxia, the biological function of Sca1high ASCs in these stress conditions may contribute to the metabolic phenotypes, such as obesity and diabetes (Oñate et al., 2012).
The Sca1high ASCs display distinct expressions of the proteins found in extracellular space and on the plasma membrane. The proteins expressed in extracellular space and on the plasma membrane appear to regulate the fat depot-specific collagen remodeling. The visceral adipose tissue adapts to nutritional challenge more rapidly than the subcutaneous adipose tissue. It is speculated that dynamic ECM remodeling is required during adipogenesis and adipose tissue expansion (Chun, 2012; Chun et al., 2006; Chun et al., 2010). As such, visceral adipose tissues may require enhanced collagenolysis at higher levels than subcutaneous adipose tissues in adult mice. On the other hand, high-fat diet feeding induces higher expression of collagens than control diet (Chun et al., 2010). We speculate that visceral adipose tissues are more susceptible to high-fat diet-induced adipose tissue fibrosis than subcutaneous adipose tissues because the delicate balance between the synthesis and the degradation of collagens needs to be maintained at higher levels in visceral adipose tissues. The imbalance in remodeling collagens and other ECM proteins may trigger pro-inflammatory macrophage infiltration that preferentially occurs in visceral adipose tissues (Inoue et al., 2013; Lumeng et al., 2008).
Collagen remodeling can be regulated not only by MMP family members, but also by lysosomal catabolism mediated by cathepsins (Arora et al., 2000) and non-proteolytic mechanism (Phillips et al., 2003; Wolf et al., 2003). Unlike the collagenolysis by iWAT-derived Sca1high cells, the collagen displacement mediated by eWAT-derived Sca-1high cells was not completely blocked in the presence of a synthetic MMP inhibitor. Further studies are needed to determine whether non-MMP proteinases, such as cathepsins, or non-proteolytic mechanism, such as cellular contraction (Phillips et al., 2003) or physical displacement (Wolf et al., 2003), mediate the collagen remodeling of eWAT-derived Sca1high ASCs in vitro and in vivo.
4. Materials and Methods
4.1. Mice and isolation of vascular stromal cells (VSCs)
C57BL/6J mice (male, 4-weeks-old) were purchased from The Jackson Laboratory. At 6-weeks-old, inguinal and epididymal adipose tissues were dissected and weighed. Part of inguinal and epididymal adipose tissues was fixed in 4% paraformaldehyde in phosphate buffered saline (PBS), pH 7.4 for 5 minutes and subjected to whole-mount immunostaining. Remaining adipose tissues were minced with scissors and digested in 5 mg/ml collagenase type 3 (derived from Clostridium histolyticum, Worthington) in Hank’s Balanced Salt Solution with calcium and magnesium, HBSS (+) (Life Technology) at 37°C for 30~45 minutes until the tissues were well dispersed. After blocking collagenase activity by adding DMEM with 10% FBS (Hyclone), cell suspensions were filtered through 100 μm cell strainer to remove debris. After spinning down, red blood cells were lysed by suspending the cell pellets in water for 2 minutes. After resuspending the pellets in a large volume of DMEM with 10% FBS, cells were passed through cell strainers. After a repeat centrifuge, the cell pellets were suspended in DMEM with 10% FBS and plated onto tissue culture plates (Falcon).
4.2. Magnetic-activated cell sorting (MACS)-based enrichment of Sca1high and Sca1low cells
Three to four hours after plating VSCs cells, plates were gently rinsed with HBSS(−) twice to remove non-adherent cells, then adherent cells were trypsinized and recovered. MACS (Miltenyi Biotec) was preformed following the manufacturer’s instruction. In this system, 50 nm superparamagnetic particles conjugated to highly specific antibodies (MACS MciroBeads) were used to magnetically label target cells. The magnetically labeled cells were retained within a MACS Column, a matrix composed of ferromagnetic spheres covered with a cell-friendly coating. The VSCs prepared from inguinal and perigonadal adipose tissues (106 cells) were suspended in 45 μl PBS with 0.5% BSA and 2 mM EDTA; 5 μl of rat anti-Sca1-FITC antibody (30 μg/ml, Miltenyi Biotec) was added, then incubated for 10 minutes at 4°C. After centrifugation and the removal of supernatant and repeated wash with 1 ml of PBS, anti-FITC MicroBeads (Miltenyi Biotec) was added and incubated for 15 minutes at 4°C. After repeated washes and the removal of supernatant, the cell suspension was passed through a MACS column attached to magnetic board; cells not trapped in the column (flow-through) were collected as Sca1low cells. After three times washes with PBS, the column was removed from the magnetic field of the separator and the target cells within the column were eluted as Sca1high cells.
4.3. Immunofluorescent imaging
After fixation in 4% paraformaldehyde/PBS, a small piece of adipose tissue (~5 mm) was permeabilized with PBS 0.5% Triton-X 100 for 5 minutes, blocked in PBS with 2% goat serum and 2% BSA, then incubated with rat anti-Ly6A/E antibody (BD Pharmingene) to detect Sca1 expression, and rabbit anti-type IV collagen antibody (Rockland) to detect adipocytes and vasculature, or polyclonal C1,2C (Col 2 3/4Cshort, IBEX Pharmaceuticals Inc, Montreal) to detect MMP-dependent collagen degradation. After repeated washing, samples were incubated with donkey Alexa Fluor 594 anti-rat IgG (Invitrogen) and/or Alexa Fluor 488 anti-rabbit IgG (Invitrogen). The nuclei were stained with Hoechst 34580 (Invitrogen). Samples were mounted in ProLong Gold Antifade Reagent (Invitrogen) and fluorescent images were obtained with Olympus FluoView 500 Laser Scanning Confocal Microscope. The detailed methods were described elsewhere (Chun and Inoue, 2014).
4.4. Flow cytometry
Vascular stromal cells were isolated as described above. Trypsinized cells were washed in HBSS (−) twice and centrifuged. The cell pellets (1.0×106 cells) were suspended in PBS with 2% goat serum and 2% BSA for 30 minutes. The isotype controls (rat IgG2a-Alexa Fluor 647 and -R-PE), anti-Sca1-Alexa Fluor 647 (Invitrogen), anti-F4/80-R-PE (Invitrogen), or both anti-Sca1-Alexa Fluor 647 and anti-F4/80-R-PE antibodies were added to each sample and incubated for 30 min at 4°C. After repeated washing, the cell pellets were suspended and passed through a cell strainer and analyzed with MoFlo Astrios (Beckman, University of Michigan Flow Cytometry Core). The gating was performed blindly by a technician at flow cytometry core based on the flow results of respective isotype controls.
4.5. In vitro adipogenesis
Adipogenesis was induced by adding 10 μg/ml porcine insulin, 0.5 mM 3-Isobutyl-1-Methyxathine, and 0.25 μM dexamethasone (all from Sigma) as described before (Sato-Kusubata et al., 2011). The lipid droplets were visualized using BODIPY 493/503 (Molecular Probe) (Listenberger and Brown, 2001).
4.6. RNA-seq
Illumina libraries were prepared at the University of Michigan DNA sequencing core using TrueSeq RNA kit (Illumina) following the manufacturer’s protocol. Samples were indexed with adapter sequences and run on Illumina HiSeq2000 for 50 bp paired-end reading. The sequenced reads were aligned to the mouse genome with Bowtie2 v2.0.6 (Schatz et al., 2010) and gene expression analyzed with Cufflinks v2.0.2 (Trapnell et al., 2010). A p-value corrected for an FDR (q-value) of 0.05 was considered statistically significant. Heatmaps were generated in R version 2.15.1 with the heatmap.2 function in the gplots package (version 2.11.0) using the hierarchical clustering method ward and the distance function euclidean. GO enrichment analysis was performed with The Database for Annotation, Visualization and Integrated Discovery (DAVID) using the default genome-wide gene list as the background list (Huang et al., 2008).
4.7. Real-time PCR
Total RNA was isolated with RNeasy kit (Qiagen) following the manufacturer’s instruction. After cDNA synthesis using Superscript III (Invitrogen), real time quantitative PCR performed in StepOnePlus (Applied Biosystems) using either SYBR Green primers or Taqman primers/probes for respective gene (Integrated DNA Technologies). The quantity of 36B4 (Rplp0) was used as a denominator to obtain the adjusted expression level of each gene. The sequences of primers and probes used are described in the supplement document.
4.8. Collagen degradation assay
Fluoresceinated collagen degradation assay was performed as described previously (Chun and Inoue, 2014; Sato-Kusubata et al., 2011). Type I collagen was extracted from aged rat tails and lyophilized. Type I collagen was solubilized in 37 mM acetic acid (2.7 mg/ml) at 4°C overnight. An 18 mm × 18 mm coverslip was placed in a well of a 6-well-plate. Eight volumes of type I collagen, 1 vol. of 10x MEM (Invitrogen), 1 vol. of 0.34N NaOH, and 25 mM HEPES were mixed on ice. Each well containing a coverslip was overlaid with 120 μl of neutralized collagen and incubated at 37°C for 1 hour. The polymerized collagen was labeled with Alexa594 carobxylic acid, succinimidyl ester (Invitrogen) following the manufacturer’s protocol. After repeated washing, cells were plated onto fluoresceinated collagen gels. 72 hours after plating, cells were fixed and coverslips were mounted onto slide glasses and observed under inverted fluorescent microscopy (Olympus IX 71).
Supplementary Material
Highlights.
Sca1high ASCs demonstrate fat depot-specific adipogenic potentials in vitro.
Sca1high ASCs display depot-specific gene expression of ECM and adhesion molecules.
Sca1high ASCs engage in MMP-mediated collagenolysis in a depot-specific manner.
Acknowledgments
We thank Carey Lumeng (University of Michigan) for scientific discussions, the University of Michigan Cancer Center Flow Cytometry Core for assistance in flow cytometry, and the DNA Sequencing Core for performing RNA-seq. This work is supported by NIH DK095137 and HL106332 (T-H.C.) and DK084079 (D.A.B). Confocal imaging was performed with the support from Michigan Diabetes Research Center funded by P30DK020572.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ailhaud G. Some new aspects on adipose tissue development. Diabetes Metabolism Reviews. 1992;8:3–7. doi: 10.1002/dmr.5610080103. [DOI] [PubMed] [Google Scholar]
- Arora PD, Manolson MF, Downey GP, Sodek J, McCulloch CAG. A Novel Model System for Characterization of Phagosomal Maturation, Acidification, and Intracellular Collagen Degradation in Fibroblasts. Journal of Biological Chemistry. 2000;275:35432–35441. doi: 10.1074/jbc.M003221200. [DOI] [PubMed] [Google Scholar]
- Chavey C, Mari B, Monthouel MN, Bonnafous S, Anglard P, Van Obberghen E, Tartare-Deckert S. Matrix Metalloproteinases Are Differentially Expressed in Adipose Tissue during Obesity and Modulate Adipocyte Differentiation. Journal of Biological Chemistry. 2003;278:11888–11896. doi: 10.1074/jbc.M209196200. [DOI] [PubMed] [Google Scholar]
- Chun TH. Peri-adipocyte ECM remodeling in obesity and adipose tissue fibrosis. Adipocyte. 2012;1:89–95. doi: 10.4161/adip.19752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TH, Hotary KB, Sabeh F, Saltiel AR, Allen ED, Weiss SJ. A pericellular collagenase directs the 3-dimensional development of white adipose tissue. Cell. 2006;125:577–591. doi: 10.1016/j.cell.2006.02.050. [DOI] [PubMed] [Google Scholar]
- Chun TH, Inoue M. 3-D Adipocyte Differentiation and Peri-adipocyte Collagen Turnover. Methods in enzymology. 2014;538:15–34. doi: 10.1016/B978-0-12-800280-3.00002-5. [DOI] [PubMed] [Google Scholar]
- Chun TH, Inoue M, Morisaki H, Yamanaka I, Miyamoto Y, Okamura T, Sato-Kusubata K, Weiss SJ. Genetic link between obesity and MMP14-dependent adipogenic collagen turnover. Diabetes. 2010;59:2484–2494. doi: 10.2337/db10-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancevic CM, Fraser FW, Smith AD, Stupka N, Ward AC, McCulloch DR. Biosynthesis and expression of a disintegrin-like and metalloproteinase domain with thrombospondin-1 repeats-15: a novel versican-cleaving proteoglycanase. The Journal of biological chemistry. 2013;288:37267–37276. doi: 10.1074/jbc.M112.418624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divoux A, Tordjman J, Lacasa D, Veyrie N, Hugol D, Aissat A, Basdevant A, Guerre-Millo M, Poitou C, Zucker JD, Bedossa P, Clément K. Fibrosis in Human Adipose Tissue: Composition, Distribution, and Link With Lipid Metabolism and Fat Mass Loss. Diabetes. 2010;59:2817–2825. doi: 10.2337/db10-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield JS, Lupher M, Thannickal VJ, Wynn TA. Host responses in tissue repair and fibrosis. Annu Rev Pathol. 2013;8:241–276. doi: 10.1146/annurev-pathol-020712-163930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English K. Mechanisms of mesenchymal stromal cell immunomodulation. Immunol Cell Biol. 2013;91:19–26. doi: 10.1038/icb.2012.56. [DOI] [PubMed] [Google Scholar]
- Gimble JM, Katz AJ, Bunnell BA. Adipose-Derived Stem Cells for Regenerative Medicine. Circulation Research. 2007;100:1249–1260. doi: 10.1161/01.RES.0000265074.83288.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Cruz RD, Darling EM. Adipose-derived stem cell fate is predicted by cellular mechanical properties. Adipocyte. 2013;2:87–91. doi: 10.4161/adip.23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee JE, Jin J, Lim JS, Oh N, Kim K, Chang SI, Shibuya M, Kim H, Koh GY. The spatiotemporal development of adipose tissue. Development. 2011;138:5027–5037. doi: 10.1242/dev.067686. [DOI] [PubMed] [Google Scholar]
- Hauner H, Entenmann G. Regional variation of adipose differentiation in cultured stromal-vascular cells from the abdominal and femoral adipose tissue of obese women. Int J Obes. 1991;15:121–126. [PubMed] [Google Scholar]
- Hausman GJ, Richardson RL. Adipose tissue angiogenesis. Journal of Animal Science. 2004;82:925–934. doi: 10.2527/2004.823925x. [DOI] [PubMed] [Google Scholar]
- Heilig C, Brosius F, Siu B, Concepcion L, Mortensen R, Heilig K, Zhu M, Weldon R, Wu G, Conner D. Implications of Glucose Transporter Protein Type 1 (GLUT1)-Haplodeficiency in Embryonic Stem Cells for Their Survival in Response to Hypoxic Stress. The American Journal of Pathology. 2003;163:1873–1885. doi: 10.1016/S0002-9440(10)63546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogduijn MJ, Roemeling-van Rhijn M, Engela AU, Korevaar SS, Mensah FK, Franquesa M, de Bruin RW, Betjes MG, Weimar W, Baan CC. Mesenchymal stem cells induce an inflammatory response after intravenous infusion. Stem Cells Dev. 2013;22:2825–2835. doi: 10.1089/scd.2013.0193. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang G, Ge G, Wang D, Gopalakrishnan B, Butz DH, Colman RJ, Nagy A, Greenspan DS. alpha3(V) collagen is critical for glucose homeostasis in mice due to effects in pancreatic islets and peripheral tissues. J Clin Invest. 2011;121:769–783. doi: 10.1172/JCI45096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Jiang Y, Barnes RH, 2nd, Tokunaga M, Martinez-Santibanez G, Geletka L, Lumeng CN, Buchner DA, Chun TH. Thrombospondin 1 mediates high-fat diet-induced muscle fibrosis and insulin resistance in male mice. Endocrinology. 2013;154:4548–4559. doi: 10.1210/en.2013-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandyba E, Leung Y, Chen YB, Widelitz R, Chuong CM, Kobielak K. Competitive balance of intrabulge BMP/Wnt signaling reveals a robust gene network ruling stem cell homeostasis and cyclic activation. Proceedings of the National Academy of Sciences. 2013;110:1351–1356. doi: 10.1073/pnas.1121312110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessenbrock K, Plaks V, Werb Z. Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan T, Muise ES, Iyengar P, Wang ZV, Chandalia M, Abate N, Zhang BB, Bonaldo P, Chua S, Scherer PE. Metabolic dysregulation and adipose tissue fibrosis: role of collagen VI. Molecular and cellular biology. 2009;29:1575–1591. doi: 10.1128/MCB.01300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listenberger LL, Brown DA. Current Protocols in Cell Biology. John Wiley & Sons, Inc; 2001. Fluorescent Detection of Lipid Droplets and Associated Proteins. [DOI] [PubMed] [Google Scholar]
- Loncar D. Development of thermogenic adipose tissue. The International journal of developmental biology. 1991;35:321–333. [PubMed] [Google Scholar]
- Lumeng CN, DelProposto JB, Westcott DJ, Saltiel AR. Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes. 2008;57:3239–3246. doi: 10.2337/db08-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macotela Y, Emanuelli B, Mori MA, Gesta S, Schulz TJ, Tseng YH, Kahn CR. Intrinsic Differences in Adipocyte Precursor Cells From Different White Fat Depots. Diabetes. 2012;61:1691–1699. doi: 10.2337/db11-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Tobita M, Uysal AC. Concise Review: Adipose-Derived Stem Cells as a Novel Tool for Future Regenerative Medicine. STEM CELLS. 2012;30:804–810. doi: 10.1002/stem.1076. [DOI] [PubMed] [Google Scholar]
- Morikawa S, Mabuchi Y, Kubota Y, Nagai Y, Niibe K, Hiratsu E, Suzuki S, Miyauchi-Hara C, Nagoshi N, Sunabori T, Shimmura S, Miyawaki A, Nakagawa T, Suda T, Okano H, Matsuzaki Y. Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. The Journal of Experimental Medicine. 2009;206:2483–2496. doi: 10.1084/jem.20091046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nho YK, Ha E, Yu KI, Chung JH, Wook NC, Chung IS, Lee MY, Shin DH. Matrix metalloproteinase-1 promoter is associated with body mass index in Korean population with aged greater or equal to 50 years. Clin Chim Acta. 2008;396:14–17. doi: 10.1016/j.cca.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Nishizuka M, Koyanagi A, Osada S, Imagawa M. Wnt4 and Wnt5a promote adipocyte differentiation. FEBS Letters. 2008;582:3201–3205. doi: 10.1016/j.febslet.2008.08.011. [DOI] [PubMed] [Google Scholar]
- Olefsky JM, Glass CK. Macrophages, Inflammation, and Insulin Resistance. Annual Review of Physiology. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- Oñate B, Vilahur G, Ferrer-Lorente R, Ybarra J, Díez-Caballero A, Ballesta-López C, Moscatiello F, Herrero J, Badimon L. The subcutaneous adipose tissue reservoir of functionally active stem cells is reduced in obese patients. The FASEB Journal. 2012;26:4327–4336. doi: 10.1096/fj.12-207217. [DOI] [PubMed] [Google Scholar]
- Phillips JA, Vacanti CA, Bonassar LJ. Fibroblasts regulate contractile force independent of MMP activity in 3D-collagen. Biochem Biophys Res Commun. 2003;312:725–732. doi: 10.1016/j.bbrc.2003.10.179. [DOI] [PubMed] [Google Scholar]
- Qian SW, Tang Y, Li X, Liu Y, Zhang YY, Huang HY, Xue RD, Yu HY, Guo L, Gao HD, Liu Y, Sun X, Li YM, Jia WP, Tang QQ. BMP4-mediated brown fat-like changes in white adipose tissue alter glucose and energy homeostasis. Proceedings of the National Academy of Sciences. 2013;110:E798–E807. doi: 10.1073/pnas.1215236110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard-Blum S. The collagen family. Cold Spring Harb Perspect Biol. 2011;3:a004978. doi: 10.1101/cshperspect.a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. Inhibition of Adipogenesis by Wnt Signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- Rupnick MA, Panigrahy D, Zhang CY, Dallabrida SM, Lowell BB, Langer R, Folkman MJ. Adipose tissue mass can be regulated through the vasculature. Proceedings of the National Academy of Sciences. 2002;99:10730–10735. doi: 10.1073/pnas.162349799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeh F, Li XY, Saunders TL, Rowe RG, Weiss SJ. Secreted versus membrane-anchored collagenases: relative roles in fibroblast-dependent collagenolysis and invasion. J Biol Chem. 2009;284:23001–23011. doi: 10.1074/jbc.M109.002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato-Kusubata K, Jiang Y, Ueno Y, Chun TH. Adipogenic Histone Mark Regulation by Matrix Metalloproteinase 14 in Collagen-Rich Microenvironments. Mol Endocrinol. 2011;25:745–753. doi: 10.1210/me.2010-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz MC, Langmead B, Salzberg SL. Cloud computing and the DNA data race. Nat Biotech. 2010;28:691–693. doi: 10.1038/nbt0710-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt BA, Horsley V. Intradermal adipocytes mediate fibroblast recruitment during skin wound healing. Development. 2013;140:1517–1527. doi: 10.1242/dev.087593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangrude G, Heimfeld S, Weissman I. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- Staszkiewicz J, Gimble JM, Dietrich MA, Gawronska-Kozak B. Diet-induced obesity in stem cell antigen-1 KO mice. Stem Cells Dev. 2012;21:249–259. doi: 10.1089/scd.2010.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staszkiewicz J, Gimble JM, Manuel JA, Gawronska-Kozak B. IFATS collection: Stem cell antigen-1-positive ear mesenchymal stem cells display enhanced adipogenic potential. STEM CELLS. 2008;26:2666–2673. doi: 10.1634/stemcells.2008-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Tordjman J, Clément K, Scherer Philipp E. Fibrosis and Adipose Tissue Dysfunction. Cell metabolism. 2013;18:470–477. doi: 10.1016/j.cmet.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, Graff JM. White Fat Progenitor Cells Reside in the Adipose Vasculature. Science. 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traurig MT, Permana PA, Nair S, Kobes S, Bogardus C, Baier LJ. Differential Expression of Matrix Metalloproteinase 3 (MMP3) in Preadipocytes/Stromal Vascular Cells From Nonobese Nondiabetic Versus Obese Nondiabetic Pima Indians. Diabetes. 2006;55:3160–3165. doi: 10.2337/db06-0373. [DOI] [PubMed] [Google Scholar]
- van de Rijn M, Heimfeld S, Spangrude GJ, Weissman IL. Mouse hematopoietic stem-cell antigen Sca-1 is a member of the Ly-6 antigen family. Proceedings of the National Academy of Sciences. 1989;86:4634–4638. doi: 10.1073/pnas.86.12.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hall G, Steensberg A, Sacchetti M, Fischer C, Keller C, Schjerling P, Hiscock N, Moller K, Saltin B, Febbraio MA, Pedersen BK. Interleukin-6 stimulates lipolysis and fat oxidation in humans. The Journal of clinical endocrinology and metabolism. 2003;88:3005–3010. doi: 10.1210/jc.2002-021687. [DOI] [PubMed] [Google Scholar]
- Vázquez F, Hastings G, Ortega MA, Lane TF, Oikemus S, Lombardo M, Iruela-Arispe ML. METH-1, a Human Ortholog of ADAMTS-1, and METH-2 Are Members of a New Family of Proteins with Angio-inhibitory Activity. Journal of Biological Chemistry. 1999;274:23349–23357. doi: 10.1074/jbc.274.33.23349. [DOI] [PubMed] [Google Scholar]
- Verma P, Dalal K. ADAMTS-4 and ADAMTS-5: Key enzymes in osteoarthritis. Journal of Cellular Biochemistry. 2011;112:3507–3514. doi: 10.1002/jcb.23298. [DOI] [PubMed] [Google Scholar]
- Wallenius V, Wallenius K, Ahren B, Rudling M, Carlsten H, Dickson SL, Ohlsson C, Jansson JO. Interleukin-6-deficient mice develop mature-onset obesity. Nature medicine. 2002;8:75–79. doi: 10.1038/nm0102-75. [DOI] [PubMed] [Google Scholar]
- Wang QA, Tao C, Gupta RK, Scherer PE. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nature medicine. 2013;19:1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welm BE, Tepera SB, Venezia T, Graubert TA, Rosen JM, Goodell MA. Sca-1pos Cells in the Mouse Mammary Gland Represent an Enriched Progenitor Cell Population. Developmental Biology. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- Wolf K, Müller R, Borgmann S, Bröcker EB, Friedl P. Amoeboid shape change and contact guidance: T-lymphocyte crawling through fibrillar collagen is independent of matrix remodeling by MMPs and other proteases. Blood. 2003;102:3262–3269. doi: 10.1182/blood-2002-12-3791. [DOI] [PubMed] [Google Scholar]
- Zabaleta J, Velasco-Gonzalez C, Estrada J, Ravussin E, Pelligrino N, Mohler MC, Larson-Meyer E, Boulares AH, Powell-Young Y, Bennett B, Happel K, Cefalu W, Scribner R, Tseng TS, Sothern M. Inverse correlation of serum inflammatory markers with metabolic parameters in healthy, Black and White prepubertal youth. Int J Obes. 2013 doi: 10.1038/ijo.2013.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.