Abstract
Regulation of hematopoietic stem cell proliferation, lineage commitment, and differentiation in adult vertebrates requires extrinsic signals provided by cells in the marrow microenvironment (ME) located within the bone marrow. Both secreted and cell-surface bound factors critical to this regulation have been identified, yet control of their expression by cells within the ME has not been addressed. Herein we hypothesize that microRNAs (miRNAs) contribute to their controlled expression. MiRNAs are small noncoding RNAs that bind to target mRNAs and downregulate gene expression by either initiating mRNA degradation or preventing peptide translation. Testing the role of miRNAs in downregulating gene expression has been difficult since conventional techniques used to define miRNA-mRNA interactions are indirect and have high false-positive and negative rates. In this report, a genome-wide biochemical technique (high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation or HITS-CLIP) was used to generate unbiased genome-wide maps of miRNA-mRNA interactions in two critical cellular components of the marrow ME: marrow stromal cells and bone marrow endothelial cells. Analysis of these datasets identified miRNAs as direct regulators of JAG1, WNT5A, MMP2, and VEGFA; four factors that are important to ME function. Our results show the feasibility and utility of unbiased genome-wide biochemical techniques in dissecting the role of miRNAs in regulation of complex tissues such as the marrow ME.
Introduction
Hematopoiesis in the adult vertebrate occurs within trabecular bone spaces where specialized functional niches are postulated to exist for different stages of hematopoietic maturation [1–3]. Many cell types populate the marrow microenvironment (ME), including endothelial cells, fibroblast-like stromal or reticular cells, osteoblasts, and macrophages. Together they produce a variety of factors that are important to hematopoietic stem cell (HSC) fate decisions including CXCL12 (stromal-derived factor 1) [4], Kit ligand (stem cell factor or KITL) [5], Angiopoietin1 (ANGPT1) [6], Notch ligands [7], Wnt proteins [8], and thrombopoietin [9]. Mechanisms that regulate expression of these factors among the various cell types of the ME remain undefined [10, 11]. Furthermore, none of these “niche associated genes” are restricted to one cell type: CXCL12 is produced by marrow stromal cells (MSCs) [12], osteoblasts [13], and endothelial cells [14]; ANGPT1 is produced by endothelial cells [6], osteoblasts [15], megakaryocytes [16], and stromal cells [17].
MicroRNAs (miRNAs) are a class of small noncoding RNAs that bind to protein-coding mRNAs in a sequence specific fashion [18]. MiRNAs regulate expression of the targeted gene by either preventing translation of the mRNA to protein or by initiating the degradation of the mRNA itself. Conventional approaches to study the role of miRNAs in regulating gene expression typically start with obtaining mRNA profiles of cell populations of interest using microarray or sequencing platforms, followed by bioinformatic prediction of potential miRNA targets. Once potentially relevant miRNAs are identified they can be overexpressed or knocked down to associate their presence or absence with levels of protein expression. Luciferase-based assays can then be used to suggest binding of miRNAs to their predicted targets. While informative, this strategy has several pitfalls [19, 20]. Most importantly, none of these experimental approaches show a direct interaction between the miRNAs and their targets in vivo. Bioinformatic prediction algorithms are heavily based on heuristics and miss those associations that do not conform to canonical descriptions using energetics, cross-species sequence conservation, and seed region pairing. Transient transfection of miRNA precursors often result in much higher levels of intracellular miRNAs which can lead to off-target effects [21]. Finally, these approaches can be used only for analysis of single miRNA-mRNA interactions and is not suitable to define functional miRNA networks. To circumvent this problem, investigators have implemented genome-wide biochemical approaches to define the miRNA-mRNA “interactome” within cells [22–24]. The technique termed HITS-CLIP (high-throughput sequencing of RNAs isolated by cross-linking immunoprecipitation or CLIP-Seq) uses ultraviolet (UV) light to cross-link RNA to adjoining proteins allowing for their stringent purification through immune-precipitation [25]. The isolated RNA is analyzed by high-throughput (or next generation) sequencing techniques. When applied to the analysis of Argonaute (Ago) proteins (Argonautes are critical components of the RNA-induced silencing complex or RISC within which miRNAs bind to their targets), unbiased genome-wide views of the miRNA-mRNA interactome can be obtained [26]. In this article, we report the implementation of a strategy based on Ago HITS-CLIP to define miRNA-mRNA interactions of functional relevance in MSCs and bone marrow endothelial cells (BMEC).
Materials and methods
Plasmids, DNA Cloning, and Viral Preparations
pCDNA-Notch1 was a kind gift of Dr. Spyros Artavanis-Tsakonas (Harvard Medical School, Boston, MA) [27]. Luciferase reporter vectors were made by cloning the 3′ UTR or miRNA responsive element (MRE), into the XhoI-NotI restriction sites of psiCheck2 vector (Promega, Madison, WI) as was reported previously [28] (primers used are detailed in Supporting Information). Retroviral constructs for stable overexpression of miR-193a were constructed by amplifying a 276 base pair fragment of genomic DNA that included the pre-miR-193a region and cloning in to the BamHI—EcoRI restriction sites of MDH1-PGK-GFP vector [29] (Addgene plasmid 11376) as previously reported [28]. Control vectors had no insert. Viral supernatants for these retroviral vectors were prepared by transient transfection of HEK-293T cells with the viral proplasmid, pVSVG, pTAT, and pJAK as previously reported [28]. Recombinant DNA research was conducted per NIH guidelines for the same.
Cell Culture, Transfection, and Sorting
Stromal cell lines HS5 and HS27A have been previously defined [30] and were maintained in RPMI-1640 supplemented with 10% fetal calf serum (FCS) and penicillin-streptomycin. Stable cell lines Hs27A-miR-193 and Hs27A-miR-Control were made by transducing HS27A with MDH-PGK-193a or MDH-PFK-Control viral vectors and selecting for green fluorescent protein (GFP)-positive single-cell clones by plating at low density and ring cloning. High expression of miR-193a in the single-cell clones was verified by quantitative RT-PCR as described previously using miR-193a specific primers [28]. Primary human mesenchymal stromal cells (hMSC) were grown from bone marrow mononuclear cells from normal donors (obtained from bone marrow “screens” as described previously) and cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS) [28, 30]. All primary MSCs were at passage number 3 or fewer. Human umbilical vein endothelial cells (HUVEC) were obtained from American Tissue Culture Association (ATCC) and cultured in recommended conditions for not more than three passages. Transformed human bone marrow endothelial cells (TrHBMEC) were a kind gift from Babette Weksler (Cornell University, New York) and cultured as described previously [31]. U937 cells were obtained from ATCC and cultured in RPMI-1640 medium supplemented with 10% FCS. Stable single-cell clones of U937-Notch1 were developed by electroporation of U937 with pCDNA-Notch1 and selection in Neomycin followed by single-cell cloning in 1.3% methyl cellulose. MiRNA mimics (miR-193a, miR-9, miR-200a, miR-185, and miR-control mimic) were from Dharmacon (Lafayette, CO) and transfected to stromal cells by “reverse transfection” (a protocol optimized for transfection of stromal cells as previously described) at a final concentration of 5 nM in six-well plates [28]. HEK-293T cells for production of viral vectors were obtained from ATCC and maintained in DMEM media supplemented with 10% FBS.
HITS-CLIP Protocol for Argonaute
The protocol was adapted from previous protocol as reported by Chi et al. [26] with minor modifications. Briefly, approximately 15–20 × 106 cells were first cross-linked with 600 mJ/cm2 of short wave UV radiation (254 nM)n in split doses. Cells were lysed and argonaute proteins (AGO1–4) along with the cross-linked RNA were immunoprecipitated (IP) with a murine monoclonal antibody that recognizes all human argonaute proteins (2A8 [32], a kind gift from Dr. Zissimos Mourelatos [University of Pennsylvania, Philadelphia, PA]). A P-32 labeled 3′ RNA linker was ligated to the RNA of the IP Ago-RNA complexes “in-bead.” The Ago-RNA complexes were resolved by SDS-PAGE, transferred to nitrocellulose, and visualized by autoradiography to determine localization of the Ago-RNA complexes. These complexes were excised, RNA extracted, and reverse transcribed to cDNA. A cDNA library suitable for sequencing on the Illumina platform was then constructed with serial amplification by PCR. Sequencing was performed on an Illumina Genome Analyzer IIx (single end 50 base pair reads). Full protocol accompanies the Supporting Information.
Western Blotting
Cell lysates were quantitated for protein content, and approximately 30 µg lysate was resolved by standard SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membrane. Blots were probed by antibodies against JAG1 (R&D, Systems, Minneapolis, MN, www.rndsystems.com), MMP2, WNT5A (both Santa Cruz, Biotechnology, Dallas, TX, www.scbt.com), VEGFA (Proteintech, Chicago, IL, www.ptglab.com), and an appropriate HRP-conjugated secondary antibody. Blots were then stripped and reprobed for alpha tubulin (Applied Biologicals) to ensure equal protein loading. The blots were then visualized using Immobilon HRP substrate (Millipore). Antibodies and blotting conditions for Western blot are summarized in Supporting Information.
U937-HS27A Coculture and Fluorescence-Activated Cell Sorting Analysis
To determine the effect of miR-193a on the function of stromal cells to support hematopoietic precursor cells in their primitive state, we first plated HS27A-193a or HS27A-Control cells 2 ml media (105 each well) in six-well adherent plates. Sixteen hours later, 105 U937-Notch1 (or control U937-pCDNA) were suspended in 0.5 ml of media and added to the adherent cells. All Trans-Retinoic Acid (ATRA) was added at 25 µM final concentration to induce differentiation. Ninety-six hours after coculture, all cells (adherent and nonadherent) were harvested by EDTA treatment and stained with APC-conjugated anti-CD11b monoclonal antibody (BD Biosciences) or appropriate isotype control. Fluorescence-activated cell sorting (FACS) analysis was performed on a FACS-Calibur on the U937 cells population by excluding the HS27A cells by GFP positivity.
Luciferase Assay
Fifty-thousand HEK-293T cells were plated in 24-well plates and cotransfected 16 hours later (cells at 40%–50% confluence) with the 0.2 µg of psicheck vectors and 5 nmol of miRNA mimic with Lipofectamine 2000 per manufacturer’s protocol. At 24 hours, the cells were lysed and assayed for both renilla and fire-fly luciferase activity using the Dual Luciferase Reporter Assay kit on a GLOMAX microplate luminometer (both from Promega) per previously described methodology [28, 33]. Ratios of renilla luciferase to fire-fly luciferase activity were calculated and statistical significance determined using Student’s t test.
Gelatin Zymography
Gelatin zymography was performed to determine MMP2 activity in cell lysates as previously described [34]. Briefly, 7.5% denaturing acrylamide gels with 3 mg/ml gelatin A was prepared and cell lysates were size-resolved under nonreducing conditions. The gel was then stained with Coomassie Blue for an hour and destained overnight and imaged.
Analysis of Next Generation Sequencing Data
Sequencing was performed on the Illumina GA IIx sequencer. Raw reads were first processed to remove adapter sequences and then aligned to the human genome 18 (hg18, UCSC) with Novoalign [35] (Selangor, Malaysia), keeping the 100 best reads of all possible mappings. Adapter sequences were trimmed from the 5′ end of read until mapped or until a read length of less than 16 bases. Statistics of aligned reads were generated using custom “peaktools” and Python scripts. Peaks were determined with a Poisson model (Yeo et al. [36]) with a moving window of 30 bp and log p-value threshold value of 5. Summits of peaks were determined by ascertaining the highest point of the peaks, and a peak footprint was determined by extending 30 bp up and downstream from the summit. Consensus peaks were assigned as those peaks present in more than one biological replicate sample. Read numbers were normalized to the total number of unique alignable reads per sample. Seed sequences of annotated mature human miRNAs (positions 2–7 and 2–8) were generated from mirbase-18 [37]. Seeds were mapped to consensus peaks and their interactions were visualized using Cytoscape [38]. Scatter plots and coefficient of determination were generated using R statistical packages (corrplot function) and Python-based data analysis toolkit Pandas. Heat maps were generated by the Partek genomics suite (Partek Inc., St. Louis, MO). Custom software packages developed for this project are available for download and use at https://github.com/jayhesselberth/peaktools.
Results
Generation and Analysis of Ago HITS-CLIP Datasets from Cells of the Marrow ME
There were two primary objectives for HITS-CLIP analysis in cells of the ME: First, to determine whether direct miRNA targeting of niche-associated genes could be defined in an unbiased fashion from the datasets. Second, to identify miRNA-regulated functional networks in the hematopoietic ME. MSCs (also called mesenchymal stem cells) and bone marrow endothelial cells (BMEC) have both been shown in both in vitro and in vivo models to be critical cellular components of the ME [10, 17, 39, 40]. Hence, we performed HITS-CLIP analysis on three human stromal populations: two cloned and functionally distinct stromal cell lines (HS5 and HS27A); and primary human MSCs or hMSC, and two endothelial populations: one derived from bone marrow endothelium (bone marrow endothelial cells or TrHBMEC) and umbilical veins (HUVEC). Human stromal cell lines HS5 and HS27A have been previously described and have been used to represent distinct components of the ME that either support maintenance of hematopoietic stem and progenitor cells in their primitive state (HS27A) or differentiation of these progenitors into the myeloid lineage (HS5) [30]. Nontransformed primary human marrow-derived stromal cells (hMSC) are arguably better for such global analysis as they are devoid of artifacts of immortalization, but hMSCs are known to be very heterogeneous in their composition, which can vary significantly among preparations [41]. Hence, we elected to use the hMSCs to confirm the overall patterns of RNA interactome isolated by Ago-HITS-CLIP from the stromal cell lines. TrHBMEC is a bone marrow endothelial cell line reported to be functionally different from endothelium derived from other vascular beds [31]. HUVEC are the most commonly available and used primary endothelial cells [42]; we performed HITS-CLIP on HUVEC to compare with marrow-derived endothelial cells. To ensure technical reproducibility, we performed biological replicate analyses using separate cDNA libraries sequenced from separate cell cultures as follows: three each of HS5, HS27A, TrHBMEC, five each of hMSC, and two of HUVEC.
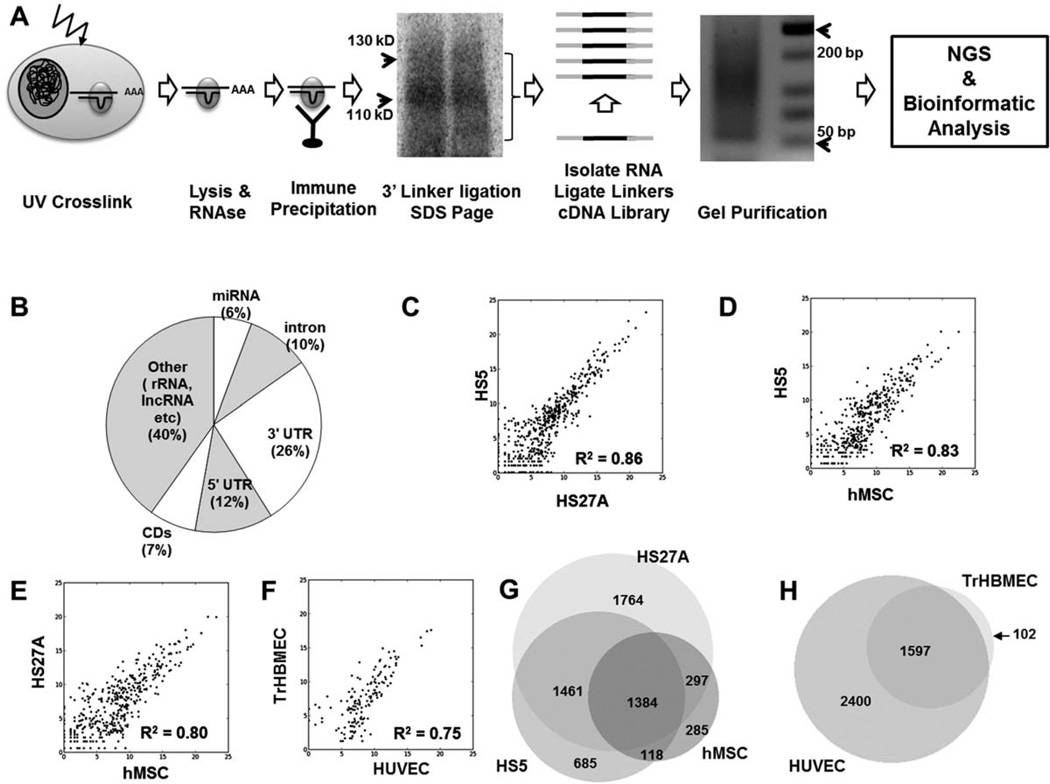
The overall scheme of HITS-CLIP analysis along with representative results from the SDS-PAGE autoradiography and PCR amplification of the CLIP cDNA library is shown in Figure 1A. The sequencing reads once processed and aligned to the human genome were first annotated as mRNA, miRNAs, other RNA (such as ribosomal RNA, lncRNA, and intergenic RNA without previously annotated transcripts). Reads within mRNA were further divided to 5′ untranslated region (5′UTR), coding regions (CD), 3′ UTR, or intronic. Distribution of aligned reads to these different kinds of transcripts for HS27A samples is shown in Figure 1B. This is comparable to results reported by Chi et al. in their initial description of HITS-CLIP [43]. Results from each of the individual samples analyzed are shown in Supporting Information Table S1. Raw data files are uploaded to the GEO short-read archive at http://www.ncbi.nlm.nlm.nih.gov/geo/query/acc.cgi?token=vdknveyqyqaucvk&acc=GSE41272. This includes the BigBed files that document the chromosomal loci of the aligned reads. For easy visualization of results, the each sample has been visualized as “tracks” visualized on the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=mmpillai&hgS_otherUserSessionName=Balakrishnan_Stem%2DCells).
Figure 1.
Ago HITS-CLIP analysis of stromal and endothelial cells. (A): Basic experimental scheme of Ago HITS-CLIP as adapted from Chi et al. UV cross-linked cells are lyzed and treated with RNase to reduce the length of mRNA fragments. Ago is immune-precipitated with monoclonal anti-Ago antibody 2A8, P32-labeled 3′ RNA linker ligated (on-bead), and resolved by SDS-PAGE electrophoresis. The protein-RNA-complex is transferred to nitrocellulose and visualized by autoradiography. In the representative autoradiograph shown, a major band is seen ~110 kD and a more diffuse band at ~130 kD. The RNA-protein complex in the 110–130 kD region is isolated and a 5′ RNA linker is ligated. The isolated RNA is reverse transcribed and PCR amplified with Illumina-compatible primers to generate a cDNA library (typical agarose gel shown). The cDNA libraries are then gel purified and sequenced on Illumina GA IIx and resultant bioinformatically analyzed. (B): Distribution of aligned reads from HS27A samples to different gene annotations. (C): Correlation of consensus peaks between HS5 and HS27A. Coefficient of determination (R2 determined by corrplot [R-package]). (D): Correlation of consensus peaks between HS5 and hMSC. (E): Correlation of consensus peaks between HS27A and hMSC. (F): Correlation of consensus peaks between TrHBMEC and HUVEC. (G): Overlap of genes enriched by consensus peaks between stromal cells (HS5, HS27A, and hMSC). (H): Overlap of genes enriched by consensus peaks between endothelial cells (TrHBMEC and HUVEC). Abbreviations: hMSC, human mesenchymal stromal cell; HUVEC, human umbilical vein endothelial cell; TrHBMEC, transformed human bone marrow endothelial cells; UV, ultraviolet.
To determine technical reproducibility, we calculated the coefficient of determination (R2) of peaks from biological replicate samples. There was a high level of correlation within biological replicates among stromal cell lines (R2 between 0.99 and 0.93), biological replicates between other cell types were more variable confirming the biological variability anticipated when analyzing primary cells (R2 between 0.95 and 0.74). Results are summarized in Supporting Information (Supporting Information Table S2 and Fig. S1). We then compared the consensus peaks to each cell type. As expected, comparison between stromal cell types showed high correlation (R2=0.86 for HS27A vs. HS5; 0.83 for HS5 vs. hMSC and 0.80 for HS27A vs. hMSC; Fig. 1C–1E). Similarly correlation between TrHBMEV and HUVEC was 0.75 (Fig. 1F). However, a comparison between stromal and endothelial cell types revealed much lesser correlation (R2=0.58–0.68, data not shown). We also determined the overlap of genes enriched in the consensus peaks among the stromal cells (Fig. 1G) and endothelial cells (Fig. 1H).
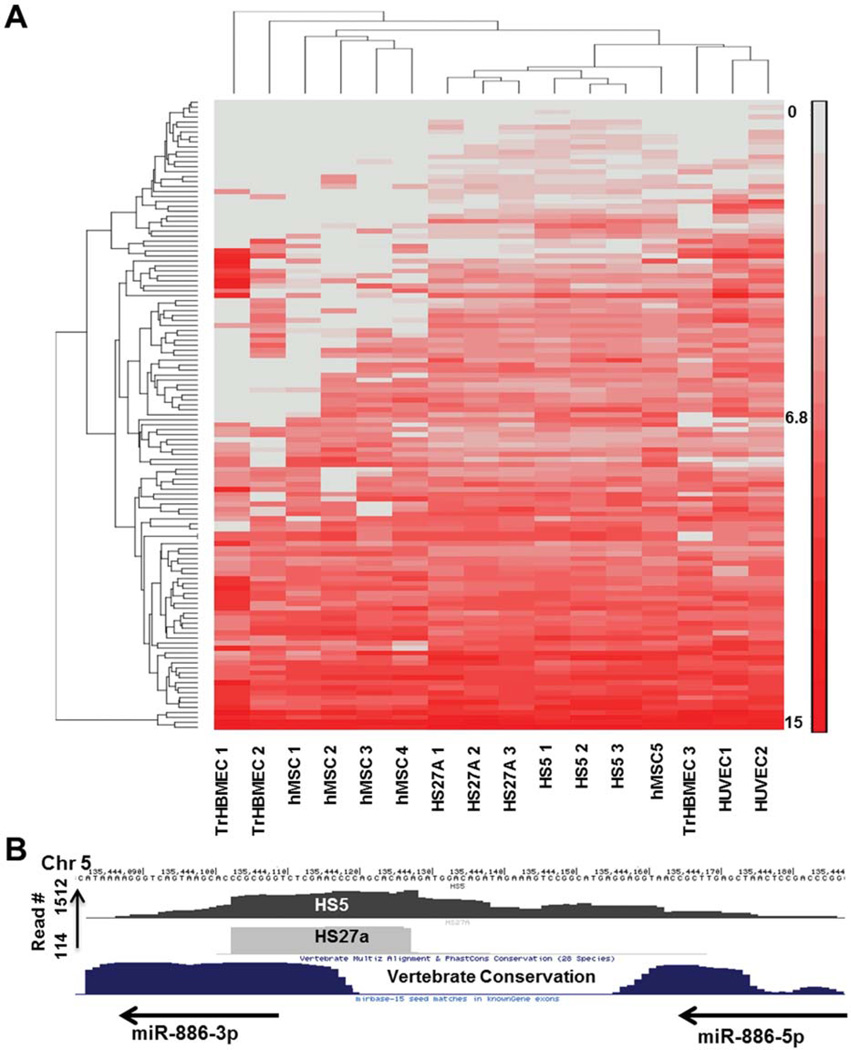
Next generation sequencing of cDNA libraries (RNA-Seq) can be used to quantitate transcript levels of coding and noncoding RNA [44]. Although similar to RNA-Seq in some technical aspects, HITS-CLIP has not been defined as a quantitative technique. We used hierarchical clustering of miRNA expression (50 miRNAs overexpressed in each of the samples) of all HITS-CLIP libraries to determine whether biological replicates would cluster together. MiRNA abundance was calculated as the number of reads per total number of alignable reads per sample. As shown in Figure 2A, stromal cell lines HS27A and HS5 samples showed tight clustering. Human MSC samples showed more variability (four of five samples clustering together while the fifth sample clustered with the stromal cell lines. Endothelial samples showed further variability. When clustering was attempted using mRNA peaks, biological replicates did not cluster together (data not shown) suggesting that miRNA isolated by HITS-CLIP is quantitative but mRNA peaks are not. When we correlated miRNA abundance from HITS-CLIP datasets abundance as determined by microarrays, no significant correlation could be obtained (data not shown). Potential reasons for this discrepancy include (a) structural bias in the 3′ adapter ligation when T4 RNA ligase is used [45] (cofolding of 3′ ends that result in poor ligation efficiency to the 3′ RNA adapter) and (b) a potential difference in the relative abundance of free versus Ago-bound miRNA in vivo.
Figure 2.
Ago-associated miRNA and vault RNAs. (A): Hierarchical clustering of miRNA abundance in all samples analyzed by HITS-CLIP. MicroRNA read numbers were normalized to total aligned reads and analyzed with PARTEK Genomics suite to generate the heat map. Fifty highly expressed miRNAs in each of the cell type were included in the analysis. (B): Hsa-miR-886 (redesignated as the vault RNA vtRNA-2) binding to AGO. Both HS5 and HS27A express Ago-bound miR-886 (precursor and mature forms). Abbreviations: hMSC, human mesenchymal stromal cell; HUVEC, human umbilical vein endothelial cell; TrHBMEC, transformed human bone marrow endothelial cells.
Vault RNAs and Other Non-miRNA Small RNAs Are Bound to Argonaute
We have previously reported on the differential expression of miR-886-3p in stromal cell lines, and the direct regulation of CXCL12 by this miRNA [10]. Later reports have since then suggested that the primary transcript for miR-886-3p is a vault RNA (vtRNA-2) [46]. vtRNAs are a family of noncoding RNAs that form parts of vault ribonucleoprotein complexes [47]. The precise function of vtRNAs is unclear; degradation of vtRNAs does not have an effect on the structure of the vaults themselves. Diverse functions have been currently proposed for vtRNAS including intracellular transport and cytoskeleton maintenance [48]. As binding to Argonaute suggests a role in RNA induced silencing, we determined if miR-886 was present in the HITS-CLIP datasets from stromal cells. As shown in Figure 2B, miR-886 was enriched in the Ago-CLIP datasets; and pattern of peaks suggested that both the pre-miR-886 and the mature miR-886-3p were bound to Ago. These results are hence in agreement with recent reports which described miRNA-like functions for small RNAs derived from vtRNAs [49]. Furthermore, other noncoding RNAs such as snoRNAs and scRNAs were also found to be enriched differentially in the different sample sets (Supporting Information Table S3). Argonaute binding of such small RNA other than classic miRNAs suggest that they too are likely functional within the context of the RISC and support other recent reports that small RNAs that have noncanonical biogenesis different from canonical miRNAs are part of the RNA silencing machinery [50, 51].
Determining miRNA Regulation of Niche-Associated Genes
We first sought to determine whether the peaks localized to the genomic coordinates of protein-coding genes (called Agom-RNA peaks) bind to specific miRNAs. HITS-CLIP provides an unbiased and genome-wide approach to determine which sequences within mature transcripts are bound by the RISC complex, but it does not allow for the precise determination of the identity of the miRNA that targets these Ago-mRNA peaks [52]. Classically, miRNAs are thought to bind to mRNAs by perfect complementarity with the seed region (nucleotides 2–7 or 2–8) of the target sequence [53]. “Non-canonical” binding that does not follow strict seed base binding is now known to exist, but is likely less common and less amenable to bioinformatic prediction [54]. Therefore, an unbiased list of all miRNAs with seed sequences complementary to the Ago-mRNA peaks was generated. This made it possible to individually assess potential miRNA binding partners associated with a particular Ago-mRNA peak. Potential binding peaks were further refined by filtering for only those miRNAs that were also present in the HITS-CLIP dataset and prioritizing those miRNAs that were differentially expressed between the two cell lines. We describe validation of miRNA regulation of four such niche-defining genes. The strategy was unsuccessful in defining downregulation of other Ago-mRNA peaks in genes such as KITLG and WNT3A.
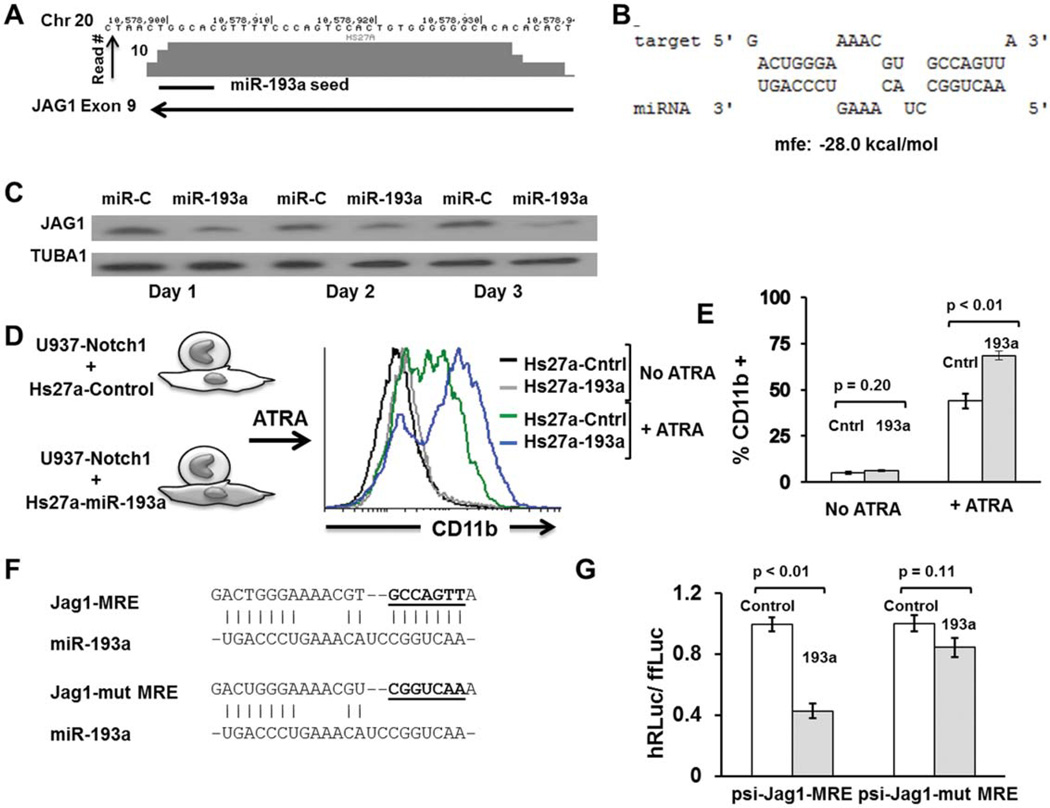
MiR-193a Regulates Expression of JAG1
Notch receptors and their ligands are a group of highly conserved proteins that mediate juxtacrine signaling across adjacent cells [55, 56]. Jagged 1 (JAG1) is one of several described notch ligands in mammals (others include JAG2, Delta-like1 or DLL1, DLL3, and DLL4). Notch signaling is considered critical to maintain HSCs in their undifferentiated state [55]. JAG1 is also known to be expressed at high levels in MSC subpopulations that support primitive hematopoietic stem and precursor cells [17, 57]. We searched for peaks of Argonaute binding in JAG1 and identified one such Ago-mRNA peak in the coding region (exon 9) with a predicted binding site to miR-193a (Fig. 3A, 3B and Supporting Information Fig. S2). To confirm that miR-193a can regulate JAG1, we first transiently transfected miR-193a mimics into HS27A cells which constitutively express high levels of JAG1. JAG1protein levels estimated by Western blotting showed significant downregulation from day 1 and lasting up to 3 days after transfection (Fig. 3C). We then used a functional assay for the Notch signaling based on the ability of JAG1 to engage Notch1 and prevent ATRA-induced differentiation as measured by CD11b expression [58]. The myeloid progenitor cell line U937 was engineered to stably overexpress Notch1. These cells were then cocultured with HS27A cells overexpressing miR-193a or a control vector, and exposed to ATRA for 4 days. FACS analysis showed significantly increased CD11b expression in U937-Notch1 cells cocultured with HS27A-miR-193a. We interpret these results to be consistent with a reduction in the ability of HS27A-193a cells to maintain hematopoietic cells in their primitive state due to a reduced level of JAG1 (Fig. 3D). Finally, luciferase assays were used to determine potential binding of miR-193a to this putative binding peak. Psi-check vectors with the MRE (a 40 bp region of the purported binding site of miR-193a on exon 9 of JAG1 transcript) or mutant MRE (with the putative seed region mutated) were cotransfected with miR-193a mimic or miR-control mimic and changes in luciferase activity measured by luminometry. As shown in Fig. 3E, 3D, miR-193a significantly reduced the luciferase activity compared to control miRNA transfection; when the seed region was mutated, this effect was abrogated. Together, our data suggest that miR-193a directly targets JAG1 expression in MSCs and reduces their ability to functionally engage the Notch1 receptor in hematopoietic cells and prevent differentiation.
Figure 3.
JAG1 is downregulated by miR-193a. (A): Genomic coordinates in chromosome 20 with a Ago-mRNA peak in the coding region of JAG1 and putative binding site of miR-193a. (B): Predicted binding and binding energy of miR-193a to its putative target on JAG1 energetics prediction performed with RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html; mfe5minimum free energy). (C): Western blot analysis of HS27A cells transfected with miR-193a or control (5 nM each). JAG1 was shown to be downregulated in miR-193a transfected cells starting 1 day after transfection with most pronounced effects by day 3. The same blots were stripped and reprobed for Tubulin 1 (TubA1) to ensure equal protein loading. (D): Functional assay to determine effect of miR-193a on ability of stromal cells to prevent hematopoietic differentiation. U937 cells stably expressing Notch1 and HS27A cells stably expressing miR-193a (or a control noinsert vector) were developed. U937-Notch1 cells were cocultured with HS27A-193a or HS27A-control in the presence of a myeloid differentiating agent (ATRA at 25 µM final concentration) for 4 days. U937 cells were then assayed for expression of CD11b by fluorescence-activated cell sorting. U937 cells cocultured with HS27A-193a had higher expression of CD11b suggesting that stromal cells expressing miR-193a had a decreased ability to retain hematopoietic cells in their primitive state. Cells without ATRA treatment did not have a difference whether they were cocultured with HS27A-Cntrl or HS27A-193a. (E): Quantitation of CD11b expression of U937-Notch cells cocultured with HS27A-Cntrl or HS27A-193a. Median fluorescence intensity from three biological replicates was averaged. Student’s t test was used to determine significance. (F): Nucleotide sequence of JAG1 MRE with the putative seed-binding region mutated; these MREs were cloned into the ψ-Check2 vector for luciferase assays. (G): Luciferase assays to confirm functional binding of miR-193a to Jag1-MRE and its disruption on mutating the seed-binding region. Psi-check Jag1-MRE or ψ-check2-Jag1-mutant MRE were transiently transfected to HEK-293T cells along with either control miRNA mimic or has-miR-193a mimic (5 nM). Cells were harvested 24 hours after transfection and assayed by luminometry serially for firefly and renella luciferase activity. Ratio of renella to firefly luciferase was calculated and normalized to control mimic transfection (ratio designated 1.0). Error bars represent SEM for triplicates and p values were calculated by Student’s t test. Abbreviations: ATRA, all trans-retinoic acid; MRE, miRNA responsive element.
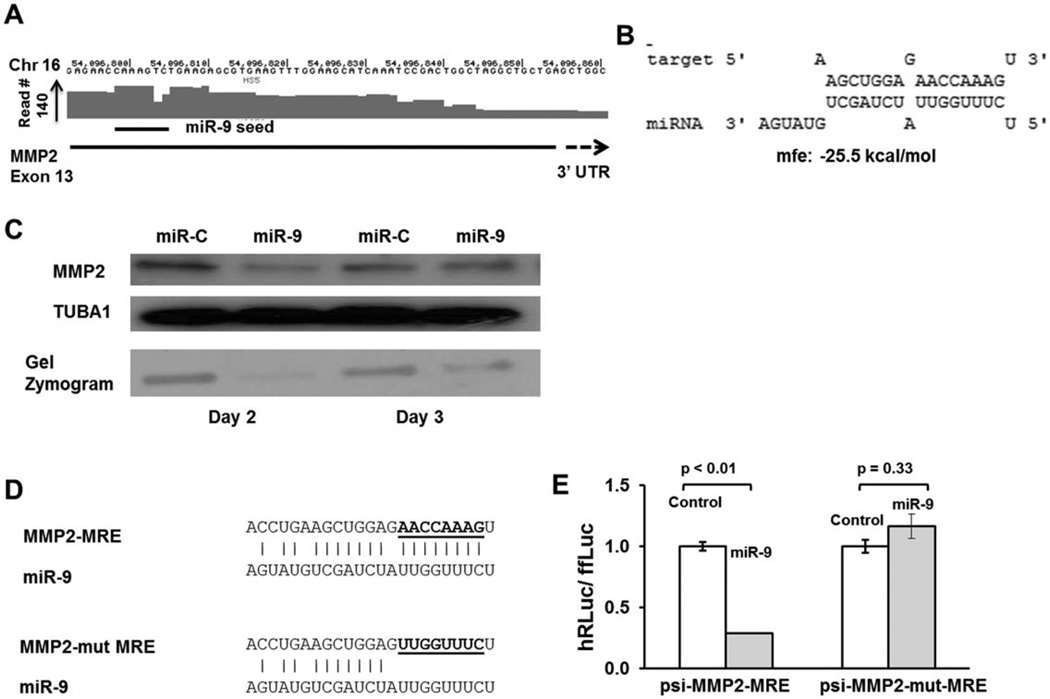
MiR-9 Regulates Expression of MMP2
Matrix metalloproteinases (MMPs) are a family of endopeptidases that cleave most proteins in the extracellular matrix thus contributing to remodeling of the extracellular space around cells and facilitating their movement [59]. MMP2 (gelatinase A) is a major stromal-derived MMP [60]. HITS-CLIP analysis showed a reproducible Ago binding peak in exon 13 close to the 3′ UTR and predicted to be targeted by miR-9 (Fig. 4A, 4B and Supporting Information Fig. S3). Western analysis of HS5 cells transfected with miR-9 mimic showed reduced MMP2 protein when compared with miR-mimic transfection. To functionally validate this protein downregulation, we also performed gelatin zymography on the cell lysates. Gelatin zymography is an electrophoretic technique that detects hydrolytic activity of enzymes on a substrate (gelatin) after size resolution on a gel that contains the substrate. Given that MMPs degrade gelatin in a dose-dependent fashion, gelatin zymography can be used to quantitate the levels of MMPs in biological samples [61]. MMP2 enzymatic activity was markedly reduced in miR-9 transfected cells when compared with controls miRNA transfected cells (Fig. 4C). Luciferase assays were then performed as described for JAG1 (by cloning the wild-type and mutant MRE of MMP2 in to psi-check2 vectors and determining relative differences in luciferase activity upon transfecting control miRNA mimic and miR-9 mimic) (Fig. 4D, 4E). Together, our results show that MMP2 is a direct target for miR-9 in marrow-derived stromal cells.
Figure 4.
MMP2 is downregulated by miR-9. (A): Genomic coordinates in chromosome 16 with an Ago-mRNA peak in the exon 13 close to stop site with a putative binding site of miR-9. (B): Predicted binding and binding energy of miR-9 to its putative target on MMP2. (C): Western blot analysis and gel zymography of HS5 cells transfected with miR-9 or control (5 nM each). MMP2 was shown to be downregulated in miR-9 transfected cells on days 2 and 3 by Western blot. Blots were stripped and reprobed for Tubulin 1 (TubA1) to ensure equal protein loading. Gel zymography was also performed using cell lysates which showed marked downregulation of matrix metalloproteinase activity. (D): Nucleotide sequence of MMP2 MRE with the putative seed-binding regions mutated; these MREs were cloned into the ψ-Check2 vector for luciferase assays to generate ψ-check-MMP2-MRE and ψ-check-mut-MMP-MRE vectors. (E): Luciferase assays to confirm functional binding of miR-9 to MMP2-MRE and its disruption on mutating the seed-binding region. Assays were performed and analyzed as described for JAG1. Abbreviations: MMP, matrix metalloproteinase; MRE, miRNA responsive element.
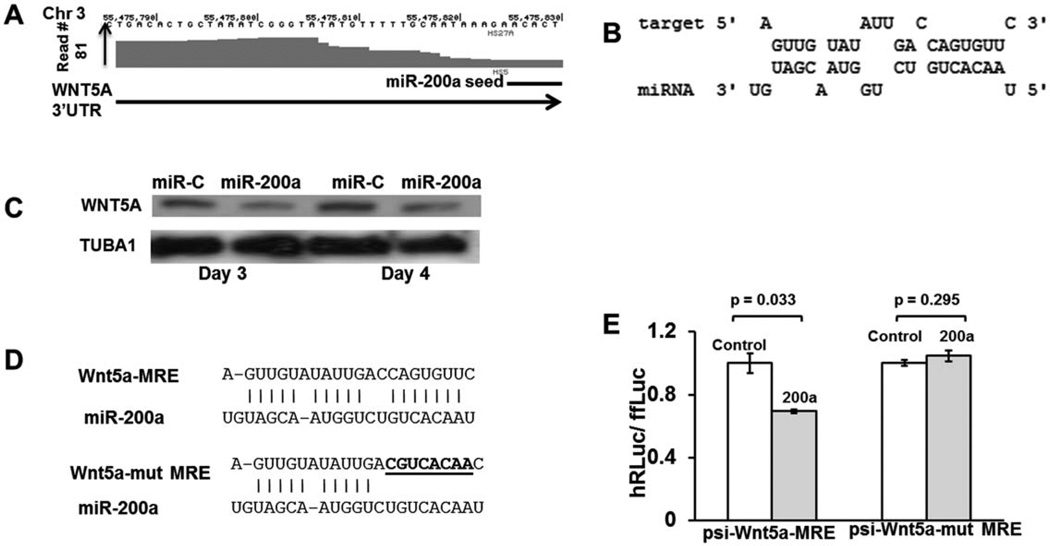
MiR-200a Regulates Expression of WNT5A
Wnt signaling is evolutionarily conserved across metazoans and is critical for embryonic and adult cell fate decisions, proliferation of stem and progenitor cells, and control of asymmetric cell division [8]. Downstream signaling by Wnt proteins is broadly classified as either canonical or noncanonical depending on the receptors that are bound by the Wnt ligands, and if β-catenin and coreceptors LRP5 and LRP6 are needed for signaling [62]. Canonical Wnt pathways are reported to be critical for the maintenance of stem cells in their undifferentiated state as well as in myeloid and lymphoid differentiation [7, 8]. WNT5A is one such ligand thought to act through the noncanonical pathway and inhibits WNT3A-mediated canonical signaling, increasing the long-term quiescence of stem cells [63].We found a Ago binding peak in the 3′ UTR of WNT5A in stromal cells and bone marrow endothelial cells with a predicted binding site for miR-200a (Fig. 5A, 5B and Supporting Information Fig. S4). Interestingly, this peak was more prominent in HS5 cells and miR-200a is overexpressed in HS5 cells compared to HS27A cells. By performing Western blot on HS5 cells transfected with miR-200a mimic or miR-control mimic, we determined that WNT5A protein levels were significantly downregulated by days 3 and 4 when miR-200a was transfected (Fig. 5C). Luciferase assays were performed as described for JAG1 by cloning the miR responsive elements which showed potential binding of miR-200a to this putative site on WNT5A (Fig. 5D, 5E). Given that WNT5A is a negative regulator of HSC quiescence while JAG1 promotes quiescence, one can speculate that miR-193a and miR-200a have opposing functional effects on the support of HSC in its niche. This hypothesis would require further experimental validation.
Figure 5.
WNT5A is downregulated by miR-200a. (A): Genomic coordinates on chromosome 3 with an Ago-mRNA peak in the 3′UTR with a putative binding site of miR-9. (B): Predicted binding and binding energy of miR-200a to its putative target on WNT5A. (C): Western blot analysis of HS27A cells transfected with miR-200a or control (5 nM each). WNT5A was shown to be downregulated in miR-200a transfected HS27A cells on days 3 and 4 by Western blot. Tubulin 1 was probed to ensure equal protein loading. (D): Nucleotide sequence of WNT5A MRE with the putative seed-binding regions mutated; these MREs were cloned into the ψ-Check2 vector for luciferase assays to generate ψ-check-WNT5A-MRE and ψ-check-mut-WNT5A-MRE vectors. (E): Luciferase assays to confirm functional binding of miR-200a to WNT5A-MRE and its disruption on mutating the seed-binding region. Assays were performed and analyzed as described for JAG1 in Figure 3. Abbreviation: MRE, miRNA responsive element.
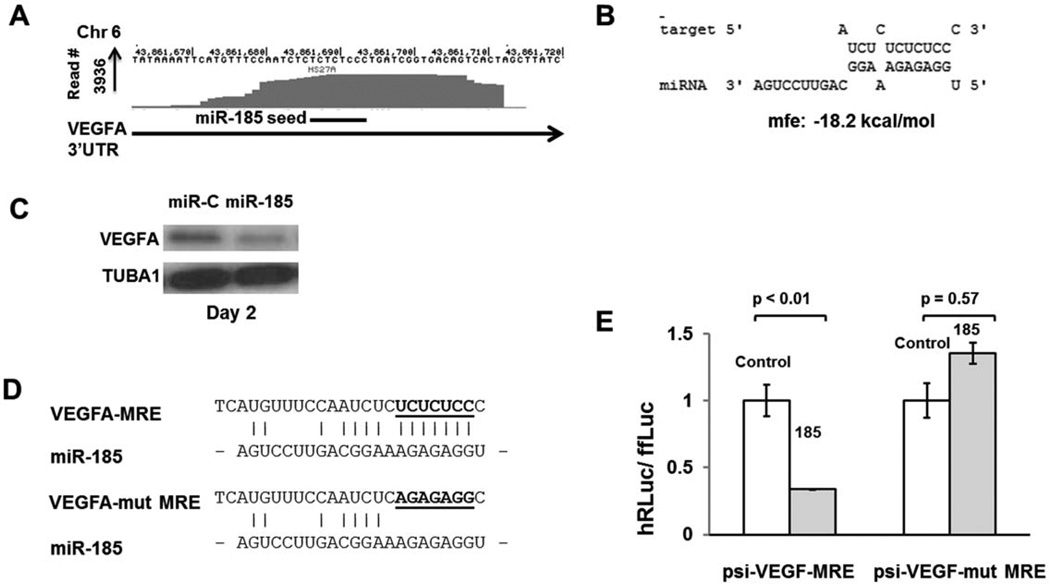
MiR-185 Regulates Expression of VEGFA
Vascular endothelial growth factor A or VEGFA is a member of the PDGF/VEGF family of endothelial mitogens [64]. The VEGF ligands along with their receptors (VEGFR1 and VEGFR2) are known to regulate formation of blood vessels as well that of blood cells. HSC survival, colony formation, and in vivo repopulation were all affected when VEGF was ablated in mice [65]. An internal autocrine loop was postulated to regulate the regulation of HSC. VEGFA has several isoforms that arise from alternative splicing of pre-mRNA [66]. We found a highly conserved Ago binding peak in the 3′ UTR of VEGFA present in stromal cells (Fig. 6A), endothelium, hematopoietic cells as well as tumor cells (data not shown). This Ago-mRNA peak was predicted to be targeted by miR-185 (Fig. 6A, 6B and Supporting Information Fig. S5). To determine whether miR-185 reduces VEGFA levels in stromal cells, we transfected HS27A cells transiently with miR-185 mimic and performed Western analyses on the cell lysates. As shown in Figure 6C, the VEGFA was significantly downregulated (a 23 kD isoform) in cells transfected with miR-185 when compared with control mimic transfections. Luciferase assays also showed potential binding of miR-185 to its putative target site (Fig. 6D, 6E). Together, our data suggest that miR-185 regulates VEGFA expression in stromal cells. Given that both miR-185 and the Ago-mRNA peak are ubiquitous, this regulation may exist across several cell types.
Figure 6.
VEGFA is downregulated by miR-185. (A): Genomic coordinates on chromosome 6 with an Ago binding peak in the 3′UTR with a putative binding site of miR-185. (B): Predicted binding and binding energy of miR-185 to its putative target on VEGFA. (C): Western blot analysis of HS27A cells transfected with miR-185 or control (5 nM each). Stromal cells produce a 23 kD isoform of VEGFA predominantly detectable by Western blot analysis. VEGFA was shown to be downregulated in miR-185 transfected HS27A cells by day 2 of transfection. TUBA1 was probed in the same blot to ensure equal protein loading. (D): Nucleotide sequence of VEGFA-MRE with the putative seed-binding regions mutated. These MREs were cloned into the ψ-Check2 vector for luciferase assays to generate ψ-check-VEGFA-MRE and ψ-check-mut-VEGFA-MRE vectors. (E): Luciferase assays to confirm functional binding of miR-185 to VEGFA-MRE and its disruption on mutating the seed-binding region. Assays were performed and analyzed as described for JAG1 in Figure 3. Abbreviations: MRE, miRNA responsive element; VEGFA, vascular endothelial growth factor A.
Discussion
Our study was designed to identify specific miRNA-mRNA interactions and attempt to predict global networks of miRNA regulation in the marrow ME using an unbiased genome-wide biochemical approach. The role of miRNAs in regulating the hematopoietic ME and HSC niche has recently received much attention with the recent report that knockout of a key miRNA processing enzyme DICER in murine stromal precursors can lead to the development of a clonal neoplastic process in unmanipulated HSCs [67]. Although several cell types are now known to functionally contribute to the marrow ME, we focused our initial efforts on two cell populations: MSCs (both immortalized lines as well as primary MSC) and bone marrow endothelial cells. Overall analysis of the resultant high-throughput sequencing data is in good agreement with previous reports using HITS-CLIP in other cell types and organ systems [26]. As expected, there was high correlation of the miRNA-RNA interactomes of similar cell types and less so between stromal and endothelial cells. We then validated this approach by directly demonstrating miRNA regulation of four genes that contribute to regulation of the HSC niche. Interestingly, the Ago-mRNA peak of VEGFA that we defined to be targeted by miR-185 was ubiquitous in all the sample types suggesting that shared mechanisms of miRNA regulation exist for at least a few of the transcripts across multiple cell types.
A few of the Ago-mRNA peaks that we pursued with a strategy based on seed region binding did not show downregulation of the protein. Two examples are shown in more detail in Supporting Information Figures S6 (KITLG) and S7 (WNT3A). In the case of KITLG, a putative binding region in the 3′ UTR for miR-188-5p was tested by transfecting HS27A with miR-188-5p or control miRNA-mimic. No change in protein levels was demonstrable by Western blot on days 2 or 3 after transfection. Similarly, a putative binding region for miR-199 in the coding region of WNT3A close to the 3′ UTR could not be confirmed as a genuine site for miRNA-based regulation. Since reproducible peaks in Ago-HITS-CLIP datasets suggest functional binding to Ago at that region, one can only speculate as to the reasons for failing to identify appropriate miRNA binding partners. Speculations include (a) noncanonical binding of miRNA to a target that cannot be predicted by seed region matching, (b) nonrepressive binding of miRNA to the target, binding by hitherto uncharacterized miRNAs, and (c) Ago binding to these mRNA regions without a corresponding miRNA partner.
Given that each miRNA can have several potential target transcripts and each transcript can be regulated by several miRNAs, it is tempting to postulate the existence of biological networks of miRNA regulation (one particular miRNA targeting several transcripts that contribute to a particular biological function). There have been reports of such networks redicted from using both bioinformatic analysis [68] as well as unbiased genome-wide biochemistry such as HITS-CLIP [43], but rigorous validation of such predictions has not been reported. Network analysis was performed for the Ago-mRNA peaks enriched in our datasets; but the results could not be conclusively validated by miRNA-overexpression approaches (results not shown). This is not entirely surprising given that network predictions based on heuristic analysis are imperfect and may not be represented in all cell types. Moreover, intracellular regulation and signaling of MSCs have not been extensively studied to date making predictions difficult.
Certain limitations were also evident from our study while using the HITS-CLIP strategy to define miRNA-mRNA interactions. One is that while the technique allows for unbiased enrichment of Ago-bound mRNA sites, bioinformatic seed-pair matching is still required to further define the exact miRNA that may be targeting a particular Ago-mRNA peak. Another limitation is that the technique itself appears to be nonquantitative for the Ago-mRNA peaks (while relative quantitation is maintained for Ago-miRNA peaks as shown in Fig. 2A). Both limitations are likely to be addressed by further refinements of the technique itself (the former by optimizing biochemistry of the HITS-CLIP protocol that can result in isolation of the mRNA peak and corresponding miRNA together as a hybrid sequence [69]; and the latter by incorporating unique molecular identifiers (UMI) at the stage of reverse transcription), to maintain relative quantitation during subsequent PCR amplifications [70]. Finally, it has also been reported that that many mRNAs that are highly regulated by miRNAs are present at a low level in HITS-CLIP datasets due to destabilization of the transcript leading to under representation of the corresponding Ago-mRNA peak [71]. This could be overcome by stabilizing particular miRNA-mRNA targets by an approach termed RISCtrap [72].
Conclusion
In summary, our results show that genome-wide biochemical approaches can be successfully used to define miRNA-mRNA interactions in cellular components of the hematopoietic ME. Unbiased genome-wide datasets of RNA-interactome will contribute to our current understanding of how the hematopoietic ME regulates different aspects of hematopoietic maturation and lineage commitment. This approach may also be used in studying miRNA-mRNA interactions in other complex functional tissue systems.
Supplementary Material
Acknowledgments
We are thankful to Dr. Zissimos Mourelatos (University of Pennsylvania, Philadelphia, PA) for the anti-argonaute monoclonal antibody 2A8, Spyros Artavanis-Tsakonas (Harvard Medical School, Boston, MA) for the pCDNA-Notch1 plasmid, Dr. Babette Weksler (Weill Cornell Medical College, New York, NY) for the TrHBMEC cells, Jill Castoe (High-Throughput Sequencing Core, UC Denver) for assistance with next generation sequencing, and Dr. Kenneth Jones (UC Denver) for logistical support of data analysis. The work was supported in part by grants from the National Institute of Health (HL104070 and DK073701 to M.M.P., CA164048 to P.K., DK082783 to A.R., and HL099993 to B.T.S.). Core facilities used were funded in part by the University of Colorado Cancer Center Grant (P30CA046934). A.R. is supported by the JP McCarthy Foundation. J.R.H. is supported by the March of Dimes and a Damon Runyon Rachleff Innovation Award.
Footnotes
Author Contributions
I.B. and XY: collection and/or assembly of data; J.B.: data analysis; A.R.: provision of study material; B.T.S.: manuscript writing; P.K.: data analysis and interpretation; J.R.H.: data analysis and interpretation and manuscript writing; M.M.P.: conception and design, financial support, data analysis and interpretation, manuscript writing, and final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
See www.StemCells.com for supporting information available online.
References
- 1.Suda T, Arai F, Hirao A. Hematopoietic stem cells and their niche. Trends Immunol. 2005;26:426–433. doi: 10.1016/j.it.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Bianco P. Bone and the hematopoietic niche: A tale of two stem cells. Blood. 2011;117:5281–5288. doi: 10.1182/blood-2011-01-315069. [DOI] [PubMed] [Google Scholar]
- 3.Papayannopoulou T, Scadden DT. Stem-cell ecology and stem cells in motion. Blood. 2008;111:3923–3930. doi: 10.1182/blood-2007-08-078147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ara T, Tokoyoda K, Sugiyama T, et al. Long-term hematopoietic stem cells require stromal cell-derived factor-1 for colonizing bone marrow during ontogeny. Immunity. 2003;19:257–267. doi: 10.1016/s1074-7613(03)00201-2. [DOI] [PubMed] [Google Scholar]
- 5.Barker JE. Sl/Sld hematopoietic progenitors are deficient in situ. Exp Hematol. 1994;22:174–177. [PubMed] [Google Scholar]
- 6.Arai F, Hirao A, Ohmura M, et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Blank U, Karlsson G, Karlsson S. Signaling pathways governing stem-cell fate. Blood. 2008;111:492–503. doi: 10.1182/blood-2007-07-075168. [DOI] [PubMed] [Google Scholar]
- 8.Austin TW, Solar GP, Ziegler FC, et al. A role for the Wnt gene family in hematopoiesis: Expansion of multilineage progenitor cells. Blood. 1997;89:3624–3635. [PubMed] [Google Scholar]
- 9.Murone M, Carpenter DA, de Sauvage FJ. Hematopoietic deficiencies in c-mpl and TPO knockout mice. Stem Cells. 1998;16:1–6. doi: 10.1002/stem.160001. [DOI] [PubMed] [Google Scholar]
- 10.Kiel MJ, Morrison SJ. Uncertainty in the niches that maintain haematopoietic stem cells. Nat Rev Immunol. 2008;8:290–301. doi: 10.1038/nri2279. [DOI] [PubMed] [Google Scholar]
- 11.Wilson A, Trumpp A. Bone-marrow haematopoietic-stem-cell niches. Nat Rev Immunol. 2006;6:93–106. doi: 10.1038/nri1779. [DOI] [PubMed] [Google Scholar]
- 12.Sugiyama T, Kohara H, Noda M, et al. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 13.Petit I, Szyper-Kravitz M, Nagler A, et al. G-CSF induces stem cell mobilization by decreasing bone marrow SDF-1 and up-regulating CXCR4. Nat Immunol. 2002;3:687–694. doi: 10.1038/ni813. [DOI] [PubMed] [Google Scholar]
- 14.Peled A, Grabovsky V, Habler L, et al. The chemokine SDF-1 stimulates integrin-mediated arrest of CD34(1) cells on vascular endothelium under shear flow. J Clin Invest. 1999;104:1199–1211. doi: 10.1172/JCI7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura Y, Arai F, Iwasaki H, et al. Isolation and characterization of endosteal niche cell populations that regulate hematopoietic stem cells. Blood. 2010;116:1422–1432. doi: 10.1182/blood-2009-08-239194. [DOI] [PubMed] [Google Scholar]
- 16.Huang YQ, Li JJ, Karpatkin S. Identification of a family of alternatively spliced mRNA species of angiopoietin-1. Blood. 2000;95:1993–1999. [PubMed] [Google Scholar]
- 17.Sacchetti B, Funari A, Michienzi S, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.Mendes ND, Freitas AT, Sagot M-F. Current tools for the identification of miRNA genes and their targets. Nucleic Acids Res. 2009;37:2419–2433. doi: 10.1093/nar/gkp145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 21.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milek M, Wyler E, Landthaler M. Transcriptome-wide analysis of protein-RNA interactions using high-throughput sequencing. Semin Cell Dev Biol. 2012;23:206–212. doi: 10.1016/j.semcdb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Ule J, Jensen K, Mele A, et al. CLIP: A method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 24.Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darnell RB. HITS-CLIP: Panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA. 2010;1:266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chi SW, Zang JB, Mele A, et al. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi H, Rand MD, Wu X, et al. Processing of the notch ligand delta by the metalloprotease Kuzbanian. Science. 1999;283:91–94. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- 28.Pillai MM, Yang X, Balakrishnan I, et al. MiR-886-3p down regulates CXCL12 (SDF1) expression in human marrow stromal cells. PLoS One. 2010;5:e14304. doi: 10.1371/journal.pone.0014304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen CZ, Li L, Lodish HF, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science (New York, NY) 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 30.Roecklein BA, Torok-Storb B. Functionally distinct human marrow stromal cell lines immortalized by transduction with the human papilloma virus E6/E7 genes. Blood. 1995;85:997–1005. [PubMed] [Google Scholar]
- 31.Schweitzer KM, Vicart P, Delouis C, et al. Characterization of a newly established human bone marrow endothelial cell line: Distinct adhesive properties for hematopoietic progenitors compared with human umbilical vein endothelial cells. Lab Invest. 1997;76:25–36. [PubMed] [Google Scholar]
- 32.Nelson PT, De Planell-Saguer M, Lamprinaki S, et al. A novel monoclonal antibody against human Argonaute proteins reveals unexpected characteristics of miRNAs in human blood cells. RNA. 2007;13:1787–1792. doi: 10.1261/rna.646007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le MTN, Xie H, Zhou B, et al. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol. 2009;29:5290–5305. doi: 10.1128/MCB.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bemis LT, Schedin P. Reproductive state of rat mammary gland stroma modulates human breast cancer cell migration and invasion. Cancer Res. 2000;60:3414–3418. [PubMed] [Google Scholar]
- 35.Keightley PD, Trivedi U, Thomson M, et al. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 2009;19:1195–1201. doi: 10.1101/gr.091231.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeo GW, Coufal NG, Liang TY, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozomara A, Griffiths-Jones S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2010;39(Database):D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopes CT, Franz M, Kazi F, et al. Cytoscape Web: An interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Méndez-Ferrer S, Michurina TV, Ferraro F, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding L, Saunders TL, Enikolopov G, et al. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phinney DG. Functional heterogeneity of mesenchymal stem cells: Implications for cell therapy. J Cell Biochem. 2012;113:2806–2812. doi: 10.1002/jcb.24166. [DOI] [PubMed] [Google Scholar]
- 42.Baudin B, Bruneel A, Bosselut N, et al. A protocol for isolation and culture of human umbilical vein endothelial cells. Nat Protoc. 2007;2:481–485. doi: 10.1038/nprot.2007.54. [DOI] [PubMed] [Google Scholar]
- 43.Chi SW, Zang JB, Mele A, et al. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhuang F, Fuchs RT, Sun Z, et al. Structural bias in T4 RNA ligase-mediated 30-adapter ligation. Nucleic Acids Res. 2012;40:e54. doi: 10.1093/nar/gkr1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee K, Kunkeaw N, Jeon SH, et al. Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA. 2011;17:1076–1089. doi: 10.1261/rna.2701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stadler PF, Chen JJ-L, Hackermuller J, et al. Evolution of vault RNAs. Mol Biol Evol. 2009;26:1975–1991. doi: 10.1093/molbev/msp112. [DOI] [PubMed] [Google Scholar]
- 48.Van Zon A, Mossink MH, Schoester M, et al. Multiple human vault RNAs. Expression and association with the vault complex. J Biol Chem. 2001;276:37715–37721. doi: 10.1074/jbc.M106055200. [DOI] [PubMed] [Google Scholar]
- 49.Persson H, Kvist A, Vallon-Christersson J, et al. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat Cell Biol. 2009;11:1268–1271. doi: 10.1038/ncb1972. [DOI] [PubMed] [Google Scholar]
- 50.Babiarz JE, Hsu R, Melton C, et al. A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus Dicer1 knockouts and small RNA sequencing. RNA. 2011;17:1489–1501. doi: 10.1261/rna.2442211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ender C, Krek A, Friedländer MR, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 52.Kishore S, Jaskiewicz L, Burger L, et al. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 53.Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee D, Shin C. MicroRNA-target interactions: New insights from genome-wide approaches. Ann N Y Acad Sci. 2012;1271:118–128. doi: 10.1111/j.1749-6632.2012.06745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kojika S, Griffin JD. Notch receptors and hematopoiesis. Exp Hematol. 2001;29:1041–1052. doi: 10.1016/s0301-472x(01)00676-2. [DOI] [PubMed] [Google Scholar]
- 56.Allman D, Aster JC, Pear WS. Notch signaling in hematopoiesis and early lymphocyte development. Immunol Rev. 2002;187:75–86. doi: 10.1034/j.1600-065x.2002.18707.x. [DOI] [PubMed] [Google Scholar]
- 57.Li L, Milner LA, Deng Y, et al. The human homolog of rat Jagged1 expressed by marrow stroma inhibits differentiation of 32D cells through interaction with Notch1. Immunity. 1998;8:43–55. doi: 10.1016/s1074-7613(00)80457-4. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka J, Iwata M, Graf L, et al. Stromal inhibition of myeloid differentiation: A possible role for hJagged1. Ann NY Acad Sci. 1999;872:171–175. doi: 10.1111/j.1749-6632.1999.tb08462.x. [DOI] [PubMed] [Google Scholar]
- 59.Janowska-Wieczorek A, Matsuzaki A, Marquez L. The hematopoietic microenvironment: Matrix metalloproteinases in the hematopoietic microenvironment. Hematology. 2000;4:515–527. [PubMed] [Google Scholar]
- 60.Clutter SD, Fortney J, Gibson LF. MMP-2 is required for bone marrow stromal cell support of pro-B-cell chemotaxis. Exp Hematol. 2005;33:1192–1200. doi: 10.1016/j.exphem.2005.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vandooren J, Geurts N, Martens E, et al. Zymography methods for visualizing hydrolytic enzymes. Nat Methods. 2013;10:211–220. doi: 10.1038/nmeth.2371. [DOI] [PubMed] [Google Scholar]
- 62.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 63.Nemeth MJ, Topol L, Anderson SM, et al. Wnt5a inhibits canonical Wnt signaling in hematopoietic stem cells and enhances repopulation. Proc Natl Acad Sci USA. 2007;104:15436–15441. doi: 10.1073/pnas.0704747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olsson A-K, Dimberg A, Kreuger J, et al. VEGF receptor signalling—In control of vascular function. Nat Rev Mol Cell Biol. 2006;7:359–371. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- 65.Gerber H-P, Malik AK, Solar GP, et al. VEGF regulates haematopoietic stem cell survival by an internal autocrine loop mechanism. Nature. 2002;417:954–958. doi: 10.1038/nature00821. [DOI] [PubMed] [Google Scholar]
- 66.Ladomery MR, Harper SJ, Bates DO. Alternative splicing in angiogenesis: The vascular endothelial growth factor paradigm. Cancer Lett. 2007;249:133–142. doi: 10.1016/j.canlet.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 67.Raaijmakers MH, Mukherjee S, Guo S, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464:852–857. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Volinia S, Galasso M, Costinean S, et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589–599. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kudla G, Granneman S, Hahn D, et al. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc Natl Acad Sci USA. 2011;108:10010–10015. doi: 10.1073/pnas.1017386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kivioja T, V�ah�arautio A, Karlsson K, et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods. 2011;9:72–74. doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
- 71.Hendrickson DG, Hogan DJ, Herschlag D, et al. Systematic identification of mRNAs recruited to argonaute 2 by specific micro-RNAs and corresponding changes in transcript abundance. PLoS One. 2008;3:e2126. doi: 10.1371/journal.pone.0002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cambronne XA, Shen R, Auer PL, et al. Capturing microRNA targets using an RNA-induced silencing complex (RISC)-trap approach. Proc Natl Acad Sci USA. 2012;109:20473–20478. doi: 10.1073/pnas.1218887109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.