Abstract
DNA mismatch repair (MMR) maintains genome stability primarily by repairing DNA replication-associated mispairs. Because loss of MMR function increases the mutation frequency genome-wide, defects in this pathway predispose affected individuals to cancer. The genes encoding essential eukaryotic MMR activities have been identified, as the recombinant proteins repair 'naked' heteroduplex DNA in vitro. However, the reconstituted system is inactive on nucleosome-containing heteroduplex DNA, and it is not understood how MMR occurs in vivo. Recent studies suggest that chromatin organization, nucleosome assembly/disassembly factors and histone modifications regulate MMR in eukaryotic cells, but the complexity and importance of the interaction between MMR and chromatin remodeling has only recently begun to be appreciated. This article reviews recent progress in understanding the mechanism of eukaryotic MMR in the context of chromatin structure and dynamics, considers the implications of these recent findings and discusses unresolved questions and challenges in understanding eukaryotic MMR.
Keywords: Mismatch repair, genetic instability, H3K36me3, histone modification, Chromatin
Introduction
DNA mismatch repair (MMR) is a highly conserved genome-maintenance system present in all species from Escherichia coli to humans. The primary function of MMR is to ensure replication fidelity by strand-specifically removing misincorporated bases and insertion-deletion mispairs in newly synthesized DNA. Mutation or hypermethylation of key MMR genes causes elevated mutation frequencies and leads to an increased incidence of certain types of cancer including hereditary non-polyposis colorectal cancer (HNPCC), also called Lynch syndrome [1–6]. Previous studies have identified essential genes and protein activities involved in MMR, including MutS family and MutL family proteins (Table 1). In eukaryotic cells, the minimal activities essential for MMR include mismatch recognition proteins MutSα (MSH2–MSH6) and MutSβ (MSH2-hMSH3), MutLα (MLH1-PMS2 in humans and Mlh1-Pms1 in yeast), proliferating cell nuclear antigen (PCNA), exonuclease 1 (EXO1), replication protein A (RPA), replication factor C (RFC), DNA polymerase δ, and DNA ligase I [2–4, 6–8].
In vitro studies have established that MMR is targeted specifically to the nicked (newly synthesized) DNA strand [9, 10]. It is generally accepted that MMR is initiated by binding of MutSα or MutSβ to a mispair (either a base-base mismatch or a small insertion-deletion loop-out). This reaction triggers concerted interactions between MutSα, MutLα, PCNA and RPA, facilitating communications between two distal sites (i.e., the mismatch and a strand break) and leading to recruitment of EXO1 to a pre-existing or MutLα-generated nick [11] 5' to the mismatch. EXO1 then excises nascent DNA from the nick toward and beyond the mismatch to generate a single-strand gap, which is filled by polymerase δ using the continuous (parental) DNA strand as template. Finally, the nick is ligated by DNA ligase I (Figure 1). These MMR proteins can efficiently process “naked” heteroduplex DNA [12–14]; however, they fail to repair DNA mismatches in the context of chromatin [15, 16]. One possible explanation for this result assumes, as proposed in one of the prevailing models for human MMR, that MutSα must slide from the DNA mismatch to an upstream nick [17, 18], and that nucleosomes physically interfere with the ability of MutSα to slide on heteroduplex DNA. These observations suggest that additional factors perform this role in vivo. In other words, the current in vitro model does not apply to how MMR occurs in eukaryotic cells. Importantly, emerging evidence suggests that chromatin remodeling/modification factors interact with both MMR proteins and the DNA replication machinery, and that epigenetic marks on histones play a role during initiation of MMR in vivo [15, 19–23]. Here, these new developments in the field of eukaryotic MMR and their implications for cancer susceptibility and therapy are described and discussed. For other recent important findings, including discovery of the potential role of ribonucleotides as a strand discrimination signal for MMR in the leading strand during DNA replication [24, 25], readers are referred to excellent recent reviews by Williams & Kunkel in this issue [26] and Jiricny [27].
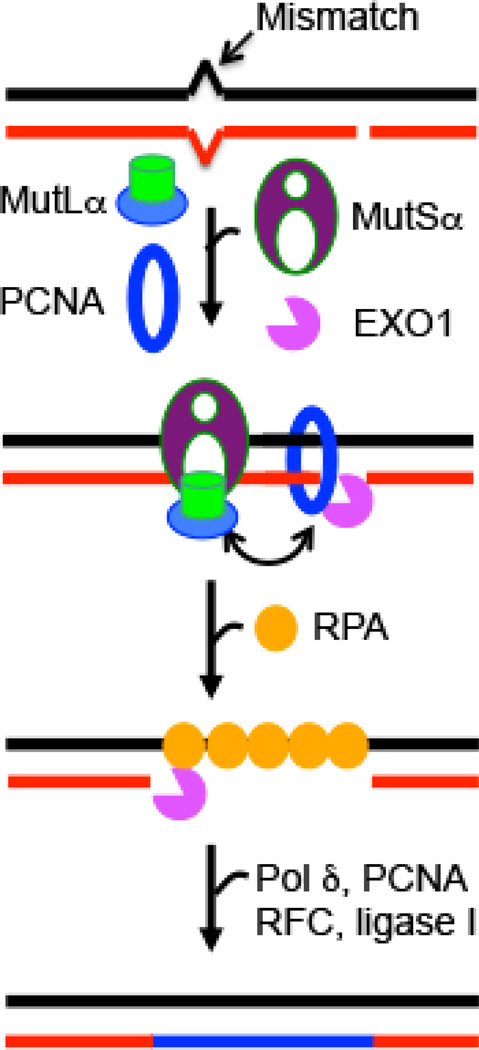
Figure 1. Eukaryotic DNA mismatch repair.
In eukaryotic cells, MMR is nick-directed and targeted to the newly-synthesized DNA strand. MMR begins with mismatch recognition by MutSα or MutSβ (not shown), which triggers concerted interactions/communications between MutSα, MutLα, PCNA and RPA, leading to the recruitment of EXO1 at a nearby nick. EXO1 then excises from the nick to the mismatch to generate a single-stranded DNA gap, which is filled by DNA polymerase (pol) δ in the presence of PCNA, RFC and RPA, followed by ligase I-catalyzed nick ligation. The image was reproduced from [87].
Role of chromatin remodeling and assembly factors in MMR
The idea that chromatin structure modulates MMR [28] and the local or regional mutation rate is not new [29]. For example, a heterotrimeric remodeling complex called RFX that regulates transcription by facilitating histone acetylation [30] also stimulates MMR in vitro [31], although a similar role in vivo has not been verified. In addition, it has been reported that hMutSα can disassemble nucleosomes on heteroduplex DNA, and this activity is enhanced by histone H3 acetylation [32]. Nevertheless, fully-modified nucleosomes from HeLa cells, which presumably carry an intact HeLa cell histone code, including H3 acetylation, inhibit MMR in vitro [15]. Therefore, the hMutSα nucleosome disassembly activity, if present, is insufficient to support MMR on chromatin, and additional factors that allow MMR to proceed in the context of chromatin have yet to be identified.
Kadyrova et al. [19] recently showed that chromatin assembly factor 1 (CAF-I), also thought to be a histone chaperone, is required during cell-free MMR to facilitate nick-dependent nucleosome assembly. Furthermore, hMutSα suppresses CAF-1-catalyzed nucleosome assembly in a mismatch-dependent manner, and nucleosome deposition by CAF-1 following mismatch removal protects the nascent DNA strand from excessive degradation by the MMR machinery. Schopf et al. [23] also demonstrated that CAF-1-catalyzed chromatin assembly occurs more slowly on heteroduplex than on homoduplex DNA. Although the detailed mechanism is not known, PCNA is thought to coordinate MMR with nucleosome loading [23], interacting with both the hMSH6 subunit of hMutSα [33–35] and CAF-1 [23]. Interestingly, hMutSα and CAF-1 also interact with each other [23]. It is possible that in the presence of a mispair, PCNA recruits hMutSα to the mismatch to promote MMR [36], and after mismatches are removed, PCNA interacts with CAF-1, triggering nucleosome assembly in nascent DNA, limiting the extent of DNA excision by the MMR machinery [19, 23]. Although evidence is lacking to support the idea, ubiquitylation, phosphorylation, or acetylation of PCNA might control the balance between its two roles, as reported for DNA polymerases during translesion DNA synthesis [37].
Role of histone modifications in MMR in vivo
Many chromatin modifying/remodeling factors contain a Pro-Trp-Trp-Pro (PWWP) domain, a member of the ‘Royal Family’ which also consists of Tudor, chromodomain and MBT domains [38]. The common feature of the “Royal Family” members is their ability to interact with methylated lysine/arginine residues in histones or other proteins through an aromatic cage [39–41]. The hMSH6 subunit of hMutSα possesses a PWWP domain [42], suggesting that it interacts with histone(s). Recent studies provide evidence to support this idea, showing that hMSH6 is a 'reader' for trimethylated Lys36 of histone H3 (H3K36me3) [43, 44]. Surprisingly, hMutSα without the hMSH6 PWWP domain is active in MMR in vitro and forms a “normal” DNA-protein co-crystal [45] as observed for other MutS family proteins lacking a PWWP motif [46–48]. The physiological function of the hMSH6 PWWP domain and its interaction with H3K36me3 were only recently discovered [15].
Using a biochemical and cellular approach, Li et al. provided evidence that the H3K36me3-hMSH6 PWWP interaction, although dispensable in vitro, is required for MMR in vivo [15]. They showed that both H3K36me3 and the hMSH6 PWWP domain are essential for localization of hMutSα to chromatin, a process that varies through the cell cycle according to the abundance of H3K36me3, This is easy to rationalize, because H3K36me3 peaks in late G1/early S and is essentially depleted in late S/G2, effectively increasing the efficiency of MMR when MMR is needed during the cell cycle to repair replication-associated misincorporation mispairs. Cells defective in H3K36 trimethyltransferase SETD2, despite being MMR-proficient in vitro, display a mutator phenotype, as if they were functionally MMR-deficient. These observations strongly suggest that the H3K36me3 histone mark plays a critical role in MMR in vivo. We now understand that H3K36me3 effectively recruits hMutSα to chromatin through its interaction with the hMSH6 PWWP domain, immediately before DNA replication initiates.
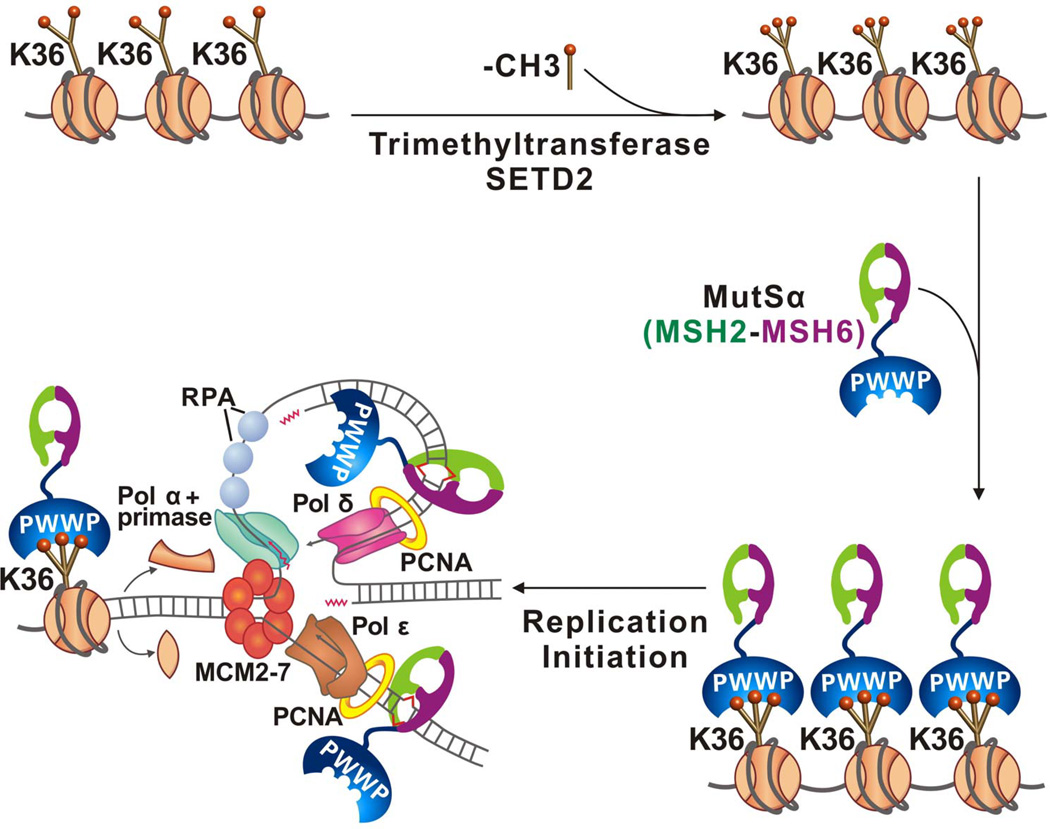
A working model for the role of H3K36me3 in MMR is presented in Figure 2. First, before cells enter S phase, SETD2 converts H3K36me2 to H3K36me3 [49]. Then, trimethylated H3K36 recruits hMutSα onto chromatin through its interaction with the hMSH6 PWWP domain. DNA replication initiates and nucleosomes are disassembled ahead of the replication fork, which also disrupts the H3K36me3-hMSH6 PWWP interaction, leading to release of hMutSα from histone octamers. hMutSα then readily binds to temporarily histone-free nascent DNA through its strong DNA binding activity and/or by interacting with PCNA via the hMSH6 PCNA-interaction protein (PIP) box. hMutSα, which possesses an ATP-dependent sliding activity [18, 50–52], then slides along the nucleosome-free DNA to locate mispairs generated during DNA replication. When hMutSα binds a mismatch, downstream MMR events ensue, such that mispaired bases are removed before mismatch-containing nascent DNA is wrapped into a nucleosome. The precise timing and sequence of events are critical, precisely because nucleosomes inhibit MMR [16, 19, 23]. The discovery of the relationship between H3K36me3 histone and the precise kinetics of MMR has been an important step in understanding how the histone code contributes to high replication fidelity in eukaryotic cells, by enhancing MMR efficiency when cells need it the most.
Figure 2. Recruitment of hMutSα to replicating chromatin.
SETD2 converts H3K36me2 to H3K36me3, which interacts with the hMSH6 PWWP domain to localize hMutSα to chromatin before DNA replication initiates. During DNA replication, nucleosomes are disassembled and the H3K36me3-PWWP interaction is disrupted, releasing hMutSα from nucleosomes. hMutSα readily binds to nascent DNA independent of PCNA, and recognizes newly-formed mismatches to initiate MMR. This image was reproduced from Ref. [15] with permission.
As with many new discoveries, this discovery has led to more questions, some of which remain unanswered at present. First, is SETD2/H3K36me3 a useful biomarker for cancer susceptibility, and might its presence correlate with microsatellite instability (MSI) in MMR-proficient cells (i.e., cells that lack mutations in MMR genes)? Could errors in the histone code explain the MSI-positive tumors, including some in HNPCC families, that do not have detectable mutations in nor hypermethylation of MMR genes [53]? Consistent with this, tumor cell lines depleted of SETD2/H3K36me3 are MMR-proficient in vitro, but display MSI [15], suggesting that loss of SETD2/H3K36me3 is responsible for the MSI phenotype in MMR-proficient tumors. In addition, a small number of gastric cancer [54], lung cancer [55, 56], leukemia [57] and renal cell carcinomas are defective in SETD2 [58–61], suggesting that SETD2-deficiency might lead to genome instability and a cancer phenotype because of its negative impact on the efficiency of MMR. Additional studies are needed to determine if this is indeed the case.
It is also worth noting that the abundance of H3K36me3 is tightly regulated by a series of histone methyltransferases (e.g., SETD2, SETD3, SETMAR, NSD1, NSD2, NSD3, ASH1L, and SMYD2) and histone demethylases (e.g., KDM2A, KDM2B, KDM4A, KDM4B, KDM4C and NO66) [62, 63]. Defects in any one of these histone methyltransferases or histone demethylases could alter the balance of H3K36 metabolism, the effects of which remain unknown. To establish whether H3K36me3 is a useful biomarker for MSI without MMR gene mutations, future studies should investigate the correlation between: 1) MSI-positive cancers, MMR status and abundance of H3K36me3; and 2) defects in H3K36me3 metabolism, MMR status and genome instability.
Second, does the local abundance of H3K36me3 influence local mutation rate? It is well recognized that some genes are more susceptible to mutations than others. For example, p53 is a mutation-prone genetic region, and >50% of human cancers carry base substitution or deletion mutations in p53 [64]. In most cases, the exact mechanism(s) that contribute to local mutation frequency are not understood. However, if the local concentration of hMutSα influences MMR efficiency, and the latter is influenced by H3K36me3 abundance, then a low level of H3K36me3 could potentially lead to an increase in local mutation rate. This hypothesis can be examined by precisely quantifying H3K36me3 in individual nucleosomes, with equally precise measurement of replication-associated mutations in cells with or without H3K36me3. Adding to the complexity, the distribution of H3K36me3 and the mutation spectrum could vary significantly in different types of cells and tissues. By analyzing the correlation between these factors, we may gain significant insight into the etiology of specific cancers, helping achieve the promise of "personalized" medicine, where cancer treatment is optimized for success in each cancer patient.
Third, how is hMutSβ recruited to chromatin/DNA? Human cells possess at least two mismatch recognition proteins, i.e., hMutSα (hMSH2–hMSH6) and hMutSβ (hMSH2–hMSH3). Unlike hMSH6, hMSH3 does not contain a PWWP domain. Is hMutSβ recruited by a different histone modification? Recent studies suggest this may be the case. For example, although yeast MutSα (yMutSα) does not contain a PWWP domain, it is localized to replicating chromatin in a mismatch-independent manner [21], possibly before DNA replication initiates. If this indicates that yMutSα recognizes and is recruited by a histone mark, then, by analogy, it is possible that hMutSβ also interacts specifically with a histone mark. Recently, Kadyrova et al. [20] showed that H3K56 acetylation (H3K56ac) regulates gross chromosomal rearrangements, base substitutions, 1-bp insertions/deletions, and complex mutations in Saccharomyces cerevisiae in a cell-cycle dependent manner. H3K56 is abundantly acetylated by Rtt109 during S phase, and deacetylated by Hst3 and Hst4 in G2/M. Strains defective in hst3 and hst4 exhibit a mutator phenotype similar to that of an msh2-deficient strain [20], suggesting that H3K56ac is involved in MMR in vivo. Additional studies are needed to establish the whether H3K56ac in fact recruits non-PWWP-containing MutS complexes (including hMutSβ, yMutSα and yMutSβ) to chromatin in a cell-cycle dependent manner.
It is worth noting that eukaryotic MSH3 and MSH6 contain a PCNA interacting protein (PIP) box [33, 34], which until now, was thought to play a major role in recruiting MutSα and MutSβ to DNA [21, 35]. However, deletion of the PIP box only moderately (~10–15%) reduces MMR activity in yeast [21, 65] and does not abolish formation of hMSH6 foci in human cells [35]; therefore, it is likely that eukaryotic MutS proteins are initially recruited to chromatin through a histone mark, after which the PIP box helps localize the chromatin-bound proteins to newly-formed mispairs via interactions with PCNA. Further investigations are needed to explore this and other possibilities, including whether MutS proteins are independently recruited by histone marks and PCNA.
The MutS-MutL interaction and MMR initiation in vivo
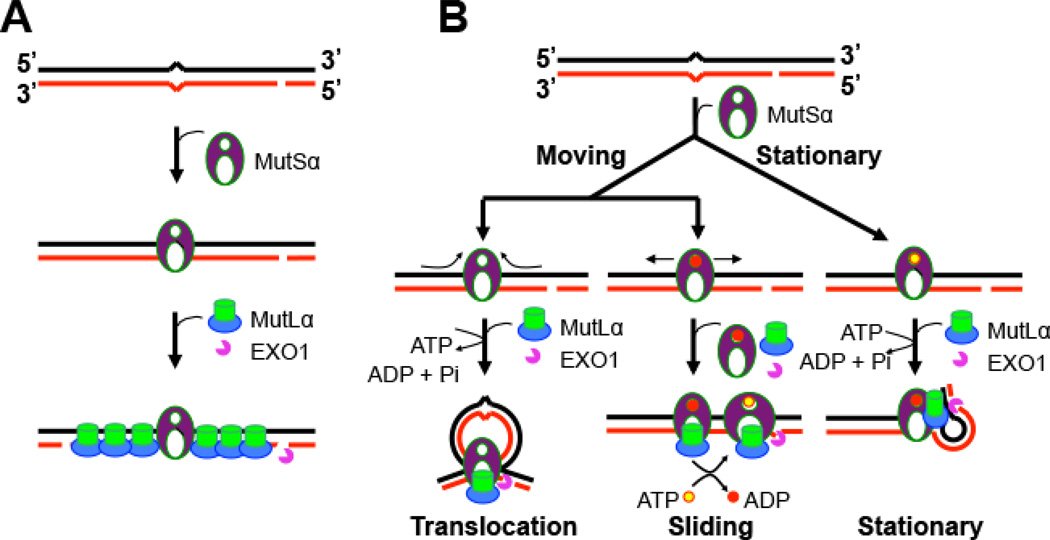
It is generally accepted that mismatch-bound MutS recruits MutL to the protein-DNA complex, and that every MutL molecule on DNA is associated with a MutS protein [66, 67]. However, this idea was brought into question, with the discovery by Hombauer et al. that yMutLα (Mlh1-Pms1 complex) and yMutSα rarely colocalize during DNA replication, and that the number of mispaired bases correlates with the number of yMutLα foci, but not with the number of yMutSα foci in yeast cells [21]. These observations led to a new model for the interaction between MutSα and MutLα (Figure 3A). In this model (referred to as the Kolodner-Hombauer model), mismatch recognition by MutSα recruits multiple molecules of MutLα to the mismatch, which activates the MutLα endonuclease [11] and EXO1, causing DNA excision to initiate at an upstream nick. It is interesting to note that this model, which does not postulate that MutSα slides away from the mismatch, is inconsistent with other current models for initiation of MMR.
Figure 3. Competing MMR models.
(A) The Kolodner-Hombauer MMR model (modified from Ref. [21]. Mismatch-bound MutSα recruits multiple molecules of MutLα to heteroduplex DNA; MutLα distributes bidirectionally from the mismatch, which activates the MutLα endonuclease, recruits EXO1 to either a pre-existing or a MutLα-generated nick, and initiates DNA excision. (B) The moving and stationary models (modified from Ref. [4]). In the stationary model (right), MutS remains bound at the mismatch. Interactions between MutS, MutL and other MMR components induce DNA bending or looping, bringing the mismatch and the nick together. There are two 'moving' models: the translocation model (left) and the sliding model (middle), in which MutS binds to the mismatch and then moves away from the mismatch in a search for the strand discrimination signal (i.e., a nick). In the translocation model, MutS creates a DNA loop as it moves from the mismatch to the nick, where it recruits EXO1 to initiate DNA excision. In the sliding model (middle), ADP-bound MutS encounters the mismatch, which triggers ADP to ATP exchange and promotes bi-directional sliding away from the mismatch, so that the mismatch is available for binding by a second incoming MutS protein. Mismatch excision begins when MutS reaches a strand break.
At least three previous models have been proposed to explain how MutS triggers DNA excision at a distant strand break after binding a mismatch (Figure 3B). Both the translocation model and the sliding model suggest that MutS loads at a mismatch and then moves away from the mismatch in an ATP-dependent manner, although the two models differ in precise stoichiometry at the mismatch and other details [17, 18, 66, 68, 69]. In contrast, the stationary model argues that MutS remains bound to a mismatch, and that the interaction between MutS, MutL and other MMR factors induces bending or looping of the DNA, bringing the mismatch and the nick into physical proximity [70, 71]. Data presented by Hombauer et al. [21] do not support active sliding of MutS on the DNA during initiation of MMR, as proposed in the translocation and sliding models. In fact, given the complicated nature of the DNA replication fork, it is difficult to imagine how MutS family proteins could freely slide along the DNA. While the Kolodner-Hombauer model and the stationary model propose a similar interaction between the mismatch and MutS proteins, they differ concerning the interaction between MutS family and MutL family proteins, and how this interaction triggers downstream events. It is hoped that future investigations will address this important question and resolve discrepancies in these three models for eukaryotic MMR.
New challenges in MMR and implications for cancer therapy
It is known that H3K36me3 promotes transcription elongation in actively transcribed genes by preventing RNA polymerase II from initiating transcription at cryptic promoters [72]. This raises the following questions. Do transcription and MMR compete with each other for the same histone mark? If they do, how is the competition regulated to avoid disruption of one or both processes? Does H3K36me3 recruit hMutSα to directly or indirectly participate in the transcription-coupled nucleotide excision repair (TC-NER) [73] that preferentially repairs bulky DNA lesions in the transcribed strand of actively transcribed genome regions [74]? Recent whole genome sequencing studies have identified histone H3.3K27M and H3.3G34R/V amino acid substitutions as the driver mutations for pediatric glioblastoma [75, 76]. Given that these mutations lead to significant decrease in H3K36me3 levels on the same and nearby nucleosomes [77–79], and that H3.3 is mainly deposited in actively transcribed genomic region [72, 80], hMutSα, which has been shown to be involved in TC-NER [81], is likely recruited to transcriptional domains for TC-NER through H3.3K36me3. However, the molecular mechanism by which hMutSα participates in the repair process remains to be investigated.
It is well established that MMR can trigger apoptosis in response to DNA damage induced by chemotherapeutic drugs, such as temozolomide and cisplatin [82]. Thus, MMR is an important target for cancer therapy. However, because MMR-deficient tumor cells are highly resistant to chemotherapeutic drugs, chemotherapy is dangerous and not a preferred treatment for patients with MSI-positive cancers. With the discovery that histone modifications regulate MMR, there may be promising new options for treating some MSI-positive cancers. This is especially important for leukemia patients, for whom chemotherapy is the only option. It is worth mentioning that many tumors lose hMSH2 or hMLH1 expression due to histone deacetylation because of the hypoxic microenvironment of the tumor [83–86]. Interestingly, the hypoxia-induced chromatin hypoacetylation and MMR deficiency can be reversed by inhibitors of histone deacetylase, such as trichostatin A [83, 86]. Thus, chemotherapeutic efficacy can be restored by normalizing the histone code, which in turn rescues the MMR dysfunction. This is an important area for future study, especially because it could ultimately provide new ways to manage difficult to treat cancers. In all cases, the molecular defect responsible for MMR-deficiency should be identified, before the course of treatment for MSI-positive cancers is selected.
Perspectives
MMR is an important mutation avoidance system found in all organisms from bacteria to man. In eukaryotic cells, chromatin structure modulates all DNA metabolic processes, including DNA replication and transcription; thus, the recent discovery that chromatin structure and the histone code play a role in coordinating MMR, DNA replication and transcription in eukaryotic cells is neither surprising nor unexpected. Looking to the future, this aspect of eukaryotic DNA metabolism and MMR warrants additional study and investigation. Such studies might identify new components or factors that influence MMR efficiency and/or timing, including histone/chromatin modifications and factors that catalyze or target such modifications. It is also possible that new roles for MMR will be discovered and that new complexities of the interaction between MMR, DNA replication and DNA transcription will emerge.
Acknowledgement
The author wishes to thank Leroy Worth for helpful comments, and acknowledge research support from the National Institutes of Health (CA167181 and GM089684) and the Kentucky Lung Cancer Research Program. The author regrets omission of citations to important studies by his colleagues, that would have been included if the publisher had authorized more space for this purpose. The author holds the James-Gardner Endowed Chair in Cancer Research.
Abbreviations
- MMR
mismatch repair
- MSI
microsatellite instability
- HNPCC
hereditary non-polyposis colorectal cancer
- PCNA
proliferating cellular nuclear antigen
- RPA
replication protein A
- RFC
replication factor C
- EXO1
exonuclease 1
- H3K36me3
histone H3 lysine 36 trimethylation
- TC-NER
transcription-coupled nucleotide excision repair
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 2.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 3.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 4.Li GM. Mechanisms and functions of DNA mismatch repair. Cell research. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 5.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 6.Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 8.Jiricny J. The multifaceted mismatch-repair system. Nature reviews. Molecular cell biology. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 9.Holmes J, Jr, Clark S, Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas DC, Roberts JD, Kunkel TA. Heteroduplex repair in extracts of human HeLa cells. The Journal of biological chemistry. 1991;266:3744–3751. [PubMed] [Google Scholar]
- 11.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 12.Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: Reconstitution of a nick-directed bidirectional reaction. The Journal of biological chemistry. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5'-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 14.Bowen N, Smith CE, Srivatsan A, Willcox S, Griffith JD, Kolodner RD. Reconstitution of long and short patch mismatch repair reactions using Saccharomyces cerevisiae proteins. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:18472–18477. doi: 10.1073/pnas.1318971110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li F, Mao G, Tong D, Huang J, Gu L, Yang W, Li GM. The Histone Mark H3K36me3 Regulates Human DNA Mismatch Repair through Its Interaction with MutSalpha. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li F, Tian L, Gu L, Li GM. Evidence that nucleosomes inhibit mismatch repair in eukaryotic cells. The Journal of biological chemistry. 2009;284:33056–33061. doi: 10.1074/jbc.M109.049874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fishel R. Mismatch repair, molecular switches, and signal transduction. Genes & development. 1998;12:2096–2101. doi: 10.1101/gad.12.14.2096. [DOI] [PubMed] [Google Scholar]
- 18.Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2–hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 19.Kadyrova LY, Blanko ER, Kadyrov FA. CAF-I-dependent control of degradation of the discontinuous strands during mismatch repair. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2753–2758. doi: 10.1073/pnas.1015914108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadyrova LY, Mertz TM, Zhang Y, Northam MR, Sheng Z, Lobachev KS, Shcherbakova PV, Kadyrov FA. A reversible histone H3 acetylation cooperates with mismatch repair and replicative polymerases in maintaining genome stability. PLoS genetics. 2013;9:e1003899. doi: 10.1371/journal.pgen.1003899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 2011;147:1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hombauer H, Srivatsan A, Putnam CD, Kolodner RD. Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science. 2011;334:1713–1716. doi: 10.1126/science.1210770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schopf B, Bregenhorn S, Quivy JP, Kadyrov FA, Almouzni G, Jiricny J. Interplay between mismatch repair and chromatin assembly. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1895–1900. doi: 10.1073/pnas.1106696109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghodgaonkar MM, Lazzaro F, Olivera-Pimentel M, Artola-Boran M, Cejka P, Reijns MA, Jackson AP, Plevani P, Muzi-Falconi M, Jiricny J. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Molecular cell. 2013;50:323–332. doi: 10.1016/j.molcel.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Molecular cell. 2013;50:437–443. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams JS, Kunkel TA. Ribonucleotides in DNA: origins, repair and consequences. DNA repair. 2014 doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiricny J. Postreplicative mismatch repair. Cold Spring Harbor perspectives in biology. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hawk JD, Stefanovic L, Boyer JC, Petes TD, Farber RA. Variation in efficiency of DNA mismatch repair at different sites in the yeast genome. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8639–8643. doi: 10.1073/pnas.0503415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012 doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 30.Reith W, Mach B. The bare lymphocyte syndrome and the regulation of MHC expression. Annu Rev Immunol. 2001;19:331–373. doi: 10.1146/annurev.immunol.19.1.331. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Yuan F, Wang D, Gu L, Li GM. Identification of RFX as a novel mismatch repair stimulatory factor. The Journal of biological chemistry. 2008;283:12730–12735. doi: 10.1074/jbc.M800460200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Javaid S, Manohar M, Punja N, Mooney A, Ottesen JJ, Poirier MG, Fishel R. Nucleosome remodeling by hMSH2–hMSH6. Molecular cell. 2009;36:1086–1094. doi: 10.1016/j.molcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional interaction of proliferating cell nuclear antigen with MSH2–MSH6 and MSH2–MSH3 complexes. The Journal of biological chemistry. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- 34.Flores-Rozas H, Clark D, Kolodner RD. Proliferating cell nuclear antigen and Msh2p–Msh6p interact to form an active mispair recognition complex. Nat Genet. 2000;26:375–378. doi: 10.1038/81708. [DOI] [PubMed] [Google Scholar]
- 35.Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes & development. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lau PJ, Kolodner RD. Transfer of the MSH2.MSH6 complex from proliferating cell nuclear antigen to mispaired bases in DNA. The Journal of biological chemistry. 2003;278:14–17. doi: 10.1074/jbc.C200627200. [DOI] [PubMed] [Google Scholar]
- 37.Lehmann AR. Ubiquitin-family modifications in the replication of DNA damage. FEBS letters. 2011;585:2772–2779. doi: 10.1016/j.febslet.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain 'Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends in biochemical sciences. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 39.Adams-Cioaba MA, Min J. Structure and function of histone methylation binding proteins. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2009;87:93–105. doi: 10.1139/O08-129. [DOI] [PubMed] [Google Scholar]
- 40.Liu K, Chen C, Guo Y, Lam R, Bian C, Xu C, Zhao DY, Jin J, MacKenzie F, Pawson T, Min J. Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18398–18403. doi: 10.1073/pnas.1013106107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu H, Wang JY, Huang Y, Li Z, Gong W, Lehmann R, Xu RM. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes & development. 2010;24:1876–1881. doi: 10.1101/gad.1956010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laguri C, Duband-Goulet I, Friedrich N, Axt M, Belin P, Callebaut I, Gilquin B, Zinn-Justin S, Couprie J. Human mismatch repair protein MSH6 contains a PWWP domain that targets double stranded DNA. Biochemistry. 2008;47:6199–6207. doi: 10.1021/bi7024639. [DOI] [PubMed] [Google Scholar]
- 43.Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Gottgens B, Bycroft M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nature structural & molecular biology. 2010;17:617–619. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- 44.Wu H, Zeng H, Lam R, Tempel W, Amaya MF, Xu C, Dombrovski L, Qiu W, Wang Y, Min J. Structural and histone binding ability characterizations of human PWWP domains. PLoS One. 2011;6:e18919. doi: 10.1371/journal.pone.0018919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the human MutSalpha DNA lesion recognition complex. Molecular cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 46.Gupta S, Gellert M, Yang W. Mechanism of mismatch recognition revealed by human MutSbeta bound to unpaired DNA loops. Nature structural & molecular biology. 2012;19:72–78. doi: 10.1038/nsmb.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 48.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 49.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. The EMBO journal. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blackwell LJ, Martik D, Bjornson KP, Bjornson ES, Modrich P. Nucleotide-promoted release of hMutSalpha from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. The Journal of biological chemistry. 1998;273:32055–32062. doi: 10.1074/jbc.273.48.32055. [DOI] [PubMed] [Google Scholar]
- 51.Gorman J, Chowdhury A, Surtees JA, Shimada J, Reichman DR, Alani E, Greene EC. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2–Msh6. Molecular cell. 2007;28:359–370. doi: 10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mendillo ML, Mazur DJ, Kolodner RD. Analysis of the interaction between the Saccharomyces cerevisiae MSH2–MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. The Journal of biological chemistry. 2005;280:22245–22257. doi: 10.1074/jbc.M407545200. [DOI] [PubMed] [Google Scholar]
- 53.Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. Journal of clinical oncology. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 54.Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, Chan TL, Kan Z, Chan AS, Tsui WY, Lee SP, Ho SL, Chan AK, Cheng GH, Roberts PC, Rejto PA, Gibson NW, Pocalyko DJ, Mao M, Xu J, Leung SY. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43:1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 55.Govindan R, Ding L, Griffith M, Subramanian J, Dees N, Walker KL, Maher CA, Fulton R, Fulton L, Wallis J, Chen K, Watson M, Mardis ER, Wilson RK. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, Sougnez C, Auclair D, Lawrence MS, Stojanov P, Cibulskis K, Choi K, de Waal L, Sharifnia T, Brooks A, Greulich H, Banerji S, Zander T, Seidel D, Leenders F, Ansén S, Ludwig C, Engel-Riedel W, Stoelben E, Wolf J, Goparju C, Thompson K, Winckler W, Kwiatkowski D, Johnson BE, Jänne PA, Miller VA, Pao W, Travis WD, Pass HI, Gabrie SB, Lander ES, Thomas RK, Garraway LA, Getz G, Meyerson M. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, Lu C, Chen SC, Wei L, Collins-Underwood JR, Ma J, Roberts KG, Pounds SB, Ulyanov A, Becksfort J, Gupta P, Huether R, Kriwacki RW, Parker M, McGoldrick DJ, Zhao D, Alford D, Espy S, Bobba KC, Song G, Pei D, Cheng C, Roberts S, Barbato MI, Campana D, Coustan-Smith E, Shurtleff SA, Raimondi SC, Kleppe M, Cools J, Shimano KA, Hermiston ML, Doulatov S, Eppert K, Laurenti E, Notta F, Dick JE, Basso G, Hunger SP, Loh ML, Devidas M, Wood B, Winter S, Dunsmore KP, Fulton RS, Fulton LL, Hong X, Harris CC, Dooling DJ, Ochoa K, Johnson KJ, Obenauer JC, Evans WE, Pui CH, Naeve CW, Ley TJ, Mardis ER, Wilson RK, Downing JR, Mullighan CG. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, Teague J, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Forbes S, Jia M, Jones D, Knott H, Kok CY, Lau KW, Leroy C, Lin ML, McBride DJ, Maddison M, Maguire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, O'Meara S, Pleasance E, Rajasingham A, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turrell K, Dykema KJ, Khoo SK, Petillo D, Wondergem B, Anema J, Kahnoski RJ, Teh BT, Stratton MR, Futreal PA. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duns G, van den Berg E, van Duivenbode I, Osinga J, Hollema H, Hofstra RM, Kok K. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 2010;70:4287–4291. doi: 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- 60.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, Bignell G, Butler A, Cho J, Dalgliesh GL, Galappaththige D, Greenman C, Hardy C, Jia M, Latimer C, Lau KW, Marshall J, McLaren S, Menzies A, Mudie L, Stebbings L, Largaespada DA, Wessels LF, Richard S, Kahnoski RJ, Anema J, Tuveson DA, Perez-Mancera PA, Mustonen V, Fischer A, Adams DJ, Rust A, Chan-on W, Subimerb C, Dykema K, Furge K, Campbell PJ, Teh BT, Stratton MR, Futreal PA. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Albert M, Helin K. Histone methyltransferases in cancer. Semin Cell Dev Biol. 2010;21:209–220. doi: 10.1016/j.semcdb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 63.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 65.Shell SS, Putnam CD, Kolodner RD. The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Molecular cell. 2007;26:565–578. doi: 10.1016/j.molcel.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Molecular cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 67.Blackwell LJ, Wang S, Modrich P. DNA chain length dependence of formation and dynamics of hMutSalpha.hMutLalpha.heteroduplex complexes. The Journal of biological chemistry. 2001;276:33233–33240. doi: 10.1074/jbc.M105076200. [DOI] [PubMed] [Google Scholar]
- 68.Allen DJ, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith JD. MutS mediates heteroduplex loop formation by a translocation mechanism. Embo J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 70.Guarne A, Ramon-Maiques S, Wolff EM, Ghirlando R, Hu X, Miller JH, Yang W. Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair. Embo J. 2004;23:4134–4145. doi: 10.1038/sj.emboj.7600412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Junop MS, Obmolova G, Rausch K, Hsieh P, Yang W. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Molecular cell. 2001;7:1–12. doi: 10.1016/s1097-2765(01)00149-6. [DOI] [PubMed] [Google Scholar]
- 72.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nature reviews. Molecular cell biology. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmidt CK, Jackson SP. On your mark, get SET(D2), go! H3K36me3 primes DNA mismatch repair. Cell. 2013;153:513–515. doi: 10.1016/j.cell.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 74.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nature reviews. Molecular cell biology. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 75.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jager N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Fruhwald MC, Roggendorf W, Kramm C, Durken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 76.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Zhang J, Baker SJ. P. St. Jude Children's Research Hospital-Washington University Pediatric Cancer Genome, Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, Sarkaria J, Zhang Z. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes & development. 2013;27:985–990. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuen BT, Knoepfler PS. Histone H3.3 mutations: a variant path to cancer. Cancer cell. 2013;24:567–574. doi: 10.1016/j.ccr.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Delbarre E, Jacobsen BM, Reiner AH, Sorensen AL, Kuntziger T, Collas P. Chromatin environment of histone variant H3.3 revealed by quantitative imaging and genome-scale chromatin and DNA immunoprecipitation. Molecular biology of the cell. 2010;21:1872–1884. doi: 10.1091/mbc.E09-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mellon I, Rajpal DK, Koi M, Boland CR, Champe GN. Transcription-coupled repair deficiency and mutations in human mismatch repair genes. Science. 1996;272:557–560. doi: 10.1126/science.272.5261.557. [DOI] [PubMed] [Google Scholar]
- 82.Li GM. The role of mismatch repair in DNA damage-induced apoptosis. Oncol Res. 1999;11:393–400. [PubMed] [Google Scholar]
- 83.Edwards RA, Witherspoon M, Wang K, Afrasiabi K, Pham T, Birnbaumer L, Lipkin SM. Epigenetic repression of DNA mismatch repair by inflammation and hypoxia in inflammatory bowel disease-associated colorectal cancer. Cancer Res. 2009;69:6423–6429. doi: 10.1158/0008-5472.CAN-09-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kondo A, Safaei R, Mishima M, Niedner H, Lin X, Howell SB. Hypoxia-induced enrichment and mutagenesis of cells that have lost DNA mismatch repair. Cancer Res. 2001;61:7603–7607. [PubMed] [Google Scholar]
- 85.Koshiji M, To KK, Hammer S, Kumamoto K, Harris AL, Modrich P, Huang LE. HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Molecular cell. 2005;17:793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 86.Mihaylova VT, Bindra RS, Yuan J, Campisi D, Narayanan L, Jensen R, Giordano F, Johnson RS, Rockwell S, Glazer PM. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol. 2003;23:3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li GM. Decoding the histone code: Role of H3K36me3 in mismatch repair and implications for cancer susceptibility and therapy. Cancer Res. 2013;73:6379–6383. doi: 10.1158/0008-5472.CAN-13-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]