Abstract
Purpose
Typically, low molecular weight cationic peptides or polymers exhibit poor transfection efficiency due to an inability to condense plasmid DNA into small nanoparticles. Here, efficient gene delivery was attained using TAT/pDNA complexes containing calcium crosslinks.
Methods
Electrostatic complexes of pDNA with TAT or PEI were studied with increasing calcium concentration. Gel electrophoresis was used to determine DNA condensation. The morphology of the complexes was probed by transmission electron microscopy. Transfection efficiency was assessed using a luciferase reporter plasmid. The accessibility of phosphate and amine groups within complexes was evaluated to determine the effect of calcium on structure.
Results
TAT/pDNA complexes were condensed into small, 50–100 nm particles by optimizing the concentration of calcium. Complexes optimized for small size also exhibited higher transfection efficiency than PEI polyplexes in A549 cells. TAT and TAT complexes displayed negligible cytotoxicity up to 5 mg/mL, while PEI exhibited high cytotoxicity, as expected. Probing the TAT-Ca/pDNA structure suggested that calcium interacted with both phosphate and amine groups to compact the complexes; however, these “soft” crosslinks could be competitively disrupted to facilitate DNA release.
Conclusion
Small and stable TAT-Ca/pDNA complexes were obtained via “soft” calcium crosslinks leading to sustained gene expression levels higher than observed for control PEI gene vectors. TAT-Ca/pDNA complexes were stable, maintaining particle size and transfection efficiency even in the presence of 10% of FBS. TAT-Ca complexes offer an effective vehicle offering potential for translatable gene delivery.
Keywords: A549 cells, gene delivery, plasmid DNA, polyethylenimine, TAT
INTRODUCTION
Intensive effort has been devoted to develop gene therapy systems capable of overcoming a variety of limitations, including low gene transfection efficiency, toxicity, and in vivo instability (1–7). Nonviral vectors have been given considerable attention as gene delivery vehicles because of their presumed safety, ease of synthesis, relatively unrestricted vector size, low cost, and low degree of immunogenicity in comparison to viral vectors (8). Plasmid DNA complexed with cationic lipids (lipoplexes) and polymers (polyplexes) are the most commonly employed nonviral gene delivery vehicles (9–17). Because naked plasmid DNA does not easily penetrate cellular membranes (18), nonviral gene delivery systems may include agents to improve intracellular delivery in an effort to promote transfection. Finally, gene delivery vehicles are subject to dilution, degradation and elimination in vivo, which amplify the need for safe and efficient gene delivery at high doses of DNA.
New strategies have been put forward to enhance cellular uptake of gene delivery vehicles, among which peptides enhancing cell adhesion and internalization have reached a prominent position (19). Peptide sequences, also designated as protein transduction domains (PTD) or membrane translocalization signals (MTS), were identified as potentially useful labels for intracellular delivery of peptides, proteins, oligonucleotides, and plasmid DNA (20–24). By modifying the surface of gene delivery vectors with cell-penetrating peptides (CPPs), vectors have been shown to traverse the membranes of biological cells within seconds to minutes (25). Polycationic CPPs have even been reported to enhance cell permeability and facilitate the intracellular delivery of nanoparticles (26).
The mechanism of cell entry of CPPs alone or with their cargoes still remains somewhat of an enigma. Some reports indicating that cellular translocation of CPPs is energy as well as endocytosis independent and that there is direct transfer of the peptides through the lipid bilayer by inverted micelle formation (27–32). Another report, however, proposed an energy dependent mechanism of cell entry of CPPs (33), which may also involve extracellular heparan sulfate and various endocytosis and macropinocytosis pathways (34–41). It was also suggested that classical and nonclassical endocytosis pathways may be associated simultaneously with CPP translocation, depending upon the biophysical properties of CPPs and their cargo (32,42,43).
One particular CPP of interest is the HIV-1 TAT peptide. This peptide, which represents a protein transduction domain (44,45) and a nuclear localization sequence (NLS) (46), has been reported to show unusual translocation abilities by directly crossing biological membranes independent of receptors and temperature (47). In addition, the NLS function of TAT peptide could facilitate nuclear localization of a therapeutic agent due to interaction with the endogenous cytoplasmic nuclear transport machinery. The cationic nature of the TAT sequence arising from several arginine residues has already been utilized in gene delivery either by covalent coupling of this CPP to the gene delivery vehicle (48–50) or by simple mixing of plasmid DNA with TAT to form TAT/pDNA complexes via noncovalent electrostatic interactions (51–54). However, the transfection efficiency of such complexes remains quite low and requires improvement.
Obviously, a strong affinity between TAT peptide and plasmid DNA is required to stabilize the resulting polyplex and to achieve an optimized transfection yield. On the other hand, a sufficiently low affinity between TAT peptide and plasmid DNA is desired to facilitate the release of the cargo after cellular uptake. Thus, the critical balance between the TAT/pDNA binding affinities has important consequences for enhancing the transfection efficiency. Our objective was to design a more efficient and less toxic means of gene delivery using TAT/pDNA complexes containing “soft” crosslinks. Calcium was found to control the delicate balance between binding affinities within TAT/pDNA polyplexes. The addition of CaCl2 to TAT/pDNA complexes directly affected particle size and transfection efficiency in a concentration-dependent manner. The optimum calcium concentration (0.3 M) resulted in a 1,000-fold enhancement in TAT/pDNA polyplex transfection efficiency and showed no detectable cytotoxicity. Gene transfection levels were as high as that observed for PEI polyplexes, suggesting the possible translation of the TAT-Ca/pDNA complexes.
MATERIALS AND METHODS
Materials
Plasmid DNA encoding firefly luciferase (pGL3, 4.8 kbp) was obtained from Promega (Madison, WI, USA) and transformed into Escherichia coli DH5ά (Invitrogen, Carlsbad, CA). A single transformed colony picked from an agar plate was cultured in LB Broth Base (Invitrogen) liquid for plasmid DNA preparation. Plasmid DNA was purified with Plasmid Giga Kit 5 (Qiagen, Germantown, MD) following the manufacturer’s instructions. All pDNA had purity levels of 1.8 or greater as determined by UV/Vis inspection (A260/A280). TAT peptide (RKKRRQRRR; Mw=1338.85 Da, TFA salt=2632.9 Da) was synthesized in house. Branched polyethylenimine (PEI, 25 kDa) was obtained from Aldrich (Milwaukee, WI). Calcium chloride (CaCl2. 2H2O) and agarose medium were purchased from Fisher Scientific (Pittsburgh, PA). A549 cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD). The cell culture medium (Ham’s F-12 Nutrient Mixture, Kaighn’s modified with L-glutamine) was purchased through Fisher Scientic. Fetal bovine serum (FBS) was purchased from Hyclone. Penicillin-streptomycin was purchased from MB Biomedical, LLC. Trypsin-EDTA was purchased through Gibco. MTS reagent [tetrazolium compound; 3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] was purchased from Promega. Heparin Sodium was obtained from Spectrum (Gardena, CA).
Preparation of TAT-Ca/pDNA Complexes
TAT/pDNA complexes were synthesized by rapidly adding 10µL (0.1µg/µL) of pDNA to 15µL (N/P ratio of 25) TAT solution while pipetting. To this solution, 15µL of known molarity (e.g. 0.3 M) CaCl2 was added and mixed by vigorous pipetting followed by 15–20 min incubation at 4°C prior to use.
Formation of PEI/pDNA Complexes
PEI-DNA complexes were prepared by adding 10µl (0.1 µg/µL) of pDNA solution to 15µL (N/P ratio of 5 or 10) PEI solution dropwise while stirring. Complexes were incubated at room temperature for 20–30 min before dilution 1.7 times (15µL) with the appropriate buffer (e.g. nuclease-free water or CaCl2 solution). Complexes were freshly prepared before each individual experiment.
Size and Zeta Potential Measurement
Suspensions containing complexes with TAT or PEI were prepared as described earlier using a DNA concentration of 0.1µg/µL. All samples intended for light scattering analyses were prepared using 10 mM Tris buffer, pH 7.4, which was prefiltered with a 0.22µm filter to remove any trace particulates. Particle sizes were measured by dynamic light scattering (DLS) using a Brookhaven (Holtsville, NY) instrument equipped with a 9000AT autocorrelator, a 50 mW HeNe laser operating at 532 nm (JDS Uniphase), an EMI 9863 photomultiplier tube, and a BI 200 M goniometer. The light scattered at 90° from the incident light was fit to an autocorrelation function using the method of cumulants. Zeta potential measurements were obtained by phase analysis light scattering using a Brookhaven Zeta PALS instrument. The electrophoretic mobility of the samples was determined from the average of 10 cycles of an applied electric field. The zeta potential was determined from the electrophoretic mobility from the Smoluchowski approximation. Zeta potential was determined in 1 mM KCl solution.
Agarose Gel Electrophoresis Assays
The pDNA binding ability of the TAT, TAT-Ca, CaCl2 and PEI complexes was analyzed by agarose gel electrophoresis. The TAT-Ca/pDNA and PEI/DNA complexes containing 1µg luciferase reporter gene were prepared as described at various N/P ratios. The N/P ratio refers to the molar ratio of amine groups in the cationic polymer, which represent the positive charges, to phosphate groups in the plasmid DNA, which represent the negative charges. The DNA complex suspensions (i.e. 25µL) at various N/P ratios were diluted by adding 4µL of 10× Tris-acetate-EDTA (TAE) gel running buffer (Promega) and 4µL of 100× SYBR Green (Invitrogen) solutions. The DNA loading buffer (7µL of 6×) was added to the complex suspensions. The mixtures were allowed to incubate at room temperature for 40 min to ensure labeling of the DNA with the SYBR Green dye. Thereafter, the complexes were loaded into individual wells of 1% agarose/ 1× TAE gel buffer, and subjected to electrophoresis at 110 V for 30 min. Uncomplexed DNA diluted with an identical volume of solution was used as a control. The resulting DNA migration patterns were revealed using an AlphaImager® Imaging System (Alpha Innotech, San Leandro, CA).
Cell Culture
Culturing of human epithelial lung cell line A549 was performed according to the protocol provided by the American Type Culture Collection. A549 cells were grown in F-12K supplemented with 10% v/v FBS and 1% v/v Penicillin/streptomycin at 37° C in a humidified air atmosphere containing 5% CO2.
In Vitro Cell Transfection Studies
A549 cells were trypsinized, counted and diluted to a concentration of approximately 80,000 cells/ mL. Then 0.1 mL of that dilution was added to each well of a 96-well plate, and the cells were incubated in a humidified atmosphere at 5%CO2 and 37°C for 24 h. Immediately before transfection, the cells were washed once with PBS and 100µl sample (20% of complex to 80%of serum-free cell culture medium) was added to each well. Cells were incubated with the complexes for 5 h. The transfection agent was then removed by aspiration, and 100µL of fresh serum medium was added followed by further incubation. The Luciferase Assay System from Promega was used to determine gene expression following the manufacturer’s recommended protocol. The light units were normalized against protein concentration in the cells’ extracts, which were measured using the Coomassie Plus™ Protein Assay (Thermo Scientific). The transfection results were expressed as Relative Light Units (RLU) per mg of cellular protein.
Assessment of Cytotoxicity (MTS Assay)
Cytotoxicity of polymers was determined by the Cell-Titer 96® Aqueous Cell Proliferation Assay (Promega). A549 cells were grown as described in the transfection experiments. Cells were treated with the samples for ~24 h. The media were then removed and replaced with a mixture of 100µL fresh culture media and 20µL MTS reagent solution. The cells were incubated for 3 h at 37°C in the 5% CO2 incubator. The absorbance of each well was then measured at 490 nm using a microtiter plate reader (SpectraMax, M25, Molecular Devices Corp., CA) to determine cell viability.
SYBR Green Assay of TAT and PEI Complexes
The degree of pDNA accessibility following complexation with TAT or PEI was assessed by the double-stranded-DNA-binding reagent SYBR Green (Invitrogen). Briefly, 10 µL (0.1 mg/mL) of pDNA was mixed with 15 µL of TAT or PEI solution, then 15 µL deionized water or metal solution was added. Complexes were allowed to form for 30 min at room temperature prior to use. After incubation, 120 µL deionized water and 160 µL 10× SYBR Green solutions were added. And then 80 µL of sample was added to three wells of a 96-well cell culture plate. The fluorescence was measured using a fluorescence plate reader (SpectraMax M5; Ex., 250 nm; Em, 520 nm).
TNBS Assay of TAT and PEI Complexes
The accessibility of free amine groups of TAT or PEI following complexation with pDNA was measured using a colorimetric assay with 2,4,6-trinitro-benzenesulphonic acid (TNBS) as an assay reagent (Pierce). Briefly, 10 µL of complex solution was added to 190 µL deionized water and then 200 µL of 0.02 % TNBS solution in 0.1 M sodium bicarbonate buffer (pH 8.5) was added. The solution was rapidly mixed. After incubation at 37°C for 2 h, 80 µL of sample was added to three wells of a 96-well cell culture plate. The absorption at 335 nm was determined using a plate reader.
The Effect of Heparin on the Stability of TAT and PEI Complexes
The effect of heparin on the stability of complexes was evaluated by the means of the change in fluorescence intensity obtained with the fluorescent probe SYBR Green. TAT and PEI complexes were freshly prepared in absence and the presence of CaCl2 (0.3 M) as above described. 120 µL of heparin solution was added to these various complexes to yield final diverse heparin concentrations up to 50µg/µL, incubated for 30 min at room temperature and then 160 µL 10× SYBR Green was added. In triplicate, 80 µL of each sample was added to the well of 96-well plate and the fluorescence was measured as indicated in SYBR Green assay.
Statistical Analysis
Statistical evaluation of comparing the significance of the difference in expression between the means of two groups was performed using the t-test, a value of p <0.05 was accepted as significant.
RESULTS
Formation of TAT-Ca/pDNA and PEI/pDNA Complexes
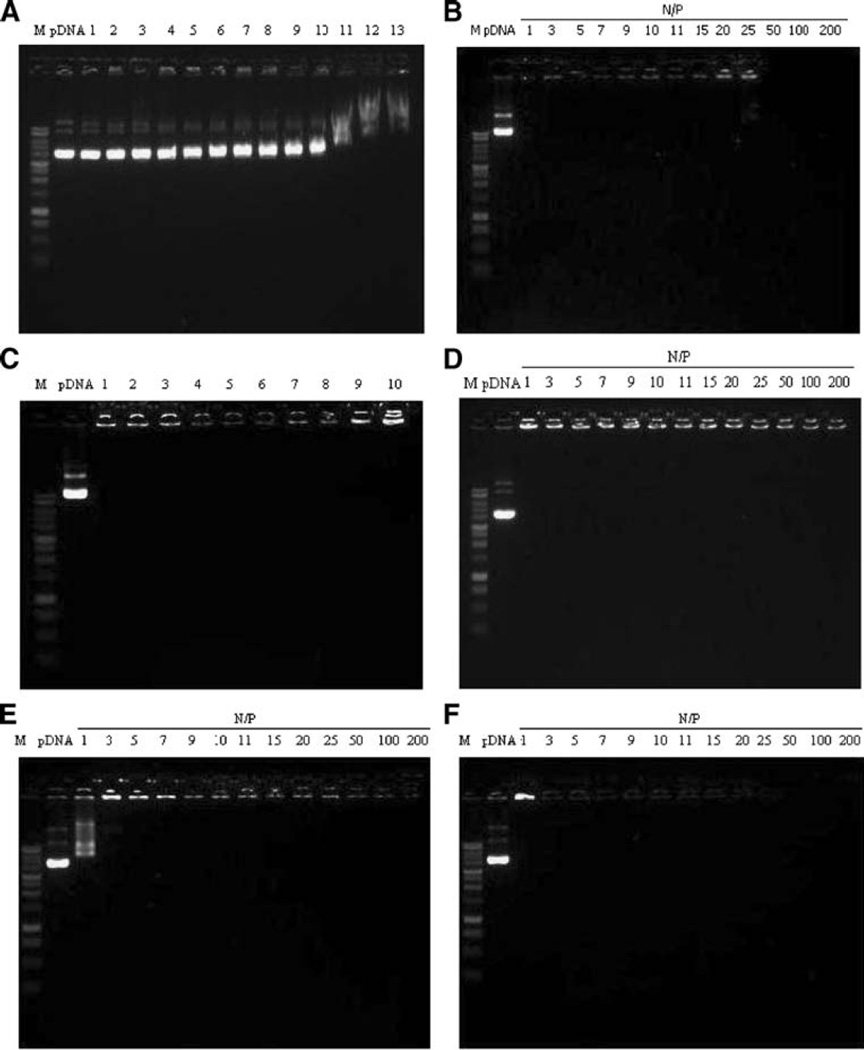
TAT and PEI complexes were prepared by mixing pDNA with each polycation at various N/P ratios as described. In order to demonstrate complex formation, a gel electrophoresis assay was performed using 1% agarose gel. Uncomplexed pDNA was used as a control. Gel electrophoresis indicated that pDNA complexed with polycations could be retained in the loading wells. Above a certain N/P ratio, no bands were observed during electrophoresis, indicating that TAT and PEI completely complexed the pDNA. Under these conditions, CaCl2 showed negligible ability to complex the plasmid DNA even at high concentration (1 M) (Fig. 1).
Fig. 1.
Gel electrophoresis study of (a) CaCl2/pDNA complexes; 1=0.1 M, 2=0.2 M, 3=0.3 M, 4=0.4 M, 5=0.5M, 6=0.6M, 7=0.7 M, 8=0.8M, 9=0.9 M, 10=1.0 M, 11=3 M, 12=5 M and 13=7 M of CaCl2. (b) TAT/pDNA complexes. (c) TAT-Ca/pDNA N/P 25 complexes; 1=0 mM, 2=62.5 mM, 3=125 mM, 4=250 mM, 5=300 mM, 6=350 mM, 7=400 mM, 8=500 mM, 9=1 M and 10=2 M of CaCl2. (d) TAT/pDNA complexes with 0.3 M CaCl2. (e) Branched PEI/pDNA complexes. (f) Linear PEI/pDNA complexes.
Morphological and Physical Characterization of TAT-Ca/pDNA and PEI/pDNA Complexes
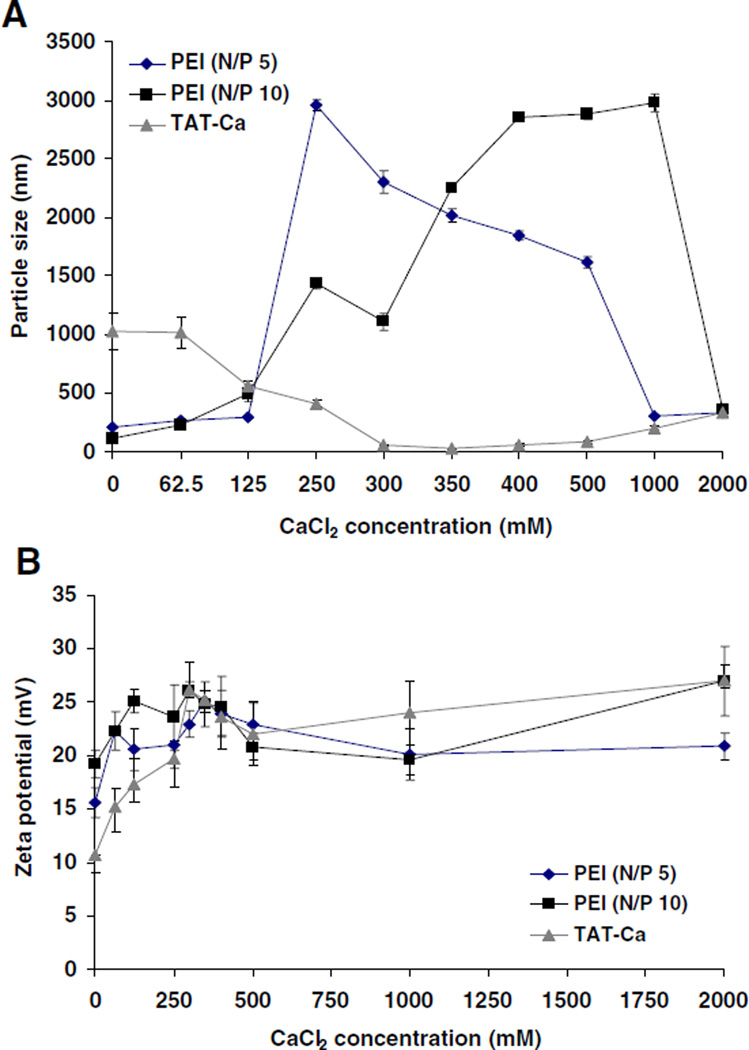
The effect of calcium chloride concentration on the particle size and surface charge of TAT/pDNA and PEI/pDNA complexes was investigated. Over a certain concentration range, calcium addition to TAT/pDNA complexes induced a substantial decrease in the particle size. In comparison, PEI/pDNA complexes showed an increase in particle size (Fig. 2A). The added CaCl2 concentration range of 0.3–0.5 M (113–188 mM final CaCl2 concentration) produced small (50–100 nm) and stable TAT/pDNA complexes with relatively narrow polydispersity (< 0.15). In general, the zeta potential of TAT and PEI complexes increased significantly from 11 to 27 mV with increasing concentration of CaCl2 (Fig. 2B).
Fig. 2.
The effect of CaCl2 concentration on (A) particle size and (B) charge of PEI/pDNA complexes (N/P 5, N/P 10) and TAT-Ca/pDNA (N/P 25) complexes. Data are presented as mean±SD (n=3).
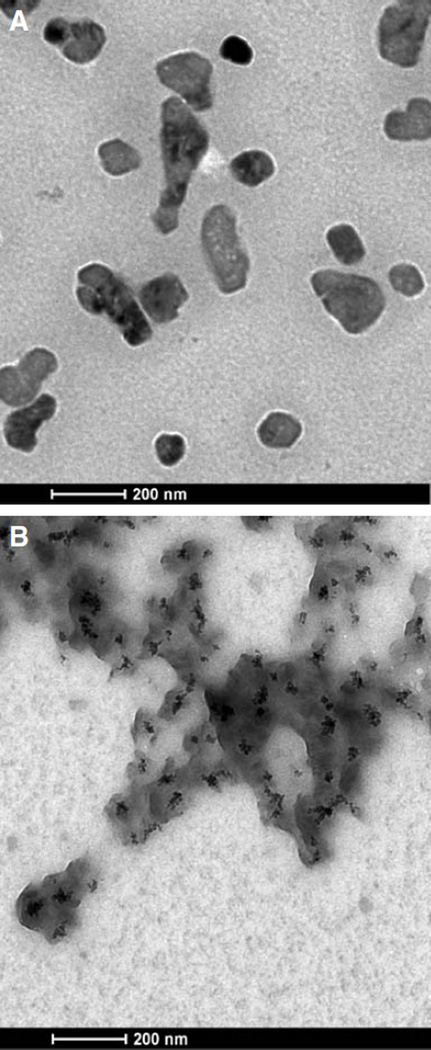
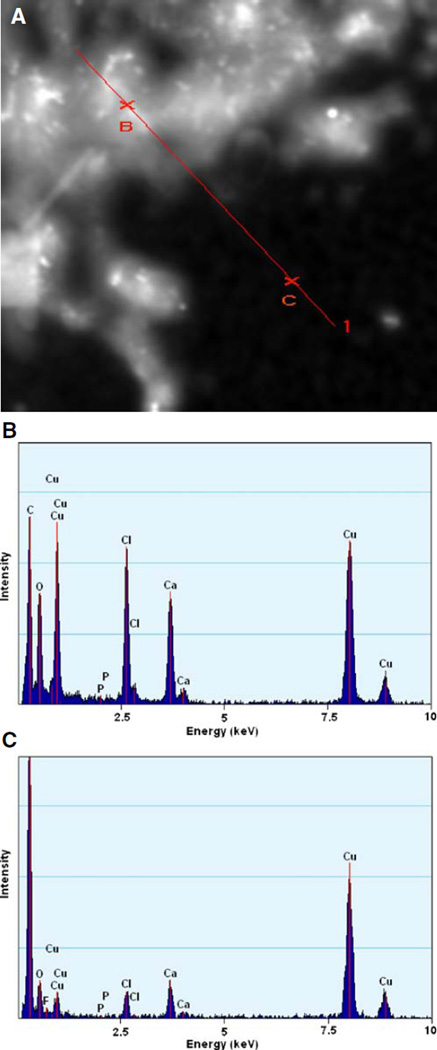
A transmission electron microscope (FEI field emission transmission electron microscope, Tecnai G2 at 200 kV) equipped with an energy-dispersive analytical X-ray (EDAX) was used to image the morphology of the TAT/pDNA and TAT-Ca/pDNA complexes and to characterize elements in these complexes, respectively. TEM samples were prepared by depositing a drop of the complex suspension on a copper carbon grid and allowing it to dry in a dessicater overnight. Transmission electron micrographs of TAT/pDNA complexes revealed an asymmetric morphology. The darker areas in TAT-Ca/pDNA seemed to suggest calcium trapped inside the particles (Fig. 3A and B). Samples were imaged using scanning transmission electron microscopy and analyzed using EDAX spectroscopy to detect the location of calcium. EDAX spectrum for two labeled areas in the scanning transmission electron micrograph (on the particles and substrate) revealed significantly higher calcium concentration in the particles compared to free calcium that would have dried on the substrate (Fig. 4A, B and C).
Fig. 3.
Transmission electron micrograph of (A) TAT/pDNA and (B) TAT-Ca/pDNA.
Fig. 4.
(A) Scanning transmission electron micrograph and EDAX spectrum of TAT-Ca/pDNA. The characterized areas (B) on particles and (C) on substrate are indicated in the scanning transmission electron micrograph. EDAX spectra showed the elemental composition of particles and substrate (Cu peaks result from the copper grid).
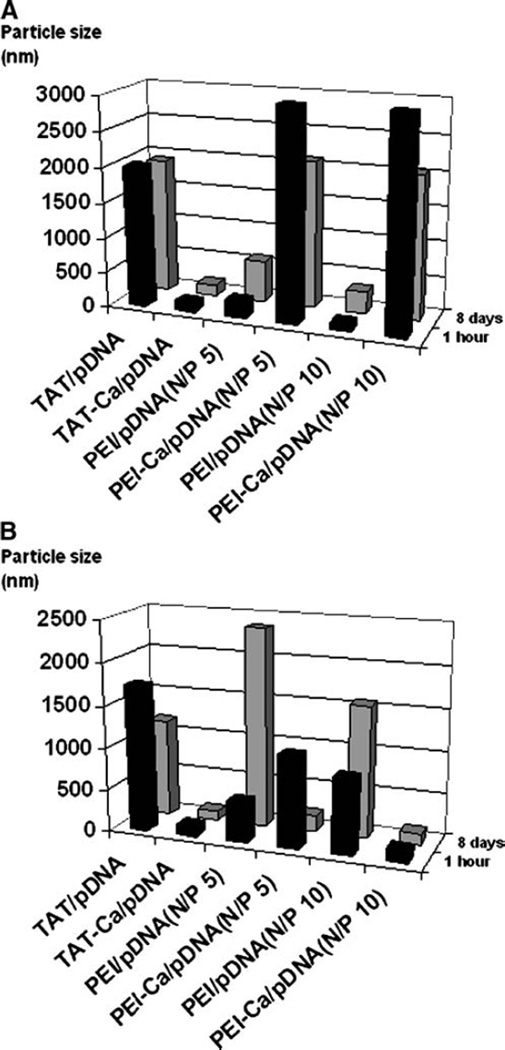
The stability of TAT and PEI complexes was investigated as a function of time in the absence and presence of 0.3 M CaCl2 and 10% FBS (Fig. 5A and B). TAT/pDNA complexes including 0.3 M CaCl2 remained stable in the absence and presence of 10% FBS over a period of 8 days. Conversely, PEI/pDNA complexes showed a marked increase in size.
Fig. 5.
The stability of TAT-Ca/pDNA and PEI/pDNA complexes over time in (A) the absence and (B) presence of 10%FBS. Results are presented as mean±SD (n=3).
Cytotoxicity of TAT and PEI Complexes
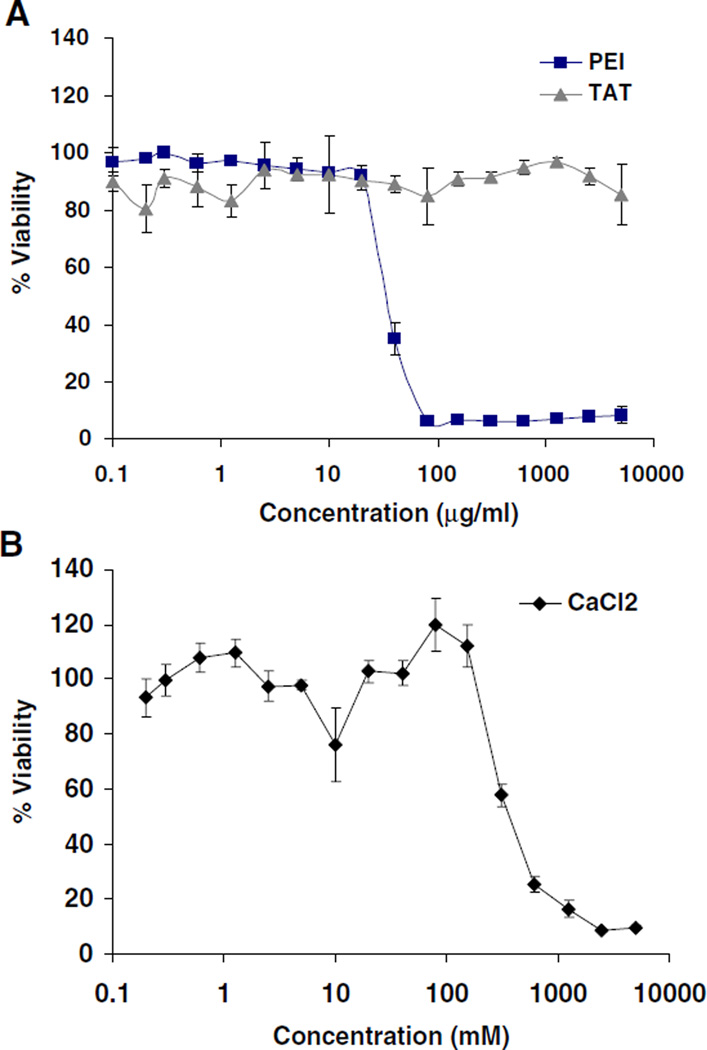
Low cytotoxicity together with high transfection efficiency are extremely important attributes for nonviral gene vectors. The cytotoxicity of free TAT, PEI, and CaCl2 was studied by incubating A549 cells with up to 5 mg/mL of TAT or PEI and with up to 0.5 M CaCl2 (Fig. 6A and B). TAT peptide revealed no evidence of cytotoxicity, and cells maintained high viability. Branched PEI induced significant cytotoxicity (IC50 ~35µg/mL). CaCl2 alone showed modest cytotoxicity (IC50 ~210 mM).
Fig. 6.
Cytotoxicity profiles of (A) PEI, TAT and (B) CaCl2. Viability is expressed as a function of polymer concentration. Results are presented as mean±SD (n=3).
In Vitro Transfection Efficiency of TAT-Ca/pDNA and PEI/pDNA Complexes
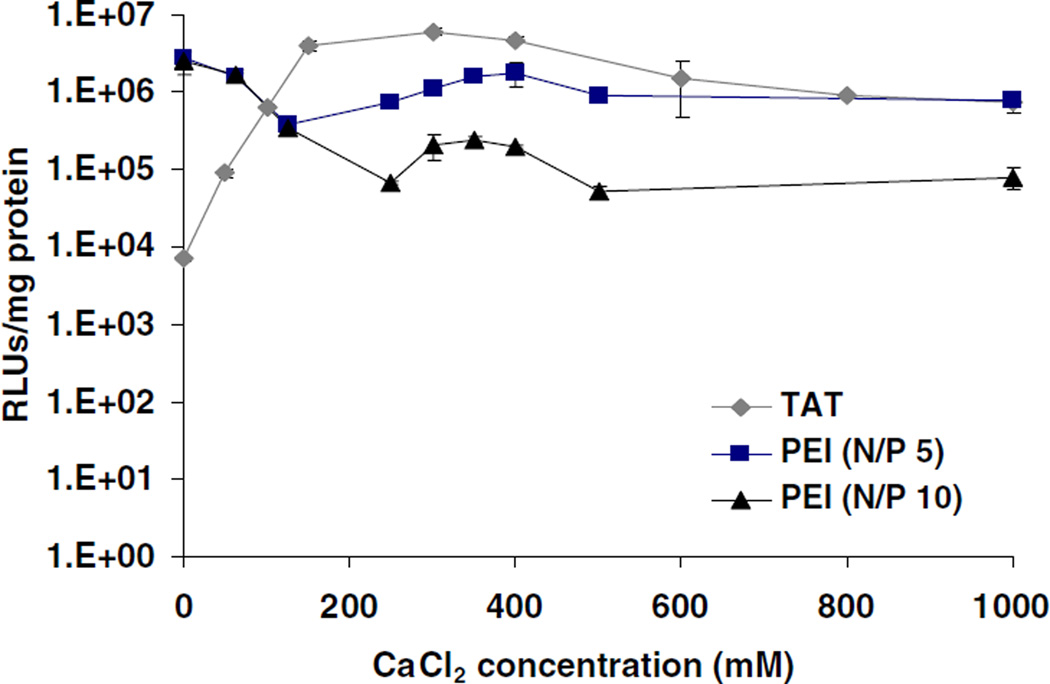
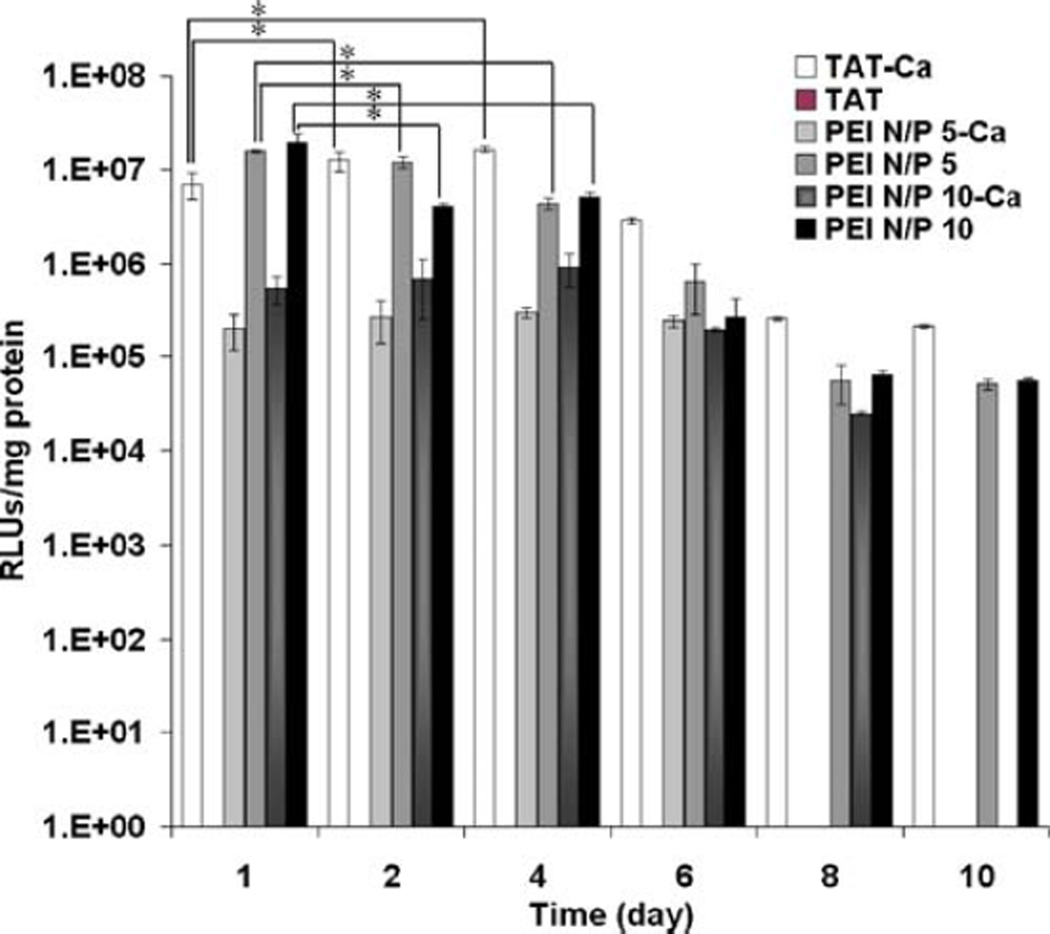
The in vitro transfection efficiency of these complexes was studied using the human lung carcinoma cell line A549. Luciferase gene expression was evaluated on day 1 of transfection using the TAT or PEI polyplexes including different concentrations of CaCl2 during the complex formation. TAT complexes showed a higher level of gene expression at 0.3 M CaCl2 when compared to PEI, which had excellent transfection efficiency in the absence of CaCl2 (Fig. 7). It was interesting that the optimized level of gene expression of TAT-Ca/pDNA complexes was similar to the transfection efficiency of branched PEI and increased over the first four days. Conversely, the gene expression of PEI/pDNA complexes showed a marked decrease during the same time-frame. The gene expression was sustained for at least 10 days, and TAT-Ca/pDNA complexes appeared to be superior to PEI/DNA complexes at day 8 and 10 (Fig. 8). Strikingly, no expression was observed for TAT/pDNA complexes without CaCl2. In addition, PEI-Ca/pDNA complexes exhibited lower levels of gene expression compared to PEI/pDNA complexes. It is important to note that the optimum gene transfection for TAT-Ca/pDNA complexes occurred when 0.3 M of CaCl2 was added, which yields a final CaCl2 concentration of 113 mM, well below the CaCl2 IC50 value of 210 mM.
Fig. 7.
The transfection efficiency of TAT-Ca/pDNA and PEI/pDNA polyplexes with different concentration of CaCl2. Results are presented as mean±SD (n=3).
Fig. 8.
The transfection efficiency of TAT-Ca/pDNA and PEI/pDNA polyplexes with and without 0.3 M CaCl2. Results are presented as mean±SD (n=3). * p<0.001.
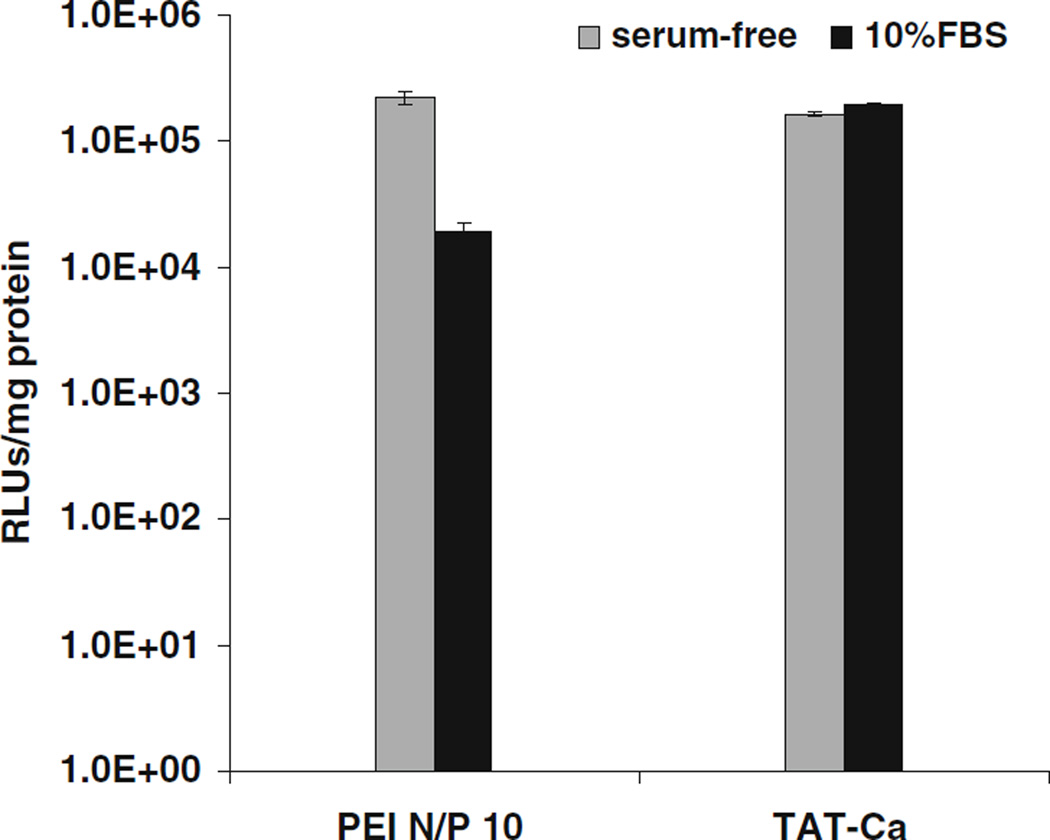
The Effect of Serum on Transfection Efficiency and Particle Size
It has been previously reported that the presence of serum may have a significant influence on the transfection efficiency of nonviral gene delivery vehicles (55–57). To determine the effect of serum on gene expression, A549 cells were transfected with optimized TAT-Ca/pDNA (0.3 M CaCl2) or PEI/pDNA complexes in the absence and presence of 10% FBS. Results indicated that serum did not significantly inhibit the transfection efficiency mediated by TAT-Ca/pDNA complexes. In contrast, PEI complexes showed slightly decreased transfection efficiency in the presence of 10% FBS (Fig. 9). The effect of serum on transfection efficiency should be considered in light of the stability of the size of TAT and PEI complexes which were investigated as a function of time. TAT/pDNA complexes without CaCl2 exhibited some agglomeration behavior in the absence and presence of 10% of FBS after 8 days (Fig. 5A and B). However, TAT-Ca/pDNA complexes showed good stability in serum-free and supplemented culture media during the same time-frame. On the other hand, PEI/pDNA complexes remained stable in the absence of serum and CaCl2 over a period of 8 days and retained their size, whereas the particle size of PEI-Ca/pDNA showed a marked decrease during the same time-frame in the presence of serum.
Fig. 9.
Transfection efficiency of TAT-Ca/pDNA and PEI/pDNA complexes in A549 cells in the absence or presence of 10% FBS. Results are presented as mean±SD (n=3).
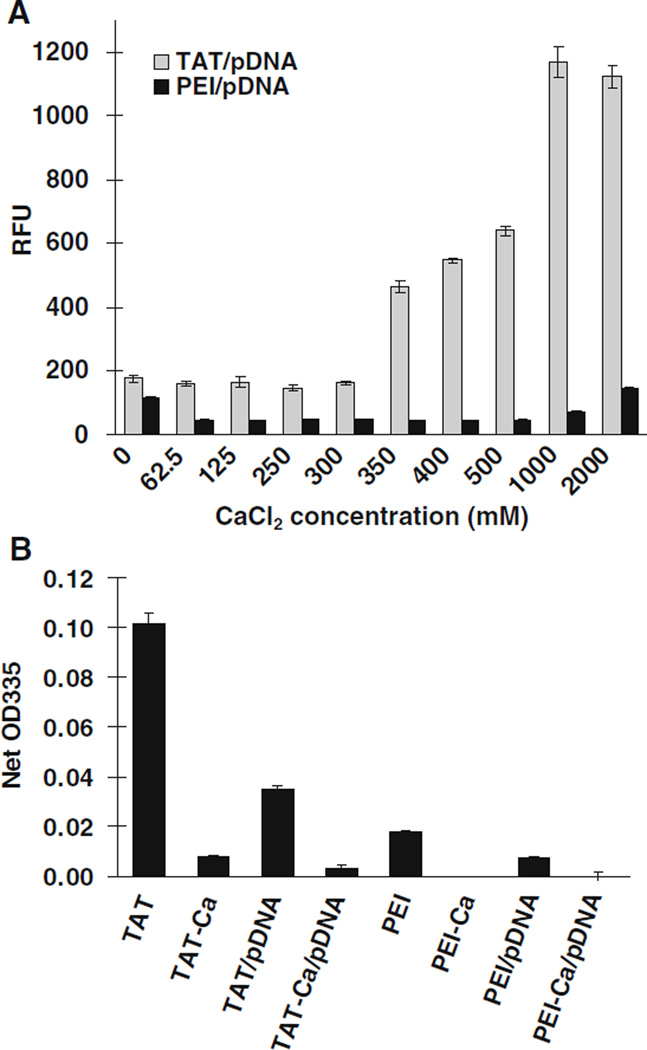
SYBR Green Assay
Dye displacement assays provide a simple, nondestructive, and high throughput method for investigating the pDNA accessibility within complexes. Various concentrations of CaCl2 were examined to identify the effect on pDNA packaging. SYBR green, which binds double-stranded DNA, was used for the assay (58). TAT/pDNA complexes showed weak fluorescence intensity. The addition of different CaCl2 concentrations up to 0.3 M likely condensed the TAT complexes, which resulted in lowering the accessibility of pDNA (Fig. 10A). The fluorescence intensity transitioned into a gradual increase with increasing concentration of CaCl2. In contrast, PEI/pDNA complexes exhibited a negligible fluorescence for all concentrations of CaCl2 except at 0 and 2 M CaCl2, where slight fluorescence intensities were observed.
Fig. 10.
(A) Effect of CaCl2 concentration on the TAT/pDNA and PEI/pDNA complexes by using the SYBR Green assay to assess DNA accessibility and (B) condensation of TAT and PEI complexes assessed using the TNBS assay as a probe for amine accessibility. Results are presented as mean±SD (n=3).
TNBS Assay
A TNBS assay was also used to determine the accessibility of the primary amine groups within the TAT/pDNA or PEI/pDNA complexes. The TNBS assay revealed that the free TAT had more primary amine groups than PEI, which decreased to negligible levels when adding the designated volume of 0.3 M CaCl2 to them (Fig. 10B). Control studies indicated that adding 62.5 mM CaCl2 decreased the absorption intensity of TNBS by less than 10%. Increasing CaCl2 concentration did not affect the absorption intensity further. These results supported the hypothesis that the efficiency of DNA compaction was increased by the introduction of CaCl2. Therefore, calcium interaction with amines also facilitated the observed stabilization and decrease in particle size for TAT/pDNA complexes. PEI complexes including CaCl2 showed low binding of the dye as well, but this may result from particle agglomeration or precipitation as suggested by the particle size studies.
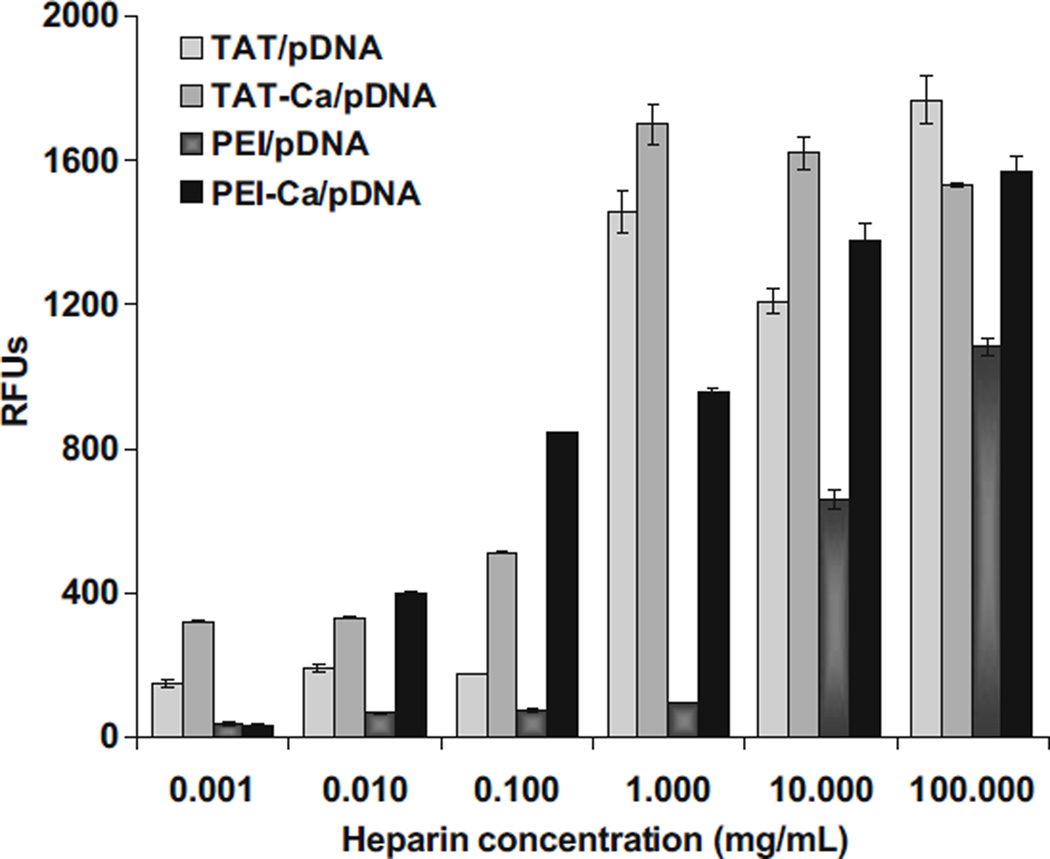
Stability of Complexes Exposed to Heparin
The stability of TAT and PEI complexes when exposed to the biological polyanion heparin was studied by determining changes in the SYBR Green-pDNA fluorescence. Exposing the complexes to heparin yielded an increase in the fluorescence signal as the heparin concentration increased (Fig. 11). TAT and PEI complexes including 0.3 M CaCl2 showed release of the plasmid DNA from the complexes at lower heparin concentrations than without calcium. These results indicated that calcium may control the delicate balance between binding affinities within polycations and pDNA complexes.
Fig. 11.
Unpackaging of TAT and PEI complexes by heparin displacement of DNA. Results are presented as mean±SD (n=3).
DISCUSSION
Improvement in the field of gene therapy is currently hindered by the lack of translatable gene delivery vectors. Synthetic, nonviral, vehicles based on polycations are promising vectors for gene delivery (59–64). The toxicity of these materials may be reduced or eliminated by reducing polycation molecular weight. However, the levels of gene expression mediated by low molecular weight polycations are typically low compared to higher molecular weight polycations (e.g. 25 kDa PEI) (65,66). As a result, many researchers have aimed to improve upon the deficiency of either system.
Among nonviral vehicles, cell-penetrating peptides (CPPs) have been used to deliver a variety of therapeutics. For example, experimental anti-cancer cargo including small molecules, proteins and nucleic acids, were delivered into cells in vitro using CPPs and have been explored to treat preclinical tumor models in vivo (67,68). Complexes of plasmid DNA with TAT peptides have been used in gene delivery either by covalent coupling of the peptide to the vectors, or by simple mixing of the plasmid DNA with the TAT peptide to form TAT/pDNA complexes via electrostatic interactions. However, the transfection efficiency for these complexes was quite low in comparison to PEI.
In our studies, TAT/pDNA complexes alone also exhibited no gene expression. The size of these complexes was quite large (~1,000 nm) for gene delivery. Therefore, calcium was explored as a condensing agent to reduce the size of TAT/pDNA complexes and to improve transfection efficiency. Calcium was found to form tight and compact complexes (~60 nm) through “soft” crosslinks that could be competitively disrupted in order to increase transfection efficiency. The precise balance between binding affinities within TAT/pDNA polyplexes was adjusted by controlling CaCl2 concentration as a means to optimize transfection efficiency. Previous studies have shown that both small and larges particles may provide efficient levels of gene expression (69–72). However, the condensed size of TAT-Ca/pDNA complexes in this study appeared to correlate with enhanced transfection efficiency.
The ability of calcium to condense TAT/pDNA complexes occurred as a result of calcium interactions with both amines (polycations) and phosphates (DNA). The binding affinity of calcium to double-stranded DNA was observed as indicated by gel electrophoresis of the TAT-Ca/pDNA complexes. A specific CaCl2 concentration range was necessary to achieve small particles. Dye displacement studies provided some indication of the extent of compaction of DNA when bound to TAT or PEI. Nearly complete exclusion of TNBS from both TAT/pDNA and PEI/pDNA complexes was achieved when 0.3 M calcium was added to the polyplexes, suggesting that the primary amine groups within these complexes were blocked. Moreover, pDNA was mostly inaccessible to SYBR green when TAT/pDNA complexes were formulated with up to 0.3 M CaCl2, suggesting that pDNA was also extensively condensed within the complexes. The accessibility of pDNA increased at higher concentrations of CaCl2 which demonstrated that calcium may also competitively inhibit the amine/phosphate interaction.
The stability of TAT and PEI complexes upon exposure to the polyanion heparin revealed that complex dissociation depended on the inclusion of calcium as well. Heparin and heparan sulfate have been previously reported to bind to PEI and release pDNA from complexes (73,74). Complex stability is significant because sufficiently low affinity between polycation and plasmid DNA may be desirable to facilitate the release of the DNA after cellular uptake. The inclusion of calcium in the formulation of TAT and PEI complexes facilitated the dissociation of these complexes by heparin. Therefore, the release of pDNA from the complexes suggested that a critical concentration of calcium can condense the complexes into small particles yet facilitate the release of DNA.
A successful gene delivery system should be able to deliver DNA to the cell without negatively affecting the viability of the host cell. A cytotoxicity study in A549 human lung carcinoma cells indicated that TAT peptide provided substantially higher cell viability than PEI. Further studies will be necessary to determine if this gene vector is benign in primary human cells.
CONCLUSION
Drug delivery strategies using CPPs such as TAT have been widely explored to improve the intracellular delivery of a large number of cargo molecules. Electrostatic complexation of pDNA using TAT has been less explored due to the relatively low levels of gene expression observed when using such low molecular weight polycations as DNA condensing agents. We have found that the binding affinity of calcium for TAT peptide and pDNA can be used to effectively mediate the charge balance within these complexes. In this study, it was shown that 0.3 M CaCl2 produced small and stable TAT/pDNA complexes via “soft” crosslinks leading to gene expression levels higher than observed for control PEI gene vectors in A549 lung epithelial cells. TAT-Ca/pDNA complexes were stable, maintaining particle size in the absence and presence of 10% of FBS over a period of 8 days. Gene expression of TAT-Ca/pDNA complexes was sustained for at least 10 days and tended to increase over the first four days of the study. Conversely, gene expression levels for PEI/pDNA complexes showed high initial gene expression that dropped to low levels after day 4. Moreover, the transfection efficiency of TAT-Ca/pDNA complexes was not significantly influenced by the presence of serum. The TAT peptide also showed negligible cytotoxicity up to 5 mg/mL. In comparison, PEI was very cytotoxic (IC50 ~35µg/mL). Thus, these data suggest that TAT-Ca complexes are a novel and effective vehicle offering some potential for translatable gene delivery.
ACKNOWLEDGMENTS
We would like to acknowledge support for this work from the Coulter Foundation, the Higuchi Biosciences Center, and the Cystic Fibrosis Foundation as well as additional lab funding from the American Heart Association, the NIH (R03 AR054035, P20 RR016443 and T32 GM08359-11) and the Department of Defense. In addition, we acknowledge the support of the NSF (CHE 0719464). We also thank Prof. C. Russ Middaugh for the use of laboratory equipment and the Microscopy Lab for assistance with electron microscopy.
REFERENCES
- 1.Check E. A tragic setback. Nature. 2002;420:116–118. doi: 10.1038/420116a. [DOI] [PubMed] [Google Scholar]
- 2.Felgner PL. Nonviral strategies for gene therapy. Sci Am. 1997;276:102–106. doi: 10.1038/scientificamerican0697-102. [DOI] [PubMed] [Google Scholar]
- 3.Hope MJ, Mui B, Ansell S, Ahkong QF. Cationic lipids, phosphatidylethanolamine and the intracellular delivery of polymeric, nucleic acid-based drugs (Review) Mol Membr Biol. 1998;15:1–14. doi: 10.3109/09687689809027512. [DOI] [PubMed] [Google Scholar]
- 4.Marshall E. Clinical trials: gene therapy death prompts review of adenovirus vector. Science. 1999;286:2244. doi: 10.1126/science.286.5448.2244. [DOI] [PubMed] [Google Scholar]
- 5.Peeters M, Patijn GA, Lieber A, Meuse L, Kay MA. Adenovirus-mediated hepatic gene transfer in mice: comparison of intravascular and biliary administration. Hum Gene Ther. 1996;7:1693–1699. doi: 10.1089/hum.1996.7.14-1693. [DOI] [PubMed] [Google Scholar]
- 6.Thomasand M, Klibanov AM. Non-viral gene therapy: polycation-mediated DNA delivery. Appl Microbiol Biotechnol. 2003;62:27–34. doi: 10.1007/s00253-003-1321-8. [DOI] [PubMed] [Google Scholar]
- 7.Yei S, Mittereder N, Tang K, O’Sullivan C, Trapnell BC. Adenovirus-mediated gene transfer for cystic fibrosis: quantitative evaluation of repeated in vivo vector administration to the lung. Gene Ther. 1994;1:192–200. [PubMed] [Google Scholar]
- 8.Huang L, Hung M, Wagner E. Nonviral Vectors for Gene Therapy. Academic; 1999. [Google Scholar]
- 9.Davis ME. Non-viral gene delivery systems. Curr Opin Biotechnol. 2002;13:128–131. doi: 10.1016/s0958-1669(02)00294-x. [DOI] [PubMed] [Google Scholar]
- 10.Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, et al. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofland HEJ, Nagy D, Liu JJ, Spratt K, Lee YL, Danos O, et al. In vivo gene transfer by intravenous administration of stable cationic Lipid/DNA complex. Pharm Res. 1997;14:742–749. doi: 10.1023/a:1012146305040. [DOI] [PubMed] [Google Scholar]
- 12.Hortobagyi GN, Ueno NT, Xia W, Zhang S, Wolf JK, Putnam JB, et al. Cationic liposome-mediated E1A gene transfer to human breast and ovarian cancer cells and its biologic effects: a Phase I clinical trial. J Clin Oncol. 2001;19:3422. doi: 10.1200/JCO.2001.19.14.3422. [DOI] [PubMed] [Google Scholar]
- 13.Med JG. Lipid-mediated siRNA delivery down-regulates exogenous gene expression in the mouse brain at picomolar levels. J Gene Med. 2005;7:198–207. doi: 10.1002/jgm.659. [DOI] [PubMed] [Google Scholar]
- 14.Ogrisand M, Wagner E. Targeting tumors with non-viral gene delivery systems. Drug Discov Today. 2002;7:479–485. doi: 10.1016/s1359-6446(02)02243-2. [DOI] [PubMed] [Google Scholar]
- 15.Templeton NS, Lasic DD, Frederik PM, Strey HH, Roberts DD, Pavlakis GN. Improved DNA: liposome complexes for increased systemic delivery and gene expression. Nat Biotechnol. 1997;15:647–652. doi: 10.1038/nbt0797-647. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Russell DW. Molecular cloning: A laboratory manual. 3rd ed. Vol. 16. Cold Spring Harbor: Cold Spring Harbor Lab; 2001. Introducing cloned gene into cultured mammalian cells; pp. 16.14–16.19. [Google Scholar]
- 17.Tang MX, Redemann CT, Szoka FC. In vitro gene delivery by degraded polyamidoamine dendrimers. Bioconjug Chem. 1996;7:703–714. doi: 10.1021/bc9600630. [DOI] [PubMed] [Google Scholar]
- 18.Godbey WT, Wu KK, Mikos AG. Poly (ethylenimine) and its role in gene delivery. J Control Release. 1999;60:149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- 19.Lundberg M, Wikström S, Johansson M. Cell surface adherence and endocytosis of protein transduction domains. Molec Ther. 2003;8:143–150. doi: 10.1016/s1525-0016(03)00135-7. [DOI] [PubMed] [Google Scholar]
- 20.Tungand CH, Weissleder R. Arginine containing peptides as delivery vectors. Adv Drug Deliv Rev. 2003;55:281–294. doi: 10.1016/s0169-409x(02)00183-7. [DOI] [PubMed] [Google Scholar]
- 21.Cao G, Pei W, Ge H, Liang Q, Luo Y, Sharp FR, et al. In vivo delivery of a Bcl-xL fusion protein containing the TAT protein transduction domain protects against ischemic brain injury and neuronal apoptosis. J Neurosci. 2002;22:5423. doi: 10.1523/JNEUROSCI.22-13-05423.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dietzand GPH, Bdhr M. Delivery of bioactive molecules into the cell: the Trojan horse approach. Mol Cell Neurosci. 2004;27:85–131. doi: 10.1016/j.mcn.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 23.Guptaand B, Torchilin VP. Transactivating transcriptional activator-mediated drug delivery. Expert Opin Drug Deliv. 2006;3:177–190. doi: 10.1517/17425247.3.2.177. [DOI] [PubMed] [Google Scholar]
- 24.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 25.Schwarze SR, Hruska KA, Dowdy SF. Protein transduction: unrestricted delivery into all cells? Trends Cell Biol. 2000;10:290–295. doi: 10.1016/s0962-8924(00)01771-2. [DOI] [PubMed] [Google Scholar]
- 26.Koch AM, Reynolds F, Merkle HP, Weissleder R, Josephson L. Transport of surface-modified nanoparticles through cell mono-layers. ChemBioChem. 2005;6:337–345. doi: 10.1002/cbic.200400174. [DOI] [PubMed] [Google Scholar]
- 27.Derossi D, Calvet S, Trembleau A, Brunissen A, Chassaing G, Prochiantz A. Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J Biol Chem. 1996;271:18188. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 28.Deshayes S, Heitz A, Morris MC, Charnet P, Divita G, Heitz F. Insight into the mechanism of internalization of the cell-penetrating carrier peptide Pep-1 through conformational analysis. Biochemistry. 2004;43:1449–1457. doi: 10.1021/bi035682s. [DOI] [PubMed] [Google Scholar]
- 29.Henriques ST, Costa J, Castanho M. Translocation of β-galactosidase mediated by the cell-penetrating peptide Pep-1 into lipid vesicles and human HeLa cells is driven by membrane electrostatic potential. Biochemistry. 2005;5:9. doi: 10.1021/bi0502644. [DOI] [PubMed] [Google Scholar]
- 30.Mano M, Teodosio C, Paiva A, Simoes S, de Lima MCP. On the mechanisms of the internalization of S413-PV cell-penetrating peptide. Biochem J. 2005;390:603. doi: 10.1042/BJ20050577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel LN, Zaro JL, Shen WC. Cell penetrating peptides: intracellular pathways and pharmaceutical perspectives. Pharm Res. 2007;24:1977–1992. doi: 10.1007/s11095-007-9303-7. [DOI] [PubMed] [Google Scholar]
- 32.Vives E, Richard JP, Rispal C, Lebleu B. TAT peptide internalization: seeking the mechanism of entry. Current Protein and Peptide Science. 2003;4:125–132. doi: 10.2174/1389203033487306. [DOI] [PubMed] [Google Scholar]
- 33.Console S, Marty C, Garcia-Echeverria C, Schwendener R, Ballmer-Hofer K. Antennapedia and HIV transactivator of transcription (TAT) “Protein transduction domains” promote endocytosis of high molecular weight cargo upon binding to cell surface glycosaminoglycans. J Biol Chem. 2003;278:35109–35114. doi: 10.1074/jbc.M301726200. [DOI] [PubMed] [Google Scholar]
- 34.Foerg C, Ziegler U, Fernandez-Carneado J, Giralt E, Rennert R, Beck-Sickinger AG, et al. Decoding the entry of two novel cell-penetrating peptides in HeLa cells: lipid raft-mediated endocytosis and endosomal escape. Biochemistry. 2005;44:72–81. doi: 10.1021/bi048330+. [DOI] [PubMed] [Google Scholar]
- 35.Gerbal-Chaloin S, Gondeau C, Aldrian-Herrada G, Heitz F, Gauthier-Rouviere C, Divita G. First step of the cell-penetrating peptide mechanism involves Rac1 GTPase-dependent actin-network remodelling. Biol Cell. 2007;99:223–238. doi: 10.1042/BC20060123. [DOI] [PubMed] [Google Scholar]
- 36.Jones AT. Macropinocytosis: searching for an endocytic identity and role in the uptake of cell penetrating peptides. J Cell Mol Med. 2007;11:670–684. doi: 10.1111/j.1582-4934.2007.00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakase I, Tadokoro A, Kawabata N, Takeuchi T, Katoh H, Hiramoto K, et al. Interaction of arginine-rich peptides with membrane-associated proteoglycans is crucial for induction of actin organization and macropinocytosis. Biochemistry. 2007;46:492–501. doi: 10.1021/bi0612824. [DOI] [PubMed] [Google Scholar]
- 38.Richard JP, Melikov K, Brooks H, Prevot P, Lebleu B, Chernomordik LV. Cellular uptake of unconjugated TAT peptide involves clathrin-dependent endocytosis and heparan sulfate receptors. J Biol Chem. 2005;280:15300. doi: 10.1074/jbc.M401604200. [DOI] [PubMed] [Google Scholar]
- 39.Silhol M, Tyagi M, Giacca M, Lebleu B, Vives E. Different mechanisms for cellular internalization of the HIV-1 Tat-derived cell penetrating peptide and recombinant proteins fused to Tat. Eur J Biochem. 2002;269:494–501. doi: 10.1046/j.0014-2956.2001.02671.x. [DOI] [PubMed] [Google Scholar]
- 40.Thorén PEG, Persson D, Isakson P, Goksor M, Onfelt A, Nordén B. Uptake of analogs of penetratin, Tat (48–60) and oligoarginine in live cells. Biochem Biophys Res Commun. 2003;307:100–107. doi: 10.1016/s0006-291x(03)01135-5. [DOI] [PubMed] [Google Scholar]
- 41.Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 42.Jarver P, Langel Ü. Cell-penetrating peptides—a brief introduction. BBA-Biomembranes. 2006;1758:260–263. doi: 10.1016/j.bbamem.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 43.Mae M, Myrberg H, Jiang Y, Paves H, Valkna A, Langel Ü. Internalisation of cell-penetrating peptides into tobacco protoplasts. BBA-Biomembranes. 2005;1669:101–107. doi: 10.1016/j.bbamem.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, et al. Tat-mediated delivery of heterologous proteins into cells. Proc Natl Acad Sci. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frankeland AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 46.Truantand R, Cullen BR. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct Importin β-dependent nuclear localization signals. Mol Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vives E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 48.Eguchi A, Akuta T, Okuyama H, Senda T, Yokoi H, Inokuchi H, et al. Protein transduction domain of HIV-1 Tat protein promotes efficient delivery of DNA into mammalian cells. J Biol Chem. 2001;276:26204–26210. doi: 10.1074/jbc.M010625200. [DOI] [PubMed] [Google Scholar]
- 49.Torchilin VP, Rammohan R, Weissig V, Levchenko TS. TAT peptide on the surface of liposomes affords their efficient intracellular delivery even at low temperature and in the presence of metabolic inhibitors. Proc Natl Acad Sci. 2001;98:8786. doi: 10.1073/pnas.151247498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tungand CH, Stein S. Preparation and applications of peptide-oligonucleotide conjugates. Bioconjug Chem. 2000;11:605–618. doi: 10.1021/bc0000334. [DOI] [PubMed] [Google Scholar]
- 51.Futaki S, Ohashi W, Suzuki T, Niwa M, Tanaka S, Ueda K, et al. Stearylated arginine-rich peptides: a new class of transfection systems. Bioconjug Chem. 2001;12:1005–1011. doi: 10.1021/bc015508l. [DOI] [PubMed] [Google Scholar]
- 52.Ignatovich IA, Dizhe EB, Pavlotskaya AV, Akifiev BN, Burov SV, Orlov SV, et al. Complexes of plasmid DNA with basic domain 47–57 of the HIV-1 Tat protein are transferred to mammalian cells by endocytosis-mediated pathways. J Biol Chem. 2003;278:42625–42636. doi: 10.1074/jbc.M301431200. [DOI] [PubMed] [Google Scholar]
- 53.Sandgren S, Cheng F, Belting M. Nuclear targeting of macromolecular polyanions by an HIV-Tat derived peptide. Role for cell-surface proteoglycans. J Biol Chem. 2002;277:38877–38883. doi: 10.1074/jbc.M205395200. [DOI] [PubMed] [Google Scholar]
- 54.Tung CH, Mueller S, Weissleder R. Novel branching membrane translocational peptide as gene delivery vector. Bioorg Med Chem. 2002;10:3609–3614. doi: 10.1016/s0968-0896(02)00248-1. [DOI] [PubMed] [Google Scholar]
- 55.Haberland A, Knaus T, Zaitsev SV, Buchberger B, Lun A, Haller H, et al. Histone H1-mediated transfection: serum inhibition can be overcome by Ca2+ Ions. Pharm Res. 2000;17:229–235. doi: 10.1023/a:1007581700996. [DOI] [PubMed] [Google Scholar]
- 56.Nchinda G, Uberla K, Zschornig O. Characterization of cationic lipid DNA transfection complexes differing in susceptability to serum inhibition. BMC Biotechnol. 2002;2:12. doi: 10.1186/1472-6750-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zelphati O, Uyechi LS, Barron LG, Szoka FC. Effect of serum components on the physico-chemical properties of cationic lipid/oligonucleotide complexes and on their interactions with cells. Biochim Biophys Acta (BBA)/Lipids Lipid Metab. 1998;1390:119–133. doi: 10.1016/s0005-2760(97)00169-0. [DOI] [PubMed] [Google Scholar]
- 58.Kwon DS, Lin CH, Chen S, Coward JK, Walsh CT, Bollinger JM., Jr Dissection of glutathionylspermidine synthetase/amidase from Escherichia coli into autonomously folding and functional synthetase and amidase domains. J Biol Chem. 1997;272:2429. doi: 10.1074/jbc.272.4.2429. [DOI] [PubMed] [Google Scholar]
- 59.Abdallah B, Hassan A, Benoist C, Goula D, Behr JP, Demeneix BA. A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: polyethylenimine. Hum Gene Ther. 1996;7:1947–1954. doi: 10.1089/hum.1996.7.16-1947. [DOI] [PubMed] [Google Scholar]
- 60.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang CY, Ma SS, Lee S, Radhakrishnan R, Braun CS, Choosakoonkriang S, et al. Enhancements in gene expression by the choice of plasmid DNA formulations containing neutral polymeric excipients. J Pharm Sci. 2002;91:1371–1381. doi: 10.1002/jps.10130. [DOI] [PubMed] [Google Scholar]
- 62.Lobo BA, Vetro JA, Suich DM, Zuckermann RN, Middaugh CR. Structure/function analysis of peptoid/lipitoid: DNA complexes. J Pharm Sci. 2003;92:1905–1918. doi: 10.1002/jps.10450. [DOI] [PubMed] [Google Scholar]
- 63.Tiyaboonchai W, Woiszwillo J, Middaugh CR. Formulation and characterization of DNA–polyethylenimine–dextran sulfate nanoparticles. Eur J Pharm Sci. 2003;19:191–202. doi: 10.1016/s0928-0987(03)00102-7. [DOI] [PubMed] [Google Scholar]
- 64.Wiethoff CM, Koe JG, Koe GS, Middaugh CR. Compositional effects of cationic lipid/DNA delivery systems on transgene expression in cell culture. J Pharm Sci. 2004;93:108–123. doi: 10.1002/jps.10519. [DOI] [PubMed] [Google Scholar]
- 65.Choosakoonkriang S, Lobo BA, Koe GS, Koe JG, Middaugh CR. Biophysical characterization of PEI/DNA complexes. J Pharm Sci. 2003;92:1710–1722. doi: 10.1002/jps.10437. [DOI] [PubMed] [Google Scholar]
- 66.Forrest ML, Koerber JT, Pack DW. A degradable polyethylenimine derivative with low toxicity for highly efficient gene delivery. Bioconjug Chem. 2003;14:934–940. doi: 10.1021/bc034014g. [DOI] [PubMed] [Google Scholar]
- 67.Lindsay MA. Peptide-mediated cell delivery: application in protein target validation. Curr Opin Pharmacol. 2002;2:587–594. doi: 10.1016/s1471-4892(02)00199-6. [DOI] [PubMed] [Google Scholar]
- 68.Snyderand EL, Dowdy SF. Cell penetrating peptides in drug delivery. Pharm Res. 2004;21:389–393. doi: 10.1023/B:PHAM.0000019289.61978.f5. [DOI] [PubMed] [Google Scholar]
- 69.Pouton CW, Lucas P, Thomas BJ, Uduehi AN, Milroy DA, Moss SH. Polycation-DNA complexes for gene delivery: a comparison of the biopharmaceutical properties of cationic polypeptides and cationic lipids. J Control Release. 1998;53:289–299. doi: 10.1016/s0168-3659(98)00015-7. [DOI] [PubMed] [Google Scholar]
- 70.Simberg D, Danino D, Talmon Y, Minsky A, Ferrari ME, Wheeler CJ, et al. Phase behavior, DNA ordering, and size instability of cationic lipoplexes. Relevance to optimal transfection activity. J Biol Chem. 2001;276:47453–47459. doi: 10.1074/jbc.M105588200. [DOI] [PubMed] [Google Scholar]
- 71.Turek J, Dubertret C, Jaslin G, Antonakis K, Scherman D, Pitard B. Formulations which increase the size of lipoplexes prevent serum-associated inhibition of transfection. J Gene Med. 2000;2:32–40. doi: 10.1002/(SICI)1521-2254(200001/02)2:1<32::AID-JGM78>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 72.Wagner E, Cotten M, Foisner R, Birnstiel ML. Transferrinpolycation-DNA complexes: the effect of polycations on the structure of the complex and DNA delivery to cells. Proc Natl Acad Sci. 1991;88:4255–4259. doi: 10.1073/pnas.88.10.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moret I, Esteban Peris J, Guillem VM, Benet M, Revert F, Dasi F, et al. Stability of PEI–DNA and DOTAP–DNA complexes: effect of alkaline pH, heparin and serum. J Control Release. 2001;76:169–181. doi: 10.1016/s0168-3659(01)00415-1. [DOI] [PubMed] [Google Scholar]
- 74.Ruponen M, Yla-Herttuala S, Urtti A. Interactions of polymeric and liposomal gene delivery systems with extracellular glycosaminoglycans: physicochemical and transfection studies. BBA-Biomembranes. 1999;1415:331–341. doi: 10.1016/s0005-2736(98)00199-0. [DOI] [PubMed] [Google Scholar]