Abstract
The uncharacterized α/β-hydrolase protein OLEI01171 from the psychrophilic marine bacterium Oleispira antarctica belongs to the PF00756 family of putative esterases, which also includes human esterase D. In the present paper we show that purified recombinant OLEI01171 exhibits high esterase activity against the model esterase substrate α-naphthyl acetate at 5 – 30°C with maximal activity at 15–20°C. The esterase activity of OLEI01171 was stimulated 3–8-fold by the addition of chloride or several other anions (0.1–1.0 M). Compared with mesophilic PF00756 esterases, OLEI01171 exhibited a lower overall protein thermostability. Two crystal structures ofOLEI01171 were solved at 1.75 and 2.1 Å resolution and revealed a classical serine hydrolase catalytic triad and the presence of a chloride or bromide ion bound in the active site close to the catalytic Ser148.Both anions were found to co-ordinate a potential catalytic water molecule located in the vicinity of the catalytic triad His257. The results of the present study suggest that the bound anion perhaps contributes to the polarization of the catalytic water molecule and increases the rate of the hydrolysis of an acyl-enzyme intermediate. Alanine replacement mutagenesis of OLEI01171 identified ten amino acid residues important for esterase activity. The replacement of Asn225 by lysine had no significant effect on the activity or thermostability of OLEI01171, but resulted in a detectable increase of activity at 35–45°C. The present study has provided insight into the molecular mechanisms of activity of a cold-active and anion-activated carboxyl esterase.
Keywords: anion activation, carboxyl esterase, cold-active enzyme, crystal structure, Oleispira antarctica, protein thermostability
INTRODUCTION
On Earth, microbial life is not limited to moderate environments, but can also be found in various extreme conditions including high and low temperatures (thermophiles and psychrophiles), acid or alkaline pH (acidophiles and alkaliphiles), high salt (halophiles), and hydrostatic pressure (barophiles), as well as high levels of heavy metals and radioactivity [1]. The ability of these micro-organisms (extremophiles) to survive and thrive in extreme environments is the result of various physiological adaptations and the molecular evolution of enzymes with selective pressure directed towards the increased stability and activity under extreme conditions. Biochemical and structural studies of extremophilic enzymes provide fundamental information about the molecular mechanisms of enzyme stability, activity, adaptation and evolution. Unique properties of these enzymes have already resulted in several applications in industrial processes, and there is a growing demand for novel enzymes in biotechnology [2].
Cold-adapted or psychrophilic micro-organisms occupy the largest ecological niche on Earth which includes oceans, polar regions and mountain areas with average temperatures of −1–5°C [3]. The lowest temperature limit for life seems to be around −20°C as found in bacteria living in permafrost or in sea ice [4]. It is estimated that around 90%of the biosphere exists at temperatures below 10°C, and that the largest proportion of biomass on Earth is produced at low temperatures [5].A diverse range of microbes have been discovered in cold environments including representatives of bacteria, archaea and eukarya [4, 6, 7]. Given the facts that cold completely penetrates microbial cells and that they are at complete thermal equilibrium with the environment, all of the structural and functional components of the cell, including membranes, transport systems, intracellular solutions and proteins, must be suitably adapted. The known structural and physiological adaptations in psychrophiles include increased membrane fluidity (associated with a decrease in fatty acid saturation and in fatty acid chain length), freeze protection (compatible solutes, antifreeze proteins and ice nucleation) and gene expression (cold-shock responses) [5, 6, 8]. In addition, to survive at such low temperatures, psychrophilic organisms have evolved enzymes which are able to perform efficiently under these conditions.
Although the ability of some micro-organisms to grow well at 0°C was first mentioned in 1887 [9], the first work on the biochemical characterization of a psychrophilic enzyme (the alkaline phosphatase from an Antarctic bacterium) was published only in 1984 [10], and the first crystal structure of a cold-active enzyme (the α-amylase from Alteromonas haloplanctis) was determined in 1998 [14]. Although recent advances in the structural and biochemical characterization of psychrophilic enzymes have provided significant information about the molecular mechanisms of cold adaptation [11, 12], much less is known about psychrophilic enzymes compared with their thermophilic counterparts. The commonly observed features of cold-active enzymes are their high catalytic efficiencies at low temperatures and increased thermolability and flexibility [12, 13]. The α-amylase from A. aloplanctis represents the best biochemically and structurally characterized cold-adapted enzyme [14, 15]. The three-dimensional structures for several other cold-active enzymes have also been published including the Pseudoalteromonas haloplanktis xylanase and formylglutathione hydrolase [16, 17]. Comparative studies of homologous proteins from psychrophiles, mesophiles and thermophiles have revealed several changes in psychrophilic enzymes including a reduction in core hydrophobicity, decreased ionic interactions, reduced charge of surface residues, additional surface loops, substitution of proline residues by glycine residues, a decreased arginine/lysine ratio, fewer aromatic interactions or hydrogen bonds, an increased exposure of hydrophobic residues to solvent, and larger catalytic cavities [4–6, 11, 12]. In general, the cold-active enzymes and their active sites are characterized by an increased flexibility of the molecular structure and decreased conformational stability. It has been demonstrated that each psychrophilic enzyme adopts its own strategy for cold adaptation and uses one or several of the abovementioned structural adjustments [13].
Oleispira antarctica is a psychrophilic hydrocarbonoclastic bacterium isolated from Antarctic coastal waters, which has the optimal growth temperature of 2–4°C and requires NaCl (3– 5%, w/v) for growth [18]. The recently sequenced O. antarctica genome has revealed the presence of many genes encoding potential psychrophilic enzymes (M. Kube, T. N. Chernikova, A. Beloqui, Y. Al-Ramahi, M. E. Guazzaroni, N. Lopez-Cortez, O. R. Kotsyrbenko, T. Y. Nechitaylo, M. Fernández, S. Juárez, S. Ciordia, A. Singer, O. Kagan, O. Egorova, P. A. Petit, P. Stogios, Y. Kim, A. Tchigvintsev, R. Flick, R. Denaro, M. Genovese, H. J. Heipieper, J. P. Albar, K. N. Timmis, O. N. Reva, H. Tran, M. Ferrer, A. Savchenko, A. F. Yakunin, M. M. Yakimov, O. V. Golyshina, R. Reinhardt and P. N. Golyshin, unpublished work). To provide further biochemical and structural insight into the activity of cold-adapted enzymes, we have biochemically characterized and determined the crystal structure of the unknown protein OLEI01171 from O. antarctica, a predicted esterase from the PF00756 protein family. We have found that OLEI01171 is a cold-active carboxyl esterase with high activity at 5°C, which is stimulated by Cl− and other anions. The structure of OLEI01171 revealed the presence of one Cl− or Br− ion bound close to the catalytic Ser148. Site-directed mutagenesis identified ten residues (including the catalytic triad) important for activity with many residues located in the acyl- or alcohol-binding pockets. The replacement of Asn225 by lysine noticeably increased the activity of OLEI01171 at higher temperatures (35–45°C).
EXPERIMENTAL
Gene cloning, protein purification and mutagenesis
The OLEI01171 gene (GenBank® accession number JN986750) was amplified by PCR using the O. antarctica genomic DNA and cloned into a modified pET15b vector as described previously [19]. The recombinant His6-tagged OLEI01171 was overexpressed in the Escherichia coli BL21(DE3) Codon-Plus strain and purified using metal-chelate affinity chromatography on Ni-NTA (Ni2+ -nitrilotriacetate; Qiagen) and gel filtration on a Superdex 200 16/60 column {in 10 mM Hepes (pH 7.5), 0.3 M NaCl and 1 mM TCEP [tris-(2-carboxyethyl)phosphine]} [19]. Site-directed mutagenesis of OLEI01171 was performed using the QuikChange® site-directed mutagenesis kit (Stratagene) according to the manufacturer’s protocol. The amino acids selected to be mutated were changed to alanine or other amino acids. DNA encoding wild-type OLEI01171 cloned into the modified pET15b vector was used as a template for mutagenesis. All of the mutations were verified by DNA sequencing, and the mutant proteins were overexpressed and purified in the same manner as the wild-type OLEI01171.
Enzymatic assays
Carboxyl esterase activity was measured spectrophotometrically using pNP (p-nitrophenyl) or α-naphthyl esters of various fatty acids (0.25–2.0 mM) in a reaction mixture containing 50 mM potassium Hepes buffer (pH 8.0) and salt (if indicated; 0.1– 1.1 M) at the standard temperatures of 20°C for OLEI01171, 30°C for Atu1476 and 37°C for the E. coli esterases YeiG [FGH (S-formylglutathione hydrolase) yeiG) and BioH [pimelyl-(acyl-carrier protein) methyl ester esterase] (0.1–2.0 µg of protein/assay). Thioesterase activity was assayed using acetyl- CoA and palmitoyl-CoA [20], feruloyl esterase activity with ethyl ferulate and rosmarinic acid [21], and activity against the glutathione derivatives (S-lactoylglutathione and S-formylglutathione) was measured as described in [19]. For determination of the Km and kcat values, the assays contained substrates at concentrations of 0.01–2.0 mM. Kinetic parameters were determined by non-linear curve fitting from the Lineweaver–Burk plot using GraphPad Prism software (version 4.00 for Windows).
Protein thermodenaturation assays
Temperature-dependent aggregation was measured using static light scattering on a StarGazer instrument as described previously [22]. The protein samples (50 µl aliquots, 0.4 mg/ml) were heated from 27°C to 80°C at a rate of 1°C per min in a clear-bottom 384-well plate (Nunc). Protein aggregation (thermodenaturation) was monitored by measuring the intensity of the scattered light every 30 s with a CCD (charge-coupled device) camera. The pixel intensities in each well were integrated, plotted against temperature and fitted to the Boltzman equation by non-linear regression. The resulting point of inflection of each resulting curve was defined as the temperature of aggregation Tagg [22].
Protein crystallization and data collection
Crystals of the OLEI01171 esterase were grown at 22°C by sitting-drop vapour diffusion with 2 µl of protein sample and an equal volume of reservoir buffer using an optimized sparse matrix crystallization screen [23]. Crystals of OLEI01171 were grown using a reservoir solution of 0.2 Mdiammonium hydrogen citrate and 20%PEG [poly(ethylene glycol)] 3350, pH 5.0. The crystals were cryoprotected with Paratone-N oil (Hampton Research) prior to flash-freezing in liquid nitrogen. For the structure of the OLEI01171–Br− complex, the crystals were soaked for 30 s in the crystallization solution supplemented with 0.8 M KBr and then extensively washed in crystallization solution containing 20% glycerol before flash-freezing in liquid nitrogen.
Structure determination
For the OLEI01171–Cl− complex, diffraction data were collected at λ=1.54178 Å (Cu Kα radiation; 1 Å=0.1 nm) on a home-source Rigaku Micromax-007 generator equipped with osmic mirrors and a Raxis4++ detector. The data were scaled and merged using HKL2000 [24]. The structure was solved by molecular replacement using a model of the protein generated using the program SWISS-MODELLER [25] using the coordinates from the PDB entry 1PV1. The final model contained two molecules of OLEI01171 in the unit cell, both found by molecular replacement using PHASER as part of the CCP4 program suite [26]. Subsequent manual model building and automated water-picking using COOT [27] and refinement using REFMAC 5.1 and TLS (Translation–Liberation–Screw rotation) [28] were used to refine the model. The final model, refined to 1.75 Å, yielded an Rwork and Rfree of 19.1% and 22.9% respectively and contained 5088 atoms, and each molecule of the asymmetric unit contains a single chain containing all residues of a 279-residue protein, except for the initiator methionine, which could not be modelled owing to poor density, probably due to conformational flexibility, for this residue. The final model showed a high quality stereochemistry [29], with 99.2% of the protein residues being found in the most or additionally favoured regions of the Ramachandran plot.
For the OLEI01171–Br− structure, the data were collected at 1.54 Å on a Rigaku FR-E superbright home diffractometer equipped with an image R-AXIS IV HTC detector. The data were integrated with XDS and scaled with Scala from the CCP4 suite [26]. The structure was solved by molecular replacement using the Mol Rep program and the OLEI01171 apo-chain A (PDB code 3I6Y) as the model. After several refinement cycles using Phenix, water molecules were built iteratively by COOT [27], and one Br− ion was added to the model. The final refinement using TLS was performed with Phenix and with the Br− ion occupancy reduced to 0.75 for an optimized B-factor. A total of 97.1% of the final model residues were in the most-favoured regions, and 2.9%were in allowed regions of the Ramachandran plot. Data collection and refinement statistics are summarized in Table 1. Co-ordinates and structure factors of the OLEI01171 complexes with Cl− or Br− have been deposited in the Protein Data Bank with the accession codes 3I6Y and 3S8Y respectively.
Table 1. Crystallographic data collection and model refinement statistics.
Values in parentheses are for the highest resolution shell.
| OLEI01171–Cl− | OLEI01171–Br− | |
|---|---|---|
| Parameters | (PDB code 3I6Y) | (PDB code 3S8Y) |
| Data collection | ||
| Space group | P21 | P22121 |
| Cell dimensions* | ||
| a, b, c (Å), β (°) | 44.8, 88.2, 84.1, 94.8 | 44.53, 84.45, 88.05 |
| Wavelength | 1.54 | 1.54 |
| Resolution (Å) | 27.9–1.7 (1.76–1.70) | 19.73–2.10 (2.21–2.10) |
| Unique reflections | 61 811 (4594) | 19 720 (2775) |
| Rmerge | 0.103 (0.313) | 0.090 (0.300) |
| I/σI | 16.3 (5.8) | 17.0 (6.3) |
| Completeness (%) | 86.9 (64.8) | 98.6 (97.0) |
| Redundancy | 3.8 (3.3) | 6.2 (6.3) |
| Refinement | ||
| Resolution (Å) | 27.9–1.75 | 19.7–2.1 |
| Number of reflections† | 60 917 (3072) | 19 117 (959) |
| Rwork/Rfree (%) | 18.8/22.9 | 15.4/18.4 |
| Number of atoms | 5088 | 2343 |
| Protein | 4547 | 2161 |
| Solvent | 508 | 181 |
| Other | 33 | 1 |
| Rmsd | ||
| Bond lengths (Å) | 0.016 | 0.008 |
| Bond angles (°) | 1.48 | 1.043 |
| Average B-factors | ||
| Overall mean | 16.0 | 17.7 |
| Wilson Plot | 15.8 | 16.6 |
| Ramachandran plot | ||
| Percentage in most favoured regions | 89.6 | 97.1 |
| Percentage in additionally allowed regions | 9.5 | 2.9 |
| Percentage in disallowed regions | 0.4 | 0 |
All of the other unit cell angles are 90° as defined by their symmetry.
Number of reflections overall and in the test set.
RESULTS AND DISCUSSION
Sequence analysis of OLEI01171
On the basis of its gene sequence, OLEI01171 belongs to the family PF00756 of putative esterases (5530 sequences and IPR000801 in the InterPro database) present in bacteria, archaea and eukaryotes. A BLAST search of the GenBank® sequences using the OLEI01171 sequence as a query revealed the presence of a large group of proteins (over 900 sequences) with high similarity with OLEI01171 (46–83%sequence identity) found in bacteria and eukaryotes, several of which have been characterized biochemically as FGHs involved in formaldehyde detoxification. A smaller group of homologous proteins or protein domains (over 400 sequences) present in bacteria, archaea and eukaryotes shares low sequence similarity with OLEI01171 (< 28%sequence identity) and includes a few proteins with demonstrated or predicted feruloyl or acetyl esterase activity involved in plant cell wall degradation. The experimentally characterized PF00756 enzymes include human esterase D [30] and FGHs from yeast (P40363) [31], E. coli [FrmB (FGH frmB) and YeiG] [19], Agrobacterium tumefaciens (Atu1476) [32] and Ps. haloplanktis [17].
Microbial esterases have been classified into eight families on the basis of their amino acid sequences [33]. Although by their size (280–320 amino acids) the PF00756 esterases are similar to the family VI esterases (23–26 kDa), their sequences show no presence of known esterase signature motifs, suggesting that these enzymes represent an independent esterase family. Sequence alignment of several microbial PF00756 esterases, including OLEI01171 and human esterase D, revealed the presence of five blocks containing five or more conserved residues (Figure 1). The two C-terminal blocks contain the predicted catalytic serine and histidine residues (Ser148 and His257 in OLEI01171), whereas the functional role of the other three blocks of conserved residues remains unknown. The sequence alignment also shows that, in the PF00756 esterases, the catalytic histidine residue (His257 in OLEI01171) has a more conserved neighbourhood than the catalytic aspartic acid residue (Asp224 in OLEI01171) (Figure 1).
Figure 1. Structure-based sequence alignment of OLEI01171 and homologous esterases from bacteria and humans.
Residues conserved in all aligned proteins are boxed, and the residues constituting the serine hydrolase catalytic triad are highlighted in grey. The five large regions with conserved sequences are labelled ‘block-1’ to ‘block-5’ below the alignment. The OLEI01171 residues mutated in the present study are marked with black triangles above the alignment and numbered. The secondary structure elements derived from structures of OLEI01171 and human EstD are shown above and below the alignment respectively. The compared proteins are OLEI01171 (GenBank® accession number D0VWZ4), Ps. haloplanktis PhEst (GenBank® accession number Q3IL66), A. tumefaciens Atu1476 (GenBank® accession number A9CJ11), E. coli FrmB (EcFrmB; GenBank® accession number P51025) and YeiG (EcYeiG; GenBank® accession number P33018) and human EstD (HsEstD; GenBank® accession number P10768).
Esterase activity and anion stimulation of OLEI01171
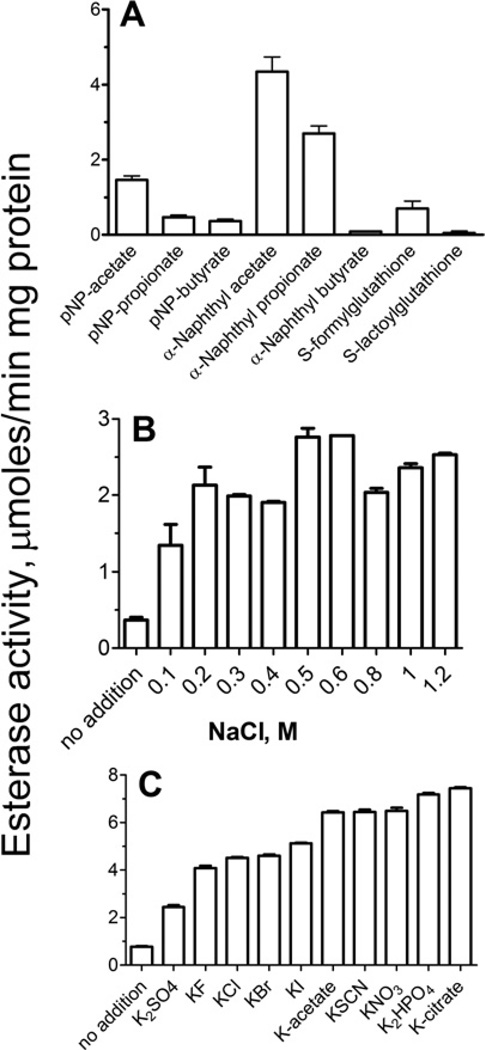
Purified OLEI01171 showed high esterase activity at 15 °C against the model substrate α-naphthyl acetate (Figure 2A), which was maximal at pH 8.0 (Supplementary Figure S1A at http://www.BiochemJ.org/bj/445/bj4450193add.htm) and was stimulated (3–6-fold) by the addition of NaCl to the reaction mixture (0.1–1.2 M) (Figure 2B). This is consistent with the growth requirement of O. antarctica cells for NaCl with the optimal growth observed at 0.5–0.9 M NaCl [18]. Interestingly, the purified esterase OLEI03762, another α/β-hydrolase from O. antarctica (PF00561), was also activated by NaCl (0.2– 0.8 M; results not shown) indicating that in this organism the activation by chloride is not limited to the PF00756 family of esterases. However, the addition of NaCl (0.1–1.6 M) had a small stimulating effect on the activity of mesophilic esterases from the PF00756 (Atu1476, E. coli YeiG or SMc01273 from Sinorhizobium meliloti) or PF00561 (E. coli BioH) protein families (24–73% increase; Supplementary Figure S2 at http://www.BiochemJ.org/bj/445/bj4450193add.htm). Strong stimulation of the OLEI01171 esterase activity was also observed after the addition of KCl (0.1–1.2 M) or other halides including KF, KBr and KI (Figure 2C and Supplementary Figures S1B–S1D). In addition, the OLEI01171 activity was stimulated by several other inorganic (SO42−, NO3− HPO42− and SCN−) or organic (acetate and citrate) anions (Figure 2C). Thus OLEI01171 is an anion-activated carboxyl esterase.
Figure 2. Esterase activity of OLEI01171.
(A) Activity profile with various substrates (measured in the presence of 0.4 M NaCl). (B) Effect of NaCl concentration. (C) Effect of various anions (50 mM). Experimental conditions were as described in the Experimental section (15 min incubation at 15°C). The reaction mixtures contained 1 µg of OLEI01171 and 2 mM substrate (1 mM for S -formylglutathione and S-lactoylglutathione). α-Naphthyl acetate (2 mM) was used as the substrate in (B) and (C). Results are means± S.D. from at least two independent determinations.
OLEI01171 also exhibited significant activity toward α-naphthyl propionate and pNP-acetate and low activity against α-naphthyl butyrate, pNP-propionate and pNP-butyrate indicating that this enzyme prefers the short acyl-chain substrates (Figure 2A). With all of the substrates, OLEI01171 showed saturation kinetics with a hyperbolic saturation curve and exhibited higher affinity with the α-naphthyl substrates (Table 2). The addition of NaCl or KCl to OLEI01171 had a stronger effect on the kcat value (4–8-fold increase) compared with the counterproductive effect on the Km value (1.8–3-fold increase), suggesting that the anions are more probable to enhance substrate hydrolysis (Table 2) than substrate binding. In addition, OLEI01171 showed no thioesterase activity (against acetyl-CoA or palmitoyl-CoA), no feruloyl esterase activity (tested with ethyl ferulate and rosmarinic acid), negligible esterase activity against S-lactoylglutathione (0.03± 0.007 µmol/min per mg of protein), and significant activity toward S-formylglutathione (0.6± 0.06 µmol/min per mg of protein). It has been previously shown that two other PF00756 esterases FrmB and YeiG from E. coli exhibited high activity against S-formylglutathione and S-lactoylglutathione, which are intermediates of the intracellular detoxification of formaldehyde or methylglyoxal respectively [19]. The ability to hydrolyse S-formylglutathione has also been demonstrated in other PF00756 enzymes, including the human esterase D, yeast FGH (P40363), A. tumefaciens Atu1476 and Ps. haloplanktis PhEst [17, 31, 32].
Table 2. Kinetic parameters of the wild-type and mutant OLEI01171 proteins.
The salts used were 0.4 M NaCl and 0.1 M KCl.
| Protein | Variable substrate | Km (mM) | kcat (s−1) |
kcat/Km (s−1 · M−1) |
|---|---|---|---|---|
| Wild-type | α-Naphthyl acetate (no salt) | 0.40 ± 0.03 | 0.8 ± 0.02 | 0.2×104 |
| Wild-type | α-Naphthyl acetate (+NaCl) | 0.70 ± 0.04 | 3.3 ± 0.1 | 0.5×104 |
| Wild-type | α-Naphthyl acetate (+KCl) | 1.2 ± 0.1 | 6.5 ± 0.2 | 0.6×104 |
| Wild-type | α-Naphthyl propionate (+NaCl) | 0.20 ± 0.03 | 1.5 ± 0.1 | 0.8×104 |
| Wild-type | α-Naphthyl butyrate (+NaCl) | 0.20 ± 0.04 | 0.06 ± 0.01 | 0.3×103 |
| Wild-type | pNP-acetate (no salt) | 0.7 ± 0.1 | 0.2 ± 0.03 | 0.3×103 |
| Wild-type | pNP-acetate (+NaCl) | 1.0 ± 0.2 | 0.8 ± 0.1 | 0.2×103 |
| Wild-type | pNP-propionate (+NaCl) | 0.7 ± 0.2 | 0.30 ± 0.03 | 0.4×103 |
| Wild-type | pNP-butyrate (+NaCl) | 0.9 ± 0.3 | 0.20 ± 0.03 | 0.2×103 |
| C57A | α-Naphthyl acetate (+NaCl) | 0.20 ± 0.01 | 0.90 ± 0.02 | 0.5×104 |
| S58A | α-Naphthyl acetate (+NaCl) | 0.40 ± 0.01 | 0.8 ± 0.1 | 0.2×104 |
| N61A | α-Naphthyl acetate (+NaCl) | 0.5 ± 0.1 | 1.8 ± 0.2 | 0.4×104 |
| K65A | α-Naphthyl acetate (+NaCl) | 0.40 ± 0.01 | 1.2 ± 0.1 | 0.3×104 |
| Y97A | α-Naphthyl acetate (+NaCl) | 1.7 ± 0.3 | 2.9 ± 0.3 | 0.2×104 |
| S172A | α-Naphthyl acetate (+NaCl) | 0.6 ± 0.1 | 5.3 ± 0.3 | 0.9×104 |
| A223K | α-Naphthyl acetate (+NaCl) | 0.30 ± 0.04 | 1.1 ± 0.1 | 0.4×104 |
| N225A | α-Naphthyl acetate (+NaCl) | 0.5 ± 0.1 | 1.7 ± 0.1 | 0.3×104 |
| N225K | α-Naphthyl acetate (+NaCl) | 0.4 ± 0.1 | 3.0 ± 0.2 | 0.8×104 |
Effect of temperature on the OLEI01171 activity
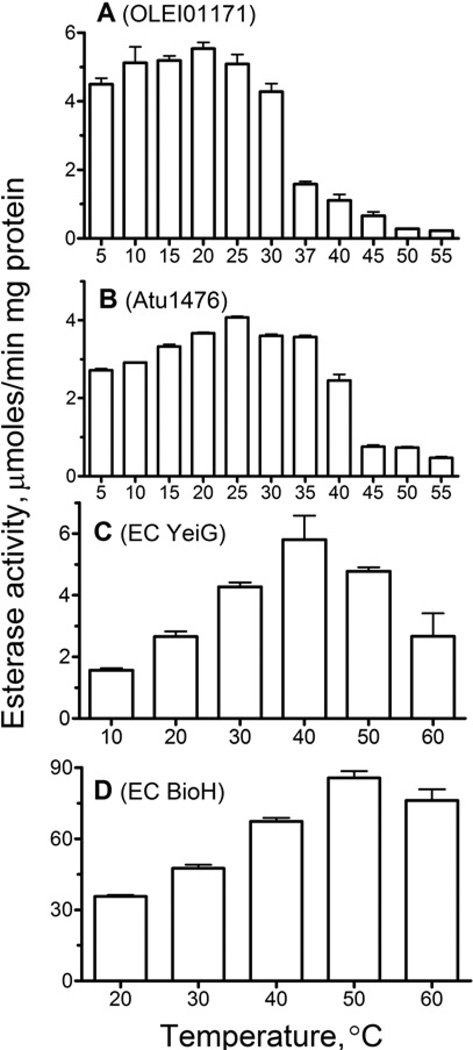
The temperature profile of esterase activity of OLEI01171 measured with α-naphthyl acetate as the substrate shows that the protein exhibits high activity within a broad range of low temperatures from 5 to 30°C with the maximal activity at 15– 20°C (Figure 3). At 5 °C, this enzyme retained 82% of its maximal activity (at 20°C). The biochemically characterized cold-active esterases from Ps. haloplanktisPs. arctica and Acinetobacter sp. strain 6 showed a 30–90%decrease in activity at 5°C [34–36]. Interestingly, the homologous esterase Atu1476 from the mesophilic bacterium A. tumefaciens also showed a relatively small decrease (~35%) in activity at 5°C (compared with its maximal activity at 25°C), whereas the E. coli YeiG (also from PF00756) and BioH (a non-homologous esterase from PF00561) retained 30–40% of maximal activity at 5°C (Figure 3). Thus the temperature profiles of the esterase activity of OLEI01171, Atu1476 and YeiG correlate with the optimal growth temperatures of these organisms (4 °C, 28°C and 37°C respectively), so the enzyme from the organism with lower optimal growth temperature retained higher residual activity at 5°C.
Figure 3. Effect of temperature on the esterase activity of OLEI01171 (A), Atu1476 (B), E. coli YeiG (C) and BioH (D).
Esterase activity of purified proteins was measured at the indicated temperatures under the optimal conditions using α-naphthyl acetate (A), pNP-butyrate (B and D) and pNP-propionate (C) as substrates (see the Experimental section for details).
At higher temperatures, the activity of OLEI01171 was slightly reduced at 30°C and became negligible at 60°C (Figure 3A). As expected, the activity of mesophilic esterases Atu1476, YeiG and BioH showed a higher resistance to increasing temperature, as they were inhibited at 40°C, 50°C and 60°C respectively (Figures 3B–3D). Thus, whereas the activity of all three PF00756 esterases showed the increased sensitivity to high temperatures, the mesophilic esterase Atu1476 retained a significant activity at low temperatures. Comparison of the temperature profiles shown in Figure 3 indicates that OLEO01171 belongs to the group-1 of psychrophilic enzymes, which show similar levels of activity and reduced thermostability compared with their mesophilic homologues [11].
Thermostability of OLEI01171
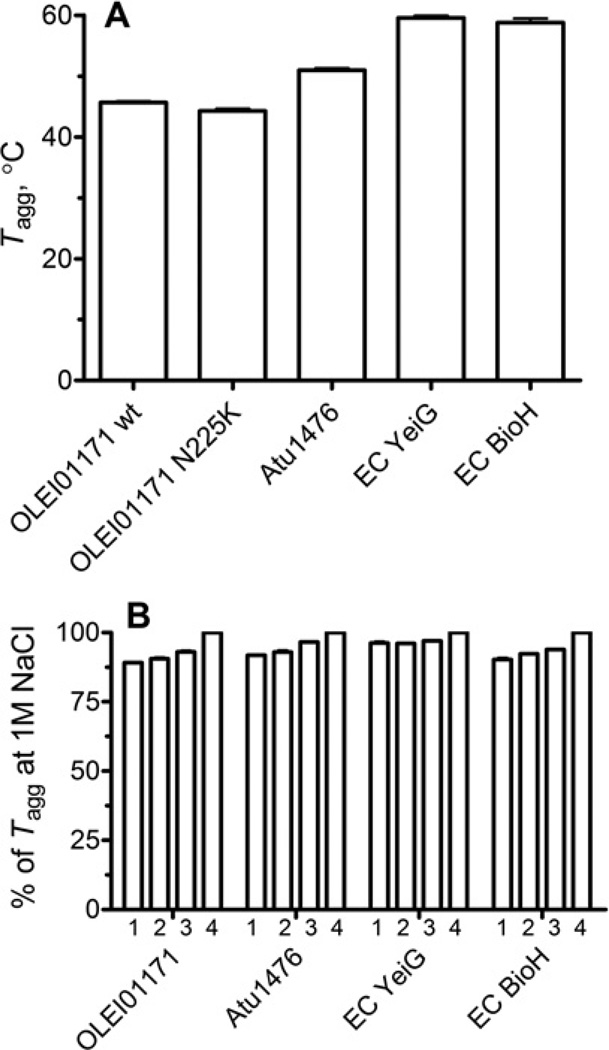
To compare the temperature activity profiles of the PF00756 esterases with their overall thermostability, we have determined the thermodenaturation (aggregation) profiles (Tagg) of these proteins. These experiments revealed that OLEI01171 has the lowest thermostability (Tagg =45.7± 0.5 °C), whereas Atu1476, YeiG and BioH exhibited higher Tagg (51.0 °C, 59.6 °C and 58.8 °C respectively) (Figure 4A). These results indicate that the homologous esterases from the PF00756 family exhibit different overall protein thermostability, although they are structurally similar and have the same structural fold. On the other hand, two non-homologous esterases YeiG and BioH showed similar thermodenaturation profiles. This suggests that protein thermostability is mainly determined by small differences in the protein structure or sequence. In contrast with the stimulating effect of chloride on the OLEI01171 esterase activity, the addition of increasing concentrations of NaCl or KCl (0.05–1 M) produced only a small increase in Tagg of OLEI01171 and other esterases (Figure 4B, only the NaCl effect is shown). Therefore the anion stimulation of the OLEI01171 activity is not due to overall protein stabilization.
Figure 4. Thermostability of OLEI01171 and other esterases (StarGazer experiments).
(A) The temperatures of aggregation (Tagg) measured under standard conditions (0.3 M NaCl). (B) The effect of increasing concentrations of NaCl on the Tagg of indicated esterases: 50 mM (1), 150 mM (2), 500 mM (3) and 1 M NaCl (4). EC, E. coli.
The comparison of the protein thermodenaturation temperatures (Tagg) and the activity temperature profiles of the studied enzymes indicates that the structurally similar esterases (OLEI01171, Atu1476 and YeiG) have similar differences between the maximal activity temperature and Tagg (20–25 °C), whereas for BioH (PF00561) this difference was almost two times smaller (9°C) (Figures 3 and 4). This suggests that the active sites of OLEI01171 and other PF00756 esterases are more heat-labile than their overall protein structures which denature at temperatures 10–15 °C higher than the temperature of activity inhibition (35–50°C) (Figure 3A). In contrast, BioH displays similar temperatures of activity inhibition (60°C) and protein denaturation (58.8 °C), indicating that protein unfolding is the main determinant for the loss of activity at high temperatures in this enzyme. Like the previous work on α-amylases [37], the results of the present study support the model of ‘localized flexibility’ of psychrophilic enzymes, which proposes that the low stability (high flexibility) of the active site is a key determinant of enzyme activity at low temperatures [38].
Crystal structure and active site of OLEI01171
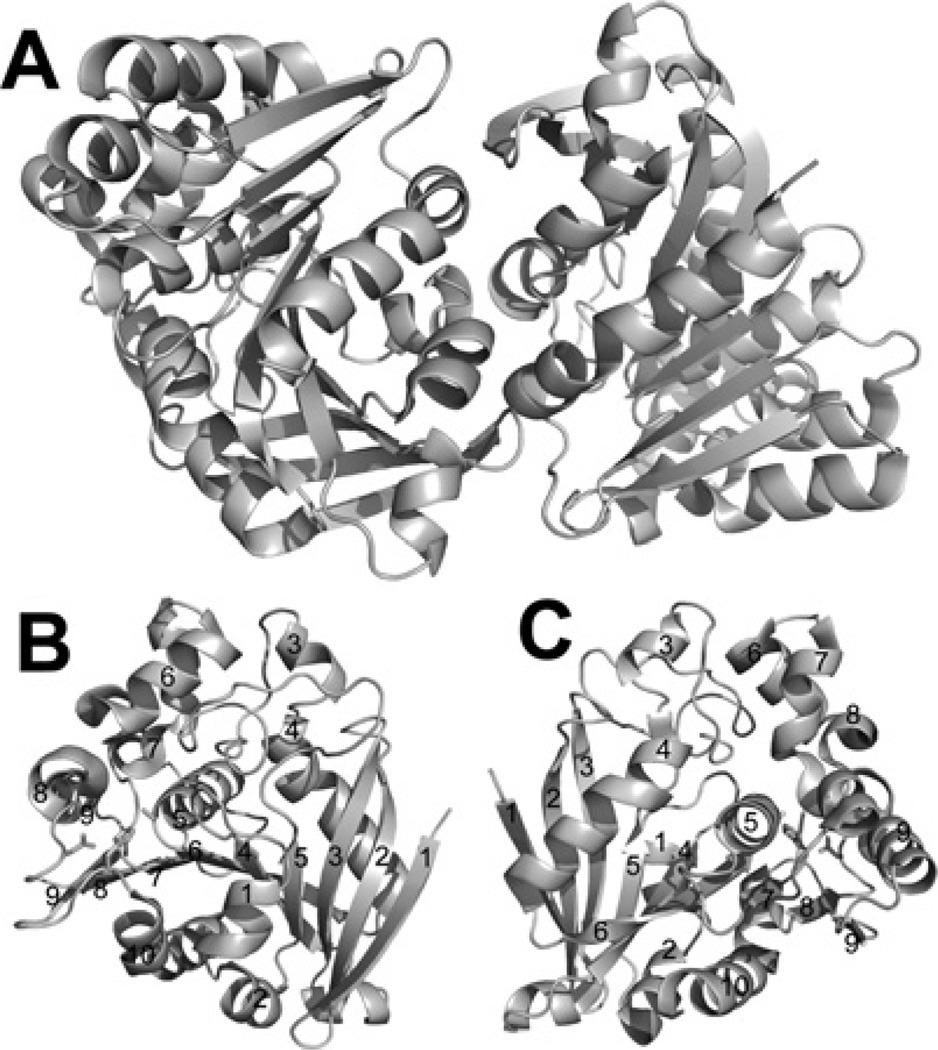
Purified wild-type OLEI01171 was crystallized (see the Experimental section for details) and its structure was solved to 1.75 Å resolution by molecular replacement using the structure of the yeast FGH (PDB code 1PV1) as a model (Table 1). Gel-filtration experiments with the purified recombinant OLEI01171 revealed that at neutral pH (7.5) it exists as a dimer in solution (51.1 kDa; predicted monomer molecular mass is 33.4 kDa). Consistent with the results of our gel-filtration experiments, the structure revealed that two OLEI01171 protomers form a dimer through multiple interactions between the residues located on three α-helices (α1, α2 and α10), one β-strand (β1) and the β9– α10 strand (Figure 5A). The core structure of the OLEI01171 protomer is formed by a nine-stranded hydrophobic β-sheet packed between two layers of amphiphilic α-helices (α/β/α sandwich) and covered by four α-helices and the long β5–α3 loop on the top (Figures 5B and 5C). A Dali (http://ekhidna.biocenter.helsinki.fi/dali_server/start) search for homologous structures in PDB identified four PF00756 esterase structures with the FGH from the psychrophilic bacterium Ps. haloplanktis (PDB code 3LS2) as the best match [Z-score=49.1 and rmsd (root mean square deviation)=0.8 Å]. The other three top hits were the human esterase D (PDB code 3FCX, Z-score= 46.5 and rmsd=0.9 Å), the FGH Atu1476 from A. tumefaciens (PDB code 3E4D, Z-score=46.5 and rmsd=1.0 Å) and the yeast FGH (esterase D) (PDB code 1PV1, Z-score= 42.2 and rmsd=1.4 Å). This rather high overall structural and sequence conservation (44–65% sequence identity) between the enzymes from the evolutionarily distant organisms suggests a recent evolutionary origin of the PF00756 esterase family.
Figure 5. Crystal structure of OLEI01171.
(A) Overall structure of the dimer. (B and C) Two views of the OLEI01171 monomer (related by a 180° rotation). The secondary structure elements are labelled and the position of the active site is indicated by the side chains of the catalytic triad (shown as sticks).
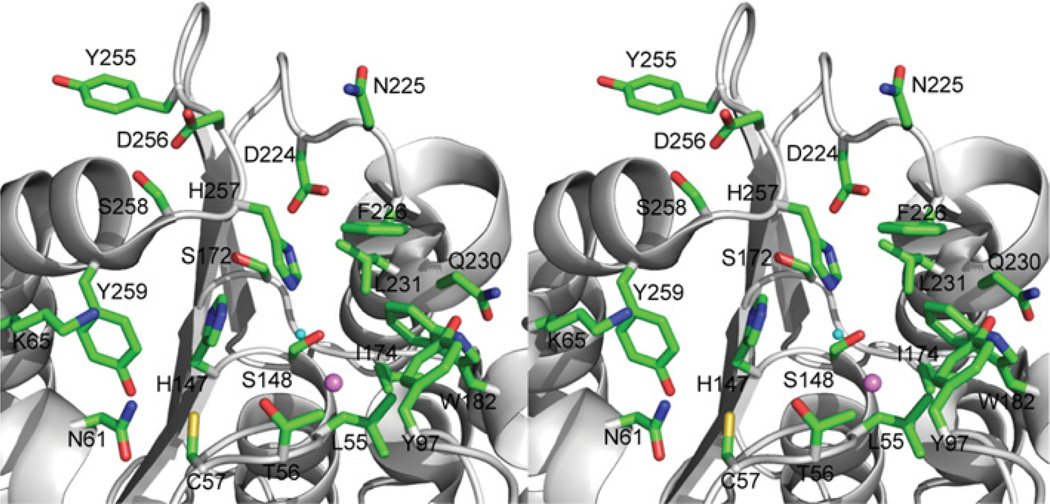
In OLEO01171, the active site is placed on the top of the β-sheet and, like in other psychrophilic enzymes [13], its catalytic cavity is open and quite shallow with the catalytic residues fairly exposed to solvent (Figure 6). The conserved Ser148 is located on the classical nucleophilic elbow (a short loop between the β6-strand and α6-helix) and makes a hydrogen bond with the side chain of the conserved His257 (3.3 Å) (Figure 6). Together with Asp224 (2.7 Å from His257), these three residues comprise the OLEI01171 catalytic serine hydrolase triad (Figure 6). In a classical serine hydrolase mechanism, the serine-histidine-aspartic acid triad catalyses the hydrolytic cleavage of the carboxyl ester substrate with the formation of a covalent acylenzyme intermediate with the active site Ser148 and two tetrahedral intermediates [39, 40]. The ester hydrolysis proceeds in five steps: after binding of the substrate, a first tetrahedral intermediate is formed by nucleophilic attack of the catalytic serine residue, with the oxyanion stabilized by two or three hydrogen bonds with the main chain nitrogens (an oxyanion hole). The ester bond is cleaved and the alcohol product leaves the active site. In a last step, the acyl-enzyme intermediate is hydrolysed by the nucleophilic attack of the catalytic water molecule activated by the catalytic histidine residue. The oxyanion hole plays an important role in ester hydrolysis through the activation of the substrate carbonyl group for attack and stabilization of the negative charge of the tetrahedral intermediate [39, 40]. On the basis of the crystal structure of OLEI01171, its oxyanion hole appears to be constituted by the main chain nitrogens of the conserved Leu55 and Met149 (5.8 Å and 3.4 Å to the Ser148 side chain hydroxy group respectively) (Figure 6).
Figure 6. Close-up stereo view of the OLEI01171 active site.
The Figure shows the bound Cl− and potential catalytic water molecule (shown as violet and cyan balls respectively). The OLEI01171 residues are shown as sticks along an OLEI01171 ribbon.
In serine esterases, the substrate-binding site is composed of two subsites involved in the binding of the acyl or alcohol part of a substrate, which are located on the top of the central β-sheet and connected at the catalytic serine residue [41]. Like a typical carboxyl esterase, OLEI01171 has a small acyl-binding site located on the right from the Ser148 side chain and filled with several hydrophobic or polar residues including Leu55, Tyr97, Ile174, Trp182, Phe226, Gln230 and Leu231 (Figure 6). The small size of the acyl-binding site is consistent with the OLEI01171 preference for the ester substrates with short acyl chains (acetyl or formyl) (Figure 2A). On the left-hand side from the catalytic Ser148, there is a larger cleft sitting on the top of the β-sheet with thewalls formed by two α helices (α1 and α10) and β4–α1 loop accommodating the OLEI01171 alcohol-binding site (Figure 6). This pocket is filled with several polar or charged residues (Cys57, Asn61, Lys65, His147, Ser172 and Tyr259), which are conserved in PF00756 esterases (except for Ser172). The comparison of the active sites of PF00756 esterases and mesophilic esterases from other protein families indicates that the former have more open active sites with the histidine residue loop more exposed to solvent (Supplementary Figure S3 at http://www.BiochemJ.org/bj/445/bj4450193add.htm) that might facilitate the maintenance of higher activity of the PF00756 esterases at low temperatures.
Anion binding in the OLEI01171 active site
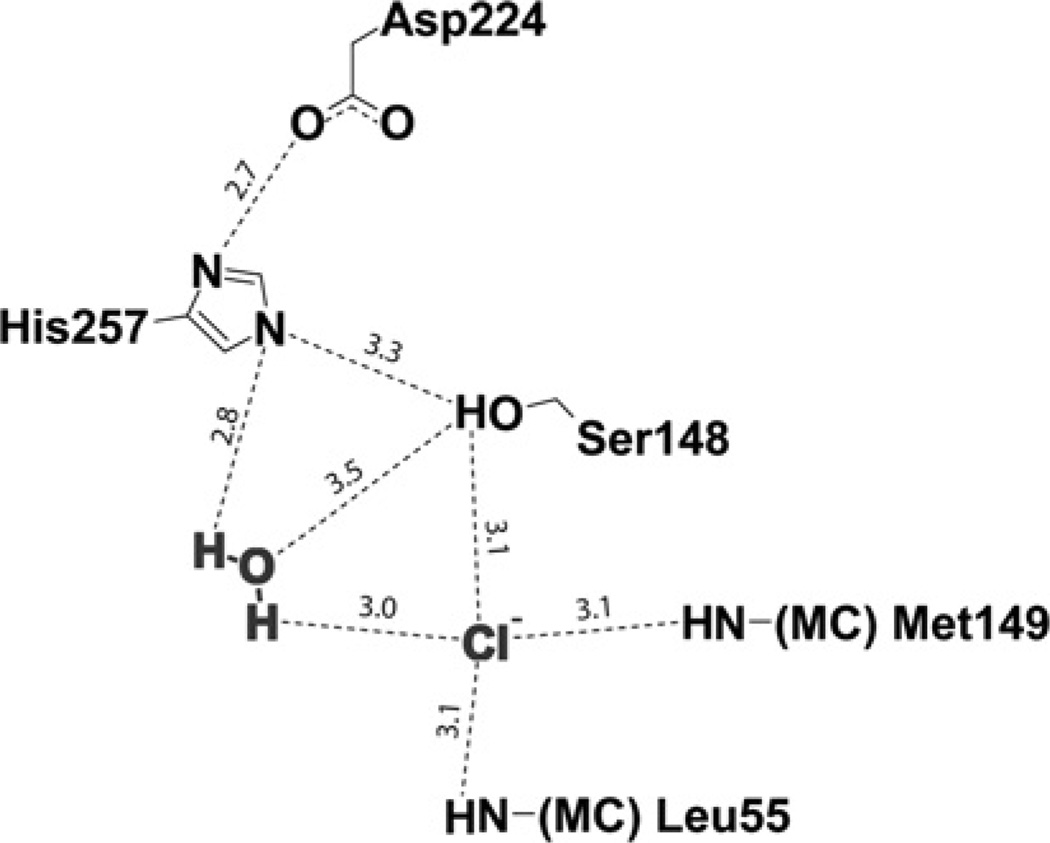
The OLEI01171 structure revealed the presence of a small density close to the predicted catalytic Ser148 (3.1 Å), which was annotated as a chloride anion (Figure 6). This chloride anion is hydrogen bonded to the main chain nitrogens of Leu55 (3.1 Å) and Met149 (3.1 Å), and therefore it might mimic the position of the substrate carbonyl oxygen bound to the oxyanion hole. However, the structure of the Est30 (thermostable carboxylesterase Est30) esterase acyl-enzyme intermediate shows that the substrate carbonyl oxygen is located closer to the catalytic serine residue hydroxyl oxygen (2.4 Å) [42]. In addition, close to the chloride ion there is a water molecule (3.1 Å), which is also co-ordinated by the catalytic His257 side chain (2.7 Å) and might represent a catalytic water molecule positioned for nucleophilic attack on the acyl-enzyme intermediate (Figure 6). Therefore this chloride ion might contribute to the polarization of the catalytic water molecule enhancing the rate of the hydrolysis of the acyl-enzyme intermediate. The OLEI01171 structure suggests that the presence of the chloride ion in the active site could interfere with substrate binding, and therefore the anion is more likely to bind to the acyl-enzyme intermediate, whose hydrolysis might represent a rate-limiting step in the OLEI01171 reaction. The potential catalytic role of the chloride ion might provide a molecular mechanism for the stimulating effect of NaCl and other anions on the activity of OLEI01171 (Figure 2). Previously, it has been shown that chloride ions stimulate activity of the α-amylases found in Ps. haloplanktis and animals, which belong to the large superfamily of the (β/α)8 barrel (TIM barrel) hydrolases, specifically to the glycoside hydrolase family 13 [43]. In the OLEI01171 structure, the chloride ion is co-ordinated by the side chain of one catalytic residue and by two main-chain nitrogens resembling the mainchain anion-binding site called the nest [44] (Figure 6). The typical anion-binding site nest is formed by the three main-chain NH groups of the adjacent residues, which point inwards and form a depression that can accommodate an anion. Another difference is that in OLEI01171 the chloride ion directly interacts with the potential catalytic water molecule (Wat280), which is also bound to the catalytic His257 and is likely to function as an attacking nucleophile in the hydrolysis of the acyl-enzyme intermediate (Figures 6 and 7). Thus, although OLEI01171 binds chloride ions in a way different from that in α-amylases, this anion might also contribute to the activation of the catalytic water molecule thereby increasing the rate of the second reaction step (hydrolysis of the acyl-enzyme intermediate).
Figure 7. Diagram showing the OLEI01171 catalytic triad and the c-ordination of bound chloride ion and potential catalytic water in the active site.
Mutational studies of OLEI01171
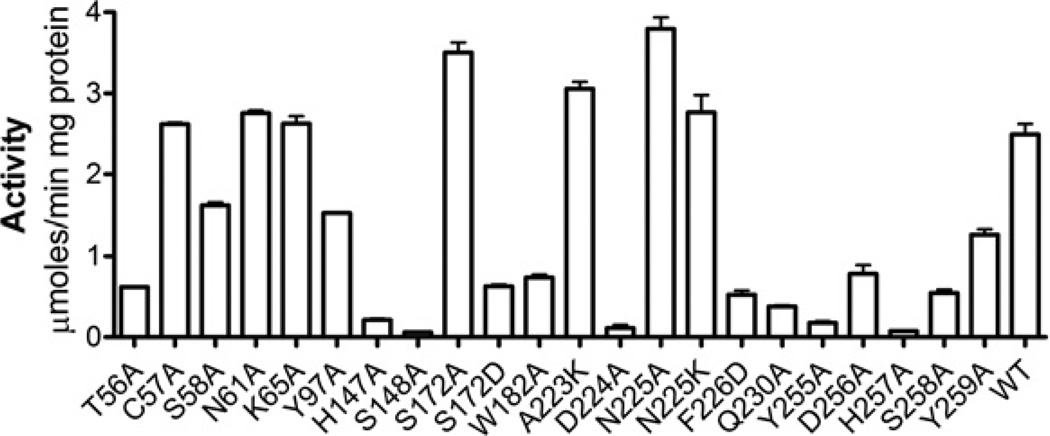
In order to probe the role of active-site residues in the activity of the OLEI01171 esterase, we mutated them to alanine or other residues. The mutated proteins were purified and tested for esterase activity using α-naphthyl acetate as a substrate. As expected, the alanine replacement of the three residues of the OLEO01171 catalytic triad (Ser148, His257 and Asp224) abolished the enzymatic activity of this protein (Figure 8). In addition, we observed a strong negative effect on the activity of seven mutant proteins with mutated conserved residues located in the substrate-binding site including T56A, H147A, W182A, F226D, Y255A, D256A and S258A (Figure 8). However, several other mutant proteins with the mutated conserved or semi-conserved residues showed a significant or wild-type activity (C57A, S58A, N61A, K65A, Y97A, S172A, A223K, N225A and Y259A) suggesting that these residues are not playing a critical role in the OLEI01171 activity under optimal conditions. Most of these mutant proteins (except for Y97A) also showed the wild-type level Km value, whereas S172A had a higher kcat value (Table 2). The sequences of the three PF00756 esterases (Atu1476, YeiG and human esterase D) contain an alanine residue at the position occupied by Ser172 in OLEI01171 (Figure 1). Accordingly, the OLEI01171 S172A mutant protein showed a wild-type level of activity, which was greatly decreased in the S172D mutant protein (Figure 8), probably owing to the strong inhibitory effect of a negatively charged residue introduced close to the catalytic His257 (Figure 6). Thus our mutagenic studies show that most of the active-site residues of OLEI01171 are important for its catalytic activity. In addition, the results of the present study show that the activity of OLEI01171 is more sensitive to mutations (structural perturbations) introduced to the histidine loop than to the aspartic acid loop since both mutations in the His257 neighbourhood (D256A and S258A) produced a strong negative effect whereas the mutations around Asp224 (A223K and N225A) had no effect on activity. Thus the structural and mutational analyses of OLEI01171 indicated that the protein residues from the conserved block-1 (Figure 1) contribute to the oxyanion hole (Leu55) and alcohol-binding site (Thr56 and Cys57), block-3 to acyl binding (Phe105), block-4 to catalysis (Ser148) and the oxyanion hole (Met149), and block-5 to catalysis (His257) and the alcohol-binding site (Ser258 and Tyr259). The role of the conserved residues from block-2 is presently unclear.
Figure 8. Site-directed mutagenesis of OLEI01171: esterase activity of purified mutant proteins.
The reaction mixtures contained 2 mM α-naphthyl acetate and 1 µg of purified protein (15 min incubation at 15°C). WT, wild-type.
Activity and thermostability of the OLEI01171 N225K mutant protein
The reduction in the content of charged residues in psychrophilic proteins is considered to be one of the mechanisms of cold adaptation [45, 46]. Accordingly, the protein surface analysis of OLEI01171 and three other PF00756 esterases with solved structures revealed that two psychrophilic enzymes (OLEI01171 and Ps. haloplanktis FGH) have a lower content of charged residues (aspartic acid, glutamic acid, arginine and lysine) on their surfaces compared with two mesophilic esterases (Atu1476 and yeast FGH) (39–49 residues against 60–61 residues respectively) (Supplementary Table S1 at http://www.BiochemJ.org/bj/445/bj4450193add.htm). This is in line with the previous analyses of the psychrophilic proteins, which demonstrated a significant decrease in charged amino acid content in these proteins [8, 47].
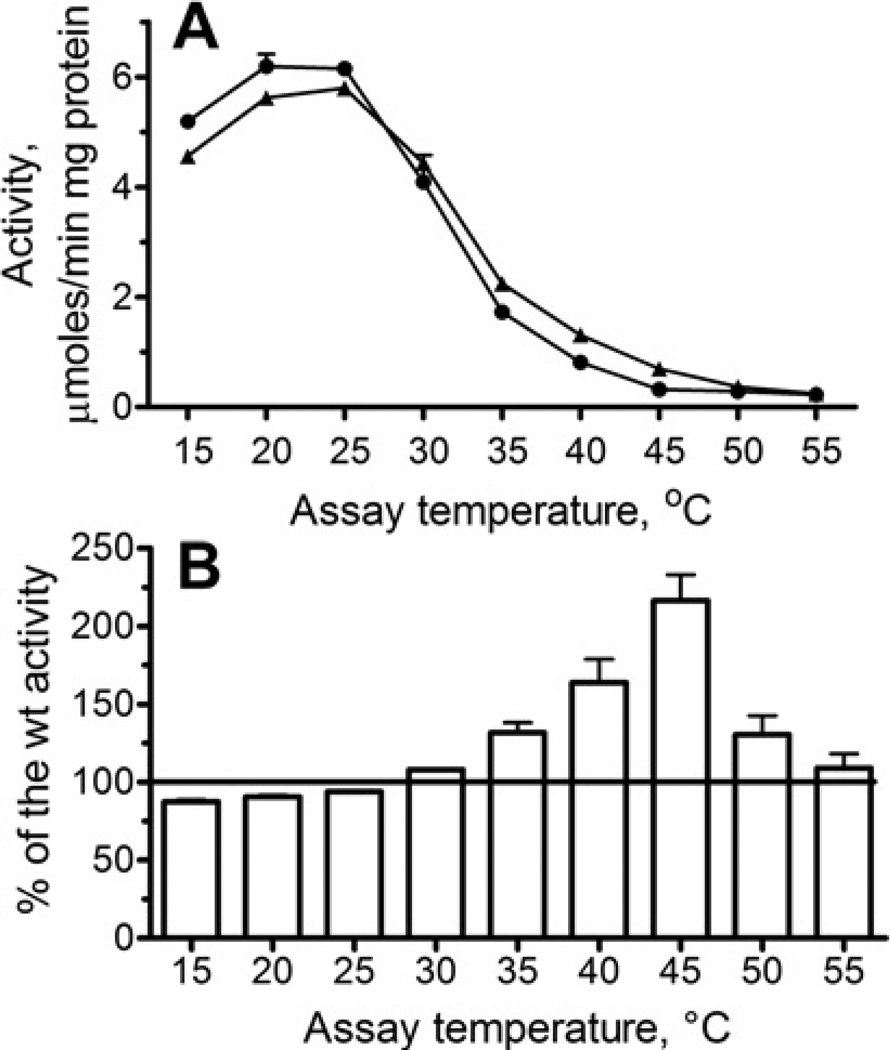
As the active site area is supposed to be the most sensitive to amino acid replacements, we wanted to determine if the introduction of a single charged residue (lysine) into the OLEI01171 aspartic acid loop (close to the catalytic Asp224)would enhance the resistance of its activity to increased temperatures. Asparagine is known to be a thermolabile amino acid residue, whose reduced frequency in thermophylic proteins has been previously reported [48]. Therefore, we mutated the OLEI01171 Asn225, as well as the nearby Ala223, to lysine and checked the effect of temperature on the protein stability (Tagg) and esterase activity of purified mutant proteins with α-naphthyl acetate as the substrate. These mutations had no significant effect on the overall protein thermostability (Tagg =44.5 °Cand 45.5 °Cfor A223K and N225K respectively) (Figure 4A, only wild-type and N225K are shown), or on the loss of its esterase activity during incubation at 45°C (Supplementary Figure 1E). However, the protein containing the N225K mutation exhibited a slightly lower activity at optimal temperatures (15–20 °C) and retained higher activity at 35–45°C compared with the wild-type OLEI01171 (Figure 9), whereas the activity of the A223K protein was inhibited by increased temperatures similar to the wild-type enzyme (results not shown). The crystal structure of the N225K mutant protein was solved and revealed no interaction of the Lys25 side chain with the histidine loop residues (results not shown), suggesting that its increased activity at elevated temperatures is probably due to the interaction of the Lys225 side chain with solvent. Thus even a single amino acid change near the active site of a psychrophilic enzyme can result in a detectable change in the thermal profile of its activity. In addition, the results of the present study suggest that the resistance of a cold-active enzyme to increased temperature can be enhanced without a reduction in its catalytic activity.
Figure 9. Replacement of Asn225 by lysine enhances the resistance of OLEI01171 to high temperatures.
(A) The effect of temperature on esterase activity of the wild-type (●) and N225K (▲) mutant proteins. (B) Esterase activity of the N225K protein shown as a percentage of the wild-type (wt) activity (100%) at the same temperature (data from A).
Thus the biochemical and structural characterization of the α/β-hydrolase protein OLEI01171 from the psychrophilic marine bacterium O. antarctica revealed a cold-active and anion-activated carboxylesterase. This enzyme retains over 80% of its catalytic activity at 5°C, and its activity is stimulated by various anions. The OLEI01171 esterase has a preference for short acyl-chain substrates and shows significant activity against several esterase substrates including α-naphthyl acetate, α-naphthyl propionate, pNP-acetate and S-formylglutathione. The crystal structure of OLEI01171 revealed the presence of a chloride or bromide ion bound in the active site close to its catalytic Ser148 and to a potential catalytic water molecule suggesting that anions might contribute to the hydrolysis of the acyl-enzyme intermediate. The potential catalytic role of the chloride ion might provide a molecular mechanism for the stimulating effect of NaCl and other anions on the activity of OLEI01171. Future biochemical and structural studies will provide a further insight into the molecular mechanisms of anion activation of cold-active enzymes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the Ontario Centre for Structural Proteomics in Toronto (SPiT) for help in conducting experiments and discussions.
FUNDING
This work was supported by the government of Canada through Genome Canada and the Ontario Genomics Institute [grant number 2009-OGI-ABC-1405] (to A.Y. and A.S.)], the National Institutes of Health [grant number GM074942 (to A.S.)], the European Science Foundation project and the European Union.
Abbreviations used
- BioH
pimelyl-(acyl-carrier protein) methyl ester esterase
- FGH
S-formyl-glutathione hydrolase
- FrmB
FGHfrmB
- pNP
p-nitrophenyl
- rmsd
root mean square deviation
- TLS
Translation-Liberation-Screw rotation
- YeiG
FGH yeiG
Footnotes
Co-ordinates and structure factors of the OLEI01171 complexes with Cl− or Br− have been deposited in the PDB under the accession codes 3I6Y and 3S8Y respectively.
AUTHOR CONTRIBUTION
Sofia Lemak, Anatoli Tchigvintsev, Robert Flick, Greg Brown, Olga Egorova, Claudio Gonzalez and Tatyana Chernikova carried out the biochemical studies; Pierre Petit, Alexander Singer, Elena Evdokimova and Olga Egorova performed the crystallization experiments; Pierre Petit, Alexander Singer and Alexei Savchenko solved and refined the crystal structures; Michail Yakimov, Michael Kube, Richard Reinhardt and Peter Golyshin sequenced and analysed the Oleispira antarctica genome and the OLEI01171 gene; and Peter Golyshin, Alexei Savchenko and Alexander Yakunin designed the research, analysed the data and wrote the paper.
REFERENCES
- 1.Rothschild LJ, Mancinelli RL. Life in extreme environments. Nature. 2001;409:1092–1101. doi: 10.1038/35059215. [DOI] [PubMed] [Google Scholar]
- 2.Antranikian G, Vorgias CE, Bertoldo C. Extreme environments as a resource for microorganisms and novel biocatalysts. Adv. Biochem. Eng. Biotechnol. 2005;96:219–262. doi: 10.1007/b135786. [DOI] [PubMed] [Google Scholar]
- 3.Morita RY. Psychrophilic bacteria. Bacteriol. Rev. 1975;39:144–167. doi: 10.1128/br.39.2.144-167.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Amico S, Collins T, Marx JC, Feller G, Gerday C. Psychrophilic microorganisms: challenges for life. EMBO Rep. 2006;7:385–389. doi: 10.1038/sj.embor.7400662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feller G, Gerday C. Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Microbiol. 2003;1:200–208. doi: 10.1038/nrmicro773. [DOI] [PubMed] [Google Scholar]
- 6.Casanueva A, Tuffin M, Cary C, Cowan DA. Molecular adaptations to psychrophily: the impact of ‘omic’ technologies. Trends Microbiol. 2010;18:374–381. doi: 10.1016/j.tim.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Cavicchioli R, Thomas T. Extremophiles. In: Lederberg J, editor. Encyclopedia of Microbiology. San Diego: Academic Press; 2000. pp. 317–337. [Google Scholar]
- 8.Methe BA, Nelson KE, Deming JW, Momen B, Melamud E, Zhang X, Moult J, Madupu R, Nelson WC, Dodson RJ, et al. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10913–10918. doi: 10.1073/pnas.0504766102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forster J. Ueber einige Eigenschaften leuchtender Bakterien. Centr. Bakteriol. Parasitenk. 1887;2:337–340. [Google Scholar]
- 10.Kobori H, Sullivan CW, Shizuya H. Heat-labile alkaline phosphatase from Antarctic bacteria: rapid 5′ end-labeling of nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 1984;81:6691–6695. doi: 10.1073/pnas.81.21.6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavicchioli R, Siddiqui KS, Andrews D, Sowers KR. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 2002;13:253–261. doi: 10.1016/s0958-1669(02)00317-8. [DOI] [PubMed] [Google Scholar]
- 12.Siddiqui KS, Cavicchioli R. Cold-adapted enzymes. Annu. Rev. Biochem. 2006;75:403–433. doi: 10.1146/annurev.biochem.75.103004.142723. [DOI] [PubMed] [Google Scholar]
- 13.Georlette D, Blaise V, Collins T, D’Amico S, Gratia E, Hoyoux A, Marx JC, Sonan G, Feller G, Gerday C. Some like it cold: biocatalysis at low temperatures. FEMS Microbiol. Rev. 2004;28:25–42. doi: 10.1016/j.femsre.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Aghajari N, Feller G, Gerday C, Haser R. Crystal structures of the psychrophilic α-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor. Protein Sci. 1998;7:564–572. doi: 10.1002/pro.5560070304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aghajari N, Feller G, Gerday C, Haser R. Structural basis of α-amylase activation by chloride. Protein Sci. 2002;11:1435–1441. doi: 10.1110/ps.0202602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Petegem F, Collins T, Meuwis MA, Gerday C, Feller G, Van Beeumen J. The structure of a cold-adapted family 8 xylanase at 1.3Å resolution. Structural adaptations to cold and investgation of the active site. J. Biol. Chem. 2003;278:7531–7539. doi: 10.1074/jbc.M206862200. [DOI] [PubMed] [Google Scholar]
- 17.Alterio V, Aurilia V, Romanelli A, Parracino A, Saviano M, D’Auria S, De Simone G. Crystal structure of an S-formylglutathione hydrolase from Pseudoalteromonas haloplanktis TAC125. Biopolymers. 2010;93:669–677. doi: 10.1002/bip.21420. [DOI] [PubMed] [Google Scholar]
- 18.Yakimov MM, Giuliano L, Gentile G, Crisafi E, Chernikova TN, Abraham WR, Lunsdorf H, Timmis KN, Golyshin PN. Oleispira antarctica gen. nov., sp. nov., a novel hydrocarbonoclastic marine bacterium isolated from Antarctic coastal sea water. Int. J. Syst. Evol. Microbiol. 2003;53:779–785. doi: 10.1099/ijs.0.02366-0. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez CF, Proudfoot M, Brown G, Korniyenko Y, Mori H, Savchenko AV, Yakunin AF. Molecular basis of formaldehyde detoxification. Characterization of two S-formylglutathione hydrolases from Escherichia coli, FrmB and YeiG. J. Biol. Chem. 2006;281:14514–14522. doi: 10.1074/jbc.M600996200. [DOI] [PubMed] [Google Scholar]
- 20.Berge RK, Farstad M. Long-chain fatty acyl-CoA hydrolase from rat liver mitochondria. Methods Enzymol. 1981;71:234–242. doi: 10.1016/0076-6879(81)71030-9. [DOI] [PubMed] [Google Scholar]
- 21.Lai KK, Lorca GL, Gonzalez CF. Biochemical properties of two cinnamoyl esterases purified from a Lactobacillus johnsonii strain isolated from stool samples of diabetes-resistant rats. Appl. Environ. Microbiol. 2009;75:5018–5024. doi: 10.1128/AEM.02837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vedadi M, Niesen FH, Allali-Hassani A, Fedorov OY, Finerty, Jr PJ, Wasney GA, Yeung R, Arrowsmith C, Ball LJ, Berglund H, et al. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimber MS, Vallee F, Houston S, Necakov A, Skarina T, Evdokimova E, Beasley S, Christendat D, Savchenko A, Arrowsmith CH, et al. Data mining crystallization databases: knowledge-based approaches to optimize protein crystal screens. Proteins. 2003;51:562–568. doi: 10.1002/prot.10340. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of X-ray data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 26.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta. Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 29.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 30.Wu D, Li Y, Song G, Zhang D, Shaw N, Liu ZJ. Crystal structure of human esterase D: a potential genetic marker of retinoblastoma. FASEB. J. 2009;23:1441–1446. doi: 10.1096/fj.08-125286. [DOI] [PubMed] [Google Scholar]
- 31.Legler PM, Kumaran D, Swaminathan S, Studier FW, Millard CB. Structural characterization and reversal of the natural organophosphate resistance of a D-type esterase, Saccharomyces cerevisiae S-formylglutathione hydrolase. Biochemistry. 2008;47:9592–9601. doi: 10.1021/bi8010016. [DOI] [PubMed] [Google Scholar]
- 32.van Straaten KE, Gonzalez CF, Valladares RB, Xu X, Savchenko AV, Sanders DA. The structure of a putative S-formylglutathione hydrolase from Agrobacterium tumefaciens. Protein Sci. 2009;18:2196–2202. doi: 10.1002/pro.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arpigny JL, Jaeger KE. Bacterial lipolytic enzymes: classification and properties. Biochem. J. 1999;343:177–183. [PMC free article] [PubMed] [Google Scholar]
- 34.Aurilia V, Parracino A, Saviano M, Rossi M, D’Auria S. The psychrophilic bacterium Pseudoalteromonas halosplanktis TAC125 possesses a gene coding for a cold-adapted feruloyl esterase activity that shares homology with esterase enzymes from gamma-proteobacteria and yeast. Gene. 2007;397:51–57. doi: 10.1016/j.gene.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki T, Nakayama T, Kurihara T, Nishino T, Esaki N. A cold-active esterase with a substrate preference for vinyl esters from a psychrotroph, Acinetobacter sp. strain no. 6: gene cloning, purification, and characterization. J. Mol. Catalysis B: Enzymatic. 2002;16:255–263. [Google Scholar]
- 36.Al Khudary R, Venkatachalam R, Katzer M, Elleuche S, Antranikian G. A cold-adapted esterase of a novel marine isolate, Pseudoalteromonas arctica: gene cloning, enzyme purification and characterization. Extremophiles. 2010;14:273–285. doi: 10.1007/s00792-010-0306-7. [DOI] [PubMed] [Google Scholar]
- 37.D’Amico S, Marx JC, Gerday C, Feller G. Activity–stability relationships in extremophilic enzymes. J. Biol. Chem. 2003;278:7891–7896. doi: 10.1074/jbc.M212508200. [DOI] [PubMed] [Google Scholar]
- 38.Fields PA, Somero GN. Hot spots in cold adaptation: localized increases in conformational flexibility in lactate dehydrogenase A4 orthologs of Antarctic notothenioid fishes. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11476–11481. doi: 10.1073/pnas.95.19.11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, et al. The α/β hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 40.Cygler M, Grochulski P, Kazlauskas RJ, Schrag JD, Bouthillier F, Rubin B, Serrequi AN, Gupta AK. A structural basis for the chiral preferences of lipases. J. Am. Chem. Soc. 1994;116:3180–3186. [Google Scholar]
- 41.Pleiss J, Fischer M, Schmid RD. Anatomy of lipase binding sites: the scissile fatty acid binding site. Chem. Phys. Lipids. 1998;93:67–80. doi: 10.1016/s0009-3084(98)00030-9. [DOI] [PubMed] [Google Scholar]
- 42.Liu P, Wang YF, Ewis HE, Abdelal AT, Lu CD, Harrison RW, Weber IT. Covalent reaction intermediate revealed in crystal structure of the Geobacillus stearothermophilus carboxylesterase Est30. J. Mol. Biol. 2004;342:551–561. doi: 10.1016/j.jmb.2004.06.069. [DOI] [PubMed] [Google Scholar]
- 43.D’Amico S, Gerday C, Feller G. Structural similarities and evolutionary relationships in chloride-dependent α-amylases. Gene. 2000;253:95–105. doi: 10.1016/s0378-1119(00)00229-8. [DOI] [PubMed] [Google Scholar]
- 44.Watson JD, Milner-White EJ. A novel main-chain anion-binding site in proteins: the nest. A particular combination of phi, psi values in successive residues gives rise to anion-binding sites that occur commonly and are found often at functionally important regions. J. Mol. Biol. 2002;315:171–182. doi: 10.1006/jmbi.2001.5227. [DOI] [PubMed] [Google Scholar]
- 45.Feller G, Gerday C. Psychrophilic enzymes: molecular basis of cold adaptation. Cell. Mol. Life Sci. 1997;53:830–841. doi: 10.1007/s000180050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marshall CJ. Cold-adapted enzymes. Trends Biotechnol. 1997;15:359–364. doi: 10.1016/S0167-7799(97)01086-X. [DOI] [PubMed] [Google Scholar]
- 47.Gianese G, Argos P, Pascarella S. Structural adaptation of enzymes to low temperatures. Protein Eng. 2001;14:141–148. doi: 10.1093/protein/14.3.141. [DOI] [PubMed] [Google Scholar]
- 48.Villeret V, Clantin B, Tricot C, Legrain C, Roovers M, Stalon V, Glansdorff N, Van Beeumen J. The crystal structure of Pyrococcus furiosus ornithine carbamoyltransferase reveals a key role for oligomerization in enzyme stability at extremely high temperatures. Proc. Natl. Acad. SciU.SA. 1998;95:2801–2806. doi: 10.1073/pnas.95.6.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.