Abstract
Proteins are often characterized in terms of their primary, secondary, tertiary, and quaternary structure. Algorithms such as DSSP can automatically assign protein secondary structure based on the backbone hydrogen-bonding pattern. However, the assignment of secondary structure elements becomes a challenge when only the Cα coordinates are available. In the current work, we present PCASSO (Protein C-Alpha Secondary Structure Output), a fast and accurate program for assigning protein secondary structure elements using only the Cα positions. PCASSO achieves ~95% accuracy with respect to DSSP and takes ~0.1 s using a single processor to analyze a 1,000 residue system with multiple chains. Our approach was compared with current state-of-the-art Cα-based methods and was found to outperform all of them in both speed and accuracy. A practical application is also presented and discussed.
Introduction
The basic protein secondary structure elements (SSEs), namely, α-helices and β-sheets, were first described by Pauling and Corey in 1951 (1, 2) and have since provided a foundation for comparing, classifying, and visualizing three-dimensional (3-D) protein folds. Traditionally, protein SSEs were manually designated through visual inspection of the polypeptide chain, which often resulted in assignments that were subjective and, at times, incomplete. Today, this tedious process is made more efficient and reproducible through automated tools such as Structural Identification (STRIDE) (3) and Define Secondary Structure of Proteins (DSSP) (4, 5). DSSP, one of the oldest and most popular SSE assignment programs available, assigns SSEs by first identifying all backbone carbonyl (C=O) and amide (N-H) hydrogen bonds based on a purely electrostatic criterion. Then, depending on the hydrogen bonding patterns, each residue is classified as a helix, strand, or loop. However, the assignment of SSEs becomes problematic when insufficient information is available (e.g., Protein Data Bank (PDB) structures with unresolved backbone atoms, Cα-only models originating from cryo-electron microscopy (cryo-EM), and coarse-grained protein models used in multiscale simulations). While the positions of the missing backbone atoms that are required for SSE assignment can be estimated from reduced models (6–11), the reconstruction methodology is imperfect and often requires some level of refinement or energy minimization through molecular dynamics simulations in order to optimize the backbone hydrogen bonding networks before being processed through DSSP. Furthermore, this time consuming process can become prohibitive when reconstructing a large number of structures from long coarse-grained MD simulations. Thus, it is advantageous to develop a fast and efficient method that avoids the reconstruction process altogether and yet can still provide reliable SSE assignments that can be generally and consistently applied across multiple scales.
Several Cα-based assignment methods such as P-SEA (12), VoTAP (13), and, more recently, SABA (14) have been reported. P-SEA utilizes a combination of distances, angles, and dihedrals for secondary structure analysis while VoTAP generates contact matrices derived from 3-D Voronoï tessellation, which are then used for assigning SSEs. SABA uses a similar approach to P-SEA but instead of directly computing the Cα coordinates SABA shifts the coordinates of the ith Cα atom to its pseudo-center (PC) position (defined as the center-of-geometry between Cα(i) and Cα(i+1)) and then assigns SSEs based on an optimized set of PC-dependent geometric criteria. This is thought to better represent the location of the backbone N-H/C=O atoms involved in secondary structure formation. While these methods appear to agree reasonably well with DSSP, P-SEA and VoTAP are no longer being maintained and SABA is available only as a web server that is limited to analyzing individually uploaded PDB files.
In the current work, we present PCASSO (Protein C-Alpha Secondary Structure Output), a fast and efficient program for assigning protein SSEs that only requires Cα atoms as input. By employing the well-known random forest (RF) (15) approach, PCASSO achieves high accuracy compared to DSSP and offers fast processing times even for large systems. PCASSO can be used for, but not limited to, evaluating individual PDBs, batch processing, and analyzing molecular dynamics (MD) simulation trajectories. The source code (licensed under the GNU General Public License v3.0) and web server are made freely available at http://brooks.chem.lsa.umich.edu/software.
Methods
Random forest (RF) is an ensemble machine learning methodology that achieves high accuracy by aggregating classifications from independent random decision trees and reporting the mode vote (15). To ensure that the trees within the forest are uncorrelated, each tree is trained on a bootstrap sample of the original data set (with replacement) and only a small, randomly chosen subset of features/variables is used to determine the best split at a given node. To compare our results with previous methods, we utilized the same protein training and test sets published by Mornon and coworkers (13) (see Table S1, Table S2, and Table S3). All structural coordinates were obtained from the Protein Data Bank (PDB) (16) and analyzed with DSSP (4, 5). The Cα atoms were then extracted from each PDB and 258 basic geometric features (see below) were computed for each residue of the reduced model.
For a given residue, i, a set of features, fCα (i) and fPC (i), were calculated from the Cα coordinates and the pseudo-center (PC) coordinates, respectively (see Table S4). The jth and kth residues form non-bonded interactions with the ith residue and help to identify interactions between strands that are separated in sequence. The jth residue has the shortest distance from residue i and, when i and j are from the same chain/segment, j must be at least i+6 residues away. Similarly, the kth residue has the shortest distance from residue i and, when i and k are from the same chain/segment, k must be at least i−6 residues away. The coordinates of the ith PC was previously defined as the center-of-geometry between Cα(i) and Cα(i+1) (14) and so the PC coordinates for the last residue of each chain/segment is undefined as are the features that reference the ultimate C-terminal residue. The feature vector, V(i), for the ith residue is made up by features from the ith, i−1th, and i+1th residues (i.e. V (i) = (fCα (i), fPC (i), fCα(i−1), fPC (i −1), fCα (i +1), fPC (i +1)}) which results in a total of (2 × 43) × 3 = 258 feature elements.
From the training set, a total of 50 trees were generated using the RF implementation found in the Open Source Computer Vision (OpenCV) library (17) and default parameters were used unless otherwise specified. At each node, 16 out of 258 features/variables were selected at random to find the best split. Node splitting was ceased either when: (i) all members of the node were of the same class (i.e. helix, strand, or loop); (ii) the maximum depth allowed (25) was reached; or (iii) the minimum sample count required for a split (10) was not satisfied. Changes in the RF parameters (i.e. number of random features used for each split, maximum tree depth, minimum sample count, total number of trees, etc) did not result in a significant increase in accuracy. Since the tree growing procedure is completely independent of the classification process, the resulting ensemble of trees was extracted from the OpenCV output, serialized as a string in pre-order, and hardcoded into PCASSO for speed and efficiency. Thus, PCASSO is a standalone program that takes either PDB structures or MD simulation trajectories as input, deserializes the tree ensemble into independent binary decision trees, calculates the full feature vector for each Cα atom and processes it through each tree, aggregates the SSE classifications, and returns the mode vote for each residue of each structure or simulation snapshot. To compare the speed and accuracy of PCASSO with the reconstruction scheme, the missing backbone atoms for each Cα model from the test set were rebuilt using the rebuild program from the Multiscale Modeling Tools for Structural Biology (MMTSB) Tool Set (6) and subsequently analyzed using DSSP. Finally, the protein test set was analyzed using PCASSO and the accuracy (relative to DSSP) was compared with the SSE assignments from P-SEA, VoTAP, and DSSP (using the reduced models with reconstructed backbone atoms as input). To demonstrate the value and applicability of PCASSO, we analyzed a previously published 58 µs MD folding trajectory of a human Pin1 WW domain variant called FiP35 (18). Simulation snapshots (n = 2900) were assessed every 20 ns and the SSE classifications were used in constructing conformation space networks. All molecular graphics were generated in PyMOL (19) and SSE time series plots were created using in-house tools.
Results and Discussion
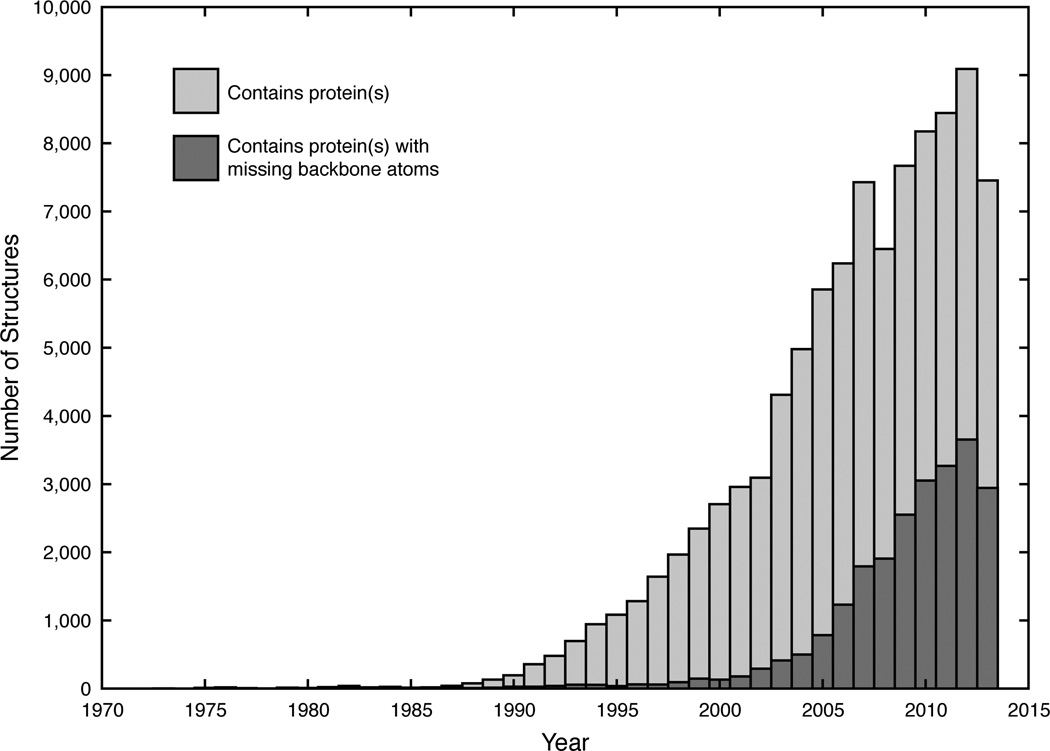
As the number of protein structures being deposited into the PDB grows, the number of X-ray, NMR, and cryo-EM structures with missing or incomplete backbone atoms also experiences a concomitant increase. For example, approximately 40% of the protein structures deposited in 2013 contained at least one or more missing backbone atoms (Figure 1). Concurrently, the number of publications that include the terms “coarse”, “grained”, “protein”, and “simulation” has also been on the rise (20). Since DSSP (4, 5), the current gold standard for assigning SSEs, depends solely upon backbone hydrogen bonding patterns, residues with only Cα coordinates are generally ignored or neglected. While the backbone atoms for a single protein can be reconstructed from the Cα atoms with reasonable accuracy, this time-consuming process, as we will demonstrate below, becomes infeasible for much larger systems and/or for rapidly rebuilding a large ensemble of structures from coarse-grained/multiscale simulations. As scientists continue to push the size of systems that can be experimentally determined (21, 22) or computationally simulated (23), the demand for faster and more efficient analysis tools that can complement these larger systems will also rise. Thus, PCASSO has been developed to provide quick and reliable SSE classifications directly from the Cα coordinates (i.e., without backbone reconstruction) with the analogous aim of being to Cα-containing structures what DSSP is to all-atom structures.
Figure 1.
The number of protein-containing structures deposited in the PDB between 1971–2013 (non-cumulative). A total of 96,286 PDB structures were analyzed and 23,295 PDB structures (~24%) were found to have incomplete/missing backbone atoms.
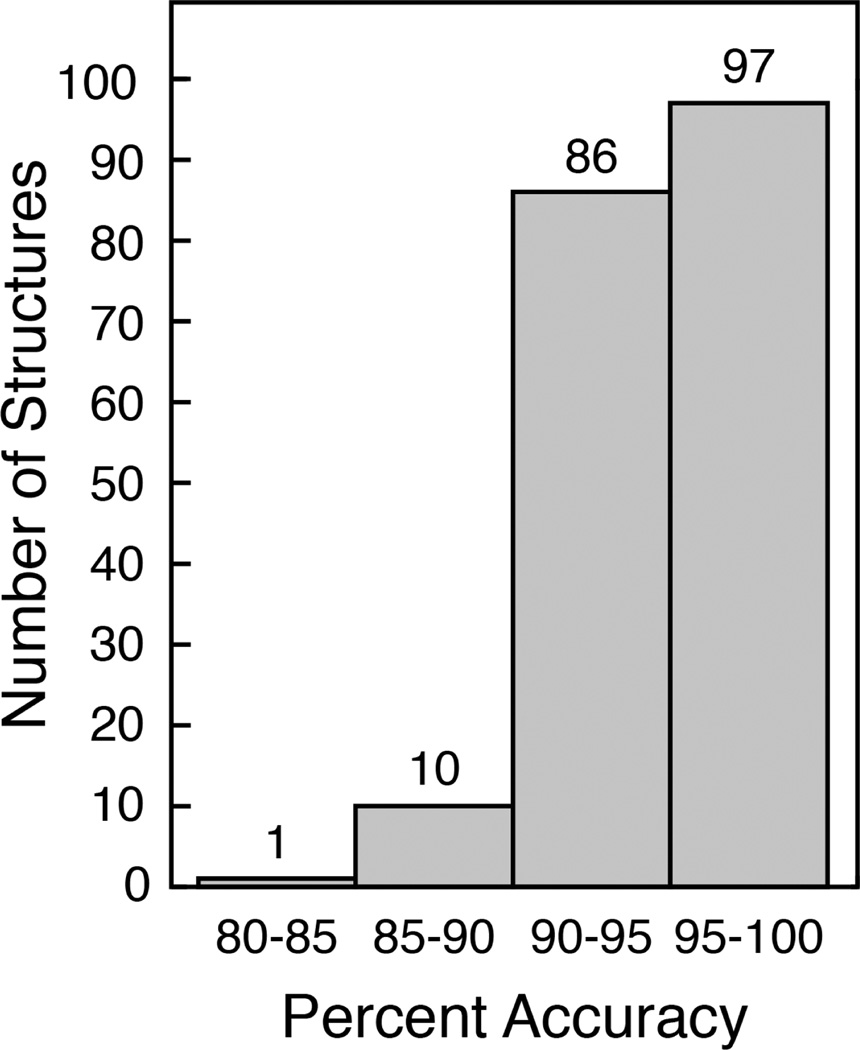
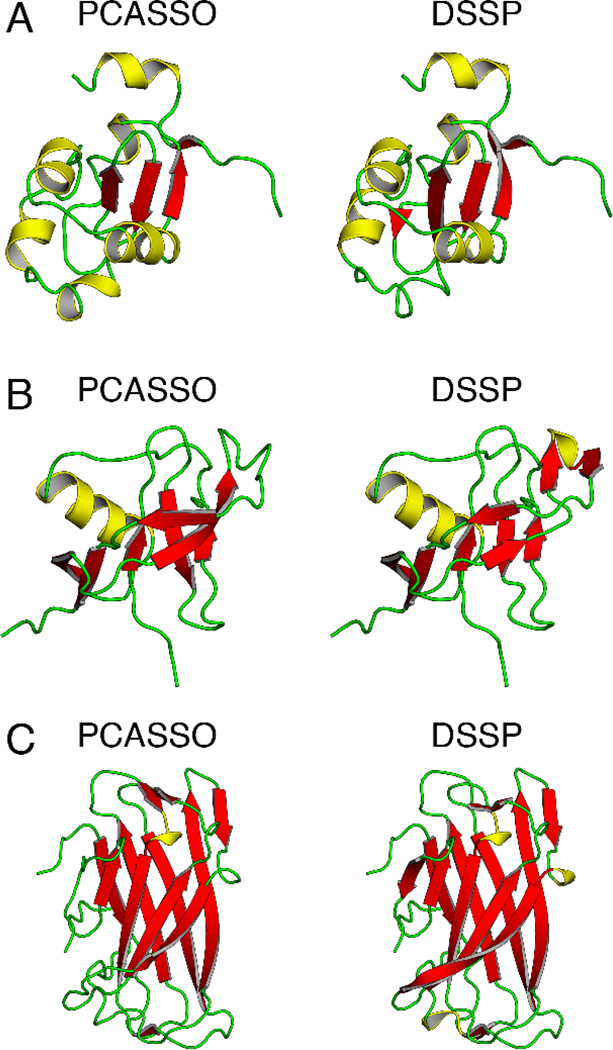
To judge the performance of PCASSO, we compared our SSE assignment accuracy relative to DSSP with assignments from P-SEA and VoTAP (Table 1). Overall, PCASSO demonstrated ~95% accuracy, which is more than an 11% increase over P-SEA and VoTAP. PCASSO showed a substantial improvement in classifying strands and loops and a moderate enhancement in classifying helices. More importantly, PCASSO was found to be equally as accurate as the reconstruction scheme (i.e., the backbone atoms were reconstructed from the Cα coordinates and then evaluated using DSSP) and exhibited a high level of precision and sensitivity for each SSE class (i.e., low false positives and low false negatives). Over 94% of the structures in the test set had a greater than 90% classification accuracy and over 99% of the structures had a greater than 85% accuracy (Figure 2). The three lowest accuracy structures (Table S5) only showed minor differences in their assignments and are displayed in Figure 3. Furthermore, since PCASSO was trained on DSSP SSE assignments, we also assessed the accuracy of PCASSO relative to STRIDE (Table S6). Remarkably, even without recalibrating PCASSO to match STRIDE, the overall accuracy was only slightly reduced to ~93% which can be attributed to a small decrease in accuracy for classifying helices and strands. It is logical that the accuracy results can somewhat vary when PCASSO is compared to different reference methods since STRIDE and DSSP are based on different approaches. In fact, it has been previously reported that STRIDE is in ~95% agreement with DSSP (13). Additionally, it has been demonstrated that these minor discrepancies can be attenuated by the use of a ternary consensus method (TCM) (12, 13, 24). However, considering the generally high level of agreement with the aforementioned all-atom-based assignment methods, we contend that TCM would not be practical or necessary.
Table 1.
PCASSO accuracy comparison
Figure 2.
Histogram of structures with varying SSE assignment accuracies
Figure 3.
SSE assignment comparison for the three lowest accuracy structures
To assess the scalability of PCASSO, we evaluated its processing time for systems of increasing size using a single CPU (Table 2). We found that PCASSO was at least 24 times faster than P-SEA and at least 11 times faster than the reconstruction scheme. In fact, by extrapolation, as the number of residues (and/or structures) increases, it becomes infeasible to use any of the pre-existing Cα-based methods for assigning SSEs due to their much longer processing times. While in all cases, multiple structures or simulation snapshots can be divided amongst multiple CPUs in an “embarrassingly parallel” manner in order to boost the speed performance, only PCASSO is amenable to further parallelization. For example, unlike P-SEA and VoTAP, which both assign helices first followed by strands and then loops (i.e., there is a residue assignment order dependency), PCASSO treats the assignment of each residue completely independently, which makes it perfectly suited for parallel processing. Additional speed improvements can also be made by distributing the evaluation of each independent decision tree to a different CPU or by removing redundant and/or highly correlated features. Thus, PCASSO is not only able to accomplish more with limited resources but its underlying implementation also allows room for future improvement and scalability.
Table 2.
Comparison of SSE processing times
| PDBID | Residues | Chains | Time (s) |
P-SEA PCASSO |
Reconstruction+DSSP PCASSO |
||||
|---|---|---|---|---|---|---|---|---|---|
| PCASSO | P-SEA | VoTAP | Reconstruction | DSSP | |||||
| 1PUC | 101 | 1 | 0.01 | 0.34 | - | 0.11 | 0.04 | 34.00 | 15.00 |
| 1NBA | 1011 | 4 | 0.11 | 2.74 | - | 1.04 | 0.17 | 24.91 | 11.00 |
| 1RVV | 4620 | 30 | 1.25 | 51.39 | - | 12.75 | 1.17 | 41.11 | 11.14 |
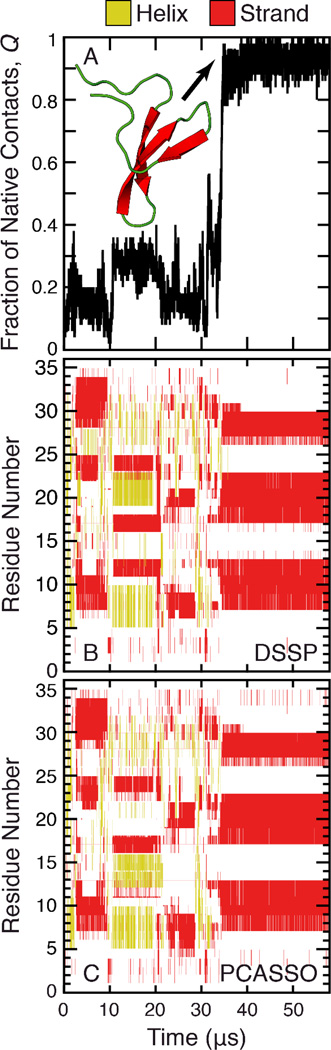
The number of coarse-grained protein simulations has experienced a steady increase over the past decade as scientists seek to understand protein structure and dynamics on much longer timescales (20). In the case of protein folding, the fraction of native amino acid contacts, Q (25), is typically used as a progress variable for monitoring the folding process. However, Q can fail to identify important nonnative contacts or protein misfolding that would have otherwise been captured through SSE analysis. To illustrate this point and to demonstrate a practical application of PCASSO, we analyzed a previously published all-atom MD folding trajectory of a human Pin1 WW domain variant called FiP35 (18), which consists of a three-stranded β-sheet connected by two β hairpins (Figure 4). Using Q as the reaction coordinate, initially, FiP35 is only partially folded but after ~35 µs the peptide forms over 80% of its native contacts and is considered fully folded (Figure 4A). However, both DSSP and PCASSO, which yield essentially the same results, reveal that FiP35 can form stable nonnative interactions at the onset and parts of the peptide actually misfold to a helix (Figure 4B–C). Thus, this example clearly demonstrates the value of SSE assignments and how this information can be complementary to Q. Furthermore, PCASSO offers a fast and reliable alternative to DSSP for analyzing protein secondary structure that can be applied to any Cα-containing multiscale model.
Figure 4.
Analyses of the FiP35 folding trajectory
In conclusion, PCASSO outperformed pre-existing programs in both accuracy and speed. Given this, PCASSO can also be used in network analysis through SSE clustering (26), high-throughput SSE studies, universal SSE assignments, SSE-based alignments (27), renormalization of Gō-like models for intrinsically disordered proteins (28), and to analyze coarse-grained simulation models that do not incorporate any native contact information (29, 30) or where the native contacts are not known a priori (e.g., to examine cooperative folding of multimers or large multi-subunit complexes). Ultimately, we hope that the work presented here will motivate the development of better and faster tools to complement the ever-growing challenges of big data.
Supplementary Material
Acknowledgements
The authors would like to acknowledge valuable scientific discussions with Bin Zhang, Junjie Zou, Shanshan Cheng, Shuai Wei, Logan Ahlstrom, Alex Dickson, Afra Panahi, Garrett Goh, and Karunesh Arora. We also thank D. E. Shaw Research for providing access to the FiP35 MD Trajectory. This work was supported by NIH through GM037554 and NSF through funding from the Center for Biological Physics (PHY0216576). ATF was supported through the University of Michigan President’s Postdoctoral Fellowship.
References
- 1.Pauling L, Corey RB. Configurations of Polypeptide Chains with Favored Orientations around Single Bonds - 2 New Pleated Sheets. Proc. Natl. Acad. Sci. U. S. A. 1951;37(11):729–740. doi: 10.1073/pnas.37.11.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pauling L, Corey RB, Branson HR. The Structure of Proteins - 2 Hydrogen-Bonded Helical Configurations of the Polypeptide Chain. Proc. Natl. Acad. Sci. U. S. A. 1951;37(4):205–211. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frishman D, Argos P. Knowledge-Based Protein Secondary Structure Assignment. Proteins-Structure Function and Genetics. 1995;23(4):566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 4.Joosten RP, et al. A Series of PDB Related Databases for Everyday Needs. Nucleic Acids Res. 2011;39:D411–D419. doi: 10.1093/nar/gkq1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kabsch W, Sander C. Dictionary of Protein Secondary Structure - Pattern-Recognition of Hydrogen-Bonded and Geometrical Features. Biopolymers. 1983;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 6.Feig M, Karanicolas J, Brooks CL., III MMTSB Tool Set: Enhanced Sampling and Multiscale Modeling Methods for Applications in Structural Biology. J. Mol. Graphics Modell. 2004;22(5):377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Li YQ, Zhang Y. Remo: A New Protocol to Refine Full Atomic Protein Models from C-Alpha Traces by Optimizing Hydrogen-Bonding Networks. Proteins. 2009;76(3):665–674. doi: 10.1002/prot.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holm L, Sander C. Database Algorithm for Generating Protein Backbone and Side-Chain Coordinates from a C-Alpha Trace Application to Model-Building and Detection of Coordinate Errors. J. Mol. Biol. 1991;218(1):183–194. doi: 10.1016/0022-2836(91)90883-8. [DOI] [PubMed] [Google Scholar]
- 9.Petrey D, et al. Using Multiple Structure Alignments, Fast Model Building, and Energetic Analysis in Fold Recognition and Homology Modeling. Proteins-Structure Function and Genetics. 2003;53(6):430–435. doi: 10.1002/prot.10550. [DOI] [PubMed] [Google Scholar]
- 10.Rotkiewicz P, Skolnick J. Fast Procedure for Reconstruction of Full-Atom Protein Models from Reduced Representations. J. Comput. Chem. 2008;29(9):1460–1465. doi: 10.1002/jcc.20906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sali A, Blundell TL. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 12.Labesse G, Colloch N, Pothier J, Mornon JP. P-SEA: A New Efficient Assignment of Secondary Structure from C Alpha Trace of Proteins. Comput Appl Biosci. 1997;13(3):291–295. doi: 10.1093/bioinformatics/13.3.291. [DOI] [PubMed] [Google Scholar]
- 13.Dupuis F, Sadoc JF, Mornon JP. Protein Secondary Structure Assignment through Voronoi Tessellation. Proteins. 2004;55(3):519–528. doi: 10.1002/prot.10566. [DOI] [PubMed] [Google Scholar]
- 14.Park SY, Yoo MJ, Shin J, Cho KH. SABA (Secondary Structure Assignment Program Based on Only Alpha Carbons): A Novel Pseudo Center Geometrical Criterion for Accurate Assignment of Protein Secondary Structures. Bmb Rep. 2011;44(2):118–122. doi: 10.5483/BMBRep.2011.44.2.118. [DOI] [PubMed] [Google Scholar]
- 15.Breiman L. Random Forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 16.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradski G. The Opencv Library. Dr. Dobb's Journal of Software Tools. 2000 [Google Scholar]
- 18.Shaw DE, et al. Atomic-Level Characterization of the Structural Dynamics of Proteins. Science. 2010;330(6002):341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 19.Schrödinger L. (Schrödinger, LLC) The Pymol Molecular Graphics System [Google Scholar]
- 20.Takada S. Coarse-Grained Molecular Simulations of Large Biomolecules. Curr. Opin. Struct. Biol. 2012;22(2):130–137. doi: 10.1016/j.sbi.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Volkmann N. Putting Structure into Context: Fitting of Atomic Models into Electron Microscopic and Electron Tomographic Reconstructions. Curr. Opin. Cell Biol. 2012;24(1):141–147. doi: 10.1016/j.ceb.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fridman K, Mader A, Zwerger M, Elia N, Medalia O. Advances in Tomography: Probing the Molecular Architecture of Cells. Nat. Rev. Mol. Cell Biol. 2012;13(11):736–742. doi: 10.1038/nrm3453. [DOI] [PubMed] [Google Scholar]
- 23.Feig M, Sugita Y. Reaching New Levels of Realism in Modeling Biological Macromolecules in Cellular Environments. Journal of Molecular Graphics and Modelling. 2013;45(0):144–156. doi: 10.1016/j.jmgm.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colloch N, Etchebest C, Thoreau E, Henrissat B, Mornon JP. Comparison of 3 Algorithms for the Assignment of Secondary Structure in Proteins - the Advantages of a Consensus Assignment. Protein Eng. 1993;6(4):377–382. doi: 10.1093/protein/6.4.377. [DOI] [PubMed] [Google Scholar]
- 25.Shakhnovich E, Farztdinov G, Gutin AM, Karplus M. Protein Folding Bottlenecks - a Lattice Monte-Carlo Simulation. Phys. Rev. Lett. 1991;67(12):1665–1668. doi: 10.1103/PhysRevLett.67.1665. [DOI] [PubMed] [Google Scholar]
- 26.Rao F, Caflisch A. The Protein Folding Network. J. Mol. Biol. 2004;342(1):299–306. doi: 10.1016/j.jmb.2004.06.063. [DOI] [PubMed] [Google Scholar]
- 27.Fontana P, et al. The Ssea Server for Protein Secondary Structure Alignment. Bioinformatics. 2005;21(3):393–395. doi: 10.1093/bioinformatics/bti013. [DOI] [PubMed] [Google Scholar]
- 28.Ganguly D, Chen JH. Topology-Based Modeling of Intrinsically Disordered Proteins: Balancing Intrinsic Folding and Intermolecular Interactions. Proteins: Struct., Funct., Bioinf. 2011;79(4):1251–1266. doi: 10.1002/prot.22960. [DOI] [PubMed] [Google Scholar]
- 29.Gopal SM, Mukherjee S, Cheng YM, Feig M. Primo/Primona: A Coarse-Grained Model for Proteins and Nucleic Acids That Preserves near-Atomistic Accuracy. Proteins. 2010;78(5):1266–1281. doi: 10.1002/prot.22645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kar P, Gopal SM, Cheng Y-M, Predeus A, Feig M. Primo: A Transferable Coarse-Grained Force Field for Proteins. J. Chem. Theory Comput. 2013;9(8):3769–3788. doi: 10.1021/ct400230y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.