Abstract
Rhodopsin transgenes carrying mutations that cause autosomal dominant retinitis pigmentosa in humans have been used to study rod photoreceptor degeneration in various model organisms including Xenopus laevis. To date, the only transgenes shown to cause rod photoreceptor degeneration in Xenopus laevis have been either mammalian rhodopsins or chimeric versions of rhodopsin based mainly on Xenopus laevis rhodopsin sequences but with a mammalian C-terminus. Since the C-terminal sequence of rhodopsin is highly conserved in mammals and divergent in Xenopus laevis, and mammalian and epitope-tagged rhodopsins may have unexpected properties as transgenes, we decided to test whether a Xenopus laevis rhodopsin transgence carrying only the P23H mutation could also cause rod photoreceptor degeneration. Xenopus laevis tadpoles expressing these transgenes indeed had shortened outer segments and, in severely affected animals, the loss of rod photoreceptors but not the loss of cone photoreceptors. RT-PCR analyses showed that less than 10% of mutant transgenic rhodopsin relative to wild-type endogenous rhodopsin mRNA was sufficient to produce severe rod photoreceptor degeneration. As observed in other animal models as well as humans carrying this particular rhodopsin mutation, the rod photoreceptor degeneration was most severe in the ventral retina and was modified by light. Thus, the rod photoreceptor degeneration produced in Xenopus laevis by the P23H mutation in an otherwise untagged Xenopus laevis rhodopsin is generally similar to that seen with mammalian rhodopsins and epitope-tagged versions of Xenopus laevis rhodopsin, though some differences remain to be explained.
Keywords: P23H, rhodopsin, Xenopus laevis, retinitis pigmentosa
1. Introduction
Retinitis pigmentosa (RP) is a group of photoreceptor degenerations with incidence of approximately one in 3,500 individuals (Boughman, Conneally et al. 1980; Bunker, Berson et al. 1984). RP exhibits extensive genetic heterogeneity and can show autosomal recessive, autosomal dominant, or X-linked patterns of inheritance (Heckenlively, Yoser et al. 1988). Clinical features in RP patients often include early night blindness, followed later by visual field loss (Heckenlively, Yoser et al. 1988; Weleber and Gregory 2006), suggesting that in these patients rod photoreceptors degenerate first followed later by the degeneration of cone photoreceptors.
Missense mutations within the rhodopsin sequence account for approximately one quarter of autosomal dominant retinitis pigmentosa (ADRP) cases. The first such pathogenic rhodopsin mutation that was found, a replacement of the proline at position 23 with a histidine, or P23H, (Dryja, McGee et al. 1990), is also the most common pathogenic rhodopsin mutation found among Americans. The severity of symptoms that result from the P23H mutation can vary considerably between patients (Kemp, Jacobson et al. 1992; To, Adamian et al. 2002), though the disease generally produces a degeneration that is both milder and progresses less quickly than some other missense mutations, especially those that cluster at the C-terminus of rhodopsin. P23H belongs to a subset of rhodopsin mutations that produces a photoreceptor degeneration that is more severe in the ventral half of the retina, thus leading to a preferential loss of the superior visual field (Heckenlively, Rodriguez et al. 1991; Stone, Kimura et al. 1991; Cideciyan, Hood et al. 1998).
Studies of transgenic mice expressing in rod photoreceptors human rhodopsin carrying the P23H mutation were the first to show that mutant rhodopsin transgenes could cause photoreceptor degeneration even when expressed at low levels (Olsson, Gordon et al. 1992). Similar studies have been done in transgenic mice with a mouse rhodopsin transgene carrying multiple amino acid substitutions, including the P23H mutation (Naash, Hollyfield et al. 1993). Transgenic expression of P23H rhodopsin transgenes in rats also produce a photoreceptor degeneration resembling retinitis pigmentosa (Lewin, Drenser et al. 1998; Machida, Kondo et al. 2000). More recently, rod photoreceptor degenerations have also been produced in Xenopus laevis by expressing mammalian rhodopsin transgenes carrying the P23H mutation (Tam and Moritz 2007) or the P23H mutation within a Xenopus laevis sequence that contains two mammalian-like epitope tags, a single amino acid substitution near the P23H mutation, namely M13F, as well as a several amino acid deletion/substitution in the C-terminus of rhodopsin (Tam, Xie et al. 2006; Tam and Moritz 2007). Curiously, in these studies the degeneration caused by the P23H mutation was described as being independent of light. However, since the rod photoreceptor degeneration produced by the P23H mutation within mammalian rhodopsins was modulated by light, this suggested that the P23H Xenopus rhodopsin was behaving differently from the mammalian counterparts when it comes to producing rod photoreceptor degeneration. In this study, we decided to study the rod photoreceptor degeneration caused by the P23H mutation in the context of an otherwise unmodified Xenopus laevis rhodopsin transgene, and to determine whether this rod photoreceptor degeneration was modulated by light.
2. Materials and methods
2.1. Constructs used for Xenopus laevis transgenesis
The control construct in which GFP is driven by 1.3 kb of the Xenopus laevis rhodopsin promoter, XOP1.3_GFP, was made by excising a 4.2 kb BglII fragment from pXOP(−5500/+41)GFP (Knox, Schlueter et al. 1998). To generate the XOP1.3 vector into which to clone rhodopsin cDNAs, the EGFP fragment in XOP1.3_GFP was replaced with the multiple cloning site from the pCS2 plasmid (Turner and Weintraub 1994) using BamHI and NotI. The Xenopus laevis rhodopsin cDNA clone was obtained from ATCC (IMAGE clone ID: 4740890) and the cDNA sequence was mutated using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, Cedar Creek, TX) using primers GGTGGTACGAAGCacaTTCGATTACCCTCAGTATTAC and GTAATACTGAGGGTAATCGAAtgtGCTTCGTACCACC. The mutated bases, shown in lower case, correspond to codon 23 and convert a proline to a histidine. To make constructs expressing the wild-type and mutant rhodopsin under the control of the rhodopsin promoter, XOP1.3_Wt_XRho and XOP1.3_P23H_XRho respectively, the Xenopus laevis rhodopsin cDNA constructs were digested with EcoRV and NotI, and cloned into the StuI and NotI sites of the XOP1.3 vector. For experiments to determine the expression level of transgenes, the XOP1.3_P23H_XRho plasmid was further mutated into XOP1.3_P23H_XRho(ΔHincII) using primers GCCTGAGgtcaatAATGAATCCTTTG and CAAAGGATTCATTattgacCTCAGGC, deleting a HincII site that is present in the rhodopsin cDNA without altering the encoded protein sequence.
2.2. Generation and genotyping of transgenic Xenopus laevis
All plasmids were linearized by BglII digestion and purified using the Geneclean II kit (Qbiogene, Inc., Carlsbad, CA). Transgenesis was performed essentially as described by Kroll and Amaya (Kroll and Amaya 1996) with few modifications (Marsh-Armstrong, Huang et al. 1999). Animals were made with each plasmid alone, or with equal amounts of the XOP1.3_EGFP and either XOP1.3_Wt_XRho, XOP1.3_P23H_XRho, XOP1.3_P23H_XRho(ΔHincII), or XOP1.3_M13F_P23H_XRhod plasmids. Animals were raised for the first week in petri dishes on laboratory bench-tops. After that, they were transferred to a tadpole raising facility in which they were grown at 24 o C at a density of 2 tadpoles per liter or less in 5 L tanks containing de-chlorinated tap water under a slow drip with an average 1-tank volume exchange per day. Animals were in a 12/12 hour light/dark cycle with fluorescent light illumination of ~600 lux average daylight room intensity, unless otherwise noted. For the analysis of the effect of light on rod photoreceptor degeneration, tadpoles with similar rates of degeneration were obtained by breeding a female frog generated with both XOP1.3_ P23H_XRhod and XOP1.3_EGFP transgenes. Five days post-fertilization, animals were screened for GFP fluorescence and GFP positive and negative animals were housed separately four weeks in 5-liter tanks that were set up in 12 hour l/d cycle under high, medium, and low lighting conditions: ~600, ~300, and ~10 Lux, respectively.
2.3. Determination of transgene expression level in rod photoreceptors
Tadpoles generated to express the XOP1.3_P23H (ΔHincII) transgene were raised to 5 to 7 weeks of age. Total RNA was isolated from one eye using the RNeasy Mini Kit (QIAGEN, Inc., Valencia, CA). Reverse transcription primed by random hexamers was generated with the SuperScript II RT kit (Invitrogen Life Technologies, Inc., Carlsbad, CA). Competitive PCR (Becker-Andre and Hahlbrock 1989) was used to determine the relative ratio of transgene to endogenous wild-type rhodopsin gene using 1:10 of the eye cDNA for each competitive PCR reaction and the same primers used in genotyping. For Southern hybridization, half of the final PCR product was electrophoresed through a 1% agarose gel, blotted onto Hybond-N+ nylon membrane (Amersham Pharmacia Biotech, Buckinhamsire, England), and hybridized using an oligonucleotide, AGGGAATGCAATGCTCATGCGGAGTA, labeled with [ 32P-ATP] by use of T4 polynucleotide kinase. Hybridized blots were exposed to FUJI imaging plate, read with a Bio-Imaging Analyzer BAS-2500 (Fuji Photo Film Co. Ltd) and analyzed with Image Gauge Version 3.2 software.
2.4. Histochemistry, in situ hybridization and immunohistochemistry
For all analyses, eyes were fixed in 4% paraformaldehyde in phosphate buffer overnight at 4°C. Some 8-µm sections were stained with hematoxylin/eosin. Other 8-µm sections were permeabilized with 0.1% Triton-X 100 in 1% BSA, blocked with 10% normal goat serum, incubated with a 1:200 dilution of the monoclonal antibody 4D2 (gift of Dr. Robert Molday, University of British Columbia) overnight at 4 ° C, and, after washing, a 1:50 dilution of a FITC-labeled goat anti-mouse IgG (Boehringer Mannheim Biochemicals, Indianapolis, IN). Control slides without incubation with the primary antibody generated no detectable signal. For blinded scoring of phenotype severities, tadpole eyes were post-fixed in 1% of OsO4/ KFeCN in 0.1 M cacodylate buffer (pH 7.4) on ice for 30 minutes, dehydrated, and embedded in Spurs resin. Plastic sections, 1 µm in thickness, were stained with toluidine blue and analyzed under the light microscope. Four observers blinded as to genotype scored disease severity on a 1–5 scale and noted whether there was any degeneration asymmetry along the dorsoventral axis. Average scores between 2 and 3.5 were considered mild or moderate degeneration, while those above 3.5 as severe.
For in situ hybridization analyses, digoxigenin (DIG) and fluorescein (FITC) labeled probes were generated for Xenopus laevis red cone opsin (IMAGE 6955813) and rhodopsin (IMAGE 4740890) clones (Open Biosystems, Huntsville, AL), respectively, and used to label 10µM thick cryosections of retinas. Slides were post fixed in 4% PFA 10 minutes and acetylated in 0.1M triethanolamine 0.25% acetic anhydride for 10 minutes with interceding washes in PBS. After the final PBS wash, sections were hybridized for 16–18 hours at 65°C with 1ng/uL of DIG and FITC labeled probes diluted in hybridization solution (50% Formamide, 1X Hybe solution (Sigma, St. Louis, MO),1mg/ml yeast RNA). After hybridization, slides were washed briefly in 5xSSC at 72°C followed by 1 hour incubation in 0.2xSSC at 72°C. Sections were then blocked for 1 hour at room temperature in 1% Blocking solution (Roche, Manneheim, Germany) and incubated at 4°C in either 1:100 α FITC-POD (Roche) antibody or 1:500 α DIG-POD antibody overnight. Antibodies were detected using a Cy5 tyramide amplification system (Perkin Elmer, Boston, MA). After in situ hybridization, sections were blocked in 10% NGS/PBT (PBS, 1% 100x–Triton), labeled with 1:10 4D2 and 1:500 of an anti-GFP antibody (Torrey Pines, San Diego, CA). Primary antibodies were detected with Cy3 (1:250, Jackson Immunoresearch, West Grove, PA) and Alexa-488 (1:250 Invitrogen, Carlsbad, CA) conjugated antibodies followed by a DAPI counterstain. For semiquantitative imaging studies, retina sections were imaged with a Zeiss 200M inverted microscope and Roper Scientific Coolsnap FX digital camera, using IPlab 4.0 custom scripts. Each retina was imaged at 20x magnification using non-saturating exposure times for each of four channels. Quantification was done with a custom script for Mac IPLab3.9.5 that measured intensity levels of each channel after manual selection of a region of interest. Three different sections were measured and averaged for each retina, and the averages of three retinas per group are presented +/‒ SEM.
3. Results
3.1. Xenopus laevis expressing a P23H rhodopsin transgene undergo rod photoreceptor degeneration
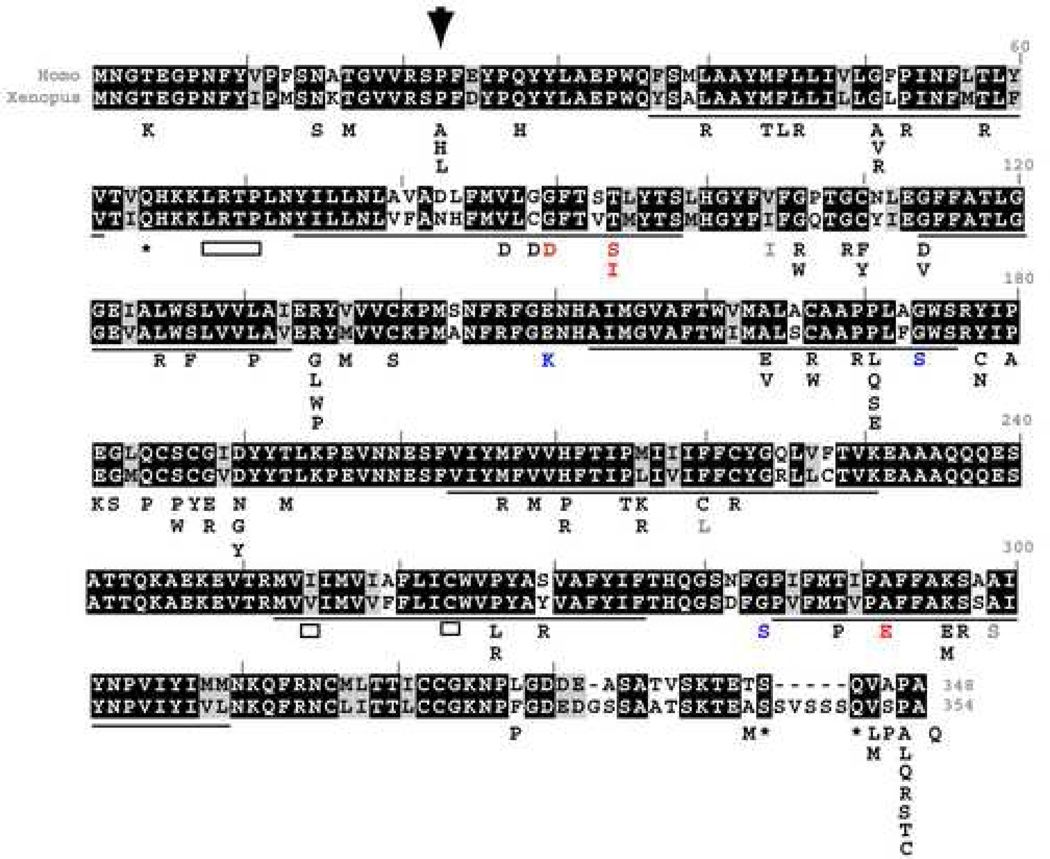
In order to test whether Xenopus laevis can be used to model human retinitis pigmentosa, we sought to generate a Xenopus laevis rhodopsin transgene carrying a mutation that causes autosomal dominant retinitis pigmentosa (ADRP) in humans. To this end, the human rhodopsin sequence (GenBank accession no. P08100) was aligned to the Xenopus laevis rhodopsin sequence (Genbank accession no. L07770) (Figure 1). Across the entire amino acid sequence, there is high conservation (82% identity with 92% similarity and 1% gaps). Among rhodopsin amino acids that when mutated cause ADRP (Briscoe, Gaur et al. 2004), the conservation is only slightly higher (87% identity and 91% similarity). One amino acid substitution that causes ADRP in humans, V137M (Ayuso, Trujillo et al. 1996), is found to occur in the wild-type Xenopus laevis sequence as in teleosts and skates (Briscoe, Gaur et al. 2004). Note that the C-termini are the most divergent part of the sequences. In order to determine whether other mutations that cause ADRP in humans can cause rod photoreceptor degeneration in Xenopus laevis, we chose to start with the single mutation in rhodopsin that accounts for the highest number of ADRP cases in the United States and which is the one most studied in various mammalian animal models, a substitution of a proline for a histidine at position 23 (P23H). The P23H mutation was introduced into the Xenopus laevis rhodopsin sequence by site directed mutagenesis. In order to make a construct that would express specifically in Xenopus laevis rod photoreceptors, wild-type and P23H Xenopus laevis rhodopsin cDNAs were cloned downstream of a 1.3 kb fragment of the Xenopus laevis rhodopsin promoter. Similar rhodopsin promoter fragments have been shown previously to drive specific expression of reporter genes in rod photoreceptors within the Xenopus laevis retina (Knox, Schlueter et al. 1998; Mani, Batni et al. 2001). Transgenic Xenopus laevis were generated by the REMI-sperm nuclear transplantation method (Kroll and Amaya 1996). All primary transgenic animals used in this study were genotyped at about four-weeks of age using PCR primers on different exons of the rhodopsin gene, which amplify the cDNA encoded by the transgene but not the endogenous rhodopsin gene (data not shown).
Fig. 1.
Human rhodopsin mutations superimposed on an alignment of Xenopus laevis and human rhodopsin sequences. In the aligned sequences, blackened boxes highlight identical amino-acids, while grey boxes highlight conserved amino-acid substitutions. Underlined are the predicted transmembrane spanning regions of rhodopsin. Underneath the aligned sequences are listed mutations occurring in human rhodopsin according to (Briscoe, Gaur et al. 2004). Mutations that cause autosomal dominant retinitis pigmentosa are shown in black: missense mutations as letters, nonsense mutations as asterisks and deletions as boxes. In blue are shown mutations that cause autosomal recessive retinitis pigmentosa. In red are shown mutations that cause congenital stationary night blindness. In grey are shown mutations found in humans that are presumed to be non-deleterious polymorphisms. An inverted arrowhead marks the position of the P23H mutation for which transgenic Xenopus laevis were generated and characterized.
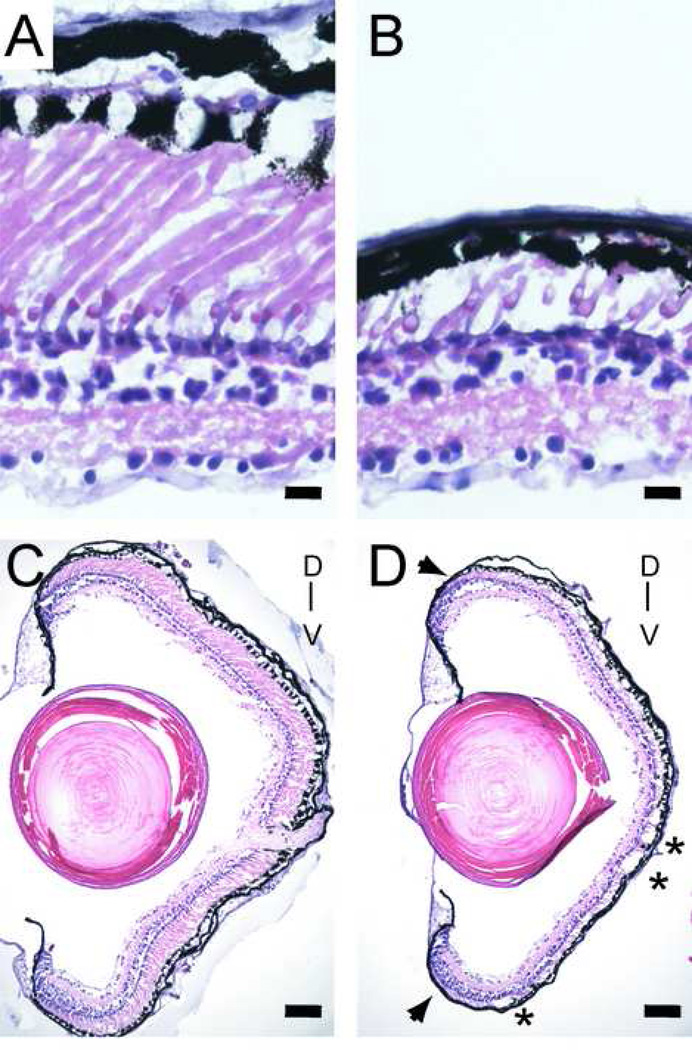
In this part of the study, the retinas from 87 animals generated by sperm-nuclear transplantation were analyzed when the animals were between 4 and 7 weeks of age, between stages 58 and 66 according to Nieuwkoop and Faber (Nieuwkoop and Faber 1956). Among these animals, 33 carried the P23H rhodopsin transgene, 6 the wild-type rhodopsin transgene, 16 only an EGFP transgene, all expressed under the same rhodopsin promoter. Additional controls included 32 aged matched animals found upon genotyping not to carry the transgenes. Hematoxylin/eosin stained cryosections showed that in control animals the outer nuclear layer of control Xenopus laevis retinas has one or two rows of photoreceptor nuclei, and nearly equal numbers of cone and rod photoreceptors (Figure 2 A). Cone photoreceptors can be identified both by the cone-shaped outer segments and the presence of an oil droplet within the inner segments. Among the tadpoles carrying the P23H transgene, a range of phenotypes was observed, ranging from outer nuclear layers that were indistinguishable from that of wild-type animals, to those manifesting severe rod photoreceptor degeneration (Figure 2 B). Both eyes of individual animals always showed very similar degrees of degeneration (data not shown). In many of the animals, the rod photoreceptor degeneration was more pronounced in the ventral half of the retina, and, occasionally, was characterized by patches manifesting more severe degeneration (Figure 2 C , D). The degeneration was most apparent in the central part of the retina, where photoreceptors are the oldest, but was also apparent as shortened outer segments near the ciliary margin, where photoreceptors are the youngest. The retinas of tadpoles expressing the wild-type rhodopsin or EGFP transgenes were undistinguishable from those of non-transgenic tadpoles (data not shown).
Fig. 2.
Rod photoreceptor degeneration in 4 week-old Xenopus laevis tadpoles expressing a P23H Xenopus laevis rhodopsin transgene, shown by hematoxylin eosin stain of cryosectioned retinas. (A) Retina of control tadpole has comparable numbers of rods and cones. (B) Retina of P23H transgenic tadpole has cones but a near absence of rods. (C) Low-power view of the same control retina shown in A. (D) Low-power view of the same P23H transgenic retina shown in B, which shows patchy degeneration (asterisks) preferentially in the ventral half of the retina. The outer segments of rods in P23H transgenic tadpoles are shorter than those in control animals, even in young rod photoreceptors near the ciliary marginal zone (arrows). D and V mark the dorsoventral axis. Scale bars: 10 µm in A-B, and 80 µm in C-D.
In order to rule out sectioning artifacts and to determine the fraction of animals with a ventral degeneration bias, plastic sections were examined (data not shown) and scored by blinded observers. Among the 13 animals in this group that carried the P23H transgene, 3 (23 %) were classified as severe, 4 (31 %) as mild to moderate, and 6 (46 %) as unaffected. All 3 of the severely affected animals had a ventral bias to the degeneration, while in none of the 4 mild to moderate was such ventral bias apparent. In 6 animals that carried the transgene and were scored as unaffected there was no evidence of any degeneration or of any photoreceptor asymmetry along the dorsoventral axis. Similarly, in an additional 9 animals scored as unaffected found subsequently not to carry the transgene, there also was no degeneration or photoreceptor asymmetry.
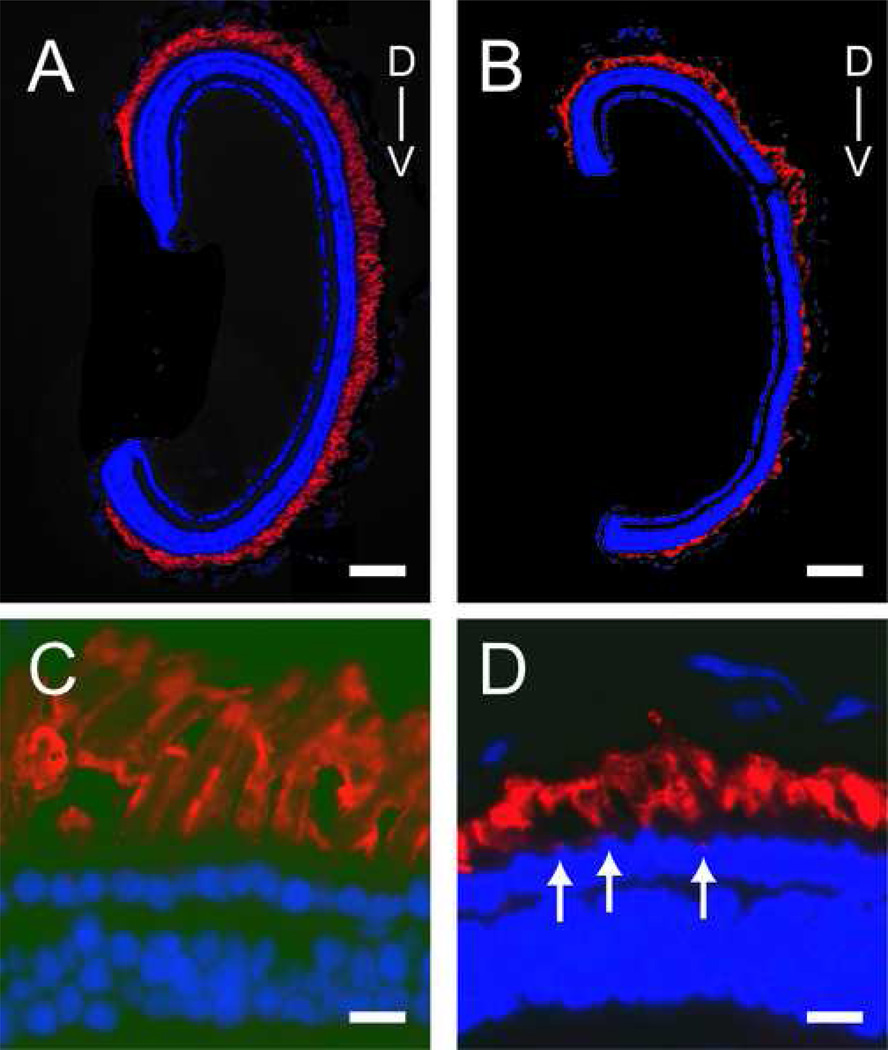
In order to examine the effect of the P23H transgene on outer segments, retina sections were stained with 4D2, an antibody that recognizes a highly conserved epitope near the carboxyl terminus of vertebrate rhodopsins (Molday. 1989). Low-power views of control (Figure 3 A) and severely affected P23H transgenic animals (Figure 3 B) show that some P23H rhodopsin transgenic animals have shorter outer segments throughout the retina and more severe degeneration preferentially in the ventral retina. When examined at higher power, the majority of the rhodopsin protein labeling can be found in the outer segments; however, some rhodopsin labeling is observed within photoreceptor inner segments of animals expressing the P23H rhodopsin transgene (Figure 3 C–D) .
Fig. 3.
Rhodopsin protein immunoreactivity in the retinas of control tadpoles and tadpoles expressing the P23H rhodopsin transgene. (A) Low power view of retina of a control tadpole stained with the 4D2 rhodopsin monoclonal antibody and the nuclear dye DAPI (blue). (B) Low power view of similarly stained retina from a P23H rhodopsin transgenic tadpole, showing preferential degeneration in the ventral retina. (C) Higher power view that shows photoreceptors of a control tadpole. (D) Higher power view from a P23H rhodopsin transgenic tadpole; note the presence of rhodopsin immunoreactivity near outer nuclear layer (arrows). D and V mark the dorsoventral axis in A-B. Scale bars 10 µm in A-B, and 100 µm in C-D.
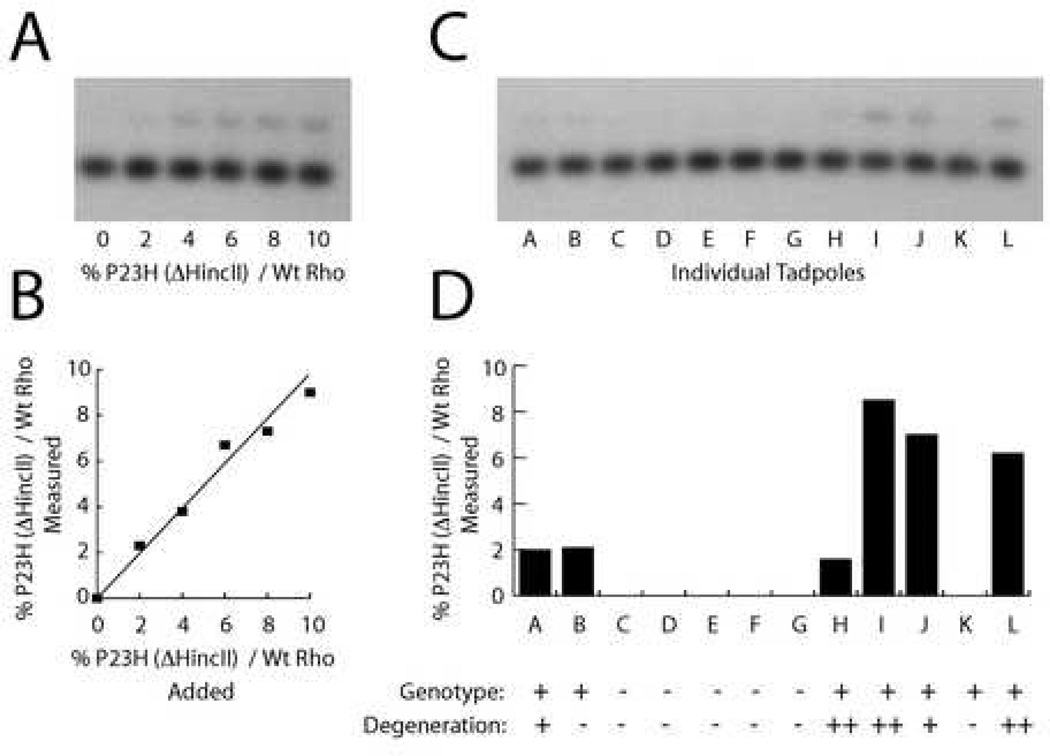
3.2. Low transgene expression causes rod photoreceptor degeneration
In order to determine whether the variability in the degeneration was caused by the amount of transgene expression, a second transgene was generated in which the Xenopus laevis P23H rhodopsin cDNA sequence was changed by a single base in order to delete a HincII restriction enzyme recognition site without altering the sequence of the encoded protein. A standard curve using varied ratios of cDNA templates with and without the HincII restriction site showed that competitive PCR (Becker-Andre and Hahlbrock 1989) may be used to determine relative cDNA levels (Figure 4 A–B). Tadpoles made to express the P23H(HincII) transgene were examined at 5 weeks of age. One eye was used for competitive PCR determination of the relative expression of transgenic to endogenous rhodopsin cDNA, while the other eye was used for a histological determination of rod photoreceptor degeneration severity. In all of the transgenic animals examined, transgenic rhodopsin expression was below 10% of endogenous rhodopsin expression (Figure 4 C–D). In animals with transgene expression between 2 and 10 % of endogenous rhodopsin, the degeneration was scored as mild to severe, with one exception of an animal with expression around 2 % in which no degeneration was apparent. In all of the animals that were subsequently found not to carry the transgene, as well as one animal that did carry the transgene, transgene expression level was found to be below 2%, the detection limit of the assay, and, in these animals no rod photoreceptor degeneration was evident. Thus, the animals with higher expression of the P23H rhodopsin transgene have more severe degeneration, and transgene expression of ~2% relative to endogenous rhodopsin is sufficient to cause a mild rod photoreceptor degeneration phenotype.
Fig. 4.
Expression of transgenic P23H rhodopsin mRNA as a percent relative to endogenous Xenopus laevisrhodopsin. (A) Competitive PCR using cDNA templates for P23H(?HincII) rhodopsin and wild-type rhodopsin in 0–10 % ratios. (B) Relationship between added and measured rhodopsin ratios from the blot shown in A. (C) Competitive PCR using cDNAs generated from individual eyes from tadpoles generated during the injection of the P23H(?HincII) transgene. (D) Expression ratio of P23H((?HincII) transgenic rhodopsin relative to wildtype endogenous rhodopsin for the tadpole eyes shown in C, calculated using standard curve shown in B. Below the graph are genotyping results showing whether the tadpoles carried the transgene, as well as the extent of photoreceptor degeneration observed in the other eye of the same tadpoles.
3.3. Rod photoreceptor degeneration resulting from the Xenopus P23H rhodopsin transgene is modified by light
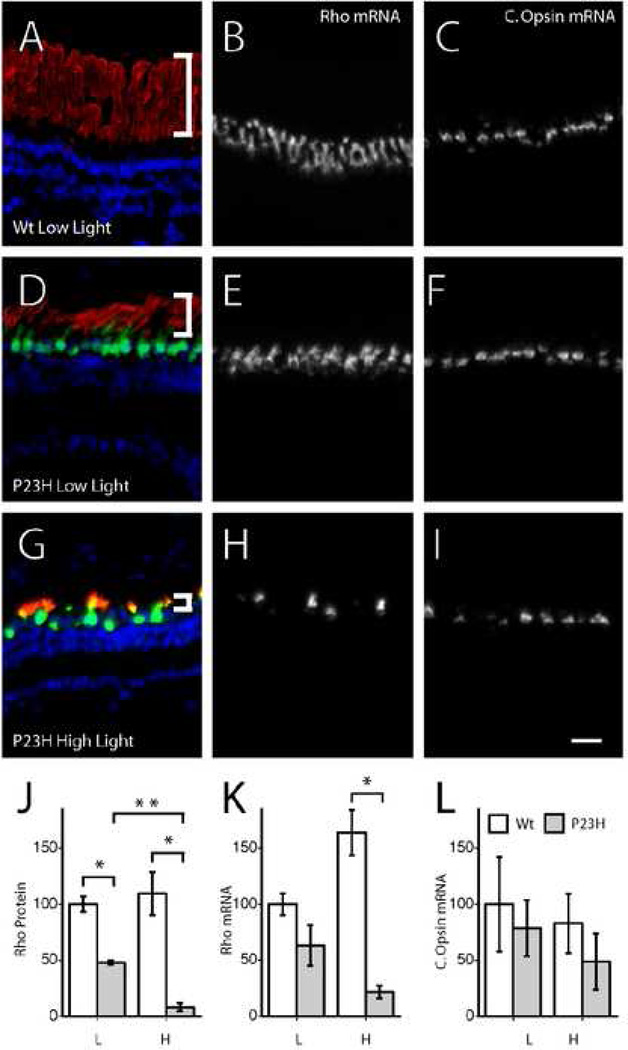
Previous studies had shown that a Xenopus laevis P23H rhodopsin transgene that also has a M13F mutation and a mammalian rhodopsin C-tail, appeared to cause rod photoreceptor degeneration through a mechanism that is unaffected by light, since the rod photoreceptor degeneration was similar in dark reared animals and was not altered by an additional mutation, K296R, that renders rhodopsin incapable of binding chromophore (Tam, Xie et al. 2006; Tam and Moritz 2007). However, mammalian P23H transgenes expressed in mice (Naash, Hollyfield et al. 1993), rats (Organisciak, Darrow et al. 2003; Jozwick, Valter et al. 2006), frogs (Tam and Moritz 2007) and even flies (Galy, Roux et al. 2005) exhibit a rod photoreceptor degeneration that is exacerbated by light. Such light sensitivity could explain the preferential degeneration of the ventral retina we observe with our Xenopus laevis P23H rhodopsin transgenic tadpoles. To explore whether the degeneration produced by our P23H transgenes was affected by light, we generated and analyzed a transgenic line carrying a Xenopus laevis P23H rhodopsin transgene together with a GFP transgene, both under the control of the same 1.3kb rhodopsin promoter. In this line, the two transgenes must have integrated into the same genomic location since all progeny from this founder that had photoreceptor expression of GFP, and none that did not, underwent the characteristic rod photoreceptor degeneration with ventral preference (data not shown). To determine whether the rod photoreceptor degeneration phenotype was modified by light, transgenic and non-transgenic siblings were raised under constant light and constant darkness as well as under cycling (12:12) light conditions: low (~10 lux), medium (~300 lux), and high (~600 lux). Animals that were raised in either constant light or constant dark failed to thrive and could not be used in analyses. Within the animals raised in the three cycling light conditions, animals reached comparable metamorphic stages within four weeks, showing that these light rearing conditions, as opposed to constant light or constant darkness, do not affect the rate of growth of the tadpoles (data not shown). Semi-quantitative analysis of the rod photoreceptor degeneration under these light conditions based on remaining rhodopsin and GFP protein expression (Fig. 5A,D,G,J, and data not shown), as well as remaining rhodopsin mRNA expression (Fig. 5B, E, H, K), show a clear dependence of the photoreceptor degeneration on light. While rod photoreceptor degeneration was present in P23H animals raised under low light conditions, it was more severe when raised under high light intensities (Fig. 5J–K) and intermediate under medium light intensities (data not shown). The degeneration was more evident at the level of rhodopsin protein than mRNA, consistent with the observation that outer segment shortening precedes photoreceptor death. A qualitative analysis based on outer segment length also supports the same conclusion. Interestingly, even in retinas with severe rod loss, as evident by the loss of cells expressing rhodopsin mRNA, there was no immediate loss of cone photoreceptors as detected by cells expressing the red cone opsin mRNA (Fig. 5C, F, I, L). However, whether there is more subtle atrophy of cones or later death of cones will need to be addressed separately.
Fig. 5.
Rod photoreceptor degeneration caused by P23H rhodopsin transgene is modified by light. Triple label of 4D2 rhodopsin antibody labeling (red), GFP antibody labeling (green) and DAPI nuclear labeling (blue) in non-transgenic siblings raised under low light (A) and transgenic siblings raised under low (D) and high (G) light intensities. White brackets show the shortening of outer segments occurring during rod photoreceptor degeneration. Rhodopsin mRNA in non-transgenic siblings raised under low light (B) and transgenic siblings raised under low (E) and high (H) light intensities. Red cone opsin mRNA in non-transgenic siblings raised under low light (C) and transgenic siblings raised under low (F) and high (I) light intensities. Quantification of rhodopsin protein (J), rhodopsin mRNA (K) and red cone opsin mRNA (L) in transgenic and non-transgenic siblings raised under low and high light intensities. * and ** mark groups that are significantly different at p<0.05 and p<0.01 levels. Scale bar 50 µm.
4. Discussion
4.1. P23H Xenopus laevis rhodopsin transgene causes rod photoreceptor degeneration when expressed at low levels
Our study demonstrates that the P23H missense mutation alone within the Xenopus laevis rhodopsin sequence is sufficient to produce the degeneration of rod photoreceptors. Because not all amino acids that cause ADRP in humans are conserved in Xenopus laevis, and, indeed, an amino acid substitution, V137M, that causes ADRP in humans occurs naturally within the frog rhodopsin, we chose to study the P23H mutation in the context of the unaltered Xenopus laevis rhodopsin sequence, without introducing epitope-tags that would have enabled discrimination between the transgenic and the endogenous rhodopsin proteins, as others have done in studying rhodopsin mutations in transgenic frogs (Tam and Moritz 2006; Tam, Xie et al. 2006; Tam and Moritz 2007). The reluctance to include an epitope tag was also based in part on a previous study in which a mouse rhodopsin transgene encoding an epitope-tag consisting of C-terminus amino acid substitutions that are normally found in bovine rhodopsin sequence and which are not associated with ADRP in humans appears to produce rod photoreceptor degeneration in mice even when expressed at relatively low levels (Tan, Wang et al. 2001).
In this study, the rhodopsin transgenes caused photoreceptor pathology when they were expressed at levels between 2 and 10% relative to the level of endogenous rhodopsin. This value matches well previous studies with P23H rhodopsin transgenes in Xenopus laevis (Tam, Xie et al. 2006; Tam and Moritz 2007), despite that fact that their studies measured relative transgene expression at the level of protein, while ours at the level of mRNA. Since the endogenous rhodopsin protein content decreases as outer segments shorten during disease progression, and different rhodopsin mutants with or without epitope tags are likely to have different protein stabilities and propensities to aggregate, protein and mRNA measures might be expected to differ slightly. Indeed, when we measured total rhodopsin levels by semi-quantitative imaging in animals raised at low light levels, significant decreases between P23H rhodopsin transgenic relative to control siblings in the amount of protein were larger and statistically significant, while the levels of mRNA were only minimally reduced and not statistically significant. The main point is that our studies agree with previous studies (Tam and Moritz 2006; Tam, Xie et al. 2006; Tam and Moritz 2007) in showing that ADRP associated mutations are able to cause rod photoreceptor degeneration when expressed at levels well below those of the endogenous rhodopsin. That such low levels of transgene expression can produce rod photoreceptor degeneration support the long-standing view that these mutations are not exerting their pathology by acting in a dominant negative manner, but rather by acting in a gain-of-function manner. However, studying the degeneration caused by P23H rhodopsin in the context of varying amounts of the wild-type rhodopsin, as has been done in mice (Wilson and Wensel 2003), may be the best way to determine whether the mutant rhodopsin is indeed acting through a gain-of-function mechanism.
4.2 Rod photoreceptor degeneration is most pronounced in the ventral half of the retina and is sensitive to light
In many tadpoles with photoreceptor pathology, the ventral half of the retina was more severely affected than the dorsal half. Such preferential degeneration in the ventral retina has been observed in some patients with P23H as well as select other rhodopsin mutations, but curiously not in all rhodopsin mutations, suggesting heterogeneity in the mechanism by which rhodopsin mutations cause rod photoreceptor degeneration (Heckenlively, Rodriguez et al. 1991; Stone, Kimura et al. 1991; Gal, Apfelstedt-Sylla et al. 1997; Cideciyan, Hood et al. 1998; To, Adamian et al. 2002). It has been suggested that this ventral susceptibility might reflect the fact that the ventral retina is exposed to higher levels of light (Cideciyan, Hood et al. 1998). Indeed, mice expressing a mouse P23H rhodopsin transgene are more susceptible to light induced rod photoreceptor degeneration (Wang, Lam et al. 1997) and their degeneration is accelerated by light preferentially in the ventral retina (Naash, Peachey et al. 1996). We show here that in Xenopus laevis as well, the P23H mutation in the context of an otherwise unchanged Xenopus laevis rhodopsin causes a rod photoreceptor degeneration that is both more severe in the ventral retina, and is also accelerated by higher light intensities. It is interesting that the P23H mutation within a Xenopus laevis rhodopsin in the context of an altered C-tail and an additional mutation near P23H, M13F, led to a degeneration that is unaffected by rearing in darkness or the presence of an amino acid substitution, K296R, which renders the transgenic protein incapable of binding chromophore. There are multiple explanations that may account for this apparent discrepancy of results. The previous studies of P23H rhodopsin transgenic Xenopus laevis used dark-rearing rather cycling light of variable intensity to address the sensitivity to light. Since dark rearing is expected to alter circadian rythms, cycling light of variable intensity is a more reliable method to address the effect of light. Indeed, we were unable to compare dark rearing to low cycling light because of the failure of Xenopus laevis tadpoles raised under constant darkness to thrive out until the metamorphic stages at which we perform our analyses. There are many ways that light might be affecting the rate of degeneration. The deleterious effect of light is not necessarily due to increased visual transduction, but rather may be due to altered metabolism by photoreceptors, pigment epithelium, or both, or a related increased turnover rate of rhodopsin. Indeed, we show a trend to increased rhodopsin mRNA expression but not protein in wildtype animals exposed to higher light conditions that would be consistent with an increased turnover rate of rhodopsin induced by light. Another possible reason that might explain the discrepancy in terms of light sensitivity between our and previous studies has to do with the fact that previous studies of rhodopsin transgenes causing rod photoreceptor degeneration in Xenopus laevis have used either mammalian rhodopsin sequences (Tam and Moritz 2007) are alternatively epitope-tagged versions of Xenopus laevis rhodopsin constructs that had in addition to the disease causing P23H mutation, an amino acid substitution near P23H, M13F, and a several amino acid substitution/deletion in the C-terminus made so that the transgenic proteins could be distinguished from the non-transgenic rhodopsin by two antibodies (Tam and Moritz 2006; Tam and Moritz 2007).
In a study of numerous pathogenic rhodopsin constructs transfected into human embryonic kidney cells, P23H rhodopsin was shown to belong to a group designated Class-II mutations, which failed to be transported properly to membranes and instead remained in the endoplasmic reticulum/Golgi complex (Sung, Schneider et al. 1991). Analyses of transgenic mice expressing a human rhodopsin containing the P23H mutation supported the notion that the mutant rhodopsin was similarly mislocalized within photoreceptors (Olsson, Gordon et al. 1992; Roof, Adamian et al. 1994). However, studies with a mouse rhodopsin containing the P23H mutation also in transgenic mice showed proper localization to the outer segment (Naash, Hollyfield et al. 1993; Wu, Ting et al. 1998). A recent study of several mammalian P23H rhodopsins and the epitope-tagged Xenopus laevis P23H rhodopsin in transgenic Xenopus laevis showed that these mutant proteins varied greatly in their tendencies to accumulate in the endoplasmic reticulum or go to the outer segment, and that the degree of accumulation was related to several factors including the susceptibility to light probably while the protein was is in or near the endoplasmic reticulum. However, all the constructs used in these studies, including the various Xenopus laevis P23H rhodopsins, had a mammalian C-terminus. The native Xenopus laevis rhodopsin C-terminus, which is highly conserved among frogs and toads (Fyhrquist, Donner et al. 1998), is the most divergent region relative to mammalian rhodopsins. The amphibian C-terminus may be optimal for trafficking at lower temperatures, leading to the rapid export and subsequent proteolytic degradation of misfolded P23H rhodopsin, resulting in the more rapid clearing of misfolded proteins. The M13F mutation near the P23H mutation may also alter the secondary structure of the protein in a way as to make it behave differently in terms of stability or endoplasmic reticulum export. A methionine at position 13 is conserved among frogs and toads (Fyhrquist, Donner et al. 1998). While M13F is not a known ADRP-causing mutation, it is near several other ADRP-causing mutations, and there are more than one divergent amino acid in the C-tail that may have co-evolved. In the case of transgenic mice, the reason why the human but not the mouse P23H rhodopsins could be found accumulating in the endoplasmic reticulum may have been due with subtle structural changes that affect their stability or their export from the endoplasmic reticulum. Such subtle structural changes are likely to result from non-pathogenic mutations throughout the entire rhodopsin sequence. Additional studies are needed to address whether or not amino acid substitutions that do not in of themselves cause rod photoreceptor degeneration are affecting the susceptibility to, or the rate or type of rod photoreceptor degeneration.
Acknowledgements
The authors thank Don Zack and Ruben Adler for providing critical advice on the manuscript. This work was supported in part by NIH grant R21 NS053696-01, and Foundation Fighting Blindness grant C-NP-0707-0418.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ayuso C, Trujillo MJ, et al. “Novel rhodopsin mutation in an autosomal dominant retinitis pigmentosa family: phenotypic variation in both heterozygote and homozygote Val137Met mutant patients”. Hum Genet. 1996;98(1):51–54. doi: 10.1007/s004390050158. [DOI] [PubMed] [Google Scholar]
- Becker-Andre M, Hahlbrock K. “Absolute mRNA quantification using the polymerase chain reaction (PCR). A novel approach by a PCR aided transcript titration assay (PATTY)”. Nucleic Acids Res. 1989;17(22):9437–9446. doi: 10.1093/nar/17.22.9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughman JA, Conneally PM, et al. “Population genetic studies of retinitis pigmentosa”. Am J Hum Genet. 1980;32(2):223–235. [PMC free article] [PubMed] [Google Scholar]
- Briscoe AD, Gaur C, et al. “The spectrum of human rhodopsin disease mutations through the lens of interspecific variation”. Gene. 2004;332:107–118. doi: 10.1016/j.gene.2004.02.037. [DOI] [PubMed] [Google Scholar]
- Bunker CH, Berson EL, et al. “Prevalence of retinitis pigmentosa in Maine”. Am J Ophthalmol. 1984;97(3):357–365. doi: 10.1016/0002-9394(84)90636-6. [DOI] [PubMed] [Google Scholar]
- Cideciyan AV, Hood DC, et al. “Disease sequence from mutant rhodopsin allele to rod and cone photoreceptor degeneration in man”. Proc Natl Acad Sci U S A. 1998;95(12):7103–7108. doi: 10.1073/pnas.95.12.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja TP, McGee TL, et al. “A point mutation of the rhodopsin gene in one form of retinitis pigmentosa”. Nature. 1990;343(6256):364–366. doi: 10.1038/343364a0. [DOI] [PubMed] [Google Scholar]
- Fyhrquist N, Donner K, et al. “Rhodopsins from three frog and toad species: sequences and functional comparisons”. Exp Eye Res. 1998;66(3):295–305. doi: 10.1006/exer.1997.0430. [DOI] [PubMed] [Google Scholar]
- Gal A, Apfelstedt-Sylla E, et al. “Rhodopsin mutations in inherited retinal dystrophies and dysfunctions”. Prog Retin Eye Res. 1997;16:51–79. [Google Scholar]
- Galy A, Roux MJ, et al. “Rhodopsin maturation defects induce photoreceptor death by apoptosis: a fly model for RhodopsinPro23His human retinitis pigmentosa”. Hum Mol Genet. 2005;14(17):2547–2557. doi: 10.1093/hmg/ddi258. [DOI] [PubMed] [Google Scholar]
- Heckenlively JR, Rodriguez JA, et al. “Autosomal dominant sectoral retinitis pigmentosa. Two families with transversion mutation in codon 23 of rhodopsin”. Arch Ophthalmol. 1991;109(1):84–91. doi: 10.1001/archopht.1991.01080010086038. [DOI] [PubMed] [Google Scholar]
- Heckenlively JR, Yoser SL, et al. “Clinical findings and common symptoms in retinitis pigmentosa”. Am J Ophthalmol. 1988;105(5):504–511. doi: 10.1016/0002-9394(88)90242-5. [DOI] [PubMed] [Google Scholar]
- Jozwick C, Valter K, et al. “Reversal of functional loss in the P23H-3 rat retina by management of ambient light”. Exp Eye Res. 2006;83(5):1074–1080. doi: 10.1016/j.exer.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Kemp CM, Jacobson SG, et al. “Abnormal rod dark adaptation in autosomal dominant retinitis pigmentosa with proline-23-histidine rhodopsin mutation”. Am J Ophthalmol. 1992;113(2):165–174. doi: 10.1016/s0002-9394(14)71529-6. [DOI] [PubMed] [Google Scholar]
- Knox BE, Schlueter C, et al. “Transgene expression in Xenopus rods”. FEBS Lett. 1998;423(2):117–121. doi: 10.1016/s0014-5793(98)00018-0. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Amaya E. “Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation”. Development. 1996;122(10):3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- Lewin AS, Drenser KA, et al. “Ribozyme rescue of photoreceptor cells in a transgenic rat model of autosomal dominant retinitis pigmentosa”. Nat Med. 1998;4(8):967–971. doi: 10.1038/nm0898-967. [DOI] [PubMed] [Google Scholar]
- Machida S, Kondo M, et al. “P23H rhodopsin transgenic rat: correlation of retinal function with histopathology”. Invest Ophthalmol Vis Sci. 2000;41(10):3200–3209. [PubMed] [Google Scholar]
- Mani SS, Batni S, et al. “Xenopus rhodopsin promoter. Identification of immediate upstream sequences necessary for high level, rod-specific transcription”. J Biol Chem. 2001;276(39):36557–36565. doi: 10.1074/jbc.M101685200. [DOI] [PubMed] [Google Scholar]
- Marsh-Armstrong N, Huang H, et al. “Asymmetric growth and development of the Xenopus laevis retina during metamorphosis is controlled by type III deiodinase”. Neuron. 1999;24(4):871–878. doi: 10.1016/s0896-6273(00)81034-x. [DOI] [PubMed] [Google Scholar]
- Molday SR. “Monoclonal antibodies to rhodopsin and other proteins of rod outer segments”. Prog Retin Eye Res. 1989;8:173–209. [Google Scholar]
- Naash MI, Hollyfield JG, et al. “Simulation of human autosomal dominant retinitis pigmentosa in transgenic mice expressing a mutated murine opsin gene”. Proc Natl Acad Sci U S A. 1993;90(12):5499–5503. doi: 10.1073/pnas.90.12.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naash ML, Peachey NS, et al. “Light-induced acceleration of photoreceptor degeneration in transgenic mice expressing mutant rhodopsin”. Invest Ophthalmol Vis Sci. 1996;37(5):775–782. [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg Till the End of Metamorphosis. New York: Elsevier/North-Holland; 1956. [Google Scholar]
- Olsson JE, Gordon JW, et al. “Transgenic mice with a rhodopsin mutation (Pro23His): a mouse model of autosomal dominant retinitis pigmentosa”. Neuron. 1992;9(5):815–830. doi: 10.1016/0896-6273(92)90236-7. [DOI] [PubMed] [Google Scholar]
- Organisciak DT, Darrow RM, et al. “Susceptibility to retinal light damage in transgenic rats with rhodopsin mutations”. Invest Ophthalmol Vis Sci. 2003;44(2):486–492. doi: 10.1167/iovs.02-0708. [DOI] [PubMed] [Google Scholar]
- Roof DJ, Adamian M, et al. “Rhodopsin accumulation at abnormal sites in retinas of mice with a human P23H rhodopsin transgene”. Invest Ophthalmol Vis Sci. 1994;35(12):4049–4062. [PubMed] [Google Scholar]
- Stone EM, Kimura AE, et al. “Regional distribution of retinal degeneration in patients with the proline to histidine mutation in codon 23 of the rhodopsin gene”. Ophthalmology. 1991;98(12):1806–1813. doi: 10.1016/s0161-6420(91)32046-3. [DOI] [PubMed] [Google Scholar]
- Sung CH, Schneider BG, et al. “Functional heterogeneity of mutant rhodopsins responsible for autosomal dominant retinitis pigmentosa”. Proc Natl Acad Sci U S A. 1991;88(19):8840–8844. doi: 10.1073/pnas.88.19.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam BM, Moritz OL. “Characterization of rhodopsin P23H–induced retinal degeneration in a Xenopus laevis model of retinitis pigmentosa”. Invest Ophthalmol Vis Sci. 2006;47(8):3234–3241. doi: 10.1167/iovs.06-0213. [DOI] [PubMed] [Google Scholar]
- Tam BM, Moritz OL. “Dark rearing rescues P23H rhodopsin-induced retinal degeneration in a transgenic Xenopus laevis model of retinitis pigmentosa: a chromophore-dependent mechanism characterized by production of N-terminally truncated mutant rhodopsin”. J Neurosci. 2007;27(34):9043–9053. doi: 10.1523/JNEUROSCI.2245-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam BM, Xie G, et al. “Mislocalized rhodopsin does not require activation to cause retinal degeneration and neurite outgrowth in Xenopus laevis”. J Neurosci. 2006;26(1):203–209. doi: 10.1523/JNEUROSCI.3849-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E, Wang Q, et al. “The relationship between opsin overexpression and photoreceptor degeneration”. Invest Ophthalmol Vis Sci. 2001;42(3):589–600. [PubMed] [Google Scholar]
- To K, Adamian M, et al. “Histopathologic study of variation in severity of retinitis pigmentosa due to the dominant rhodopsin mutation Pro23His”. Am J Ophthalmol. 2002;134(2):290–293. doi: 10.1016/s0002-9394(02)01545-3. [DOI] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. “Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate”. Genes Dev. 1994;8(12):1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Wang M, Lam TT, et al. “Expression of a mutant opsin gene increases the susceptibility of the retina to light damage”. Vis Neurosci. 1997;14(1):55–62. doi: 10.1017/s0952523800008750. [DOI] [PubMed] [Google Scholar]
- Weleber RG, Gregory K. Retinitis pigmentosa and allied disorders. In: Ryan SJ, Hinton R, Schachat AP, Wilkinson P, editors. Retina. Philoadelphia: Elsevier Mosby; 2006. [Google Scholar]
- Wilson JH, Wensel TG. “The nature of dominant mutations of rhodopsin and implications for gene therapy”. Mol Neurobiol. 2003;28(2):149–158. doi: 10.1385/MN:28:2:149. [DOI] [PubMed] [Google Scholar]
- Wu TH, Ting TD, et al. “Opsin localization and rhodopsin photochemistry in a transgenic mouse model of retinitis pigmentosa”. Neuroscience. 1998;87(3):709–717. doi: 10.1016/s0306-4522(98)00173-0. [DOI] [PubMed] [Google Scholar]