Abstract
Low bone mineral density (BMD) is a risk factor of osteoporosis. Osteoporosis is more prevalent in the females than the males. So far, the pathophysiological mechanisms underlying osteoporosis are unclear. Peripheral blood monocytes (PBM) are precursors of bone-resorbing osteoclasts. This study aims to identify PBM-expressed proteins (genes) influencing hip BMD in humans.
We utilized three independent study cohorts (N=34, 29, 40), including premenopausal Caucasians with discordant hip BMD. We studied PBM proteome-wide protein expression profiles in Cohort 1 and identified 57 differentially expressed proteins (DEPs) between low vs. high BMD subjects. One protein gelsolin (GSN), after validation by Western blotting, was subject to follow-up. We compared GSN mRNA level in PBM between low vs. high BMD subjects in Cohorts 2 and 3. We genotyped SNPs across GSN in 2,286 unrelated Caucasians (Cohort 4) and 1,627 Chinese (Cohort 5), and tested association with hip BMD in the females and males, respectively.
We discovered and validated that GSN protein expression level in PBM was down-regulated 3.0-fold in low vs. high BMD subjects (P<0.05). Down-regulation of GSN in PBM in low BMD subjects was also observed at mRNA level in both Cohorts 2 and 3. We identified that SNP rs767770 was significantly associated with hip BMD in female Caucasians (P=0.0003) only. Integrating analyses of the datasets at DNA, RNA, and protein levels from female Caucasians substantiated that GSN is highly significant for hip BMD (P=0.0001).
We conclude that GSN is a significant gene influencing hip BMD in female Caucasians.
Keywords: Bone mineral density, Monocyte, Gelsolin, Integration analysis
Osteoporosis is a worldwide public health problem. It is characterized by low bone mineral density (BMD) and high risk to osteoporotic fractures. Hip osteoporotic fracture is the most serious consequence of osteoporosis, which is associated with significant morbidity and mortality [1–4]. White population has higher hip fracture rate than Asian and Black populations [3, 5]. Prevalence of osteoporosis and incidence of osteoporotic fracture are much higher in women than in men [6].
BMD is a gold standard for diagnosing osteoporosis [3, 7]. Heritability of BMD is estimated to be 0.5–0.9 [8–11]. Extensive genetic and genomic studies, conducted over the past decade, identified a number of genes associated with BMD variation in humans. Collectively, however, these implicated genes explain no more than 10% of BMD variation in any individual human population. So the basis for the majority of variation in BMD that is genetically determined still awaits identification.
In humans, BMD increases with age till around 25 when peak BMD is attained and maintained thereafter until around 50 (or menopause in females). Then, BMD decreases gradually with aging. Peak BMD level is a strong predictor of BMD value and osteoporosis risk in later life.
Osteoporosis is attributed to bone resorption by osteoclasts exceeding bone formation by osteoblasts [12–14]. Peripheral blood monocyte (PBM) can serve as precursors of osteoclasts, migrate from circulation to bone surface, subsequently fuse into immature multinuclear osteoclasts and be activated into mature osteoclasts to resorb bone [15–18]. PBM also secretes cytokines participating in bone metabolism, like interleukin-1, interleukin-6, tumor necrosis factor-α and transforming growth factor-α [19–21]. Therefore, studies on PBM in vivo in humans may provide novel insight into pathophysiology of osteoporosis.
Proteins are major executors of gene functions in biological organisms. Alteration of cellular protein expression levels may reflect changes in physiological conditions. Quantitative proteomic study, through identifying and quantifying proteins at a proteome-wide scale, has proved as a novel, powerful, and promising method to discover disease biomarkers in bone field [22].
Utilizing the quantitative proteomics methodology, and a strategy of multi-disciplinary and integrative studies, the present work aims to identify proteins (genes) important to osteoporosis in humans. Specifically, identification of genes important to osteoporosis was based on studies at three molecule levels (protein, RNA, and DNA) and based on evidence from multiple independent study cohorts.
MATERIALS AND METHODS
Human subjects
The study was approved by appropriate Institutional Review Boards. Signed informed-consent documents were obtained from all study participants. Basic characteristics of the five study cohorts involved in the present work was summarized in Table 1. Strict criteria were applied to exclude non-genetic factors that might affect bone metabolism and BMD determination. The exclusion criteria include chronic disorders involving vital organs (heart, lung, liver, kidney, and brain), serious metabolic diseases (such as diabetes, hypo- or hyper- parathyroidism, hyperthyroidism), other skeletal diseases (such as Paget’s disease, osteogenesis imperfecta, rheumatoid arthritis), chronic use of drugs affecting bone metabolism (such as corticosteroid therapy, anticonvulsant drugs, estrogens, thyroid hormone), and malnutrition conditions (such as chronic diarrhea, chronic ulcerative colitis). For cohorts 1–3, we adopted additional exclusion criteria to minimize effects of any known disorders or conditions that might affect gene expression of PBM. Those disorders and conditions include influenza (within one week of recruitment), autoimmune or autoimmune-related diseases (such as systemic lupus erythematosus), and immune-deficiency conditions (such as AIDS), hematopoietic, and lymphoreticular malignancies (such as leukemia), etc.
Table 1.
Basic Characteristics of the Study Cohorts
| Cohort 1 | Cohort 2 | Cohort 3 | Cohort 4 | Cohort 5 | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Ethnicity | Caucasian | Caucasian | Caucasian | Caucasian | Chinese | |||
|
| ||||||||
| Category | High BMD | Low BMD | High BMD | Low BMD | High BMD | Low BMD | Random Population | |
| *Sample Size | 17 | 17 | 15 | 14 | 20 | 20 | 2,286(798/930/558) | 1,627(608/217/802) |
| Age (years) | 51.8(2.2) | 50.2(1.9) | 51.0(1.8) | 50.1(2.1) | 41.7(1.8) | 42.3 (1.8) | 50.9(13.8) | 34.5(13.2) |
| Height (cm) | 162.7(5.9) | 167.4(6.6) | 164.0(4.8) | 167.1(7.4) | 164.9(6.3) | 163.8(7.0) | 166.3(8.5) | 164.3(8.2) |
| Weight (kg) | 76.3(16.3) | 65.3(7.7) | 83.9(19.3) | 65.1(8.4) | 83.9(17.4) | 60.9(11.4) | 75.3(17.5) | 60.1(10.5) |
| **Hip BMD | 1.3(0.5) | −1.0(0.3) | 1.6(0.5) | −0.9(0.4) | 1.3(0.7) | −1.1(0.5) | 0.968(0.175) | 0.920 (0.134) |
Note: Presented are mean (standard deviation).
The numbers of premenopausal/postmenopausal/male subjects in cohorts 4 and 5 were presented in brackets.
Hip BMD is presented in Z-score for cohorts 1–3 and in g/cm2 for cohorts 4 and 5, respectively. Z-score is defined as the number of standard deviations a subject’s BMD differs from the average BMD of their age-, gender-, and ethnicity- matched population.
Cohort 1 for protein expression study
Based on archived database of Caucasian populations in Kansas City, Missouri and Omaha, Nebraska in the Midwestern area of United States, we recruited 34 unrelated premenopausal Caucasian women with extremely discordant BMD. They were composed of 17 subjects with high hip BMD and 17 subjects with low hip BMD (Z-score: 1.3 ± 0.5 vs. −1.0 ±0.3; Mean ± S.D.), from top- and bottom- 20% of BMD distribution.
Cohorts 2 and 3 for mRNA expression study
Cohort 2
Based on an archived dataset of Caucasian population from Kansas City and its vicinity areas in the Midwest of US, we recruited 29 unrelated premenopausal Caucasian women with extremely discordant BMD. They were composed of 15 subjects with high hip BMD and 14 subjects with low hip BMD (Z-score: 1.6 ± 0.5 vs. −0.9 ± 0.4; Mean ± S.D.), from top- and bottom- 20% of BMD distribution.
Cohort 3
Based on an archived dataset of Caucasian population from Omaha, Nebraska in the US, we recruited 40 unrelated premenopausal Caucasian women with extremely discordant BMD. They were composed of 20 subjects with high hip BMD and 20 subjects with low hip BMD (Z-score: 1.3 ± 0.7 vs. −1.1 ± 0.5; Mean ± S.D.), from top- and bottom- 20% of BMD distribution.
Cohorts 4 and 5 for SNP association study
Cohort 4 is composed of 2,286 (1,728 females, 558 males) unrelated adult Caucasians recruited from Kansas City, Missouri and Omaha, Nebraska and their surrounding areas in United States. This cohort includes 798 premenopausal women and 930 postmenopausal women. The age of menopause ranged from 19–62 yrs, with an average of 48.3 (6.2, S.D.) yrs. Forty-nine percent of the postmenopausal women had not become menopausal until after 50 yrs.
Cohort 5 is composed of 1,627 (825 females, 802 males) unrelated adult Chinese recruited from the cities of Xi’an and Changsha and their surrounding areas in China. This cohort includes 608 premenopausal women and 217 postmenopausal women.
BMD measurement
Total hip BMD (g/cm2) was measured with Hologic QDR 4500 dual energy X-ray absorptiometer scanners (Hologic, Waltham, MA, USA). It is a combined value at femoral neck, trochanter, and intertrochanter. The machines were calibrated daily with a control vertebral phantom. The coefficient of variation for repeated total hip BMD measurements was about 1.34%.
Peripheral blood monocyte (PBM) isolation
For cohorts 1–3, 30–60 ml peripheral blood was drawn from each subject by certificated phlebotomist. EDTA was used as anti-coagulant. The fresh blood samples were processed instantly for PBM isolation by experienced technicians. First, peripheral blood mononuclear cells (PBMC) were isolated from whole blood using density gradient centrifugation with Histopaque-1077 (Sigma-Aldrich, Cat. No.10771). Then, PBM isolation was performed with the Monocyte Isolation Kit II (Miltenyi Biotec, Cat. No. 130-091-153) following manufacturer’s recommendation. Flow cytometry analyses showed that the purity of isolated monocyte samples is >90%.
PBM protein extraction and proteome-wide protein expression profiling
For each subject in cohort 1, PBM total proteins were extracted using ProteoExtract™ Complete Mammalian Proteome Extraction Kit (Calbiochem, Cat. No.539779). Each total protein sample (20 μg) was precipitated with ProteoExtract® Protein Precipitation Kit (Calbiochem, Cat. No. 539180), and digested to peptides by trypsin with routine procedure [23]. Peptides solution were concentrated to approximately 20μl, and mixed with 4μl of 0.5% formic acid (FA) and 16μl of 100 fmol yeast alcohol dehydrogenase I digest standard (ADH1, Waters, Cat. No.186002328).
PBM proteomes were profiled using a method of LC-nano-ESI-MSE [24], through nanoAcquity Ultra Performance Liquid Chromatographer coupled with Synapt High Definition Mass Spectrometer (HDMS) (Waters Corporation). Proteome data acquisition was controlled by MassLynx 4.1 software (Waters). Briefly, the protein digests (~ 500 ng) were injected into a BEH C18 75 μm × 150 mm analytical column (Waters), and separated by solvent A (water with 0.1% FA) and solvent B (acetonitrile with 0.1% FA) at a flow rate of 0.3 μl/min using a gradient of 2-hours as follows: 3% B initial, 10% B at 1.0 min, 30% B at 75 min, 40% B at 90 min, 95% B at 91 min, 95% B at 95 min, 3% B at 96 min, equilibrate thereafter till 120 min. The eluate was analyzed by HDMS under positive ion V-mode. The following parameters were set for data acquisition: collision energy: 5 volts for MS and ramp 15–40 volts for MSE; scan time: 0.6 second per scan. The HDMS machine was calibrated daily to ensure high accuracy (2.0 ppm for lock mass of m/z 785.8426).
For each PBM proteome digest sample, triplicate LC-nano-ESI-MSE datasets were acquired. Then, the MSE data were processed with ProteinLynx Global Server v2.3 (Waters) using default parameters. Based on the alternating low- and elevated- energy nature of MSE data, properties of each ion (mass-to-charge ratio, retention time, intensity, etc.) were determined, and a list of all precursor and product ions was produced. Specifically, the ion’s intensity was derived from the areas of both chromatographic and mass spectrometric peaks. The precursor ion intensity threshold was set to be above 1,000 counts. Human protein database International Protein Index v3.56 (153,078 protein entries) was searched by using the following parameters: enzyme specificity: trypsin; number of missed cleavages permitted: 1; fixed modification: Carbamidomethyl C; variable modifications: Acetyl N-TERM, Deamidation N, Deamidation Q, and Oxidation M; mass tolerance for precursor ions: 15 ppm; mass tolerance for product ions: 30 ppm; minimum peptide matches per protein: 1; minimum fragment ion matches per protein: 7; minimum fragment ion matches per peptide: 3; false positive rate: limited to 4% per randomized database searching.
To further control false positive protein identification, proteins identified in only one of the triplicate LC-nano-ESI-MSE analyses were filtered. Consequently, proteins identified repeatedly retained. Total ion counts of the three most intense matched peptides were used to quantify each protein. With the standard ADH1 as references, protein quantification level was exported in femtomol and nanogram. Mean values from triplicate analyses were used to represent protein expression levels in each PBM sample.
Western blotting
Western blotting experiments were conducted to validate differentially expressed proteins (DEPs) discovered by LC/MS method. Protein samples were the same as used for discovery. Due to limited amount of protein sample, only two DEPs (GSN and ITGA2B, picked by random) were subject to the validation. About 10 μg of total protein per sample was loaded. For target protein GSN specifically, mouse anti-human gelsolin primary antibody (Abnova, Cat. No. MAB0786) and horseradish peroxidase (HRP)-conjugated rabbit anti-mouse IgG-H&L polyclonal secondary antibody (Abcam, Cat. No. ab6728) was used. In addition, HRP-conjugated mouse anti-human beta actin primary antibody (Abcam, Cat. No. ab20272) was used to detect internal control protein beta-actin. Blot images were obtained through X-ray exposure developed using ChemiGlow West Chemiluminescence Substrate Kit (Cellbiosciences, Cat. No.60-12596-00). Protein bands were analyzed by a VersaDoc MP 4000 System (Bio-Rad Laboratories, Inc.), and quantified using QuantityOne software (Bio-Rad Laboratories, Hercules). Western blotting experiments were conducted in triplicate. For each sample, protein band quantity was normalized against beta-actin band quantity. Normalized data was subject to statistical analyses.
PBM total RNA extraction and GSN mRNA expression assay
For cohorts 2 and 3, PBM total RNA was extracted using Qiagen RNeasy Mini Kit (Qiagen, Inc., Valencia, CA) [25]. The mRNA expression level of GSN gene was mined from datasets acquired with Affymetrix Human Exon 1.0 ST Arrays (Cohort 2) and Affymetrix Human Genome U133A Array (Cohort 3). Robust Multiarray Average (RMA) algorithm was used to transform the probe-level data into gene expression data [26]. Compared with other algorithms, RMA provides the most reproducible results and shows the highest correlation coefficients with RT-PCR data [27, 28].
DNA extraction and GSN SNP genotyping
For cohorts 4 and 5, genomic DNA was extracted from 30 ml peripheral blood using a DNA isolation kit (Gentra systems, Minneapolis, MN, USA). DNA concentration was assessed by a DU530 UV/VIS spectrophotometer (Beckman Coulter Inc.). DNA quantification was double-checked using PicoGreen® dsDNA Reagent and Kits (Invitrogen). Genotypes for a total of 42 SNPs within 20-kb upstream and 20-kb downstream of the target gene GSN were mined from datasets acquired with Affymetrix GeneChip Human Mapping SNP 6.0 arrays.
Statistical analyses
Protein differential expression analyses in cohort 1
Based on PBM proteome profiles for cohort 1, student’s t-test was used to compare mean expression levels and to identify proteins differentially expressed between the two groups of subjects with high vs. low BMD. Specifically, raw protein expression levels were normalized against internal control protein beta-actin. Herein, only proteins detected in at least five PBM samples in both high and low BMD groups were subject to the comparative analyses. For Western blotting verification, student t-test was used to compare normalized expression levels between the high vs. low BMD groups of subjects.
mRNA differential expression analyses in cohorts 2 and 3
Based on mRNA expression data transformed by the RMA algorithm, student’s t-test was conducted to compare probe-level mean expression signals for the target gene GSN between the two groups of subjects with high BMD vs. low BMD.
SNP association analyses in cohorts 4 and 5
To correct for population stratification in association analyses in cohorts 4 and 5, respectively, EIGENSTRAT program was employed to perform principal component (PC) analyses with genome wide SNP data in subjects with genome-wide SNP call rate >95%. Covariates, including age, age2, menopause status (for the females only), height, weight, and PC1-10 generated by EIGENSTRAT[29], were tested for their significance of effect on hip BMD. Significant covariates were then used to adjust raw BMD values [30]. PLINK [31, 32] was used to perform genotypic association analyses by comparing mean hip BMD values among carriers of different genotypes at each SNP site.
In the female Caucasians from cohort 4, 33 among the total 42 examined SNPs across GSN gene passed QC check (i.e., minor allele frequency, MAF>0.01; Hardy-Weinberg equilibrium test, P>0.001). We tested the 33 SNPs for association with hip BMD in the female Caucasians. Then, SNPs associated with hip BMD in the female Caucasians were further tested for association with hip BMD in the male Caucasians, and in the female and male Chinese, respectively.
Integration analyses for GSN with DNA, RNA, and protein-level data
To evaluate the overall significance of GSN gene to hip BMD in female Caucasians, we performed integration analyses using the method proposed by Tyekucheva et al [33] to combine evidence from multi-level data (designated as X), i.e., SNP genotype, RNA expression, and protein expression data, in female Caucasians from cohorts 1–4. Briefly, the analysis procedure is divided into two stages. Stage I Individual Analysis: we carried out t-test for cohorts 1, 2 and 3 and females only for cohort 4 and computed score t (X, BMD) to capture the relationship between X and hip BMD. Stage II Integrated Analysis: scores computed in stage I for GSN gene (designated as g) were integrated into one score T (g) by:
where, d represents data at multiple levels, and ωd is the weight of d-th data in score T(g).
We have two datasets for mRNA level data. Thus, we set the weights as 1.0, 0.5, 0.5, and 1.0 for cohorts 1 to 4, respectively. Considering that different statistics were generated from different data, we adopted Wang’s method [34] to adjust for the impacts of different statistics on the integrated score. Statistical significance testing of the integrated score was done by permutation-based procedure [33].
RESULTS
GSN protein was significantly down-regulated in PBM in vivo in low BMD subjects
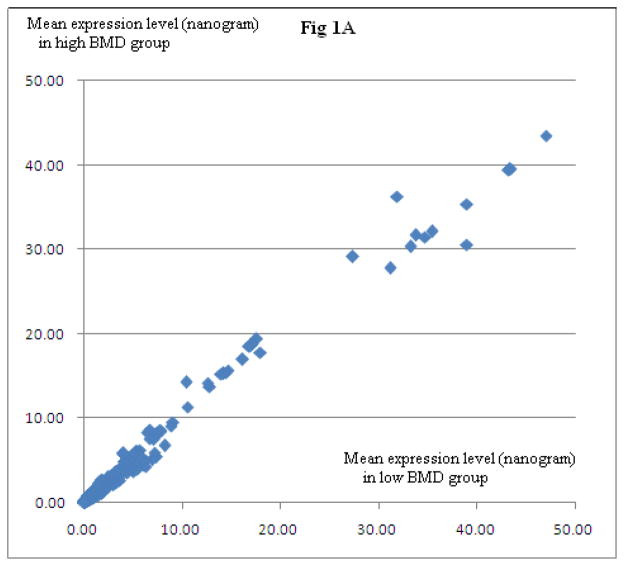
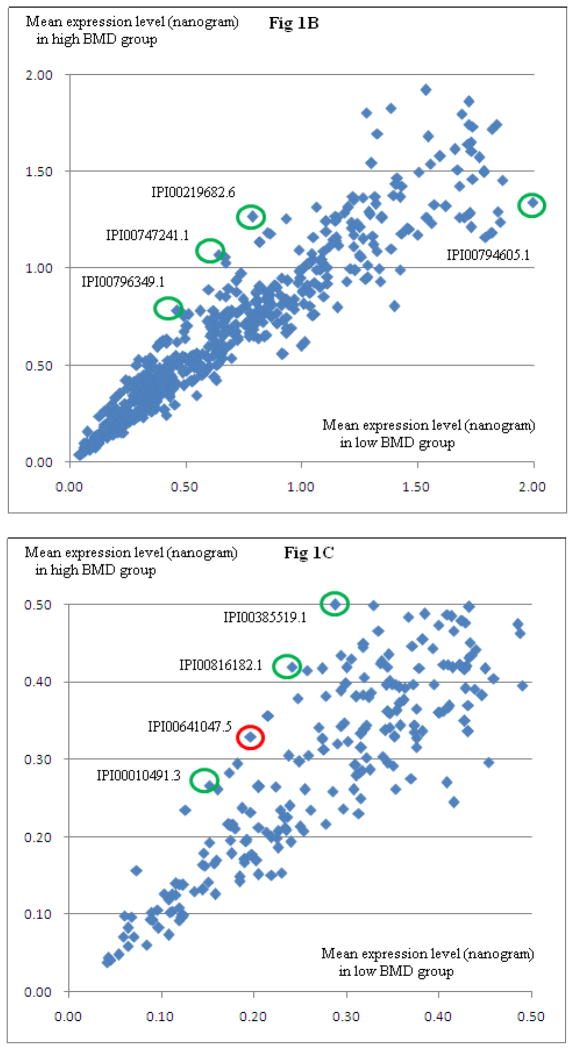
In cohort 1, we identified a total of 2,083 PBM-expressed proteins. Among the 2083 proteins, 759 proteins were detected in at least five PBM samples in both high and low BMD groups of subjects. Figure 1 illustrates the relative protein expression levels for the 759 proteins in low BMD and high BMD subjects. Based on normalized protein expression levels, we identified 57 DEPs between high vs. low BMD subjects (P≤0.05, Supplemental Table 1). Among the 57 proteins, eight proteins (IPI00641047.5, GSN; IPI00010491.3, RAB27B; IPI00816182.1, RAP1B; IPI00219682.6, STOM; IPI00385519.1 and IPI00747241.1, ITGA2B; IPI00794605.1, GAPDH; IPI00796349.1, RAP1B) showed differential expression by raw expression data analyses.
Figure 1. Relative Proteome Expression in PBM in Low vs. High BMD Subjects.
Presented are mean protein expression levels in subjects with high BMD and low BMD, respectively. Three panels are presented at different scales, for a clear view. Fig 1A is a full view of all identified proteins. Fig 1B and Fig 1C are zoomed-in presentation. Approximately 500 ng PBM total proteins were analyzed using LC-nano-ESI-MSE method. Circled in Fig 1B and Fig 1C are eight DEPs (IPI00641047.5, GSN; IPI00010491.3, RAB27B; IPI00816182.1, RAP1B; IPI00219682.6, STOM; IPI00385519.1 and IPI00747241.1, ITGA2B; IPI00794605.1, GAPDH; IPI00796349.1, RAP1B), suggested by normalized data analyses, as well as raw data analyses. Due to limited protein amount per sample, Western Blotting experiments were conducted for GSN (circled in red) and ITGA2B proteins only.
Differential expression of GSN protein was validated by Western Blotting method
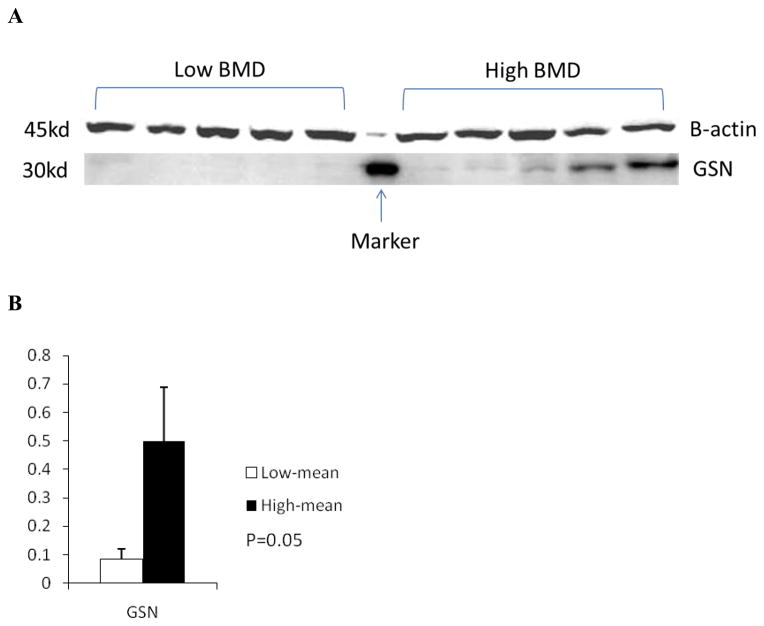
Due to limited amount of protein sample remained for subjects in cohort 1, only two DEPs (GSN and ITGA2B) were subject to validation by Western blotting. Western blotting validated differential expression for GSN (Figure 2), but not for ITGA2B. Specifically, per beta-actin normalized data, gelsolin protein was down-regulated 3.0-fold in low vs. high BMD subjects.
Figure 2. Verification of GSN Up-regulation in High BMD Subjects by Western Blot.
A: Presented are representative blot of triplicate experiments.
B: Student t-test was used to compare gelsolin expression levels in the two groups of subjects.
GSN mRNA was significantly down-regulated in PBM in vivo in low BMD subjects
As shown in Table 2, significant down-regulation of GSN gene expression in PBM in low BMD subjects was replicated at mRNA level in both cohort 2 (as reflected by probe 3187711) and cohort 3 (as reflected by both probes 200696_s_at and 214040_s_at), further implying relevance of GSN gene function to bone metabolism.
Table 2.
Differential Expression of GSN at Protein and mRNA Levels in High vs. Low BMD Subjects
| ID | P-value | Trend | Sample | |
|---|---|---|---|---|
| Protein | IPI00641047.5 | *2.2E-02 | ↑ | Cohort 1 |
| mRNA (probe set) | 3187711 | 4.5E-02 | ↑ | Cohort 2 |
| 200696_s_at | 5.6E-04 | ↑ | Cohort 3 | |
| 214040_s_at | 2.2E-04 | ↑ |
Note: The mRNA expression levels were mined from datasets acquired with Affymetrix Human Exon 1.0 ST Arrays (cohort 2) and Affymetrix Human Genome U133A Arrays (cohort 3), respectively. RMA algorithm was used to transform probe-level intensity data to gene expression data. Student’s t-test was used to compare the mean expression levels in the two groups of subjects.
P-value of student’s t-test using beta-actin-normalized data.
↑: up-regulation in high vs. low BMD subjects.
GSN SNP was associated with hip BMD in female Caucasians
In the female Caucasians, an intronic SNP rs767770 in GSN was found associated with hip BMD (P=0.0003), which is significant even after correcting for multiple testing using the highly conservative Bonferroni method. SNP rs767770 was not associated with hip BMD in the Chinese females or in the two ethnic groups of males (P>0.5). In the female Caucasians, the minor allele (A) frequency for SNP rs767770 is 0.04. Specifically, the homozygous GG carriers (N=1530), on average, have lower hip BMD than the heterozygous AG carriers (N=132) (0.94±0.14 vs. 0.97±0.31, Mean ± S.D.).
GSN was significantly associated with hip BMD in integration analysis
Integrating evidence from DNA, RNA, and protein levels, by simultaneously employing the above study datasets collected from female Caucasians in cohorts 1–4, showed that GSN is highly significantly associated with hip BMD in female Caucasians (P=0.0001).
DISCUSSION
The present work represents our continuous effort to identify osteoporosis risk genes for female Caucasians using a proteomics-based multi-disciplinary and integrative study strategy. We employed state-of-art quantitative proteomics methodology to profile proteome-wide protein expression in in vivo PBM in subjects with discordant hip BMD, hence to discover proteins thus genes that are functionally relevant to osteoporosis. Among a list of DEPs identified, we validated that PBM-expressed GSN was significantly down-regulated in premenopausal Caucasians with low vs. high hip BMD.
Furthermore, down-regulation of GSN in PBM in low BMD subjects was also replicated at mRNA level in another two independent cohorts of premenopausal Caucasian women. The findings stress functional relevance of PBM-expressed GSN gene to hip BMD in female Caucasians. Through gender-stratified SNP association analyses, we identified an intronic SNP significantly associated with hip BMD in female Caucasians, specifically. The DNA-level findings warrant the significance of GSN to hip BMD in female Caucasians. Meanwhile, the results imply that the effect of GSN on hip BMD is probably modified by gender-related factors.
We would point out that, in the discovery sample, the two group of subjects with discordant BMD have significant difference in body weight as well (P <0.05). Thus, there is a possibility that genes discovered in cohort 1 are related to body weight instead of BMD. To ascertain the relationship between GSN and body weight, we tested association between GSN SNPs and body weight in the female Caucasians. However, no evidence of association was observed (P>0.05). In contrast, we observed significant association for SNP rs767770 and hip BMD, after adjusting for the covariate effect of body weight. Therefore, GSN is most likely related to BMD itself rather than body weight. Evidence showed that the expression of gelsolin was regulated by androgen in prostate cancer cell line LNCaP [35]. Whether GSN expression in PBM is regulated by androgen or not has yet to be studied. Previous studies suggest that gelsolin protein plays an important role in regulating androgen-mediated effects on osteoclastogenesis and bone resorption. Testosterone, as a steroid hormone from androgen group, circulates and affects bone metabolism in females [36]. In purified human peripheral blood CD14+ monocytes, testosterone, via binding to androgen receptor (AR) directly, inhibit osteoclast formation and bone resorption at physiological concentrations [37]. Furthermore, testosterone exerts significant growth inhibition effect and pro-apoptotic effects on monocytes, as observed in cultured human monocytic THP-1 cells [38, 39]. It was known that the effects of androgen are mediated through AR, which regulates expression of downstream target genes [40–42]. AR expression was observed on monocytes [39] as well. It contains two transcriptional activation functions: activation function 1 (AF-1) in the NH2-terminal domain and activation function 2 (AF-2) in the ligand binding domain [43–46]. Interestingly, C terminal of gelsolin can bind to AF-2 domain, in the presence of androgen, to facilitate AR nuclear translocation [47]. Therefore, gelsolin is a co-regulator of AR transactivation [47].
Based on the above functional evidences and the findings of the present study, we proposed that gelsolin, through interaction with AR, may enhance androgen-induced AR transactivation thus promote growth inhibition and pro-apoptosis of monocytes. Reduced precursor cells of bone-resorbing osteoclasts, i.e., monocytes, may attenuate osteoclast formation and bone resorption. Consequently, higher level of gelsolin expression in PBM, through enhancing PBM growth inhibition and pro-apoptosis, may decrease osteoclast formation and bone resorption activity, and eventually lead to higher level of BMD. This proposed mechanism is consistent with and explains the associations observed in the cohorts 1–3, i.e., the higher the gelsolin expression level, the higher the BMD level. In-depth studies are needed to test the regulatory effect of gelsolin on osteoclastogenesis, to explore the interaction effect of gelsolin and androgen on BMD, and to verify the proposed mechanism.
To be mentioned, GSN is an actin filament severing and capping protein involved in actin cytoskeletal organization [48–50]. Previous studies showed that it plays an important role in osteoclasts. GSN deficiency blocks assembly and motility of podosome in mouse osteoclasts [51], reduces bone resorption in vivo and produce increased bone mass and strength in mice [49, 51]. The present study represents the first evidence showing that lower gelsolin level in human PBM in vivo is associated with DECREASED BMD. Although the above findings in mice and human look controversial, the discrepancy might be partially attributed to the following reasons. Firstly, the study cell types are different. One is osteoclast, the other is peripheral blood monocyte. These two cell types have significantly different internal environments. Secondly, the study species are different. One is mice, the other is humans. The physical conditions in the two biological systems are dramatically different. Protein functions not only depend on the protein expression levels, but also depend on protein-protein interactions, external signals, and cell-environment interaction. Therefore, it is not surprising that the same molecule will exert diverse cellular and biological functions in different cell types and biological systems. However, how GSN expression level in PBM in vivo in humans is regulated by internal environment, such as circulating hormone level, and how it interacts with other molecules to affect bone metabolism still await further investigation.
In summary, through differential expression studies in premenopausal Caucasian women with extremely discordant hip BMD, we identified, validated, and replicated that GSN is significantly down-regulated in low vs. high BMD subjects. Through SNP association studies, we identified a SNP in GSN significantly associated with hip BMD in female Caucasians, specifically. Based on the supportive evidences from both individual and integrative studies, we conclude that GSN is a significant gene influencing hip BMD, and PBM-expressed gelsolin is involved in pathogenesis of osteoporosis in female Caucasians. Additionally, our findings would await replication in other cohorts. Whether GSN is associated with osteoporotic fractures or BMD at other skeletal sites, and whether PBM-expressed gelsolin is associated with male osteoporosis still need investigation in future.
Supplementary Material
Peripheral blood monocytes (PBM) are precursor cells of bone-resorbing osteoclasts.
A proteomic study discovered that PBM-expressed gelsolin is 3.0-fold up-regulated in premenopausal Caucasians with extremely high vs. low hip BMD.
The same trend of differential expression was observed at PBM-expressed GSN mRNA level in another two independent study cohorts.
SNP rs767770 in GSN was found significantly associated with hip BMD in female Caucasians (P=0.0003), specifically.
Integrative analyses of the above DNA, RNA, and protein datasets substantiated that GSN is highly significant for hip BMD.
Acknowledgments
The study was partially supported by Soochow University Startup Fund (Q413900712), National Natural Science Foundation of China (K113931013), NSFC from Jiangsu Province (L213908513), a Project of the Priority Academic Program Development of Jiangsu Higher Education Institutions, grants from NIH (P50AR055081, R01AR057049, R01AR050496, R01AG026564, and R03TW008221), and Tulane Edward G. Schlieder Endowment.
Footnotes
All authors declare that they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sun L, Tan LJ, Lei SF, Chen XD, Li X, Pan R, Yin F, Liu QW, Yan XF, Papasian CJ, Deng HW. Bivariate genome-wide association analyses of femoral neck bone geometry and appendicular lean mass. PLoS One. 2011;6:e27325. doi: 10.1371/journal.pone.0027325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melton LJ., 3rd Adverse outcomes of osteoporotic fractures in the general population. J Bone Miner Res. 2003;18:1139–41. doi: 10.1359/jbmr.2003.18.6.1139. [DOI] [PubMed] [Google Scholar]
- 3.Jordan KM, Cooper C. Epidemiology of osteoporosis. Best Pract Res Clin Rheumatol. 2002;16:795–806. doi: 10.1053/berh.2002.0264. [DOI] [PubMed] [Google Scholar]
- 4.Pulkkinen P, Partanen J, Jalovaara P, Jamsa T. Combination of bone mineral density and upper femur geometry improves the prediction of hip fracture. Osteoporos Int. 2004;15:274–80. doi: 10.1007/s00198-003-1556-3. [DOI] [PubMed] [Google Scholar]
- 5.Riggs BL, Melton LJ., 3rd The worldwide problem of osteoporosis: insights afforded by epidemiology. Bone. 1995;17:505S–511S. doi: 10.1016/8756-3282(95)00258-4. [DOI] [PubMed] [Google Scholar]
- 6.Iki M. Difference in osteoporosis in men and women. Clin Calcium. 2011;21:1377–83. [PubMed] [Google Scholar]
- 7.Lauritzen JB. Hip fractures: incidence, risk factors, energy absorption, and prevention. Bone. 1996;18:65S–75S. doi: 10.1016/8756-3282(95)00382-7. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen TV, Howard GM, Kelly PJ, Eisman JA. Bone mass, lean mass, and fat mass: same genes or same environments? Am J Epidemiol. 1998;147:3–16. doi: 10.1093/oxfordjournals.aje.a009362. [DOI] [PubMed] [Google Scholar]
- 9.Deng HW, Stegman MR, Davies KM, Conway T, Recker RR. Genetic determination of variation and covariation of peak bone mass at the hip and spine. J Clin Densitom. 1999;2:251–63. doi: 10.1385/jcd:2:3:251. [DOI] [PubMed] [Google Scholar]
- 10.Recker RR, Deng HW. Role of genetics in osteoporosis. Endocrine. 2002;17:55–66. doi: 10.1385/ENDO:17:1:55. [DOI] [PubMed] [Google Scholar]
- 11.Jian WX, Long JR, Li MX, Liu XH, Deng HW. Genetic determination of variation and covariation of bone mineral density at the hip and spine in a Chinese population. J Bone Miner Metab. 2005;23:181–5. doi: 10.1007/s00774-004-0558-3. [DOI] [PubMed] [Google Scholar]
- 12.Horowitz MC, Xi Y, Wilson K, Kacena MA. Control of osteoclastogenesis and bone resorption by members of the TNF family of receptors and ligands. Cytokine Growth Factor Rev. 2001;12:9–18. doi: 10.1016/s1359-6101(00)00030-7. [DOI] [PubMed] [Google Scholar]
- 13.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–42. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 14.Teitelbaum SL. Bone resorption by osteoclasts. Science. 2000;289:1504–8. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 15.Quinn JM, Neale S, Fujikawa Y, McGee JO, Athanasou NA. Human osteoclast formation from blood monocytes, peritoneal macrophages, and bone marrow cells. Calcif Tissue Int. 1998;62:527–31. doi: 10.1007/s002239900473. [DOI] [PubMed] [Google Scholar]
- 16.Udagawa N, Takahashi N, Akatsu T, Tanaka H, Sasaki T, Nishihara T, Koga T, Martin TJ, Suda T. Origin of osteoclasts: mature monocytes and macrophages are capable of differentiating into osteoclasts under a suitable microenvironment prepared by bone marrow-derived stromal cells. Proc Natl Acad Sci U S A. 1990;87:7260–4. doi: 10.1073/pnas.87.18.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matayoshi A, Brown C, DiPersio JF, Haug J, Abu-Amer Y, Liapis H, Kuestner R, Pacifici R. Human blood-mobilized hematopoietic precursors differentiate into osteoclasts in the absence of stromal cells. Proc Natl Acad Sci U S A. 1996;93:10785–90. doi: 10.1073/pnas.93.20.10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bar-Shavit Z. The osteoclast: a multinucleated, hematopoietic-origin, bone-resorbing osteoimmune cell. J Cell Biochem. 2007;102:1130–9. doi: 10.1002/jcb.21553. [DOI] [PubMed] [Google Scholar]
- 19.Cohen-Solal ME, Graulet AM, Denne MA, Gueris J, Baylink D, de Vernejoul MC. Peripheral monocyte culture supernatants of menopausal women can induce bone resorption: involvement of cytokines. J Clin Endocrinol Metab. 1993;77:1648–53. doi: 10.1210/jcem.77.6.8263153. [DOI] [PubMed] [Google Scholar]
- 20.Pacifici R. Estrogen, cytokines, and pathogenesis of postmenopausal osteoporosis. J Bone Miner Res. 1996;11:1043–51. doi: 10.1002/jbmr.5650110802. [DOI] [PubMed] [Google Scholar]
- 21.Nathan CF. Secretory products of macrophages. J Clin Invest. 1987;79:319–26. doi: 10.1172/JCI112815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Recker R, Lee WN, Xiao GG. Proteomics in bone research. Expert Rev Proteomics. 2010;7:103–11. doi: 10.1586/epr.09.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng FY, Lei SF, Zhang Y, Zhang YL, Zheng YP, Zhang LS, Pan R, Wang L, Tian Q, Shen H, Zhao M, Lundberg YW, Liu YZ, Papasian CJ, Deng HW. Peripheral blood monocyte-expressed ANXA2 gene is involved in pathogenesis of osteoporosis in humans. Mol Cell Proteomics. 2011;10:M111 011700. doi: 10.1074/mcp.M111.011700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5:144–56. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Deng FY, Lei SF, Chen XD, Tan LJ, Zhu XZ, Deng HW. An integrative study ascertained SOD2 as a susceptibility gene for osteoporosis in Chinese. J Bone Miner Res. 2011;26:2695–701. doi: 10.1002/jbmr.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 27.Seo J, Hoffman EP. Probe set algorithms: is there a rational best bet? BMC Bioinformatics. 2006;7:395. doi: 10.1186/1471-2105-7-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu Y, He F, Chen Y. Different effects of the probe summarization algorithms PLIER and RMA on high-level analysis of Affymetrix exon arrays. BMC Bioinformatics. 2010;11:211. doi: 10.1186/1471-2105-11-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 30.Guo Y, Tan LJ, Lei SF, Yang TL, Chen XD, Zhang F, Chen Y, Pan F, Yan H, Liu X, Tian Q, Zhang ZX, Zhou Q, Qiu C, Dong SS, Xu XH, Guo YF, Zhu XZ, Liu SL, Wang XL, Li X, Luo Y, Zhang LS, Li M, Wang JT, Wen T, Drees B, Hamilton J, Papasian CJ, Recker RR, Song XP, Cheng J, Deng HW. Genome-wide association study identifies ALDH7A1 as a novel susceptibility gene for osteoporosis. PLoS Genet. 2010;6:e1000806. doi: 10.1371/journal.pgen.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singer JB. Candidate gene association analysis. Methods Mol Biol. 2009;573:223–30. doi: 10.1007/978-1-60761-247-6_13. [DOI] [PubMed] [Google Scholar]
- 32.Zhang LS, Hu HG, Liu YJ, Li J, Yu P, Zhang F, Yang TL, Tian Q, Zheng YP, Guo Y, Deng HW. A follow-up association study of two genetic variants for bone mineral density variation in Caucasians. Osteoporos Int. 2011 doi: 10.1007/s00198-011-1863-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyekucheva S, Marchionni L, Karchin R, Parmigiani G. Integrating diverse genomic data using gene sets. Genome Biol. 2011;12:R105. doi: 10.1186/gb-2011-12-10-r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 2007;81:1278–83. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urbanucci A, Waltering KK, Suikki HE, Helenius MA, Visakorpi T. Androgen regulation of the androgen receptor coregulators. BMC Cancer. 2008;8:219. doi: 10.1186/1471-2407-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mosekilde L, Eriksen EF, Charles P. Effects of thyroid hormones on bone and mineral metabolism. Endocrinol Metab Clin North Am. 1990;19:35–63. [PubMed] [Google Scholar]
- 37.Michael H, Harkonen PL, Vaananen HK, Hentunen TA. Estrogen and testosterone use different cellular pathways to inhibit osteoclastogenesis and bone resorption. J Bone Miner Res. 2005;20:2224–32. doi: 10.1359/JBMR.050803. [DOI] [PubMed] [Google Scholar]
- 38.Cutolo M, Capellino S, Montagna P, Ghiorzo P, Sulli A, Villaggio B. Sex hormone modulation of cell growth and apoptosis of the human monocytic/macrophage cell line. Arthritis Res Ther. 2005;7:R1124–32. doi: 10.1186/ar1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cutolo M, Sulli A, Craviotto C, Felli L, Pizzorni C, Seriolo B, Villaggio B. Modulation of cell growth and apoptosis by sex hormones in cultured monocytic THP-1 cells. Ann N Y Acad Sci. 2002;966:204–10. doi: 10.1111/j.1749-6632.2002.tb04216.x. [DOI] [PubMed] [Google Scholar]
- 40.Chang CS, Kokontis J, Liao ST. Molecular cloning of human and rat complementary DNA encoding androgen receptors. Science. 1988;240:324–6. doi: 10.1126/science.3353726. [DOI] [PubMed] [Google Scholar]
- 41.Lubahn DB, Joseph DR, Sullivan PM, Willard HF, French FS, Wilson EM. Cloning of human androgen receptor complementary DNA and localization to the X chromosome. Science. 1988;240:327–30. doi: 10.1126/science.3353727. [DOI] [PubMed] [Google Scholar]
- 42.Chang CS, Kokontis J, Liao ST. Structural analysis of complementary DNA and amino acid sequences of human and rat androgen receptors. Proc Natl Acad Sci U S A. 1988;85:7211–5. doi: 10.1073/pnas.85.19.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jenster G, van der Korput HA, Trapman J, Brinkmann AO. Identification of two transcription activation units in the N-terminal domain of the human androgen receptor. J Biol Chem. 1995;270:7341–6. doi: 10.1074/jbc.270.13.7341. [DOI] [PubMed] [Google Scholar]
- 44.Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–87. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 45.He B, Kemppainen JA, Voegel JJ, Gronemeyer H, Wilson EM. Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH(2)-terminal domain. J Biol Chem. 1999;274:37219–25. doi: 10.1074/jbc.274.52.37219. [DOI] [PubMed] [Google Scholar]
- 46.Bevan CL, Hoare S, Claessens F, Heery DM, Parker MG. The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol Cell Biol. 1999;19:8383–92. doi: 10.1128/mcb.19.12.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishimura K, Ting HJ, Harada Y, Tokizane T, Nonomura N, Kang HY, Chang HC, Yeh S, Miyamoto H, Shin M, Aozasa K, Okuyama A, Chang C. Modulation of androgen receptor transactivation by gelsolin: a newly identified androgen receptor coregulator. Cancer Res. 2003;63:4888–94. [PubMed] [Google Scholar]
- 48.Sun HQ, Yamamoto M, Mejillano M, Yin HL. Gelsolin, a multifunctional actin regulatory protein. J Biol Chem. 1999;274:33179–82. doi: 10.1074/jbc.274.47.33179. [DOI] [PubMed] [Google Scholar]
- 49.Silacci P, Mazzolai L, Gauci C, Stergiopulos N, Yin HL, Hayoz D. Gelsolin superfamily proteins: key regulators of cellular functions. Cell Mol Life Sci. 2004;61:2614–23. doi: 10.1007/s00018-004-4225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwiatkowski DJ. Functions of gelsolin: motility, signaling, apoptosis, cancer. Curr Opin Cell Biol. 1999;11:103–8. doi: 10.1016/s0955-0674(99)80012-x. [DOI] [PubMed] [Google Scholar]
- 51.Chellaiah M, Kizer N, Silva M, Alvarez U, Kwiatkowski D, Hruska KA. Gelsolin deficiency blocks podosome assembly and produces increased bone mass and strength. J Cell Biol. 2000;148:665–78. doi: 10.1083/jcb.148.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.