Abstract
Bone Mineral Density (BMD) is major index for diagnosing osteoporosis. PhosSNPs are non-synonymous SNPs that affect protein phosphorylation. The relevance and significance of phosSNPs to BMD and osteoporosis is unknown. This study aims to identify and characterize phosSNPs significant for BMD in humans. We conducted a pilot genome-wide phosSNP association study for BMD in three independent population samples, involving ~5,000 unrelated individuals. We identified and replicated three phosSNPs associated with both spine BMD and hip BMD in Caucasians. Association with hip BMD for one of these phosSNPs, i.e., rs6265 (major/minor allele: G/A) in BDNF gene, was also suggested in Chinese. Consistently in both ethnicities, individuals carrying AA genotype have significant lower hip BMD than carriers of GA and GG genotypes. Through in vitro molecular and cellular studies, we found that compared to osteoblastic cells transfected with wild-type BDNF-Val66 (encoded with allele G at rs6265), transfection of variant BDNF-Met66 (encoded with allele A at rs6265) significantly decreased BDNF protein phosphorylation (at amino acid residue T62), expression of osteoblastic genes (OPN, BMP2, and ALP), and osteoblastic activity. The findings are consistent with and explain our prior observations in general human populations. We conclude that phosSNP rs6265, via regulating BDNF protein phosphorylation and osteoblast differentiation, influence hip BMD in humans. This study represents our first endeavor to dissect the functions of phosSNPs in bone, which might stimulate extended large-scale studies on bone or similar studies on other human complex traits and diseases.
Keywords: BMD, SNP, protein phosphorylation, BDNF, osteoblast
Introduction
Genetics of BMD and Osteoporosis
Bone mineral density (BMD), as a measurable and powerful index predicting osteoporosis and OF risks, is the current “golden standard” for diagnosing osteoporosis. BMD is a complex trait largely determined by genetic factors, with heritability larger than 0.6(1). In the past decades, extensive studies, including candidate gene association studies, genome-wide linkage and association studies, have been launched to search for quantitative trait loci underlying BMD variation in humans. However, currently identified quantitative trait loci alone can only account for a small proportion of BMD variation, leaving the majority of genetic variation to be explained by novel factors to be identified. In addition, most of the genes identified in one population have yet to be tested for their significance in independent sample(s). So far, an increasing number of genome-wide association studies (GWAS) systemically explore the relationship between single nucleotide polymorphisms (SNPs) and phenotypic variation, and also established some associations. However, the mechanism(s) underlying a majority of these associations are largely unclear. Identification of significant SNPs influencing BMD and characterization of their cellular and molecular functions will contribute to elucidating pathophysiological mechanism of osteoporosis.
Proteins, Protein Phosphorylation, and Diseases
Proteins, as direct executors of genes’ function, participate in a wide variety of biological processes. As such, proteome-wide protein expression studies have been utilized to systemically search for causal proteins or protein markers which are differentially expressed in diseased vs. healthy subjects. However, such expression proteomics studies generally focused on quantitative change(s) of protein(s) expression levels and cast little attention to amino acid residue change(s) and potential impact of such sequence changes on phenotypes of interest.
As the most widely studied post-translational modification, protein phosphorylation is involved in various signaling pathways and plays fundamental roles in regulating almost all kinds of normal cellular functions and biological processes. Thus, abnormal regulation of protein phosphorylation has been found associated with pathogenesis of a serial of diseases, such as cancer (2), Alzheimer’s disease (3), osteoporosis (4), etc. In the bone field, more than a dozen of review articles highlight the importance of protein phosphorylation-regulated signaling events for osteoporosis, such as IL-6 signaling (5), TNF signaling (6), and events in regulation of CSF-1 induced osteoclast spreading (7), integrin-mediated osteoclast adhesion and activation (8), etc. However, few studies have systemically explored the relationship between protein phosphorylation and bone metabolism and/or assess the significance of phosphorylation sites in influencing BMD in humans.
SNP and Protein Phosphorylation
Protein phosphorylation is a reversible post-translational modification mainly targeting at amino acid residues serine (S), threonine (T), and tyrosine (Y). It is catalyzed by protein kinases. Ren et al. reported that ~70% of non-synonymous SNP (nsSNP) in human genome are potential phosphorylation-related SNPs, i.e., so called phosSNPs (9). They reported a total of 64,035 phosSNPs, among which 2,004 are experimentally validated. PhosSNPs could affect protein phosphorylation by either changing protein phosphorylation amino acid site(s) or changing protein kinase(s) which catalyze phosphorylation of specific amino acid site(s). Despite their potential significance, so far, whether phosSNPs are associated with variation in complex traits, such as human BMD, is unknown.
Study Purpose
Through a pilot genome-wide phosSNP association study and replication studies in three independent human populations, the present work is attempted to identify significant phosSNPs influencing BMD. Furthermore, through follow-up in vitro studies on top significant phosSNP identified, we aim to characterize their biological functions, hence to illustrate pathophysiological mechanism of osteoporosis in humans.
Materials and Methods
Human Subjects
The study was approved by Institutional Review Boards of involved institutes. Signed informed-consent documents were obtained from all study participants before enrollment in the study.
Caucasian Sample 1 (CAU1)
This sample contains 1,000 unrelated subjects (age: 50.3 ± 18.3 years) selected from our established and expanding genetic database currently containing more than 7,000 subjects and largely recruited in Midwestern U.S. in Omaha, Nebraska. All the subjects were U.S. Caucasians of European origin. CAU1 serves as a discovery cohort in this study.
Caucasian Sample 2 (CAU2)
This sample contains 2,286 unrelated subjects (age: 51.4 ± 13.8 years) recruited in Midwestern U.S. in Kansas City, Missouri and Omaha, Nebraska. All the subjects were U.S. Caucasians of European origin. This sample, independent of CAU1, serves as a replication cohort to validate findings in CAU1.
Chinese Sample (CHN)
This sample consists of 1,627 unrelated subjects (age: 34.5 ± 13.2 years) recruited from central south region of China. All the subjects were Han Chinese. This sample serves as a replication cohort for across-ethnicity validation to test ethnic- general or specific effects of phosSNPs identified and/or validated in Caucasians.
For subject recruitment, strict exclusion criteria (10) were adopted to minimize any known or potential confounding effects on variation of bone phenotype. Briefly, patients with chronic diseases/conditions that may potentially affect bone mass were excluded. These diseases/conditions included chronic disorders involving vital organs (heart, lung, liver, kidney, brain), serious metabolic diseases (diabetes, hypo- or hyperparathyroidism, hyperthyroidism), other skeletal diseases (Paget’s disease, osteogenesis imperfecta, rheumatoid arthritis), chronic use of drugs affecting bone metabolism (corticosteroid therapy, anticonvulsant drugs), and malnutrition conditions (chronic diarrhea, chronic ulcerative colitis). Bone mineral density (g/cm2) at lumbar spine (L1_4) and total hip were measured using daily calibrated dual energy X-ray absorptiometry (DXA) machines (Hologic Inc., Bedford, MA, USA).
PhosSNP Genotyping
Out of the total 64,035 phosSNPs in the phosSNP 1.0 database (9), those covered by Affymetrix SNP Arrays (Affymetrix, Inc., Santa Clara, CA, USA) were studied herein. Specifically, genomic DNA was extracted from leukocytes using Puregene DNA Isolation Kit (Gentra systems, Minneapolis, MN, USA). For the CAU1 sample, SNP genotyping with the Affymetrix Mapping 250 k Nsp and 250 k Sty arrays was performed in Vanderbilt Microarray Shared Resources (VMSR) (http://array.mc.vanderbilt.edu/) using the standard protocol recommended by Affymetrix. For CAU2 and CHN samples, SNP genotyping with Affymetrix Genome-Wide Human SNP Array 6.0 was performed using the standard protocol recommended by the manufacturer. Fluorescence intensities were quantified using an Affymetrix array scanner 30007G. Data management and analyses were performed using the Affymetrix® GeneChip® Command Console® Software. Contrast quality control (QC) threshold was set at the default value of greater than 0.4 for data QC.
After excluding SNPs with minor allele frequency (MAF) less than 0.01 and/or SNPs deviating from Hardy-Weinberg Equilibrium (HWE test, p <0.01) in individual population sample, a total of 2,474 phosSNPs retained in the three study samples, including 660, 1,797, and 1,662 phosSNPs in the CAU1, CAU2, and CHN samples, respectively. The discrepancy between the numbers of phosSNPs “retained” in CAU1 and the other two samples mainly reflects the difference in SNP coverage between the genotyping arrays used. The relatively small difference in the numbers of phosSNPs between CAU2 and CHN primarily reflects the difference in genetic background between the two ethnicities of Caucasian and Chinese. Among those retained phosSNPs, 653 phosSNPs were overlapped in all the three samples.
Association Test between PhosSNPs and BMD in Human Populations
Age, gender, height, and weight were used as covariates to adjust the raw BMD measurements. PLINK (11) was used to perform genotypic association analyses between phosSNPs and adjusted BMD. Specifically, the analyses compared the difference of mean BMD values among carriers of the three different genotypes for each phosSNP. PhosSNPs, discovered to be significant to both spine BMD and hip BMD in the CAU1 sample, were analyzed in the CAU2 sample for within-ethnicity validation purpose, and further analyzed in the CHN sample for across-ethnicity validation. Miscellaneous statistical analyses were performed using the software packages SAS (SAS Institute Inc., Cary, NC) and Minitab (Minitab Inc., State College, PA).
Predicting and Validating Impact of PhosSNP on Protein Phosphorylation
Group-based Prediction System (GPS 2.0) (12) was applied to predict the impact of phosSNPs on protein phosphorylation, i.e., to predict the resultant changes of protein phosphorylation sites and/or corresponding changes of protein kinases (12). For a specific phosSNP rs6265 that we identified as important for hip BMD in general human populations, we conducted the following studies to validate its predicted impact on protein phosphorylation (detailed in the section of Results). To be noted, rs6265 (major/minor allele: G/A) is located in BDNF gene (Entrez Gene database accession number: 627), which encodes brain-derived neurotrophic factor.
Firstly, to ascertain the substrate-kinase relationship between the protein BDNF and the predicted protein kinase CHEK2, we tested their protein-protein interaction in bone cells. Briefly, we co-transfected the human fetal osteoblastic 1.19 cell line (hFOB, ATCC, Cat CRL-11372) with expression vector pCMV6-AC-GFP-CHEK2, together with wild-type pCDNA3.1-BDNF-V66T62, variant pCDNA3.1-BDNF-M66T62, or mutant pCDNA3.1-BDNF-V66A62, respectively. Herein, pCDNA3.1-BDNF-V66A62 was to replace a suspect target site of phosphorylation (i.e., Threonine at amino acid residue 62) with an unphosphorylatable residue (i.e., Alanine, A) at the encoded BDNF protein product. After 48 hours, we collected hFOB cell lysate and quantified total proteins. We then employed Co-Immunoprecipitation (Co-IP) procedures to pull down BDNF protein from the cell lysate and to probe CHEK2 protein in the Co-IP product through Western Blotting (WB) procedures.
Secondly, for testing whether rs6265 affect BDNF protein phosphorylation, we co-transfected hFOB cells with expression vector pCMV6-AC-GFP-CHEK2, together with wild-type pCDNA3.1-BDNF-V66T62, variant pCDNA3.1-BDNF-M66T62, or mutant pCDNA3.1-BDNF-V66A62, respectively. After 48 hours, we collected hFOB cell lysate, quantified total proteins, and purified BDNF from equal amount of total proteins. Then, we conducted phosphoprotein phosphate estimation assay to quantify overall BDNF phosphoyrlation levels under different conditions.
Thirdly, to validate the prediction that BDNF-T62 is the target site of phosphorylation regulated by rs6265, we co-transfected human osteoblast-like cell line MG63 (ATCC, Cat CRL-1427) with expression vector pCMV6-AC-GFP-CHEK2, together with wild-type pCDNA3.1-BDNF-V66T62, variant pCDNA3.1-BDNF-M66T62, or mutant pCDNA3.1-BDNF-V66A62, respectively. After 48 hours, we collected MG63 cell lysate and quantified total proteins. We then analyzed BDNF-T62 site-specific protein phosphorylation levels by Western Blotting method using anti-BDNF-pT62 antibody.
Detailed procedures of the above experiments are described as follows.
Plasmid Constructs
The pCDNA3.1-BDNF-V66T62 and pCDNA3.1-BDNF-M66T62 constructs were kind gifts from Dr. Francis Lee’s lab (13). Specifically, the human BDNF cDNA was subcloned into pCDNA3.1 hygro expression vector (Invitrogen, Cat V870-20) at Hind III and Xho I restriction endonuclease sites. The pCDNA3.1-BDNF-V66T62 construct was mutated to pCDNA3.1-BDNF-V66A62 (i.e., ACT->GCT at codon 62, for amino acid substitution: T62->A62) through PCR-based method by using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, Cat 200524). Primers used were 5′-ttgacatcattggctgacgctttcgaacacgtgatag-3′ and 5′-ctatcacgtgttcgaaagcgtcagccaatgatgtcaa-3′. All the constructs were confirmed by Sanger sequencing to exclude potential PCR-introduced mutations. The pCMV6-AC-GFP-CHEK2 construct was purchased from Origene (Rockville, MD, USA, Cat RG201278).
Osteoblastic Cell Culture
Human fetal osteoblastic 1.19 cell line (hFOB) was obtained from American Type Culture Collection (ATCC, Cat CRL-11372). The hFOB has the ability to differentiate into mature osteoblasts expressing normal osteoblast phenotype (14). In this study, hFOB cells were maintained at 37°C in complete medium consisting of 1:1 Dulbecco’s modified Eagle’s medium (DMEM)/Ham’s F-12 medium (DMEM/F-12) without phenol red (Hyclone, Cat SH3027201), supplemented with 10% fetal bovine serum (Hyclone, Cat SH30070.03) and 0.3 mg/mL G418/geneticin (Invitrogen, Cat 10131-035). For osteoblastic differentiation, confluent cultures of hFOB cells were maintained at 37°C in complete medium with the addition of differentiation cocktail: 50 μg/μL ascorbic acid, 10−8 M dexamethasone, and 8mM β-glycerolphosphate (all from Sigma, St Louis, MO).
Human osteoblast-like cell line MG63 (ATCC, Cat CRL-1427) was cultured in Eagle’s Minimum Essential Medium (EMEM) (ATCC, Cat 30-2003) containing 10% fetal bovine serum and 1% penicillin and streptomycin (Sigma-Aldrich, Cat P4333) at 37°C in an atmosphere of 5% CO2.
Transient and Stable Transfection
For transient transfection, the hFOB cells or MG63 cells were seeded at 6×105 cells/well in polystyrene-coated 6-well plates. After 24 hours, cells were transfected with 1.0 μg of plasmids, 6.0 μl of Lipofectamine PLUS reagent (Invitrogen, Cat 11514-015), plus 4.0 μl of Lipofectamine™ reagent per well (Invitrogen, Cat 18324-020). The pCMV6-AC-GFP-CHEK2 was co-transfected with pCDNA3.1-BDNF-V66T62, pCDNA3.1-BDNF-M66T62, and pCDNA3.1-BDNF-V66A62, respectively. After 1–3 days, hFOB cells were harvested and lysed for downstream assays. All transfection experiments were performed three times (duplicates for each condition).
For stable transfection, hFOB cells were transfected with pCDNA3.1-BDNF-V66T62, pCDNA3.1-BDNF-M66T62, and pCDNA3.1-BDNF-V66A62, respectively. After 24 hours, 50 μg/ml selective antibiotic hygromycin B was added to the culture medium. After one week selection, 25 μg/ml hygromycin B was maintained in the culture.
BDNF-CHEK2 Protein Interaction Assay
Co-immunoprecipitation (Co-IP) and Western Blotting (WB) procedures were employed to test protein-protein interaction between protein BDNF and kinase CHEK2. Briefly, 48 hours after transient transfection, hFOB cells were lysed and clarified by centrifugation. Total proteins in supernatants were quantified using a BCA Protein Assay Kit (Pierce Chemical Co., Cat 23225). A Co-immunoprecipitation Kit (Pierce, Cat 26149) was used to pull down BDNF from hFOB total proteins. Specifically, mouse anti-human BDNF antibody (Abcam, Cat ab10505) was coupled to an amine-reactive gel. Then, the supernatants of cell lysate (50 μg total protein) were incubated with the antibody-coupled gel in spin-columns for 2.0 hours at 4 °C. After washing, proteins retained in the spin columns were eluted. The eluates were separated by electrophoresis on a 12% Tris-HCl gel and transferred to a PVDF membrane. The membrane was incubated with mouse anti-human CHEK2 monoclonal antibody (Abcam, Cat ab3292) or an IgG control antibody, and then incubated with goat anti-mouse HRP-conjugated secondary antibody (Abnova, Cat PAB0096). Protein bands were visualized using chemiluminescent detection reagents (Bio-Rad, Cat 170-5070) and imaged by using VesDoc MP 4000 system (Bio-Rad, Hercules, CA, USA).
BDNF Protein Phosphorylation Assay
a) BDNF total protein phosphorylation assay
BDNF protein was purified from transiently transfected hFOB cell lysate through IP procedure by using the Co-IP kit (Pierce, Cat 26149) and mouse anti-human BDNF antibody (Abcam, Cat ab10505). Then, BDNF protein phosphorylation level was quantitated using a Phosphoprotein Phosphate Estimation Assay Kit (Thermo Scientific, Cat 23270) according to manufacturer’s instruction. The assay is based on the alkaline hydrolysis of phosphate from seryl and threonyl residues in phosphoprotein, followed by colorimetric quantification of the released phosphate by use of malachite green and ammonium molybdate. Herein, BioTek Synergy HT Multi-Mode Microplate Reader (BioTek, Winooski, VT, USA) and BioTek Gen5 Data Analyses Software were used to collect and analyze the data.
b) BDNF T62 site-specific phosphorylation assay
Transiently transfected MG63 were lysed in 1X cell lysis buffer (62.5 mM Tris-HCl pH 6.8, 10% glycerol, and 2% SDS) and homogenized. Equal amount of cell lysate (25 μg total protein) was subjected to electrophoresis on a 12% Tris-HCl gel under reducing conditions, and transferred to a PVDF membrane (Millipore, Billerica, MA, USA). Membrane was blocked with 5% non-fat milk in phosphate buffered saline (0.1% Tween). The membrane was incubated with rabbit anti-human BDNF-pT62 polyclonal antibody (Custom-made, ProSci Incorporated, Poway, CA, USA), and a goat anti-rabbit HRP-conjugated secondary antibody (Sigma-aldrich, Cat A0545). In a parallel blot, total BDNF was incubated with mouse anti-human BDNF primary antibody (Abcam, Cat ab10505) and goat anti-mouse HRP-conjugated secondary antibody (Abnova, Cat PAB0096). Both the BDNF-pT62 and total BDNF proteins were visualized using chemiluminescence detection kit (Bio-Rad, Cat 170-5070) and imaged with VesDoc MP 4000 system (Bio-Rad, Hercules, CA, USA). Then, both blots were stripped and incubated with goat anti-rabbit actin antibody (Sigma, Cat A5060) and imaged again to detect house-keeping protein beta-actin (internal control).
Protein band intensities were analyzed with Quantity One software (Bio-Rad, Hercules, CA, USA). For each sample, the intensity of the BDNF-pT62 and total BDNF protein bands were normalized against that of beta-actin band on the two parallel blots, respectively. The ratio of the normalized BDNF-pT62 intensity to the normalized total BDNF intensity was used to represent BDNF phosphorylation level at site T62. Western blot experiments were performed twice.
Testing Effect of PhosSNP rs6265 on Osteoblastogenesis in Vitro
Osteopontin (OPN) and alkaline phosphatase (ALP) are known to be regulated during osteoblastic differentiation and are commonly used as “osteoblast markers” (15–17). Bone morphogenetic protein 2 (BMP2) is a potent osteoblastic factor stimulating osteoblastogenesis. To assess the effect of rs6265 on osteoblastogenesis, we compared mRNA expression levels (OPN and BMP2) and enzyme activity (ALP) of these osteoblastic maker genes in hFOB cells that had been transfected with wild-type vs. variant BDNF gene. Related experimental procedures are detailed as follows.
mRNA Expression Assay of Osteoblastic Genes (OPN and BMP2) by Quantitative real time PCR
24 hours after transfection of pCDNA3.1-BDNF-V66T62, pCDNA3.1-BDNF-M66T62, pCDNA3.1-BDNF-V66A62 constructs, and pCDNA3.1 empty vector individually, hFOB cells were lysed and reverse transcribed using the Power SYBR Green Cells-to-CT Kit (Applied Biosciences, Cat 4402954) according to the manufacturer’s instructions. OPN and BMP2 mRNA levels were quantified by quantitative PCR using a MiQ qPCR cycler and MiQ software (Bio-Rad). Both gene expression levels were normalized against the house-keeping gene Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH, internal control). The PCR primers used are shown in Supplemental Table 1. The quantitative RT-PCR experiments were performed in triplicates for each condition and repeated twice.
Osteoblastic ALP Enzyme Activity Assay by ALP Staining
ALP staining was performed to test ALP enzyme activity by using TRACP & ALP Double-Stain Kit (Takara, Cat MK300) following the manufacturer’s instructions. 5×104 stably transfected hFOB cells were plated on 48-well plates the day before staining. The experiments were performed in triplicates for each condition and repeated twice.
Results
Identification of PhosSNPs Significant for BMD in Humans
Basic characteristics of the three study population samples were summarized in Table 1. In the CAU1 sample, we identified nine phosSNPs associated with both spine BMD and hip BMD (p<0.05) (Table 2). Among the nine phosSNPs, three phosSNPs (rs16861032, rs2657879, and rs6265) were replicated in the CAU2 sample (p=0.05). Notably, association of phosSNP rs6265 (major/minor allele: G/A) with hip BMD was also suggested in the CHN sample (p=0.09), suggesting that rs6265 has ethnic-general effect on hip BMD. Interestingly, the direction of genotypic effect is consistent among the three studied population samples. Specifically, homozygous AA carriers have significantly decreased hip BMD than allele G carriers (GA or GG) (Figure 1).
Table 1.
Basic Characteristics of the Three Human Population Samples
| CAU1 (N = 1,000) | CAU2 (N = 2,286) | CHN (N = 1,627) | |
|---|---|---|---|
| Age (yrs) | 50.33 ± 18.31 | 51.37 ± 13.75 | 34.48 ± 13.24 |
| Height (m) | 1.71 ± 0.10 | 1.66 ± 0.08 | 1.64 ± 0.08 |
| Weight (kg) | 80.16 ± 17.79 | 75.25 ± 17.53 | 60.12 ± 10.48 |
| Spine BMD (g/cm2) | 1.03 ± 0.16 | 1.02 ± 0.15 | 0.95 ± 0.13 |
| Hip BMD (g/cm2) | 0.97 ± 0.16 | 0.96 ± 0.15 | 0.92 ± 0.13 |
Note: presented are mean and S.E.
Table 2.
PhosSNPs Associated with Spine BMD and Hip BMD in CAU1 and/or Replicated in CAU2 and CHN Samples
| SNP | Allele 1 | Allele 2 | P Value for Spine BMD | P Value for Hip BMD | Gene Symbol | ||||
|---|---|---|---|---|---|---|---|---|---|
| CAU1 | CAU2 | CHN | CAU1 | CAU2 | CHN | ||||
| rs11096957 | C* | A | 4.25E-05 | 7.33E-01 | 3.05E-01 | 1.28E-03 | 9.00E-01 | 3.42E-01 | TLR10 |
| rs11096955 | C* | A | 6.05E-05 | 6.62E-01 | 3.10E-01 | 1.81E-03 | 9.54E-01 | 3.22E-01 | TLR10 |
| rs16861032 | C | G | 2.15E-03 | 2.57E-02 | 1.59E-01 | 3.24E-02 | 1.17E-02 | 7.51E-01 | CCDC52 |
| rs2657879 | C | T | 5.62E-03 | 3.54E-02 | 6.31E-01 | 4.42E-02 | 7.27E-02 | 8.99E-01 | GLS2 |
| rs594445 | A | C | 5.97E-03 | 3.82E-01 | 3.44E-01 | 1.15E-04 | 2.02E-01 | 6.29E-01 | MOCOS |
| rs6265 | A | G | 1.33E-02 | 3.64E-03 | 3.08E-01 | 1.87E-03 | 5.12E-02 | 9.92E-02 | BDNF |
| rs16995685 | A* | C | 1.56E-02 | 3.53E-01 | 2.65E-01 | 4.92E-02 | 7.37E-01 | 5.06E-01 | DEFB127 |
| rs2071460 | C* | T | 4.17E-02 | 1.78E-01 | 7.31E-01 | 4.04E-02 | 8.68E-01 | 5.43E-01 | CSNK2A1P |
| rs185435 | G | A | 4.74E-02 | 8.64E-01 | 5.41E-01 | 6.84E-04 | 9.01E-01 | 8.00E-01 | FCHO2 |
Allele 1 is the minor allele in both CAU and CHN, except those marked in * which are minor in CAU but major in CHN.
Figure 1. Genotypic Effect of PhosSNP rs6265 on Hip BMD in Human Populations.
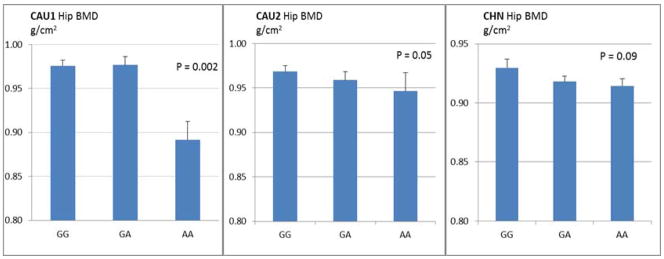
Presented are mean and S.E. Homozygous AA carriers lack wild-type protein BDNF-V66. The osteoblastic signaling pathways, regulated by BDNF-V66-mediated BDNF-T62 phosphorylation, probably are either blocked or attenuated, leading to inhibited bone formation and decreased BMD in humans.
Predictive Impact of PhosSNP on Protein Phosphorylation
The potential impacts of the above three significant phosSNPs (rs16861032, rs2657879, and rs6265) on protein phosphorylation were predicted and summarized in Supplemental Table 2. Generally speaking, the allele-specific gene sequences encode different protein isoforms, which either change (create or remove) phosphorylation site(s) of the encoded protein or change the kinases that catalyze phosphorylation of target site(s). To take phosSNP rs6265 as an example, the BDNF gene harboring major allele G and minor allele A encodes BDNF protein isoforms with amino acid residue Val (V) and Met (M) at position 66 (abbreviated as BDNF-V66 and BDNF-M66), respectively. Bioinformatics analyses predicted that compared with BDNF-M66, BDNF-V66 creates a new phosphorylation site at residue T62, which can be targeted and catalyzed by protein kinase CHEK2. CHEK2 is the only one kinase that is predicated to act on this specific phosphorylation site of BDNF. However, the substrate-kinase relationship between BDNF and CHEK2 and the effect of rs6265 on BDNF-T62 phosphorylation have not been experimentally validated previously.
Experimental Validation of rs6265 on Protein Phosphorylation
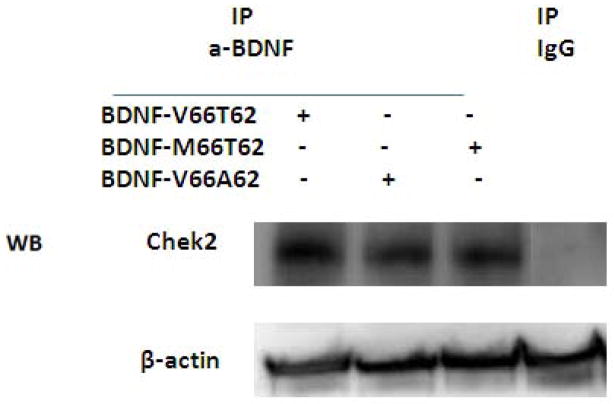
We found that CHEK2 protein kinase in hFOB cell lysates can be pulled down together with BDNF protein by anti-BDNF antibody during co-IP process, as visualized on Western blot using anti-CHEK2 antibody (Figure 2). The finding supports and validates substrate-kinase interaction between BDNF and CHEK2 proteins in bone cells. Furthermore, comparison of CHEK2 protein band intensity on Western blot, after beta-actin normalization, showed that more CHEK2 protein was pulled down with BDNF-V66T62 than BDNF-M66T62 or BDNF-V66A62, suggesting weakened interaction between protein kinase CHEK2 and the variant/mutant BDNF protein isoforms.
Figure 2. Substrate-Kinase Interaction Assay for BDNF and CHEK2 Proteins.
The protein-protein interaction assay was assessed by Co-IP and WB.
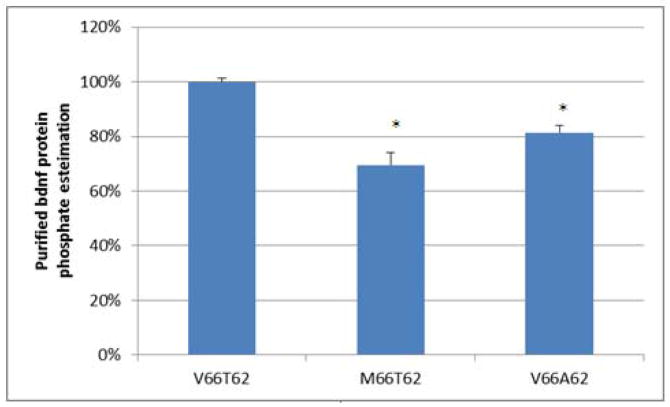
We also found that BDNF total protein phosphorylation level was significantly reduced in hFOB cells transfected with variant BDNF-M66T62, compared with that transfected with wild-type BDNF-V66T62 (Figure 3). The finding suggests that BDNF total protein phosphorylation is regulated by the amino acid substitution Val66Met, resulting from the nucleotide variation at rs6265.
Figure 3. Allele-Specific Effects of rs6265 on BDNF Total Protein Phosphorylation Level.
Presented are estimated phosphate levels in purified BDNF proteins (mean and standard error). The phosphate level under the condition of wild-type BDNF transfection (V66T62) is scaled as 100%. The Phosphoprotein Phosphate Estimation Assay is based on the alkaline hydrolysis of phosphate from seryl and threonyl residues in phosphoprotein followed by quantification of the released phosphate by use of malachite green and ammonium molybdate. hFOB cells transfected with variant or mutant BDNF (M66T62 or V66A62) showed decreased total BDNF phosphorylation levels, compared with hFOB cells transfected with wild-type BDNF (V66T62). Experiments are in duplicates. *: P<0.05 as compared with V66T62.
Furthermore, site-specific protein phosphorylation analyses showed that normalized BDNF-pT62 protein band intensities level on Western blot was significantly decreased in MG63 cells transfected with variant BDNF-M66T62, compared with that transfected with wild-type BDNF-V66T62 (Figure 4). The finding suggests that T62 is the target phosphorylation site regulated by amino acid residue at position 66, and that substitution of amino acid residue V66 to M66 attenuates BDNF phosphorylation at site T62. Consistently, when T62 is substituted with unphosphorylatable A62, BDNF total phosphorylation level was significantly decreased (Figure 3, V66A62 vs. V66T62), suggesting that T62 is a primary target site of phosphorylation in BDNF protein.
Figure 4. Allele-Specific Effects of rs6265 on BDNF Protein Phosphorylation Level at Site T62.
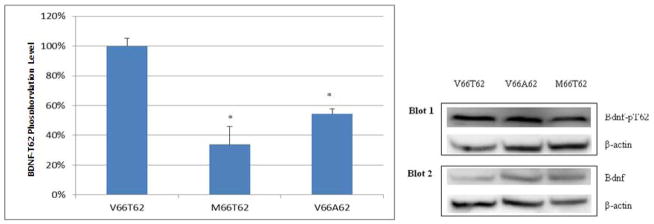
Both the BDNF-pT62 and total BDNF, under the same condition, were quantified from two parallel blots. Their intensities were normalized against internal control beta-actin protein, respectively. The ratio of normalized BDNF-pT62 intensity to normalized total BDNF intensity was used to represent BDNF T62 site-specific phosphorylation level. Presented to the right are representative western blot images. Presented to the left are BDNF T62 site-specific phosphorylation level (mean and standard error). The phosphorylation level is scaled as 100% under the condition of wild-type BDNF transfection (V66T62). MG63 cells transfected with variant or mutant BDNF (M66T62 or V66A62) showed decreased T62 phosphorylation, compared with MG63 cells transfected with wild-type BDNF (V66T62). Experiments are in duplicates. *: P<0.05, as compared with V66T62.
Effect of PhosSNP rs6265 on Osteoblast Differentiation In Vitro
Quantitative real time PCR experiments showed that, transient transfection with wild-type BDNF-V66T62 significantly increases OPN and BMP2 mRNA expression levels in hFOB cells, as compared with empty vector control (Figure 5). In contrast, the stimulatory effects on both OPN and BMP2 mRNA expression were significantly reduced by transfection of either variant BDNF-M66T62 or mutant BDNF-V66A62, as compared with that of wild-type BDNF-V66T62 transfection (Figure 5). ALP staining of hFOB cell cultures showed that, transfection with wild-type BDNF-V66T62 significantly increases ALP enzyme activity in hFOB cells, as compared with empty vector control (Figure 6, A vs. B). In contrast, the stimulatory effects on ALP activity were significantly reduced by transfection with either variant BDNF-M66T62 or mutant BDNF-V66A62, as compared with that of wild-type BDNF-V66T62 transfection (Figure 6, B vs. C and B vs. D).
Figure 5. Allele-Specific Effects of rs6265 on Osteoblastic Gene Expression (OPN and BMP2) in hFOB Cells.
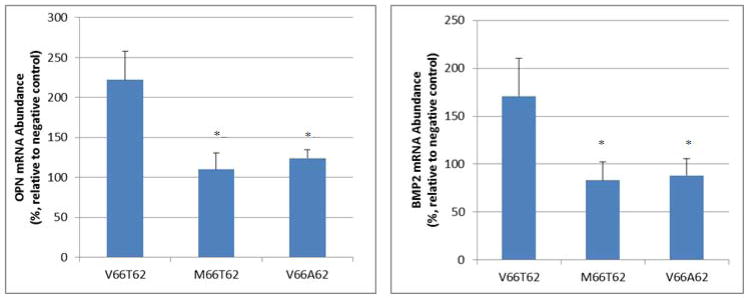
Presented are mRNA abundance in hFOB cells. Raw data were normalized against the internal control GAPDH mRNA (mean and standard error). Negative control refers to empty vector transfection. The hFOB cells transfected with variant or mutant BDNF (M66T62 or V66A62) showed decreased OPN and BMP2 mRNA expression levels, compared with hFOB cells transfected with wild-type BDNF (V66T62). Experiments are in triplicates and repeated twice. *: P<0.05, as compared with V66T62.
Figure 6. Osteoblastic Enzyme Activity in hFOB Cell Culture by ALP Staining.
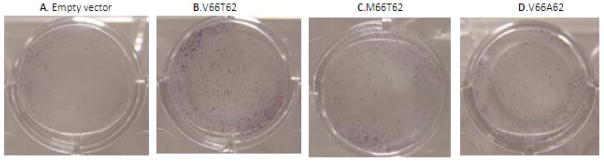
ALP is a specific osteoblast maturation marker. Compared with negative control (i.e., empty vector transfection), over-expression of wild-type BDNF (V66T62) significantly enhances ALP activity in hFOB cells (A vs. B). In contract, hFOB cells transfected with mutant BDNF (M66T62 or V66A62) show lower ALP activity, compared with hFOB cells transfected with wild-type BDNF (V66T62) (B vs. C; B vs. D).
Discussion
Through a pioneer genome-wide phosSNP association study for BMD in three independent human population samples, we identified and replicated three phosSNPs, i.e., rs16861032, rs2657879, and rs6265, which are significant for both spine BMD and hip BMD in Caucasians. Specially, one phosSNP, i.e., rs6265, was found to be associated with hip BMD across ethnicity boundary in Chinese. Through functional studies, we validated and characterized that rs6265 affects substrate-kinase interaction between BDNF protein and CHEK2 kinase, regulates BDNF phosphorylation at site T62, and influences osteoblastic gene expression and osteoblast differentiation. This study, driven by biological significance of protein phosphorylation and integrating advantage of hypothesis-free genome-wide screening, focuses on a subset of potentially functional SNPs while limiting multiple testing problem of traditional GWAS studies. Evidence gained from in vitro molecular and cellular studies supplements prior findings generated in human samples, validates and strengthens significance of the identified phosSNP to BMD.
SNP rs6265 is located in BDNF gene. BDNF is conserved in chimpanzee, dog, cow, mouse, rat, and chicken, indicating its important role through evolution. Protein BDNF promotes differentiation and survival of developing neurons and their maintenance in the adult nervous system (18,19). Previous animal studies showed that BDNF also functions in chondrocyte and osteoblast, participates in cartilage development, ossification, and osteogenesis, and plays an important role in bone growth and development, remodeling and regeneration (20–22). This study, for the first time, highlights the significance of BDNF to BMD variation in human populations.
SNP rs6265 (Val66Met) plays a very important role in determining many phenotypes in humans. Previous studies show that SNP rs6265 was associated with neuropsychiatric disorders, such as bipolar disorder (23) and eating disorder (24), weight (25), obesity (25–28), etc. To the best of our knowledge, this study firstly identified that rs6265 is associated with bone phenotype in humans. To be noted, association of rs6265 with BMD variation was also suggested in the largest meta-analysis involving 32,961 individuals of European and East Asian ancestry (i.e., GEFOS study, http://www.gefos.org/) (29). Per the released GEFOS data, rs6265 was associated with femoral neck BMD in men (p=0.0034) (29). Furthermore, the direction of its effect was consistent with that in our study samples. Specifically, homozygous minor allele A carriers (AA) have significantly decreased BMD than major allele G carriers (GA and GG).Consistently, BDNF-V66 (major allele G at rs6265) transfection significantly increases expression of osteoblast-specific markers (OPN, BMP2, and ALP) and promotes osteoblast differentiation and maturation in cell culture. In contrast, BDNF-M66 (allele A at rs6265) transfection decreased expression of osteoblast marker genes and osteoblast activity. Our findings suggest that rs6265, via regulating gene expression, regulates osteoblastogenesis and bone formation.
It has been reported that variant BDNF-M66 alters intracellular trafficking and impairs BDNF secretion (13,30). Extracellular BDNF protein induces mRNA expression of ALP, OPN, and BMP-2 in human cementoblast-like (HCEM) cells, through TrkB-c-Raf-ERK1/2-Elk-1 signaling pathway (31). The rs6265 (Val66Met) is located in the N-terminal pro-peptide (AA 19–128) of BDNF precursor protein (247 AA in total; UniProtKB database accession number: P23560). Cleaving off of the pro-domain can take place before or after secretion of the pro-BDNF (32). In this study, we presented that variant BDNF-M66 impaired BDNF-T62 phosphorylation. However, whether and how the altered BDNF-T62 phosphorylation influences BDNF processing, maturation, and activity is unclear yet. Herein, we propose that rs6265, through changing interaction between protein substrate BDNF and protein kinase CHEK2 and mediating BDNF-T62 phosphorylation, regulates osteoblastic gene expression and differentiation, hence influences bone formation and BMD.
To be noted, in this pilot phosSNP-BMD association study, due to limited coverage of phosSNPs by the current Affymetrix SNP arrays, a large number of phosSNPs has not been studied. This issue could be addressed by two means. On one hand, availability of high coverage custom-made SNP arrays with an exclusive coverage of phosSNPs may accelerate high-throughput screening and discovery of phosSNPs involved in determining variation of human complex traits or involved in pathogenesis of human complex diseases, including osteoporosis. On the other hand, based on available microarray data and through referring ethnicity-matched human genome haplotype map, genotype imputation for uncovered phosSNPs could be performed to attain a higher coverage of phosSNP at genome-wide scale, thus contributing to identification of more interesting phosSNPs for human complex traits or diseases of interest. To be noted, ~97% of the phosSNPs in the phosSNP 1.0 database were predicted by bioinformatics. Thus, their potential impacts on protein phosphorylation have yet to be validated by experiments. Meanwhile, functional mechanisms of these SNPs in human body have yet to be elucidated through in-depth investigation. This study represents our novel endeavor to dissect the functions of phosSNPs in the bone field, which might stimulate more comprehensive large-scale studies in future. Furthermore, similar strategies could be applied to studies of other human complex traits and complex diseases as well.
In summary, we have identified and characterized a functional phosSNP significant for BMD in humans, and illustrated a novel biological mechanism underlying BMD variation in population. We conclude that SNP rs6265, through affecting protein substrate-kinase interaction, and regulating BDNF protein phosphorylation and osteoblast differentiation, influences hip BMD in general human populations.
Supplementary Material
Acknowledgments
We are grateful to Drs. Yu Xue and Jian Ren for sharing the annotation information of the phosSNPs compiled in PhosSNP 1.0 database. We are thankful to Dr. Francis Lee for his gift plasmids and Dr. Tianhua Niu for editing of the manuscript. The study personnel were partially supported by Soochow University Startup Fund, a Project of the Priority Academic Program Development of Jiangsu Higher Education Institutions, and grants from National Institutes of Health (Website: http://www.nih.gov; Grant numbers: P50AR055081, R01AG026564, R01AR050496, R01AR057049, and R03TW008221). FYD: study design, data analyses, manuscript drafting and finalization; LJT: functional experiments, data analyses, manuscript revision; HS, YJL, YZL: coordinated genotyping effort for the CAU1 and CAU2 samples; JL and QT: prepared genotype and phenotype data for GWAS; XZZ and XDC: coordinated the genotyping effort for the CHN sample; MZ: functional experiment design and manuscript revision; HWD: study design, manuscript revision and finalization.
Footnotes
All authors state that they have no conflicts of interest.
Reference List
- 1.Shen H, Zhang YY, Long JR, Xu FH, Liu YZ, Xiao P, Zhao LJ, Xiong DH, Liu YJ, Dvornyk V, Rocha-Sanchez S, Liu PY, Li JL, Conway T, Davies KM, Recker RR, Deng HW. A genome-wide linkage scan for bone mineral density in an extended sample: evidence for linkage on 11q23 and Xq27. J Med Genet. 2004;41(10):743–51. doi: 10.1136/jmg.2004.020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frogne T, Laenkholm AV, Lyng MB, Henriksen KL, Lykkesfeldt AE. Determination of HER2 phosphorylation at tyrosine 1221/1222 improves prediction of poor survival for breast cancer patients with hormone receptor-positive tumors. Breast Cancer Res. 2009;11 (1):R11. doi: 10.1186/bcr2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munoz L, Ammit AJ. Targeting p38 MAPK pathway for the treatment of Alzheimer’s disease. Neuropharmacology. 58(3):561–8. doi: 10.1016/j.neuropharm.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Srivastava S, Weitzmann MN, Kimble RB, Rizzo M, Zahner M, Milbrandt J, Ross FP, Pacifici R. Estrogen blocks M-CSF gene expression and osteoclast formation by regulating phosphorylation of Egr-1 and its interaction with Sp-1. J Clin Invest. 1998;102(10):1850–9. doi: 10.1172/JCI4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374(Pt 1):1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ihnatko R, Kubes M. TNF signaling: early events and phosphorylation. Gen Physiol Biophys. 2007;26(3):159–67. [PubMed] [Google Scholar]
- 7.Insogna K, Tanaka S, Neff L, Horne W, Levy J, Baron R. Role of c-Src in cellular events associated with colony-stimulating factor-1-induced spreading in osteoclasts. Mol Reprod Dev. 1997;46(1):104–8. doi: 10.1002/(SICI)1098-2795(199701)46:1<104::AID-MRD16>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 8.Duong LT, Rodan GA. Integrin-mediated signaling in the regulation of osteoclast adhesion and activation. Front Biosci. 1998;3:d757–68. doi: 10.2741/A319. [DOI] [PubMed] [Google Scholar]
- 9.Ren J, Jiang C, Gao X, Liu Z, Yuan Z, Jin C, Wen L, Zhang Z, Xue Y, Yao X. PhosSNP for systematic analysis of genetic polymorphisms that influence protein phosphorylation. Mol Cell Proteomics. 2010;9(4):623–34. doi: 10.1074/mcp.M900273-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng HW, Shen H, Xu FH, Deng HY, Conway T, Zhang HT, Recker RR. Tests of linkage and/or association of genes for vitamin D receptor, osteocalcin, and parathyroid hormone with bone mineral density. J Bone Miner Res. 2002;17(4):678–86. doi: 10.1359/jbmr.2002.17.4.678. [DOI] [PubMed] [Google Scholar]
- 11.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue Y, Ren J, Gao X, Jin C, Wen L, Yao X. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics. 2008;7(9):1598–608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen ZY, Patel PD, Sant G, Meng CX, Teng KK, Hempstead BL, Lee FS. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci. 2004;24(18):4401–11. doi: 10.1523/JNEUROSCI.0348-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris SA, Enger RJ, Riggs BL, Spelsberg TC. Development and characterization of a conditionally immortalized human fetal osteoblastic cell line. J Bone Miner Res. 1995;10 (2):178–86. doi: 10.1002/jbmr.5650100203. [DOI] [PubMed] [Google Scholar]
- 15.Garcia T, Roman-Roman S, Jackson A, Theilhaber J, Connolly T, Spinella-Jaegle S, Kawai S, Courtois B, Bushnell S, Auberval M, Call K, Baron R. Behavior of osteoblast, adipocyte, and myoblast markers in genome-wide expression analysis of mouse calvaria primary osteoblasts in vitro. Bone. 2002;31(1):205–11. doi: 10.1016/s8756-3282(02)00781-0. [DOI] [PubMed] [Google Scholar]
- 16.Ongphiphadhanakul B, Jenis LG, Braverman LE, Alex S, Stein GS, Lian JB, Baran DT. Etidronate inhibits the thyroid hormone-induced bone loss in rats assessed by bone mineral density and messenger ribonucleic acid markers of osteoblast and osteoclast function. Endocrinology. 1993;133(6):2502–7. doi: 10.1210/endo.133.6.8243271. [DOI] [PubMed] [Google Scholar]
- 17.Zhou H, Choong P, McCarthy R, Chou ST, Martin TJ, Ng KW. In situ hybridization to show sequential expression of osteoblast gene markers during bone formation in vivo. J Bone Miner Res. 1994;9(9):1489–99. doi: 10.1002/jbmr.5650090922. [DOI] [PubMed] [Google Scholar]
- 18.Jones KR, Reichardt LF. Molecular cloning of a human gene that is a member of the nerve growth factor family. Proc Natl Acad Sci U S A. 1990;87(20):8060–4. doi: 10.1073/pnas.87.20.8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutchison MR, Bassett MH, White PC. SCF, BDNF, and Gas6 are regulators of growth plate chondrocyte proliferation and differentiation. Mol Endocrinol. 2010;24(1):193–203. doi: 10.1210/me.2009-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamashiro T, Fukunaga T, Yamashita K, Kobashi N, Takano-Yamamoto T. Gene and protein expression of brain-derived neurotrophic factor and TrkB in bone and cartilage. Bone. 2001;28(4):404–9. doi: 10.1016/s8756-3282(01)00405-7. [DOI] [PubMed] [Google Scholar]
- 22.Aiga A, Asaumi K, Lee YJ, Kadota H, Mitani S, Ozaki T, Takigawa M. Expression of neurotrophins and their receptors tropomyosin-related kinases (Trk) under tension-stress during distraction osteogenesis. Acta Med Okayama. 2006;60(5):267–77. doi: 10.18926/AMO/30739. [DOI] [PubMed] [Google Scholar]
- 23.Fan J, Sklar P. Genetics of bipolar disorder: focus on BDNF Val66Met polymorphism. Novartis Found Symp. 2008;289:60–72. doi: 10.1002/9780470751251.ch5. discussion 72–3, 87–93. [DOI] [PubMed] [Google Scholar]
- 24.Gratacos M, Gonzalez JR, Mercader JM, de Cid R, Urretavizcaya M, Estivill X. Brain-derived neurotrophic factor Val66Met and psychiatric disorders: meta-analysis of case-control studies confirm association to substance-related disorders, eating disorders, and schizophrenia. Biol Psychiatry. 2007;61(7):911–22. doi: 10.1016/j.biopsych.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 25.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, Kiemeney LA, Pedersen O, Kong A, Thorsteinsdottir U, Stefansson K. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 26.Gunstad J, Schofield P, Paul RH, Spitznagel MB, Cohen RA, Williams LM, Kohn M, Gordon E. BDNF Val66Met polymorphism is associated with body mass index in healthy adults. Neuropsychobiology. 2006;53(3):153–6. doi: 10.1159/000093341. [DOI] [PubMed] [Google Scholar]
- 27.Shugart YY, Chen L, Day IN, Lewis SJ, Timpson NJ, Yuan W, Abdollahi MR, Ring SM, Ebrahim S, Golding J, Lawlor DA, Davey-Smith G. Two British women studies replicated the association between the Val66Met polymorphism in the brain-derived neurotrophic factor (BDNF) and BMI. Eur J Hum Genet. 2009;17(8):1050–5. doi: 10.1038/ejhg.2008.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu L, Xi B, Zhang M, Shen Y, Zhao X, Cheng H, Hou D, Sun D, Ott J, Wang X, Mi J. Associations of six single nucleotide polymorphisms in obesity-related genes with BMI and risk of obesity in Chinese children. Diabetes. 2010;59(12):3085–9. doi: 10.2337/db10-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, Oei L, Albagha OM, Amin N, Kemp JP, Koller DL, Li G, Liu CT, Minster RL, Moayyeri A, Vandenput L, Willner D, Xiao SM, Yerges-Armstrong LM, Zheng HF, Alonso N, Eriksson J, Kammerer CM, Kaptoge SK, Leo PJ, Thorleifsson G, Wilson SG, Wilson JF, Aalto V, Alen M, Aragaki AK, Aspelund T, Center JR, Dailiana Z, Duggan DJ, Garcia M, Garcia-Giralt N, Giroux S, Hallmans G, Hocking LJ, Husted LB, Jameson KA, Khusainova R, Kim GS, Kooperberg C, Koromila T, Kruk M, Laaksonen M, Lacroix AZ, Lee SH, Leung PC, Lewis JR, Masi L, Mencej-Bedrac S, Nguyen TV, Nogues X, Patel MS, Prezelj J, Rose LM, Scollen S, Siggeirsdottir K, Smith AV, Svensson O, Trompet S, Trummer O, van Schoor NM, Woo J, Zhu K, Balcells S, Brandi ML, Buckley BM, Cheng S, Christiansen C, Cooper C, Dedoussis G, Ford I, Frost M, Goltzman D, Gonzalez-Macias J, Kahonen M, Karlsson M, Khusnutdinova E, Koh JM, Kollia P, Langdahl BL, Leslie WD, Lips P, Ljunggren O, Lorenc RS, Marc J, Mellstrom D, Obermayer-Pietsch B, Olmos JM, Pettersson-Kymmer U, Reid DM, Riancho JA, Ridker PM, Rousseau F, Slagboom PE, Tang NL, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44(5):491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112(2):257–69. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 31.Kajiya M, Shiba H, Fujita T, Ouhara K, Takeda K, Mizuno N, Kawaguchi H, Kitagawa M, Takata T, Tsuji K, Kurihara H. Brain-derived neurotrophic factor stimulates bone/cementum-related protein gene expression in cementoblasts. J Biol Chem. 2008;283(23):16259–67. doi: 10.1074/jbc.M800668200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lessmann V, Brigadski T. Mechanisms, locations, and kinetics of synaptic BDNF secretion: an update. Neurosci Res. 2009;65(1):11–22. doi: 10.1016/j.neures.2009.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.