Abstract
While targeted therapy based on the idea of attenuating the activity of a preselected, therapeutically relevant protein has become one of the major trends in modern cancer therapy, no truly specific targeted drug has been developed and most clinical agents have displayed a degree of polypharmacology. Therefore, the specificity of anticancer therapeutics has emerged as a highly important but severely underestimated issue. Chemical proteomics is a powerful technique combining postgenomic drug-affinity chromatography with high-end mass spectrometry analysis and bioinformatic data processing to assemble a target profile of a desired therapeutic molecule. Due to high demands on the starting material, however, chemical proteomic studies have been mostly limited to cancer cell lines. Herein, we report a down-scaling of the technique to enable the analysis of very low abundance samples, as those obtained from needle biopsies. By a systematic investigation of several important parameters in pull-downs with the multikinase inhibitor bosutinib, the standard experimental protocol was optimized to 100 µg protein input. At this level, more than 30 well-known targets were detected per single pull-down replicate with high reproducibility. Moreover, as presented by the comprehensive target profile obtained from miniaturized pull-downs with another clinical drug, dasatinib, the optimized protocol seems to be extendable to other drugs of interest. Sixty distinct human and murine targets were finally identified for bosutinib and dasatinib in chemical proteomic experiments utilizing core needle biopsy samples from xenotransplants derived from patient tumor tissue. Altogether, the developed methodology proves robust and generic and holds many promises for the field of personalized health care.
Keywords: kinase, inhibitor, drug specificity, proteomic profiling, chemical proteomics, tandem mass spectrometry, Bosutinib, Dasatinib, tumor biopsies
INTRODUCTION
Over the past few decades, intensive research into cancer has led to the identification of a large number of oncogenes and other genes that are not inherently oncogenic but can significantly contribute to cancer development and progression. Thus, our understanding of the causes of cancer and the subsequent molecular pathology has significantly broadened. Naturally, these findings in cancer fundamentals have also opened new possibilities for both diagnosis and especially treatment. In this field, one of the greatest milestones was the advent of targeted therapy that is often connected with the discovery of imatinib (Gleevec) in the late 1990s. Imatinib revolutionized the treatment of chronic myeloid leukemia (CML) and was the first of a class of small molecule compounds that modulated the activity of a desired, therapeutically relevant protein (or in accordance with the present view of targeted therapy and modulation of the activity of several selected proteins simultaneously). Thus, neoplastic cells were specifically killed.1 Since imatinib entered the market, several other small-molecule targeted drugs have been approved by the U.S. Food and Drug Administration agency and the importance of targeted therapy in modern oncology continues to grow.2
Despite the undeniable success of specific anticancer agents, recent years have shown that several issues connected with this prospective concept still remain. Undoubtedly, one of the most frequently discussed topics is drug specificity.3 Even if enormous efforts have been made to design and develop compounds affecting only given molecular nodes in cancer circuits (as documented by research programs of many pharmaceutical and biotechnological companies), no strictly selective agent has been discovered. The majority of novel, potential therapeutics displays much broader inhibitory profiles than originally planned. Affecting unexpected targets can consequently lead to unwanted toxicity and cause a number of adverse side effects. On the other hand, incidental drug promiscuity does not necessarily have to be harmful. For example, new targets can lead to an increase in potency of a drug and/or new clinical indications, as demonstrated by the aforementioned CML drug imatinib. On the basis of the fact that it is, in addition to BCR-ABL, also a potent inhibitor of c-KIT and PDGFR, it has been approved for eight other diseases.2 Hence, the determination of the specificity profile and the delineation of plausible mechanism of action should be an integral part of the modern drug discovery pipeline.
In recent years, a variety of techniques such as haploinsufficiency profiling, three hybrid system, phage display, RNAi arrays, gene expression analyses or in silico computational predictions have been developed and successfully employed in the identification of binding partners of many promising antitumor agents.4,5 Moreover, some of these methods have become available as a commercial service. This is the case especially for the assessment of inhibitors against a panel of recombinant protein kinases6 that is provided, for example, by Millipore (KinaseProfiler), Invitrogen (SelectScreen Kinase Profiling), or DiscoveRx (KINOMEscan).7
Proteomics also offers a technology for molecular target identification. An affinity approach called chemical proteomics employs immobilized drugs to isolate protein interactors from complex protein mixtures. Analysis of these specific protein sets is then achieved by modern high-end mass spectrometry (MS) and collected data are subjected to bioinformatic processing and evaluation. Together, these principles promote chemical proteomics as an unbiased method that is applicable to any type of biological sample. One of the major advantages of this particular technology is that it enables the characterization of interacting proteins in a native form (splice variant, conformation, post-translational modifications, and expression level) under physiological conditions.8–10
Naturally, as with any analytical technique, chemical proteomics also harbors some limitations.10 For instance, chemical proteomics usually requires large quantities of biological material that restricts the majority of studies to cell lines, which at least in theory can be expanded infinitely. Although these cells are accepted as a valid experimental model and possess important attributes of primary cancer cells, none entirely display the phenotype of the original tumor from which the cell lines are derived.11 This difference has been clearly demonstrated in our recent study with the BCR-ABL kinase inhibitor bosutinib (SKI- 606), in which we have observed distinct target profiles in K562 cells versus peripheral blood mononuclear cell samples derived from two CML patients.12 Furthermore, it can be hypothesized that the possibility to perform systematic chemical proteomic experiments using as limited amounts of material as can be obtained from clinically relevant samples would open a new chapter in personalized medicine, particularly considering the emerging appreciation of the heterogeneity between tumors from different patients and, in fact, within different areas of any individual tumor.13
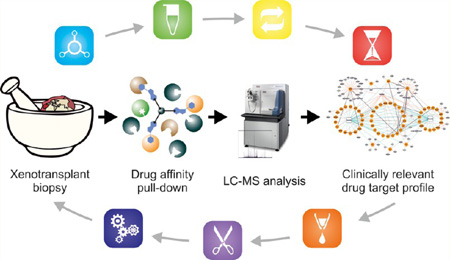
By combining acidic elution with gel-free one-dimensional proteomic analysis, we have previously achieved successful down-scaling of the pull-down procedure to 500 µg protein,14 which constituted about 10% of the amounts described in the original protocol.15 In the present study, we describe a further miniaturization of the methodology extending the range of the application to amounts obtainable from tumor biopsies. To obtain the highest number of relevant target identifications from 100 µg protein, a group of parameters affecting gel-free one-dimensional chemical proteomic experiments was addressed in pull-downs with bosutinib as a model drug. The optimized protocol allowed us to significantly reduce the input material and also the number of replicates analyzed by liquid chromatography– mass spectrometry (LC–MS), while still unambiguously identifying numerous targets of bosutinib. Additionally, successful pull-downs performed with another important clinical kinase inhibitor, dasatinib, show that the miniaturized protocol can be potentially applied to other drugs. All of these findings were finally verified in experiments using core needle biopsies from xenotransplants derived from patient lung tumor tissue.
MATERIALS AND METHODS
Chemicals
All chemicals used in this study were from commercial sources and were of analytical grade, unless stated otherwise.
Biological Material
Human CML K562 cells (ATCC, Washington, D.C.) were grown in suspension in RPMI 1640 medium supplemented with 10% fetal calf serum, l-glutamine, and penicillin/streptomycin and harvested by centrifugation. Harvested cells were washed with ice cold PBS, snap-frozen in liquid nitrogen and stored at −80 °C until lysed.
Lung tumor biopsies were obtained from patient-derived xenotransplants, as described earlier.16 In brief, primary tumor specimens were acquired at initial surgery from early stage nonsmall cell lung cancer (NSCLC) patients, cut into small pieces and immediately subcutaneously transplanted in immuno-deficient NOD/SCID mice (Taconic, Hudson, NY). Tumor growth was observed daily and at palpable tumor size, a core biopsy was performed with an 18 gauge needle. Biopsy samples were snap-frozen in liquid nitrogen and stored at −80 °C until lysed. Consent for use of human tumor tissue was obtained from all patients and the study was approved by the University of South Florida Institutional Review Board.
Compounds and Immobilization on Chromatography Media
c-Bosutinib was generated by Vichem Chemie (Budapest, Hungary) and c-dasatinib was synthesized by WuXi PharmaTech (Shanghai, China). Compounds were immobilized on NHS-activated Sepharose 4 Fast Flow (GE Healthcare Life Sciences, Uppsala, Sweden) as follows. Matrix (100, 50, or 25 µL) was washed with DMSO and incubated for at least 16 h at room temperature with a defined amount of appropriate compound and 0.1 M triethylamine. The completion of compound immobilization was verified by the analysis of the supernatant using HPLC−MS, and unreacted functional groups were blocked with an excess of ethanolamine (final concentration of 0.8 M). After blocking, the affinity resin was washed with DMSO prior to use for affinity chromatography.
Preparation of Cell and Tissue Lysate and Drug Affinity Purification Procedure
K562 cell pellets were resuspended in lysis buffer (50 mM Tris- HCl, pH 7.5; 100 mM NaCl; 0.2% NP-40; 5% glycerol; 1.5 mM MgCl2; 1 mM dithiothreitol (DTT); 25 mM NaF; 1 mM Na3VO4; 1 mM phenylmethylsulfonyl fluoride; 5 µg/µL tosyl-l-lysine chloromethyl ketone; 1 µg/µL leupeptin; 1 µg/µL aprotinin; 10 µg/µL soybean trypsin inhibitor), and cells were homogenized by passing the suspension 10 times through a 21 gauge needle. The lysate was incubated for 30 min on ice, and cell debris was removed by centrifugation (10 min at 20000g followed by 1 h at 100000g). Protein concentration was determined by Bradford assay (Bio-Rad, Hercules, CA), and the lysate was diluted to the desired concentration with the lysis buffer. The lysates were either snap-frozen in liquid nitrogen or used directly for affinity purifications.
Frozen biopsy samples were ground to a fine powder in liquid nitrogen using a precooled mortar and pestle. The homogenate was transferred to an eppendorf tube and extracted with 150 µL of the lysis buffer for 30 min on ice. After centrifugation, the lysate was then processed essentially as described above.
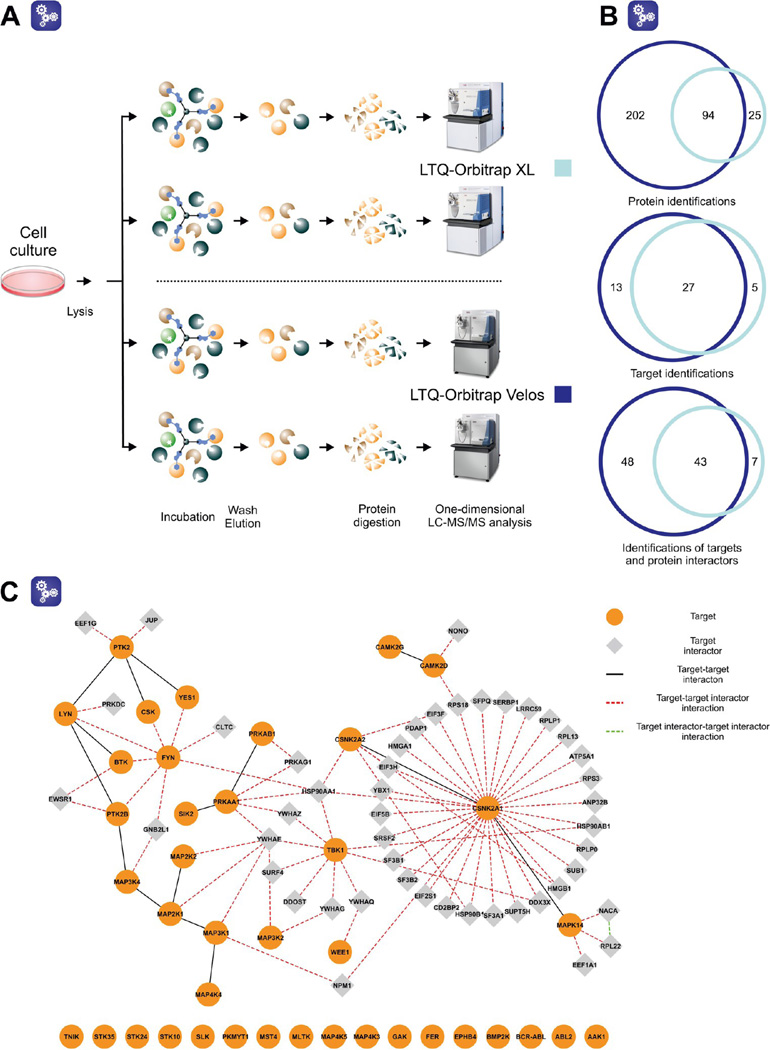
All affinity isolations were performed in duplicates, as previously described.14 Briefly, before performing a pull-down, the affinity resin was washed with precooled lysis buffer. Cell lysates were cleared by centrifugation (if frozen; 20 min at 100000g) and subjected to association with equilibrated resin at 4 °C under continuous agitation. Subsequently, the lysate−resin suspension was centrifuged (3 min at 75g) and transferred to a Micro Bio-Spin chromatography column (Bio-Rad). The resin was drained by gravity flow and washed with 5 mL lysis buffer and 2.5 mL HEPES/NaOH buffer (HEPES/NaOH, pH 7.5; 100 mM NaCl; 0.5 µM EDTA), respectively. The bound proteins were eluted directly into a glass vial containing 125, 250, or 500 µL (depending on the elution volume) 1 M triethylammonium bicarbonate buffer (SIGMA-Aldrich, St. Louis, MO) with 100 mM formic acid (FA), mixed by gentle vortexing and stored at −20 °C until further analysis. For more detailed information concerning the entire procedure, see Figure 1.
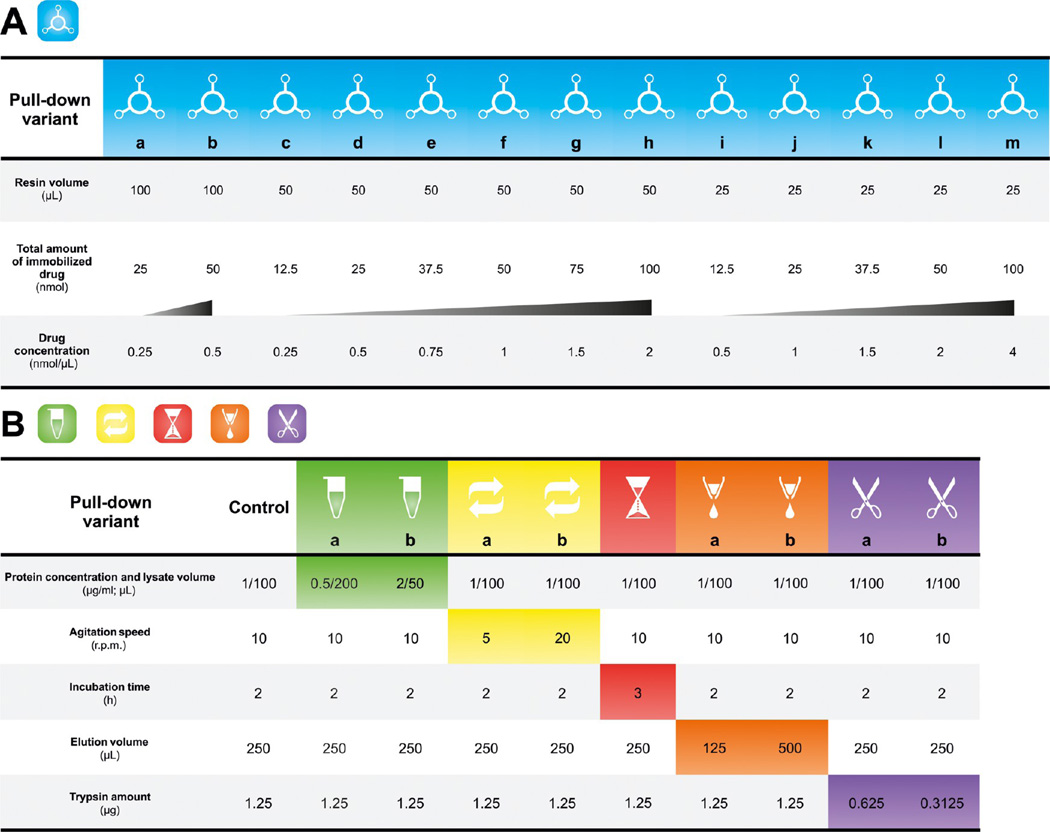
Figure 1.
Detailed procedure of bosutinib pull-down experiments. (A) Summary of bosutinib resin setups that were used for the investigation of affinity resin influences. In these experiments, the resin was incubated with 100 µL of K562 cell lysate (protein concentration of 1 µg/µL) at 4 °C for 2 h under continuous stirring (10 rpm). The elution volume was 250 µL, and the amount of trypsin used for protein digestion was 1.25 µg. For each affinity resin volume, the increasing concentration of bosutinib is marked by a shaded triangle. (B) Summary of the experimental conditions employed for the assessment of pull-down- and sample-processing effects. 50 nmol of bosutinib immobilized on 50 µL of the Sepharose resin (bosutinib concentration of 1 nmol/µL) was used for all listed pull-downs.
Solution Digestion of Eluted Proteins and Sample Preparation for Mass Spectrometry
Proteins in the neutralized eluate were reduced with 10 mM DTT at 56 °C for 1 h, alkylated with 55 mM iodoacetamide for 30 min at room temperature in the dark, and digested overnight at 37 °C with sequencing grade modified porcine trypsin (Promega, Madison, WI) (Figure 1). The resultant tryptic digest was acidified with 30% trifluoroacetic acid (TFA) and desalted C18 columns were concentrated in a vacuum centrifuge (Eppendorf, Hamburg, Germany) to a volume of approximately 2 µL and then reconstituted to 10 µL with 5% FA.
Liquid Chromatography Mass Spectrometry
Mass spectrometry was performed on either a hybrid LTQ-Orbitrap XL or an LTQ-Orbitrap Velos mass spectrometer (ThermoFisher Scientific, Waltham, MA) using Xcalibur, version 2.0.7 and 2.1.0 SP1.1160, respectively. Both instruments were coupled to Agilent 1200 HPLC nanoflow systems (dual pump with one precolumn and one analytical column) (Agilent Biotechnologies, Palo Alto, CA) via a nanoelectrospray ion source using liquid junction (Proxeon, Odense, Denmark). Solvents for LC−MS separation of the digested samples were as follows: solvent A consisted of 0.4% FA in water and solvent B consisted of 0.4% FA in 70% methanol and 20% isopropanol. From a thermostatted microautosampler, 8 µL of the tryptic peptide mixture were automatically loaded onto a trap column (Zorbax 300SB-C18 5 µm, 5 × 0.3 mm, Agilent Biotechnologies, Palo Alto, CA) with a binary pump at a flow rate of 45 µL/min. 0.1% TFA was used for loading and washing the precolumn. After washing, the peptides were eluted by back-flushing onto a 16 cm fused silica analytical column with an inner diameter of 50 µm packed with C18 reversed phase material (ReproSil-Pur 120 C18- AQ, 3 µm, Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). The peptides were eluted from the analytical column with a 27 min gradient ranging from 3 to 30% solvent B, followed by a 25 min gradient from 30 to 70% solvent B and, finally, a 7 min gradient from 70 to 100% solvent B at a constant flow rate of 100 nL/min.17 The analyses were performed in a data-dependent acquisition mode and dynamic exclusion for selected ions was 60 s. For the LTQ-Orbitrap XL and Velos, a top 6 and top 15 collision-induced dissociation (CID) method were used, respectively. No lock masses were used for the Orbitrap XL and a single lock mass at m/z 445.120024 [Si(CH3)2O)6]18 was employed for the Orbitrap Velos. Maximal ion accumulation time allowed in the CID mode was 150 ms for MSn in the LTQ and 1000 ms in the C-trap (LTQ-Orbitrap XL) and 50 ms for MSn in the LTQ and 500 ms in the C-trap (LTQ-Orbitrap Velos). Automatic gain control was used to prevent overfilling of the ion traps and was set to 5000 in the MSn mode for the LTQ and 106 ions for a full FTMS scan. Intact peptides were detected in the Orbitrap XL and Velos at 100000 and 60000 resolution at m/z 400, respectively.
Data Analysis
The acquired raw MS data files were processed with msconvert (ProteoWizard Library v2.1.2708) and converted into Mascot generic format (.mgf) files. The resultant peak lists were searched against the human Swiss-Prot database version v2011.06_20110609 or v2011.12_20111220 (35683 and 35879 sequences, respectively, including isoforms as obtained from varsplic.pl) with the search engines Mascot (v2.3.02, MatrixScience, London, U.K.) and Phenyx (v2.5.14, GeneBio, Geneva, Switzerland).19 Submission to the search engines was via a Perl script that performs an initial search with relatively broad mass tolerances (Mascot only) on both the precursor and fragment ions (±10 ppm and ±0.6 Da, respectively). High-confidence peptide identifications were used to calculate independent linear transformations for both precursor and fragment ion masses that would minimize the mean square deviation of measured from theoretical. These recalibrating transformations were applied to all precursor and fragment ion masses prior to a second search with narrower mass tolerances (±4 ppm and ±0.3 Da). One missed tryptic cleavage site was allowed. Carbamidomethyl cysteine was set as a fixed modification and oxidized methionine was set as a variable modification. To validate the proteins, Mascot and Phenyx output files were processed by internally developed parsers. Proteins with ≥2 unique peptides above a score T1, or with a single peptide above a score T2, were selected as unambiguous identifications. Additional peptides for these validated proteins with score > T3 were also accepted. For Mascot and Phenyx, T1, T2, and T3 peptide scores were set to 16, 40, and 10 and 5.5, 9.5, and 3.5, respectively (P value < 10e−3). The validated proteins retrieved by the two algorithms were merged, any spectral conflicts discarded and grouped according to shared peptides. A false discovery rate (FDR) of <1% and <0.1% (including the peptides exported with lower scores) was determined for proteins and peptides, respectively, by applying the same procedure against a reversed database.
Comparisons between analytical methods involved comparisons between the corresponding sets of identified proteins. This was achieved by an internally developed program that simultaneously computes the protein groups in all samples and extracts statistical data such as the number of distinct peptides, number of spectra, and sequence coverage. Spectral counts were further used to estimate abundance by the distributed normalized spectral abundance factor (dNSAF)20 as follows:
where
Briefly, the distributed spectral abundance factor (dSAF) of protein i is the weighted sum of spectral counts divided by protein length. Unique spectral counts (uSpC) have the weight of 1. The weight of shared spectral counts (sSpC) is a ratio of spectral counts that are unique to protein i and the sum of all unique spectral counts for M proteins that share peptides with protein i. Finally, dSAF is normalized against the sum of all dSAFs in the corresponding LC−MS analysis.
Gene Ontology Annotation Enrichment Analysis
A Cytoscape21 plug-in, ClueGo,22 was used to identify all enriched gene ontology (GO) molecular function annotations compared with a reference gene population set as the entire human genome. Enrichment analysis was based on two-sided minimal-likelihood test on the hypergeometric distribution that is equivalent to a classical Fisher’s exact test. The Benjamini− Hochberg correction was employed to adjust P values for statistically enriched GO annotations. To define an annotation functional group, a kappa score of 0.3 was required. Group leading terms were assigned according to the highest percentage of found genes per annotation.
RESULTS AND DISCUSSION
Experimental Strategy
The most delicate of all biopsy procedures, a fine needle aspiration, can produce in the order of 0.85 × 10e6 cells.23 In accordance with our experience this corresponds to approximately 200 µg of protein. Thus, the aim of this study was to achieve a drug pull-down with the highest possible number of relevant targets identified from a protein input of 100 µg. In theory, such an amount should enable a duplicate analysis from one patient sample. In the case of pull-downs with similar target numbers, higher target relative quantities were taken as the decisive criterion.
For this purpose, bosutinib (SKI-606) was chosen as the drug of reference (Figure 2A) and human K562 CML cells24 as the biological material. This decision was based on the broad kinase specificity of bosutinib12,14,25 and is advantageous because the number of observed kinases and the relative quantities should reflect all changes introduced during the procedure optimization. We have shown that elution with formic acid and gel-free one-dimensional proteomic analysis of the isolated proteins results in a comparative target profile (including all biologically relevant proteins) that can be obtained with a low protein input of 500 µg.14 Therefore, this methodology was adopted as a part of the workflow (Figure 2B). To evaluate the abundance of isolated proteins, dNSAF measure was used.20 In this study, a semiquantitative spectral counting approach was chosen because of the (i) ease of adaptability to any type of biological material, (ii) technical variations related to analyses of sample replicates, (iii) fact that larger proteins tend to produce more peptides that are detected by MS, and (iv) peptide inference problems are taken into account. Also, in contrast to stable isotope labeling methods, such as iTRAQ, TMT, or ICPL, dNSAF does not require any special sample treatment that can lead to undesirable sample losses.
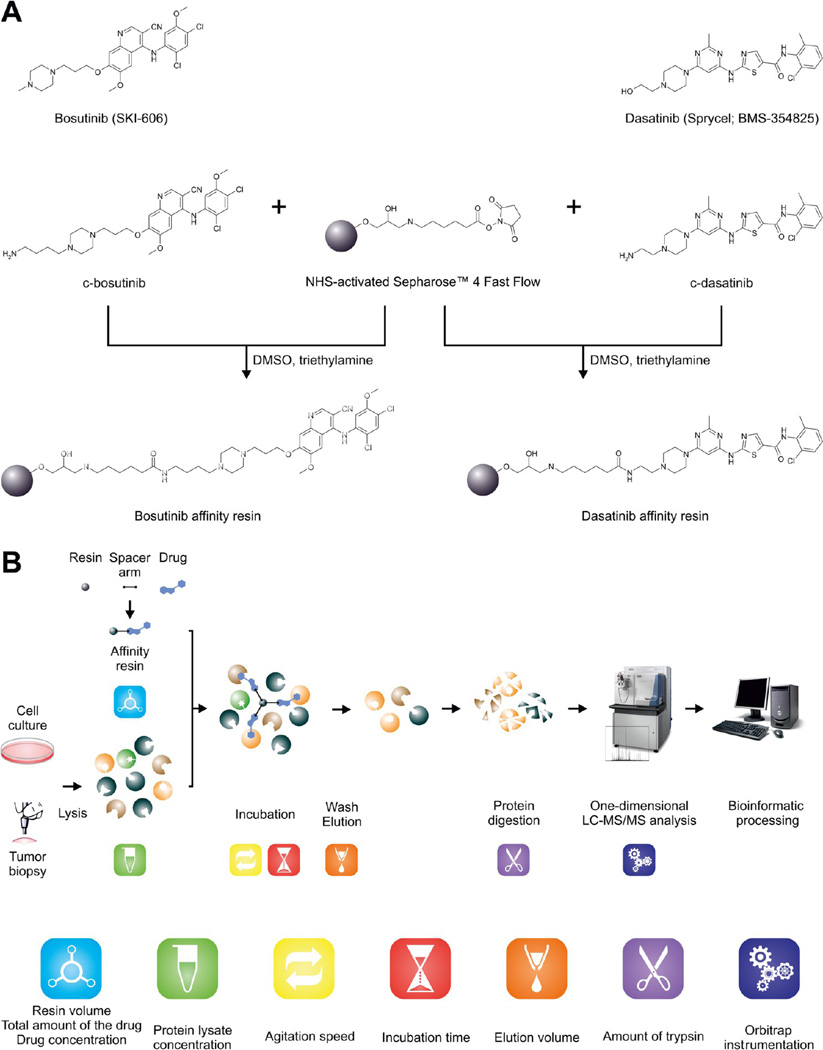
Figure 2.
Overview of the experimental strategy. (A) Chemical structures of the two clinical kinase inhibitors used in this study and the immobilization reaction to produce the desired affinity resins. (B) The overall experimental workflow. The assessed parameters are denoted by appropriate pictograms.
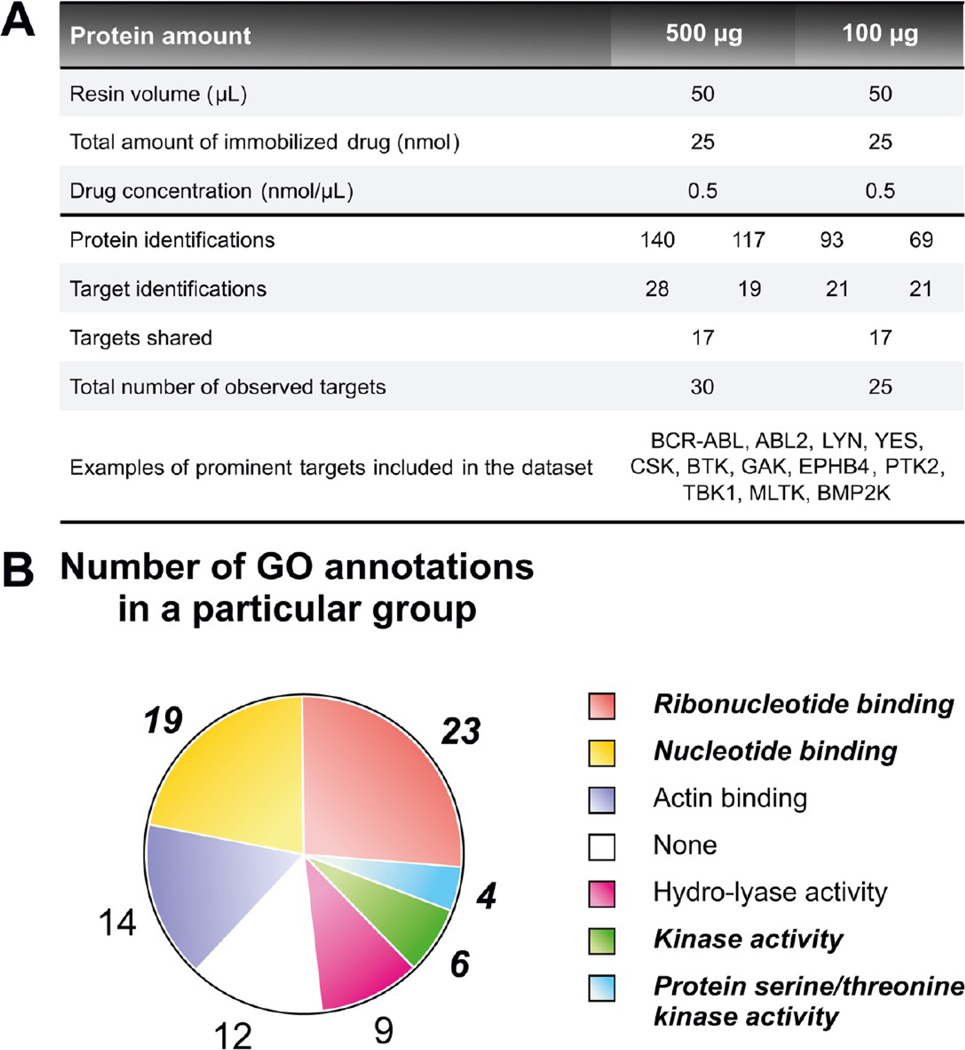
In the initial experiment of the study, the performance of the published protocol14 with 100 µg protein was examined. As a reference, a pull down with a protein input of 500 µg was performed. In average, approximately 80 proteins were unambiguously identified with 100 µg of protein. From these, the kinase targets accounted for nearly 25% of all protein identifications. In the whole group of detected specific interaction partners (25 kinases), many prominently known bosutinib targets, such as BCR-ABL, SRC kinases (LYN and YES), CSK, BTK, GAK, EPHB4 and PTK2, were present (Figure 3A). GO molecular function annotation analysis of the proteins revealed a high enrichment of gene ontology terms connected to kinase activity (Figure 3B; Figure 1 of the Supporting Information). This was in agreement with all previously mentioned findings. Interestingly, these results were very similar to those obtained from the reference pull downs with 500 µg of input protein. Furthermore, unexpectedly, the quantities of isolated kinases represented by the dNSAFs were higher when 100 µg of protein was loaded onto the affinity resin (Table 2 of the Supporting Information).
Figure 3.
Drug pull-down miniaturization: a proof-of-priniciple. (A) Summary statistics of the initial pull-downs with two different amounts of protein. The table shows protein and target identifications for both replicates performed per pull-down variant. (B) GO functional annotation groups found among the proteins identified in the pull-downs with 100 µg protein input. GO terms related to kinase functions are in bold italics.
Overall, the results obtained from these experiments with 100 µg of input material were highly encouraging as a starting point for further optimization. Although the protein input was lowered by 5-fold, only 15 targets less than in our previous gel-free one-dimensional experiments were observed (the overlap between the current and previous study was 19 kinases).14 It should also be stressed at this point that bosutinib target profiles shown in the paper by Fernbach et al.14 were compiled from 6 LC−MS analyses, whereas only 2 analyses were needed to achieve comparable kinase numbers in the present study. Finally, the method showed good reproducibility, indicated by an overlap of almost 70% of the targets that were identified in two pull-down replicates. For further optimization, several parameters that can influence the result of a gel-free one-dimensional chemical proteomic experiment were selected: (i) resin volume, (ii) concentration of the immobilized drug, (iii) total amount of drug, (iv) concentration of the protein lysate, (v) incubation time, (vi) agitation speed, (vii) elution volume, and (viii) amount of trypsin used for protein digestion (Figure 2B).
Influence of Affinity Resin-Related Parameters
At the binding equilibrium, the formation of complexes between an immobilized drug and subsequent targets depends on the concentration of the targets, the dissociation constant of the complex, and the concentration of the immobilized drug. To determine the influence of the latter, pull-downs were performed with three different resin volumes (25, 50, and 100 µL) bearing six different quantities of immobilized bosutinib (12.5, 25, 37.5, 50, 75, and 100 nmol). This combination provided seven different bosutinib concentrations (for details, see Figure 1A) and enabled an elaborate and detailed study of these interconnected parameters.
Of the 39 bosutinib kinase targets identified within this series of experiments, only 2 kinases were seen with 100 µL of the resin. On top of that, these identifications were achieved only if a relatively high amount of bosutinib (50 nmol) was immobilized. This was in sharp contrast to target numbers obtained with the other two resin volumes assessed. For instance, even if 12.5 nmol of bosutinb was used for the pull-down with 25 µL of the resin, 19 kinases were still detected (Figure 4A). On the basis of this behavior and the fact that the same amount of eluant (250 µL) was employed for all three resin volumes, we reasoned that the cause of the poor success rate observed for 100 µL of bosutinib resin was most likely inefficient elution of retained kinases.
Figure 4.
Affinity resin effects. (A) Summary statistics of pull-down experiments investigating thirteen different affinity resin designs. The table shows protein and target identifications for both replicates performed per pull-down variant. (B) Heat map showing relative quantities of all targets identified for each pull-down. Presented dNSAF values are calculated means of the original dNSAFs obtained from relevant duplicate pull-downs. The most efficient pull-down variant is highlighted in red.
As shown in Figure 3A, if the same amount of bosutinib was immobilized on either 25 or 50 µL of the resin, the number of identified proteins increased with higher drug concentrations. This increase in the protein identifications was also evident when pull-downs were performed with the same resin volume but with increasing bosutinib concentration. Notably, the number of targets did not follow the same general trend. Pull-downs performed with the same amount of bosutinib tended to provide very similar numbers of observed targets. This can be highlighted by pull-downs with 37.5 nmol of bosutinib that resulted in the same number of target identifications (21) for 25 and 50 µL. For the constant resin volume, the higher quantities of bosutinib led to a higher number of detected kinases with a maximum at 50 nmol (Figure 4A). It can be hypothesized that this interesting phenomenon was due to steric hindrances preventing the retention of additional target molecules. Other undesired effects, however, such as competition for positive charge and/or signal suppression by the other molecules during the MS analysis cannot be completely eliminated.
In summary, the most favorable results (namely, 31 and 30 kinases identified) were achieved with 50 nmol bosutinib. In a direct comparison, the pull-down variant with 50 µL of resin (and a bosutinib concentration of 1 nmol/µL) provided higher enrichment of almost all identified targets, as documented by higher dNSAFs (Figure 4B) and was thus chosen for further investigations.
Influence of Pull-Down- and Sample Processing-Related Parameters
Aside from the affinity matrix-related parameters, the formation of immobilized drug−target complexes also involves the concentration of the targets. Other important pull-down-related parameters are (i) the speed of agitation to ensure optimal binding kinetics and (ii) the incubation time that should be sufficient to reach the binding equilibrium. Of the group of sample processing-related parameters, a potential influence of the elution volume was indicated by the experiments performed with 100 µL of affinity resin described in the previous section that were not successful. As a very low amount of eluted proteins is expected in chemical proteomic experiment with 100 µg of protein input, the amount of trypsin can also play a role in the number of identified proteins (including targets). To be specific, an optimal amount of trypsin should be determined to facilitate complete digestion of the eluted protein mixture while avoiding an undesired contamination by overabundant trypsin autolysis peptides. To address all of these points, a set of pull downs were performed where the mentioned parameters were systematically altered.
In theory, an increase in the concentration of the protein lysate (i.e., the concentration of the targets) should lead to an increase in the quantity of retained targets. Contradictory to this premise, when the lysate with a doubled protein concentration was used for the bosutinib pull downs, the dNSAFs of target proteins were lower than those obtained with the control experiments. Furthermore, the use of a diluted lysate led to a moderate increase in both the number of detected kinases and the subsequent quantities (Figure 5). This discrepancy can stem from the fact that for these particular experiments, three different incubation volumes were employed. Ultimately, this could also affect the efficiency of agitation and thus alter the binding kinetics of the retained targets.
Figure 5.
Pull-down procedure and sample processing effects. (A) Summary statistics for pull-downs examining selected parameters of the pull-down procedure and subsequent sample processing. The table shows protein and target identifications for both replicates performed per pull-down variant. (B) Heat map showing relative quantities of all targets identified for each pull-down. Presented dNSAF values are calculated means of the original dNSAFs obtained from relevant duplicate pull-downs. Affinity resin parameters used for all pull-downs were as follows: (i) 50 µL of the affinity resin and (ii) bosutinib concentration of 1 nmol/µL. The pull-down with the parameters chosen for further experiments is highlighted in red.
Indeed, target binding kinetics proved to be one of the key pull-down-related parameters in the experiments, in which the influence of the agitation speed was evaluated. While 10 rpm used in the control pull-downs proved to be optimal, any changes in the speed were accompanied by a decrease in target numbers (Figure 5A). Surprisingly, in the case of lowering the speed to 5 rpm, this decrease was not connected with a decrease in target quantities. On the contrary, the dNSAFs of retained kinases were higher under these conditions (Figure 5B).
To verify whether the binding equilibrium was reached, a pull down with prolonged incubation time (3 h) was performed, and the obtained results were compared with the control. Neither the number of identified targets nor the target quantities showed any major differences (Figure 5). On the basis of this observation, it was concluded that the incubation time was already optimized. However, this is certainly a parameter that may vary somewhat with any given drug.
The importance of an optimal elution volume was demonstrated by pull downs with 125 and 500 µL of 100 mM FA. As anticipated, less kinase targets were detected if the elution volume was reduced by half (125 µL). The results from this pull-down were also compromised at the level of kinase dNSAFs. This is clearly represented, e.g., by the signal of BTK. Elution with 500 µL of formic acid did not alter any of the monitored variables (Figure 5).
The only parameter that did not show any clear trend was the amount of trypsin used for digesting the eluted proteins. No matter what amount (1.25, 0.625, or 0.3125 µg) was used, the results were almost identical (Figure 5). Therefore, it was decided to retain the original trypsin input of 1.25 µg.
Taken together, the optimal parameters elicited from the described pull-down experiments were as follows: (i) 50 nmol of a drug immobilized on (ii) 50 µL resin, (iii) a protein lysate concentration of 0.5 µg/µL (with the incubation volume of 200 µL), (iv) an agitation speed of 10 rpm, (v) an incubation time of 2 h, (vi) an elution volume of 250 µL, and (vii) 1.25 µg of trypsin for digestion of the eluted proteins.
An Aside: The Effect of Orbitrap Instrumentation
All of the results discussed above were acquired on a hybrid LTQ-Orbitrap XL mass spectrometer. Nonetheless, in the course of this project, an LTQ-Orbitrap Velos mass spectrometer was installed in our laboratory. Compared to the former LTQ-Orbitrap XL, the LTQ-Orbitrap Velos provides increased sensitivity and remarkably accelerated scanning and detection rates. This is achieved, primarily, by a combination of (i) a new, more efficient and robust atmosphere/vacuum ion guide (S-lens)26 and (ii) a fast scanning dual-pressure linear ion trap.27 As such, the LTQ-Orbitrap Velos confers a marked advantage when analyzing limited but still complex protein samples.28 For this reason, the performance of this instrument was assessed by LC−MS analysis of miniaturized drug pull-downs. Using the same protein lysate, a bosutinib pull-down following the optimized protocol was performed in duplicate, analyzed on the LTQ-Orbitrap Velos, and compared to the corresponding pull-down performed as earlier in this study (see previous sections) (Figure 6A).
Figure 6.
Miniaturized pull-downs analyzed on two different Orbitrap instruments. (A) Schematic representation of the experiment. (B) The overlap of the total protein and target identifications and combined identifications of targets and the bound interacting proteins. (C) Interaction network of targets and the annotated protein partners that were identified with the LTQ-Orbitrap Velos. The information on protein interactions were downloaded from the following databases: BioGRID,32 IntACT,33 MINT,34 InnateDB,35 CORUM,36 and DIP.37.
As expected, the number of protein identifications markedly increased with the use of the LTQ-Orbitrap Velos. In total, almost 300 proteins were detected, including about 80% of the proteins previously observed with the LTQ-Orbitrap XL. While the number of identified proteins markedly increased, there were only 8 additional targets using the LTQ-Orbitrap Velos. With a high overlap between the two target data sets (Figure 6B), it appears that by employing the optimized protocol, the majority of the targets that were retained by the affinity resin had already been identified with the LTQ-Orbitrap XL instrumentation. Therefore, it was concluded that the use of the LTQ-Orbitrap Velos in the initial phases of the study would not have influenced our subsequent optimization decisions. This is particularly pertinent if one considers that all the investigated changes that were introduced in the early phase of the study were at the level of the affinity separation performed off-line. On the other hand, the LTQ-Orbitrap Velos detected more protein interactors of direct targets than the LTQ-Orbitrap XL (Figure 6, panels B and C). It was therefore decided to employ the Orbitrap Velos instrument for all following sample analyses.
Application of the Miniaturized Pull-down Protocol to Dasatinib
Miniaturization of the drug pull-down procedure using bosutinib was successful, however, a question arose as to whether the optimized protocol was drug dependent. To shed light on this issue, the protocol was applied to dasatinib (Sprycel), another multikinase inhibitor that is approved for CML (Figure 2A).29 Importantly, the protein interaction profile of dasatinib in K562 cells has been characterized,15 and although the overall target spectrum is substantially different, several targets are shared with bosutinib. Thus, together these two drugs allow an evaluation of the applicability of the new method.
Using a previously validated dasatinib analogue as bait,15,30 22 different kinases were identified in duplicate pull-downs (22 and 18, respectively, with an overlap of 19 kinases). Among these, the cognate dasatinib targets BCR-ABL and ABL2 and SRC kinases YES and LYN were the most prominent. Other important interactors of dasatinib were the negative regulator of the SRC kinase family CSK, the TEC kinases BTK and TEC, the tyrosine kinase KIT, the serine/threonine kinase GAK, and several MAP kinases (Table 6 of the Supporting Information).
Although the number of kinases identified in the dasatanib pull-downs was lower than with bosutinib, the data obtained from the miniaturized protocol correlated well with the results from our previous studies.12,15,30 This fact, together with the observation of clinically important dasatinib targets and several unique kinase interactors (i.e., KIT and TEC) indicates that the miniaturized pull-down protocol can be extended to other drugs of interest. Nevertheless, the need for some additional fine tuning on a drug-by-drug basis may be warranted.
Miniaturized Pull-Downs in Tumor Biopsies
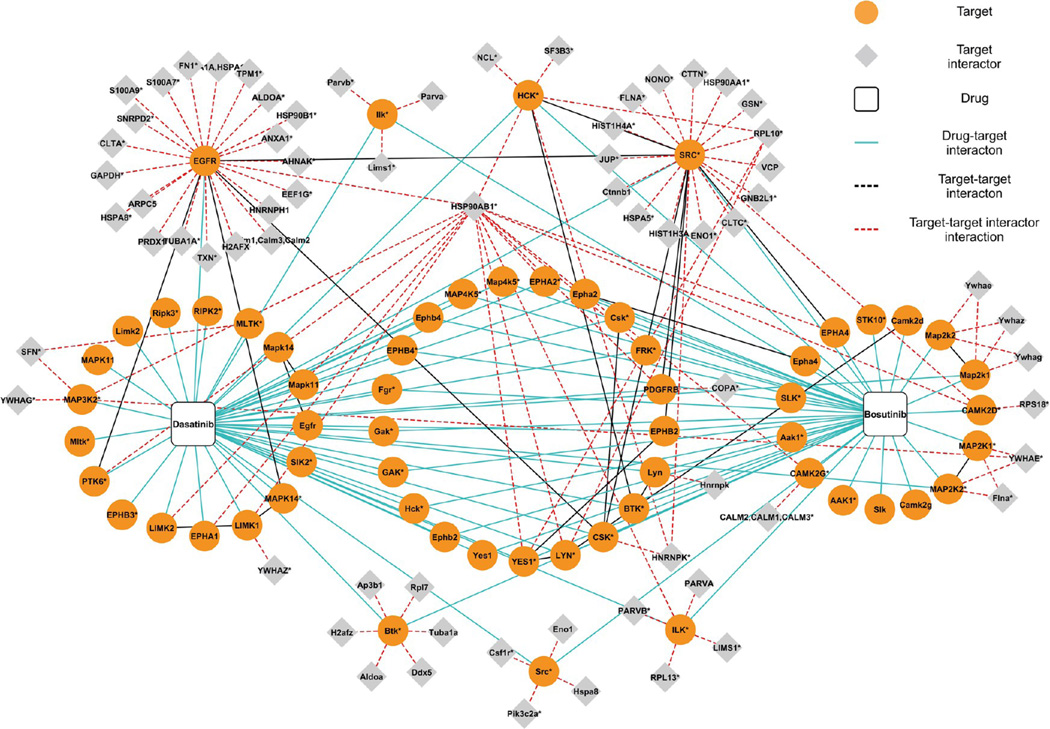
In order to validate the suitability of the optimized protocol for clinical samples at microgram levels of protein input, the scenario of a patient obtaining an image-guided needle biopsy was simulated by using primary lung cancer patient-derived xenotransplants. patients diagnosed with early stage NSCLC by transplantation of surgical tumor specimens into mice.16 When tumors became palpable, core biopsies were taken, samples lysed (Figure 2 of the Supporting Information), and duplicate pull downs performed with the protein lysates, using bosutinib and dasatinib as affinity probes.
Altogether, 60 distinct kinase targets were identified; 41 of these were found to be associated with bosutinib and 47 with dasatinib. Again, many well-known clinical targets were present in both of the drug target profiles. For example, the dasatinib target profile was almost identical to that published by us two years ago31 and included several SRC (SRC, LYN, and YES) and SRC-related kinases (FRK and PTK6). Another kinase family that was markedly enriched in the dasatinib pull-downs was the ephrin receptor family represented by EPHA1, EPHA2, EPHA4, EPHB2, EPHB3, and EPHB4. In addition to tyrosine kinases, dasatinib also retained multiple serine/threonine kinases such as GAK and various MAP kinases (Figure 7). The two replicates performed for each drug shared more than 60% of the identified targets, which is consistent with a high reproducibility of all experiments throughout this study.
Figure 7.
Interaction network of bosutinib and dasatinib in core biopsy samples. Names of human proteins are written in upper case, and murine proteins are marked in lower case with the first letter capitalized. Proteins, for which it was impossible to exclude the presence of a certain homologue or choose from two taxon-related variants due to peptide inference are labeled with an asterisk. The information on protein interactions were downloaded from BioGRID,32 IntACT,33 MINT,34 InnateDB,35 CORUM,36 and DIP37 databases.
In conclusion, the optimized protocol presented here proved to be well suited for routine target profiling from limited protein samples such as those acquired from tissue biopsies.
CONCLUSIONS AND OUTLOOK
In this article, we report a successful adaptation of the current chemical proteomic experimental protocol to protein levels that can be obtained from clinical biopsy samples. By optimization of eight preselected protocol parameters, the required protein input was downscaled to 100 µg. With the use of core needle biopsies from lung cancer xenotransplants, the identification of a large number of relevant kinase targets and secondary interactors of the two FDA-approved drugs bosutinib and dasatinib was demonstrated.
We believe this improved technology will have an important impact on personalized medicine. It is now possible to evaluate the physical interactions that a given drug can have with desired target proteins in clinical samples prior to initiation of a particular therapeutic regimen. Looking forward, the concept of combining a miniaturized chemical proteomic methodology with other techniques such as molecular imaging, deep sequencing and proteomic and metabolomic profiling is tantalizing. Such an integrative “-omics” approach could provide a wealth of valuable data on the unique genetic, biological, and therapeutic response of patient tumors, thus aiding in fine-tuning suitable treatment strategies.
ACKNOWLEDGMENTS
The authors thank Miroslav Strnad and Marek Šebela for catalyzing the collaboration that led to this work and for supporting the stay of Ivo Chamrád at the CeMM Research Center for Molecular Medicine in Vienna. Research in our laboratory at CeMM is supported by the Austrian Academy of Sciences, the Austrian Federal Ministry for Science and Research (GenAu APP and BIN), the Austrian Science Fund FWF, and the Austrian National Bank. The work was also partially funded by grants from the Moffitt Cancer Center SPORE in Lung Cancer (P50-CA119997).
ABBREVIATIONS
- CID
collision-induced dissociation
- CML
chronic myeloid leukemia
- DTT
dithiothreitol
- FA
formic acid
- FDR
false discovery rate
- GO
gene ontology
- LC−MS
liquid chromatography−mass spectrometry
- MS
mass spectrometry
- dNSAF
distributed normalized spectral abundance factor
- NSCLC
nonsmall cell lung carcinoma
- dSAF
spectral abundance factor
- sSpC
shared spectral counts
- uSpC
unique spectral counts
- TFA
trifluoroacetic acid
Footnotes
ASSOCIATED CONTENT
S Supporting Information
Raw MS data, .mgf, and protein search files for all analyzed pull-down experiments are available in the Peptide Atlas Repository (http://www.peptideatlas.org/), data set accession code PASS00205. Two supplementary figures as a .pdf file and seven supplementary tables as Excel files are attached. These files contain detailed information on the GO enrichment analysis (Figure 1), biopsy samples and corresponding protein yields (Figure 2), basic statistics for all affinity experiments (Table 1) and all identified peptides, proteins, and target dNSAFs for particular blocks of pull downs (Tables 2−7). This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The authors declare no competing financial interest.
REFERENCES
- 1.Capdeville R, Buchdunger E, Zimmermann J, Matter A. Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat. Rev. Drug Discovery. 2002;1:493–502. doi: 10.1038/nrd839. [DOI] [PubMed] [Google Scholar]
- 2.Aggarwal S. Targeted cancer therapies. Nat. Rev. Drug Discovery. 2010;9:427–428. doi: 10.1038/nrd3186. [DOI] [PubMed] [Google Scholar]
- 3.Ghoreschi K, Laurence A, O’Shea JJ. Selectivity and therapeutic inhibition of kinases: To be or not to be? Nat. Immunol. 2009;10:356–360. doi: 10.1038/ni.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terstappen GC, Schlüpen C, Raggiaschi R, Gaviraghi G. Target deconvolution strategies in drug discovery. Nat. Rev. Drug Discovery. 2007;6:891–903. doi: 10.1038/nrd2410. [DOI] [PubMed] [Google Scholar]
- 5.Chan JN, Nislow C, Emili A. Recent advances and method development for drug target identification. Trends Pharmacol. Sci. 2010;31:82–88. doi: 10.1016/j.tips.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein DM, Gray NS, Zarrinkar PP. High-throughput kinase profiling as a platform for drug discovery. Nat. Rev. Drug Discovery. 2008;7:391–397. doi: 10.1038/nrd2541. [DOI] [PubMed] [Google Scholar]
- 7.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP. A quantitative analysis of kianse inhibitor selectivity. Nat. Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 8.Kruse U, Bantscheff M, Drewes G, Hopf C. Chemical and pathway proteomics. Powerful tools for drug discovery and personalised health care. Mol. Cell. Proteomics. 2008;7.10:1887–1901. doi: 10.1074/mcp.R800006-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Bantscheff M, Scholten A, Heck AJR. Revealing promiscuous drug-target interactions by chemical proteomics. Drug Discovery Today. 2009;14:1021–1029. doi: 10.1016/j.drudis.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat. Chem. Biol. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 11.van Staveren WC, Solís DY, Hébrant A, Detours V, Dumont JE, Maenhaut C. Human cancer cell lines: Experimental model for cancer in situ? For cancer stem cells? Biochim. Biophys. Acta. 2009;1795:92–103. doi: 10.1016/j.bbcan.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Remsing Rix LL, Rix U, Colinge J, Hantschel O, Bennett KL, Stranzl T, Müller A, Baumgartner C, Valent P, Augustin M, Till JH, Superti-Furga G. Global target profile of the kinase inhibitor bosutinib in primary chronic myeloid leukemia cells. Leukemia. 2009;23:477–485. doi: 10.1038/leu.2008.334. [DOI] [PubMed] [Google Scholar]
- 13.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernbach N, Planyavsky M, Müller A, Breitweiser FP, Colinge J, Rix U, Bennett KL. Acid elution and one-dimensional shotgun analysis on an Orbitrpmss spectrometer: An application to drug affinity chromatography. J. Proteome Res. 2009;8:4753–4765. doi: 10.1021/pr900455x. [DOI] [PubMed] [Google Scholar]
- 15.Rix U, Hantschel O, Dürnberger G, Remsing Rix LL, Planyavsky M, Fernbach NV, Kaupe I, Bennett KL, Valent P, Colinge J, Köcher T, Superti-Furga G. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 16.Fichtner I, Rolff J, Soong R, Hoffmann J, Hammer S, Sommer A, Becker M, Merk J. Establishment of patient-derived non-small cell lung cancer xenografts as models for the identification of predictive biomarkers. Clin. Cancer Res. 2008;14:6456–6468. doi: 10.1158/1078-0432.CCR-08-0138. [DOI] [PubMed] [Google Scholar]
- 17.Bennett KL, Funk M, Tschernutter M, Breitwieser FP, Planyavsky M, Ubaida Mohien C, Müller A, Trajanoski Z, Colinge J, Superti-Furga G, Schmidt-Erfurth U. Proteomic analysis of human cataract aqueous humour: Comparison of one-dimensional gel LCMS with two-dimensional LCMS of unlabelled and iTRAQ-labelled specimens. J. Proteomics. 2011;74:151–166. doi: 10.1016/j.jprot.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Olsen JV, de Godoy LMF, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per million mass accuracy on an Orbitrpmss spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics. 2005;4.12:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Colinge J, Masselot A, Giron M, Dessingy T, Magnin J. OLAV: Towards high-throughput tandem mass spectrometry data identification. Proteomics. 2003;3:1454–1463. doi: 10.1002/pmic.200300485. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: How to deal with peptides shared by multiple proteins. Anal. Chem. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welker L, Akkan R, Holz O, Schultz H, Magnussen H. Diagnostic outcome of two different CT-guided fine needle biopsy procedures. Diagn. Pathol. 2007;2:31. doi: 10.1186/1746-1596-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lozzio CB, Lozzio BB. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 25.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat. Biotechnol. 2011;29:1039–1045. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim T, Tolmachev AV, Harkewicz R, Prior DC, Anderson G, Udseth HR, Smith RD. Design and implementation of a new electrodynamic ion funnel. Anal. Chem. 2000;72:2247–2255. doi: 10.1021/ac991412x. [DOI] [PubMed] [Google Scholar]
- 27.Second TP, Blethrow JD, Schwartz JC, Merrihew GE, MacCoss MJ, Swaney DL, Russell JD, Coon JJ, Zabrouskov V. Dual-pressure linear ion trpmss spectrometer improving the analysis of complex protein mixtures. Anal. Chem. 2009;81:7757–7765. doi: 10.1021/ac901278y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, Wouters ER, Senko M, Makarov A, Mann M, Horning S. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics. 2009;8.12:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lombardo LJ, Lee FY, Chen P, Norris D, Barrish JC, Behnia K, Castaneda S, Cornelius LA, Das J, Doweyko AM, Fairchild C, Hunt JT, Inigo I, Johnston K, Kamath A, Kan D, Klei H, Marathe P, Pang S, Peterson R, Pitt S, Schieven GL, Schmidt RJ, Tokarski J, Wen ML, Wityak J, Borzilleri RM. Discovery of N-(2-chloro-6-methyl-phenyl)-2-(6-(4-(2-hydroxyethyl)-piperazin-1-yl)-2-methylpyrimidin-4-ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J. Med. Chem. 2004;47:6658–6661. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 30.Hantschel O, Rix U, Schmidt U, Bürckstümmer T, Kneidinger M, Schütze G, Colinge J, Bennett KL, Ellmeier W, Valent P, Superti-Furga G. The Btk tyrosine kinase is a major target of the Bcr-Abl inhibitor dasatinib. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13283–13288. doi: 10.1073/pnas.0702654104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Rix U, Fang B, Bai Y, Edwards A, Colinge J, Bennett KL, Gao J, Song L, Eschrich S, Superti-Furga G, Koomen J, Haura EB. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat. Chem. Biol. 2010;6:291–299. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, Kohler C, Khadake J, Leroy C, Liban A, Lieftink C, Montecchi-Palazzi L, Orchard S, Risse J, Robbe K, Roechert B, Thorneycroft D, Zhang Y, Apweiler R, Hermjakob H. IntAct: Open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A, Gardy JL, Roche FM, Chan THW, Shah N, Lo R, Naseer M, Que J, Yau M, Acab M, Tulpan D, Whiteside MD, Chikatamarla A, Mah B, Munzner T, Hokamp K, Hancock REW, Brinkman FSL, Innate DB. Facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruepp A, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Stransky M, Waegele B, Schmidt T, Doudieu ON, Stümpflen V, Mewes HW. CORUM: The comprehensive resource of mammalian protein complexes. Nucleic Acids Res. 2008;36:D646–D650. doi: 10.1093/nar/gkm936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xenarios I, Rice DW, Salwinski L, Baron MK, Marcotte EM, Eisenberga D. DIP: The Database of Interacting Proteins. Nucleic Acids Res. 2000;28:289–291. doi: 10.1093/nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]