Abstract
Collagen IV is the predominant protein network of basement membranes, a specialized extracellular matrix, which underlie epithelia and endothelia. These networks assemble through oligomerization and covalent cross-linking to endow mechanical strength and shape cell behavior through interactions with cell surface receptors. A novel sulfilimine (S=N) bond between a methionine sulfur and hydroxylysine nitrogen reinforces the collagen IV network. We demonstrate that peroxidasin, an enzyme found in basement membranes, catalyzes formation of the sulfilimine bond. Drosophila peroxidasin mutants exhibit disorganized collagen IV networks and torn visceral muscle basement membranes pointing to a critical role for the enzyme in tissue biogenesis. Peroxidasin generates hypohalous acids as reaction intermediates suggesting a paradoxically anabolic role for these usually destructive oxidants. This work highlights sulfilimine bond formation as the first known physiologic function for peroxidasin, a role for hypohalous oxidants in tissue biogenesis, and a possible role for peroxidasin in inflammatory diseases.
A basic organizational unit of animal tissues is a polarized epithelium attached to an underlying basement membrane, a specialized form of extracellular matrix.1 The collagen IV protein network is the predominant constituent of basement membrane and provides structural integrity to epithelial and vascular tissues, serves as a scaffold for macromolecular assembly, and interacts with cell surface receptors such as integrins to control cell adhesion, migration, proliferation, and differentiation.1,2 The triple helical protomer is the building block which self-assembles into collagen IV networks by oligomerization. The C-terminal trimeric NC1 domains of two protomers associate with each other to form a hexameric structure.3 Importantly, the C-terminal interface between two protomers is covalently cross-linked by a sulfilimine bond (S=N) between apposed lysine and methionine residues.4
Based on collagen IV sequence homology, the sulfilimine bond appears early in animal evolution at the divergence of Placazoa and Cnidaria coinciding with the evolution of primordial basement membranes and thus representing a critical innovation for tissue biogenesis.4 The sulfilimine bond also confers immune privilege to the collagen IV auto-antigen in human Goodpasture’s disease suggesting its formation or cleavage participates in the pathogenesis of this autoimmune disease.5
Given the critical role of the collagen IV sulfilimine bond in tissue development and human disease, we endeavored to delineate the molecular mechanism of bond formation. Here, we show that peroxidasin catalyzes sulfilimine bonds directly within basement membranes using hypohalous acid intermediates. These findings represent the first known function for peroxidasin and highlight a biosynthetic role for conventionally toxic hypohalous oxidants.
Results
A Model to Study Collagen IV Sulfilimine Bond Formation
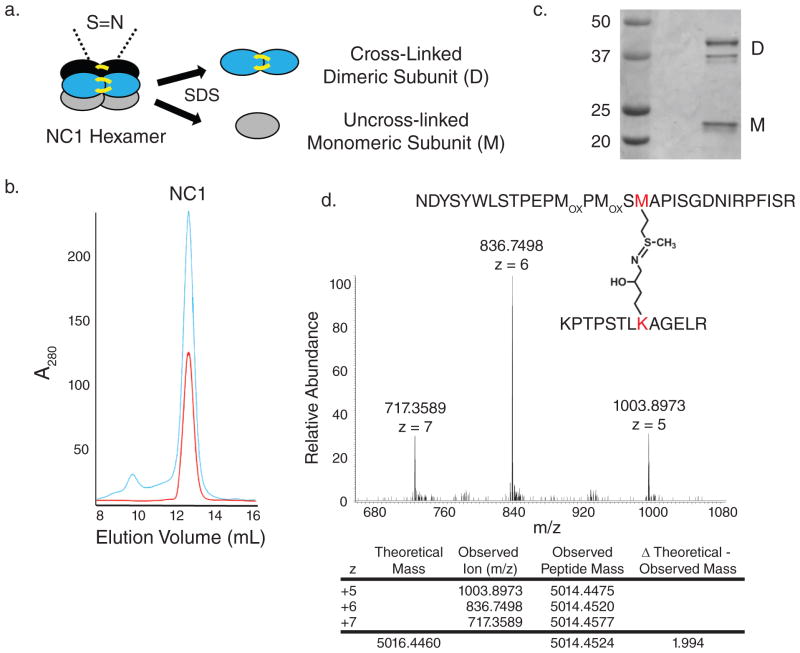
To study sulfilimine bond formation, the PFHR-9 mouse endodermal cell line was used as an experimental system, since it produces biochemically tractable quantities of collagen IV.6 When grown past confluency, PFHR-9 cells progressively accumulated basement membrane which was isolated to purify collagen IV NC1 hexamers after collagenase digestion. SDS dissociation of NC1 hexamers and gel electrophoresis revealed both cross-linked NC1 dimeric and uncross-linked monomeric subunits (Fig. 1a–c). Mass spectrometry provided chemical evidence for a sulfilimine bond joining methionine 93 (Met93) and hydroxylysine 211 (Hyl211) in adjacent protomers (Fig. 1d). We initially focused on known oxidative matrix-associated enzymes as possible mediators of sulfilimine bond formation in collagen IV. Using small molecule inhibitors during cellular deposition of basement membrane, structurally distinct peroxidase inhibitors, including phloroglucinol (IC50 = 0.5 μM),7 methimazole (IC50 = 0.8 μM for thyroid peroxidase, 3 mM inhibits myeloperoxidase by 70%),8,9 and 3-aminotriazole (near complete inhibition of thyroid peroxidase at 2 mM and of myeloperoxidase at 10 mM)10,11 universally prevented formation of collagen IV cross-links. As described in the Discussion, we initially examined iodide as a possible peroxidase substrate to form hypoiodous acid as a reactive intermediate. Unexpectedly, potassium iodide inhibited collagen IV cross-link formation and therefore used as an inhibitor in subsequent experiments (Fig. 2a). Lysyl oxidase (β-aminopropionitrile; IC50 = 3–8 μM)12 and transglutaminase inhibitors (putrescine; Km 0.026 – 0.847 mM)13 had no effect despite using concentrations exceeding published inhibitory constants (Fig. 2a). Peroxidase inhibitors did not perturb collagen IV assembly in this system, since NC1 hexamers formed quantitatively in the absence of sulfilimine cross-links (Supplementary Fig. 1). Peroxidase inhibitors also did not break cross-links after formation, but specifically prevented bond formation (Fig. 2b). These findings suggest that a peroxidase, embedded within basement membrane, forms sulfilimine bonds in collagen IV. If so, an isolated basement membrane preparation should recapitulate this biochemical event in vitro with the addition of hydrogen peroxide (H2O2), a required substrate for peroxidases. PFHR-9 cells were grown in the presence of a peroxidase inhibitor (10 mM potassium iodide) to deposit a collagen IV network devoid of sulfilimine cross-links. A basement membrane preparation was isolated and incubated without inhibitor in the absence or presence of H2O2. Sulfilimine bonds formed rapidly when peroxidase inhibitors were removed only in the presence of H2O2 pointing to a peroxidase residing within the basement membrane (Fig. 2c; Supplementary Fig. 2). Alternatively, H2O2 may chemically form sulfilimine cross-links in collagen IV. To investigate this possibility, PFHR-9 basement membrane was extracted with 2M guanidine to inactivate and/or extract the basement membrane peroxidase without affecting collagen IV. Indeed, guanidine pre-treatment of basement membrane eliminated cross-linking activity even in the presence of H2O2 consistent with the loss of an enzymatic activity rather than direct chemical oxidation by H2O2 (Supplementary Fig. 3).
Figure 1. PFHR-9 cells produce a basement membrane collagen IV network with sulfilimine cross-links.
(a) Schematic of collagen IV NC1 hexamer with sulfilimine cross-links bridging the trimer-trimer interface. Upon addition of SDS, the hexamer dissociates into cross-linked dimeric subunits (D) and uncross-linked monomeric subunits (M). (b) Gel filtration chromatography elution profile of PFHR-9 collagen IV NC1 hexamer (blue) and native, purified placental basement membrane NC1 hexamer (red) run successively. (c) SDS-PAGE of the purified NC1 hexamer with cross-linked dimeric (D) and uncross-linked monomeric subunits (M). As seen in placental and mouse Engelbreth-Holm-Swarm (EHS) tumor collagen IV, at least two and occasionally three dimeric subunit bands and one or two monomeric subunit bands were observed.46 (d) Mass spectrometry of purified PFHR-9 NC1 hexamer revealed a tryptic peptide with a mean observed mass of 5014.4524. The mass of the methionine 93 containing peptide added to the hydroxylysine 211 containing peptide provides a “theoretical” mass of 5016.446. The difference between the theoretical and observed mass of 1.994 represents the loss of two hydrogens upon sulfilimine bond formation in collagen IV.4 MOX stands for methionine sulfoxide, a common oxidation product of methionine.
Figure 2. A basement membrane peroxidase forms the collagen IV sulfilimine bond.
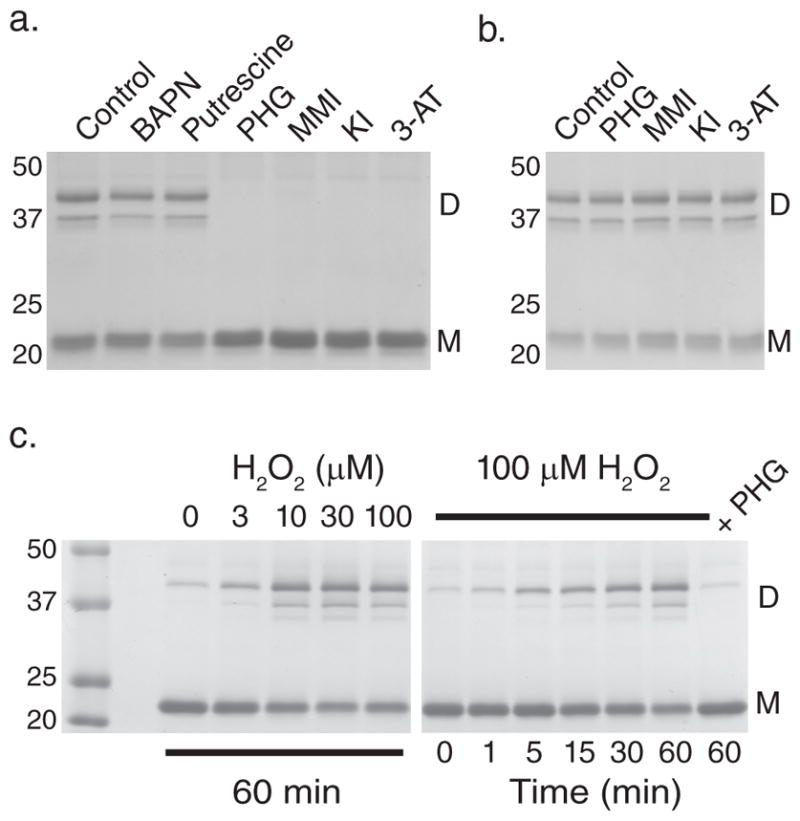
(a) Coomassie blue stained gel after SDS-PAGE of NC1 hexamers isolated from PFHR-9 cells grown in the presence of β-aminopropionitrile (BAPN; 500 μM), putrescine (2.5 mM), phloroglucinol (PHG; 50 μM), methimazole (MMI; 1 mM), 3-aminotriazole (3-AT; 10 mM), and potassium iodide (KI; 10 mM). Collagen IV NC1 hexamer from untreated cells (control) is shown for comparison. Gel is representative of 5 independent experiments. (b) PFHR-9 basement membrane was allowed to form normally, isolated, and treated with phloroglucinol (PHG; 50 μM), methimazole (MMI; 1 mM), potassium iodide (KI; 10 mM), and 3-aminotriazole (3-AT; 10 mM) for 24 hours at 37°C. Collagen IV NC1 hexamer was isolated and underwent SDS-PAGE and Coomassie blue staining to visualize sulfilimine cross-link content. (c) Coomassie blue stained gel after SDS-PAGE of NC1 hexamers after reacting uncross-linked PFHR-9 basement membrane with H2O2 at varying concentrations for 1 hour (left panel) or for varying durations with 100 μM H2O2 (right panel) in 1X PBS (phosphate buffered saline; 150 mM NaCl, 10 mM sodium phosphate, pH 7.4). The gel is representative of 8 independent experiments. D represents NC1 cross-linked dimeric subunits, while M denotes uncross-linked monomeric subunits. Full gel images are provided in Supplementary Fig. 13.
Peroxidasin Catalyzes Formation of Sulfilimine Bonds
To rapidly identify candidates, we developed a novel approach to covalently label and capture basement membrane-bound peroxidases. Inorganic azide (N3−) is a known suicide inhibitor of peroxidases. In the presence of azide and H2O2, peroxidases generate azidyl radicals which covalently attach to the peroxidase heme moiety to form an organic azide (R-N3) and eliminate enzymatic activity (KI = 1.47 mM, kinact = 0.69 min−1 for horseradish peroxidase).14 PFHR-9 basement membrane was isolated and treated with azide and H2O2 to form an organic azide conjugate with matrix peroxidases. After basement membrane proteins were solubilized with SDS, azide-peroxidase conjugates were then biotinylated using alkyne biotin to react with the organic azide in a copper catalyzed “click” chemistry reaction.15 Electrophoresed proteins were blotted with streptavidin-HRP to detect biotinylated proteins revealing a single streptavidin reactive band at about 160–200 kDa with reactivity increasing in a dose dependent manner with azide concentration (Supplementary Fig. 4). Streptavidin agarose affinity chromatography was used to purify the azide labeled peroxidase revealing a single predominant band on Coomassie stained protein gels at the same molecular weight observed with streptavidin blotting (Supplementary Fig. 4). The stained protein band was excised and digested with trypsin. Mass spectrometry of the resulting peptides revealed peroxidasin as an azide labeled peroxidase residing within PFHR-9 basement membrane (Supplementary Table 1). Recognizing the azide labeling technique as a screening tool with limitations, we next tested whether our identified candidate, peroxidasin, is truly capable of and responsible for the formation of sulfilimine cross-links in collagen IV.
To determine whether peroxidasin is biochemically able to catalyze sulfilimine bond formation, we heterologously expressed and purified human peroxidasin (Supplementary Fig. 5). When reacted with purified NC1 hexamer, prepared without crosslinks, peroxidasin led to robust formation of cross-linked dimeric subunits at low enzyme to substrate ratios (< 1:30) only in the presence of H2O2 (Fig. 3a). Mass spectrometry of the peroxidasin reacted NC1 hexamer confirmed sulfilimine bond formation at levels near native PFHR-9 hexamer (Supplementary Fig. 6). To determine whether the ability to catalyze bond formation is a universal property of animal peroxidasins, we reacted Drosophila peroxidasin with uncross-linked collagen IV and found similar cross-linking activity (Supplementary Fig. 7). Taken together, peroxidasin cross-links collagen IV NC1 hexamer in vitro.
Figure 3. Peroxidasin forms hypohalous acids and sulfilimine bonds in collagen IV.
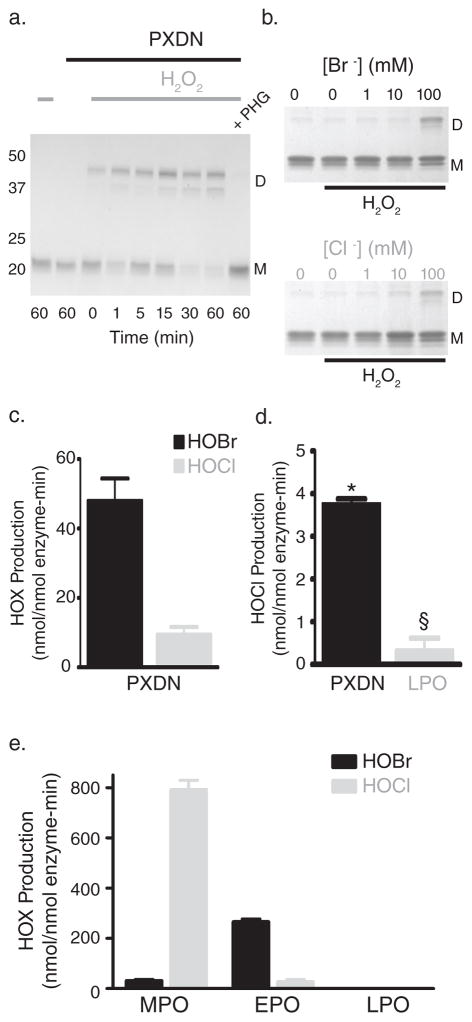
(a) SDS-PAGE of reactions consisting of 16 nM purified human peroxidasin, 500 nM monomeric NC1 hexamer (3 μM potential cross-links), and 10 μM H2O2 in 1X PBS. Control reactions without H2O2 or in the presence of the peroxidase inhibitor, phloroglucinol (PHG; 50 μM), were also conducted. D represents cross-linked dimeric NC1 subunits, while M denotes uncross-linked monomeric subunits. (b) Coomassie stained gel after SDS-PAGE of collagen IV NC1 hexamer is shown to illustrate relative amounts of sulfilimine cross-linked dimeric (D) and uncross-linked monomeric (M) subunits after incubation of uncross-linked PFHR-9 basement membranes in varying buffer halide concentrations (Br− or Cl− as K+ salt) with or without 1 mM H2O2. (c) Peroxidasin (PXDN) mediated hypohalous acid (HOX) production expressed as nmol hypohalous acid generated per nmol enzyme per minute measured in 1X PBS + 100 μM NaBr. Values represent mean ± s.e.m. (n=3). (d) HOCl production measured directly in 1X PBS without added Br−. Values denote mean ± s.e.m. (n=4). PXDN mediated HOCl generation was significantly greater than LPO (*unpaired two-tailed t-test, p < 0.05), while LPO was not statistically different from zero (§ one sample t-test, p = 0.32). (e) Hypohalous acid (HOX) production in nmol hypohalous acid generated per nmol enzyme per minute for myeloperoxidase (MPO), eosinophil peroxidase (EPO), and lactoperoxidase (LPO) in 1X PBS + 100 μM NaBr. Values represent mean ± s.e.m. (n=3). Full gel images are displayed in Supplementary Fig. 14.
Peroxidasin Forms Sulfilimine Bonds Via Hypohalous Acids
Animal heme peroxidases, such as peroxidasin, myeloperoxidase, eosinophil peroxidase, and lactoperoxidase, catalyze oxidative reactions using distinct halogenation and peroxidase cycles.16 Both begin with hydrogen peroxide oxidation of the prosthetic heme iron to form an intermediate denoted compound I.16 Compound I may oxidize halides into their respective hypohalous acids (or related oxidants in equilibrium) which may directly or indirectly halogenate susceptible moieties. Alternatively, compound I undergoes sequential reduction to form single electron free radicals of energetically favorable substrates in the peroxidase cycle. Both pathways eventually regenerate reduced, native enzyme.16 To determine whether peroxidasin forms sulfilimine bonds using a halogenation cycle, we first tested whether peroxidasin cross-links collagen IV in the absence of halides. When H2O2 was added to uncross-linked basement membrane without halides, very little cross-linked collagen IV dimeric subunits formed until halide (Cl− or Br−) concentrations approached 100 mM suggesting the involvement of a peroxidase halogenation cycle (Fig. 3b). Peroxidasin is known to iodinate proteins, but little is known about its ability to oxidize other halides such as bromide and chloride.7 Using taurine to trap hypohalous acids as stable taurine haloamines,11,17 peroxidasin formed hypobromous and hypochlorous acid at modest rates with a preference for bromide (Fig. 3c, d). Consistent with previous work, myeloperoxidase preferentially formed hypochlorous acid, eosinophil peroxidase primarily yielded hypobromous acid, and lactoperoxidase formed neither hypohalous acid (Fig. 3e).16 Taken together, peroxidasin produces hypohalous acids and requires halides (Cl− or Br−) to form sulfilimine bonds suggesting a link between the two activities.
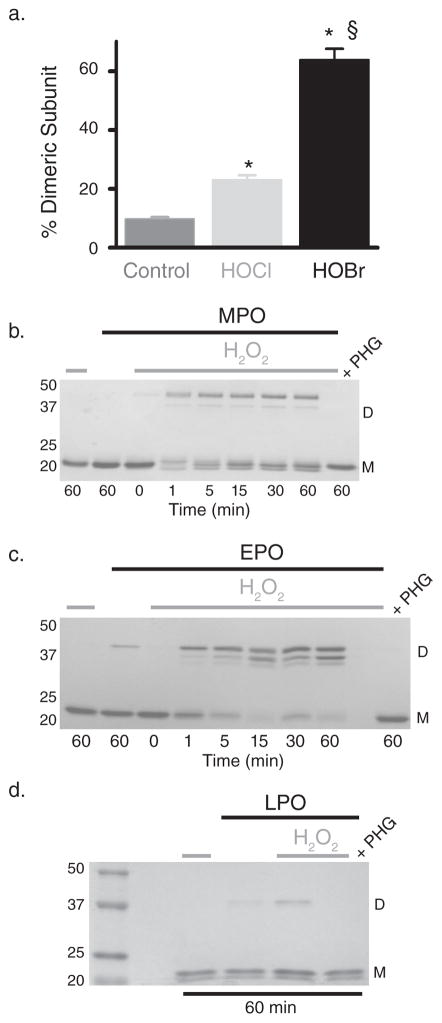
If peroxidasin utilizes hypohalous acids as intermediates to form sulfilimine bonds, these intermediates should recapitulate the reaction when directly added to purified, uncross-linked collagen IV NC1 hexamer. Indeed, reacting collagen IV with hypochlorous or hypobromous acid yielded cross-linked dimeric subunits (Fig. 4a; Supplementary Fig. 8, 9). Alternatively, other peroxidases should be able to catalyze sulfilimine bond formation when a halide is provided to form reactive hypohalous acids. Myeloperoxidase and eosinophil peroxidase formed sulfilimine cross-links in collagen IV (Fig. 4b, c), while lactoperoxidase poorly catalyzed cross-link formation since it does not efficiently form hypochlorous or hypobromous acid (Fig. 3e, 4d).16
Figure 4. Hypohalous acids form collagen IV sulfilimine bonds.
(a) 500 nM collagen IV NC1 hexamer (3 μM potential cross-links) was incubated alone (control) or with 5 μM hypochlorous (HOCl) or hypobromous acid (HOBr) for 30 minutes at 37°C. % dimeric subunit (mean ± s.e.m.) as quantified with densitometry of Coomassie stained SDS-PAGE gels (Supplementary Fig. 8) increased significantly with HOCl and HOBr treatment (control n=10, HOCl n=9, HOBr n=6; ANOVA with Tukey’s post-hoc comparison between groups, * = p < 0.05 compared to control, § = p < 0.05 HOCl versus HOBr). (b–d) 16 nM myeloperoxidase (MPO), eosinophil peroxidase (EPO), and lactoperoxidase (LPO) were reacted with 500 nM NC1 hexamer (3 μM potential cross-links) for varying time points in 1X PBS with or without 10 μM H2O2. In the case of LPO, all reactions proceeded for 60 minutes. Collagen IV sulfilimine cross-link content was visualized after SDS-PAGE and Coomassie blue staining of the reactions. Each gel is representative of 3 independent experiments. Complete gel images are provided in Supplementary Fig. 15.
Peroxidasin Cross-Links Collagen IV for Tissue Integrity
Though peroxidasin forms sulfilimine bonds in vitro, we tested whether peroxidasin catalyzes the formation of the sulfilimine bond within native insoluble collagen IV networks. HEK293 cells expressing human peroxidasin were plated on top of a PFHR-9 deposited basement membrane, which was produced in the presence of phloroglucinol to render a collagen IV network without sulfilimine cross-links (Fig. 5a). Only overlaid cells expressing human peroxidasin formed dimeric cross-linked NC1 subunits, while wildtype HEK293 cells or peroxidasin transfected cells in the continued presence of phloroglucinol failed to cross-link collagen IV (Fig. 5b). As a resident basement membrane protein,7 we hypothesized peroxidasin uniquely cross-links collagen IV networks, while other peroxidases, though capable of bond formation in solution, will not form cross-links within basement membranes. To test this hypothesis, HEK293 cells were plated on uncross-linked PFHR-9 basement membrane and transiently transfected with peroxidasin, myeloperoxidase, and lactoperoxidase cDNA or empty expression vector to determine whether peroxidasin specifically cross-links collagen IV. Only peroxidasin formed sulfilimine bonds in collagen IV even though myeloperoxidase enzymatic activity was at least 30-fold greater than peroxidasin (Fig. 5c, d). These data suggest only peroxidasin, embedded within basement membranes, generates hypohalous acid in close proximity to its collagen IV substrate. Comparatively greater, but spatially indiscriminate, generation of hypohalous acid by myeloperoxidase artificially cross-links soluble collagen IV NC1 hexamer, but fails to cross-link insoluble, basement membrane collagen IV.
Figure 5. Peroxidasin uniquely cross-links native collagen IV networks.
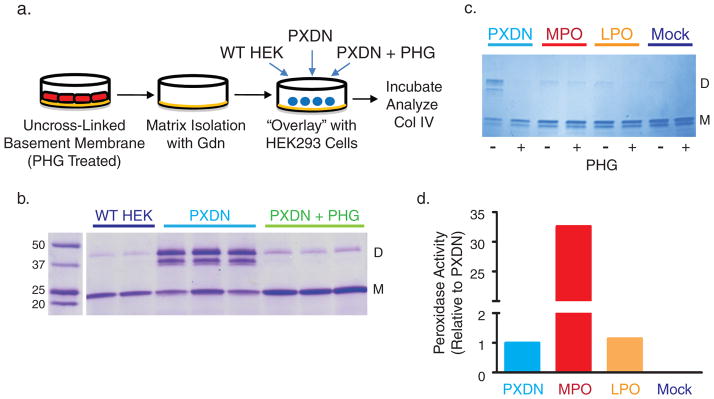
(a) Experimental design of “overlay” experiments. PFHR-9 cells were grown in the presence of phloroglucinol (PHG; 50 μM) to deposit uncross-linked collagen IV networks. The cells were then removed and the basement membrane extracted with 4M guanidine (Gdn) to inactivate endogenous peroxidasin. Human peroxidasin stably transfected cells (PXDN) or untransfected HEK293 cells (WT HEK) were plated on top of the PFHR9 basement membrane, which was subsequently analyzed for collagen IV cross-link content. (b) Collagen IV sulfilimine bond formation in the indicated experimental conditions as demonstrated by stained SDS-PAGE gel. 2 (wildtype HEK cells) or 3 (PXDN transfected with or without PHG) out of 5 independent experiments are displayed. (c) Coomassie stained gel of collagen IV NC1 hexamers isolated from uncross-linked PFHR-9 basement membrane overlaid with HEK293 cells transiently transfected with human peroxidasin cDNA, mouse myeloperoxidase cDNA (MPO), mouse lactoperoxidase cDNA (LPO), or empty vector (Mock). (d) Media from PXDN, MPO, LPO, and mock transfected cells was assayed for peroxidase activity using a tetramethylbenzidine based colorimetric assay. Activity was expressed relative to peroxidasin (A650 of given peroxidase divided by A650 for peroxidasin). Full gel images are displayed in Supplementary Fig. 16.
To further substantiate that peroxidasin functions to form sulfilimine bonds in collagen IV and to delineate the role of this function in basement membrane homeostasis, we turned to the Drosophila genetic model system where peroxidasin was first discovered.7 Using mass spectrometry of purified Drosophila collagen IV NC1 hexamer, we first experimentally determined that the collagen IV sulfilimine bond is present in Drosophila larvae as sequence conservation of methionine 93 and lysine 211 may not necessarily translate into a cross-link bridging these residues (Supplementary Fig. 10).4 With biochemical characterization of the collagen IV sulfilimine bond in hand, we examined basement membrane architecture in Drosophila larvae homozygous for a severely, hypomorphic peroxidasin (Pxn) allele (Pxnf07229/f07229; denoted as Pxn −/−) before their demise as third instar larvae. With the collagen IV GFP protein trap line Viking-GFPG454, we visualized collagen IV networks within basement membranes of the longitudinal and circumferential midgut visceral muscles.18 These networks appeared severely distorted and extensively torn in Pxn −/− mutants when compared with heterozygous Pxn +/− and wildtype Pxn +/+ larvae (Fig. 6a). Collagenase solubilization of larval basement membrane revealed Pxn −/− collagen IV NC1 content was about 20% of wildtype (Pxn +/+) based on immunoreactivity (Fig. 6b). Furthermore, Pxn −/− mutants demonstrated a shift towards uncross-linked monomer subunits with immunoreactivity rising to 42% of total band density compared to < 9% in Pxn +/− larvae (Fig. 6b). Thus, peroxidasin forms sulfilimine bonds that cross-link collagen IV to reinforce basement membranes and maintain tissue integrity.
Figure 6. Peroxidasin is critical for collagen IV and basement membrane integrity.

(a) Confocal fluorescence microscopy images of Drosophila anterior midgut using a collagen IV GFP protein trap line (Viking-GFP) to delineate collagen IV distribution. Representative sections from wildtype Pxn +/+ (Pxn +/+), heterozygote Pxn +/− (Pxn+/f07229), and mutant Pxn −/− (Pxnf07229/f07229) flies are shown. Distorted and torn collagen IV networks (arrows) with gross defects (“holes”) in the circumferential muscle layer (asterisks) typified Pxn −/− sections. Scale bar = 10 μm. (b) Immunoblot of collagenase solubilized basement membrane isolated from Drosophila Pxn +/− and Pxn −/− larvae. Pxn −/− mutants demonstrate grossly reduced collagen IV immunoreactivity at 20.4% of wildtype, while Pxn +/− flies have relatively maintained collagen IV NC1 content at 82% of wildtype (Supplementary Fig. 11). Pxn −/− mutants also demonstrated a shift in the % uncross-linked immunoreactivity with 42% of total band density in the uncross-linked form compared to < 9% total band density in Pxn +/− flies (Supplementary Fig. 11).
Discussion
In this work, we demonstrate that peroxidasin catalyzes sulfilimine bond formation in collagen IV, the first known bond of its kind in a biomolecule.4 Peroxidasin was initially discovered as a basement membrane constituent in Drosophila, but herein we establish its first bona fide function namely cross-linking collagen IV.7 Both the Drosophila mutant described in this work and C. elegans mutants of peroxidasin demonstrate defects in basement membrane integrity reminiscent of mutations in collagen IV itself.19,20 Our data provide a molecular mechanism for this phenotypic similarity. Loss of peroxidasin function leads to fewer collagen IV cross-links, destabilizes collagen IV, and reduces its content within basement membranes. Mutations in the human peroxidasin gene were recently discovered in a subset of patients with inherited anterior segment dysgenesis and cataracts. Accounting for two peroxidasin homologs in humans,21 we hypothesize that partial loss of peroxidasin activity compromises the collagen IV network of anterior eye basement membranes and again recapitulates an ocular phenotype commonly observed in patients with partial loss of function in collagen IV.22–26 Taken together, peroxidasin, collagen IV, and the sulfilimine cross-link form an important triad for basement membrane function and tissue biogenesis alongside laminin, nidogen, and proteoglycan.
Though this work identifies the first function of peroxidasin, the formation of sulfilimine cross-links in collagen IV may not be its only function. Peroxidasin is upregulated in response to transforming growth factor (TGFβ) stimulation of fibroblasts and in renal interstitial fibrosis.27 Collagen IV, a constituent primarily of basement membranes, is minimally present in fibroblast generated extracellular matrix.3 Thus, peroxidasin may form sulfilimine cross-links in other matrix proteins or execute non-catalytic functions involving protein-protein interactions with cell surface receptors and matrix proteins.
Peroxidasin generates hypohalous acids and requires halides to form sulfilimine cross-links, while hypohalous acids produce sulfilimine bonds when directly applied to collagen IV NC1 hexamer. Similarly, hypohalous acids, including HOBr and HOCl, form an intramolecular sulfilimine bond to convert methionine into dehydromethionine.28,29 We hypothesize peroxidasin, embedded within basement membranes near its collagen IV substrate, locally generates hypohalous acids, which form an intermolecular sulfilimine bond across two collagen IV protomers in a reaction mechanism akin to the formation of dehydromethionine. Specifically, HOBr and HOCl react with the sulfur of Met93 to form a halosulfonium cation intermediate which is then trapped by the Hyl211 amine to form a sulfilimine bond (Supplementary Fig. 12).30 Close proximity of the amine to the thioether creates a high effective amine concentration to prevent the halosulfonium cation from reacting with solvent water in a side reaction producing methionine sulfoxide. In collagen IV, the close apposition of Met93 and Hyl211 on separate NC1 trimers provides the required approximation of nitrogen and sulfur atoms to yield a sulfilimine bond bridging the NC1 trimer-trimer interface.29
While the parallel between the chemical synthesis and enzymatic catalysis of sulfilimine bonds suggests a mechanistic link, our data point to some differences. Iodine (I2) or hypoiodous acid (HOI) also efficiently converts methionine to dehydromethionine,28,29,31 yet iodide paradoxically inhibits cross-link formation in collagen IV. Many possible mechanisms could explain this inhibition including I− quenching of reactive hypohalous acid intermediates,32 competition between I− and H2O2 preventing compound I formation,33 or complex halide interactions at the peroxidasin catalytic site.33–35 Future work will need to address the mechanism of iodide inhibition and formally test the proposed reaction scheme for sulfilimine bond formation (Supplementary Fig. 12).
Hypohalous acids typically conjure images of microbial destruction and unintended toxicity, but this work points to a surprising, anabolic role for these highly reactive species. Peroxidasin is optimally suited to productively use hypohalous acids as its non-catalytic leucine repeat rich (LRR) and immunoglobulin (Ig) protein interaction domains presumably place peroxidasin in close proximity to its collagen IV substrate so that relatively modest amounts of hypohalous acids form sulfilimine cross-links without pathologic “collateral damage.” The use of hypohalous acids as anabolic intermediates presumably depends on coupling peroxidasin oxidant generation with sulfilimine cross-link formation and possibly on local anti-oxidant mechanisms. Excessive peroxidasin activity either due to overexpression or increased H2O2 substrate availability, may uncouple hypohalous acid generation from sulfilimine bond formation allowing free hypohalous acid oxidants to accumulate and produce intended or unintended toxicity. Indeed, mosquito gut peroxidasin is upregulated after bacterial infection and its knockdown reduces bacterial clearance and host survival.36 Invertebrate peroxidasin may generate antimicrobial hypohalous acids as a primitive form of innate immunity analogous to vertebrate myeloperoxidase and eosinophil peroxidase.37
Oxidative stress and reactive oxygen species play a central role in the pathogenesis of atherosclerosis, diabetes mellitus associated complications, and hypertensive vascular disease which are the leading causes of morbidity and mortality in developed nations.38–40 Human peroxidasin, also known as vascular peroxidase 1 (VPO1), is upregulated in cell culture models of hypertension and atherosclerosis and promotes smooth muscle proliferation and fibrosis, but the mechanistic connection between peroxidasin and downstream pathologic events is unknown.27,41–43 Since peroxidasin consumes H2O2 produced by cell surface NADPH oxidases (NOX), enhanced NOX generated H2O2 in pathologic states may promote peroxidasin mediated matrix cross-linking and stabilization eventually leading to tissue fibrosis.21,43 Alternatively, “uncoupled” peroxidasin activity may lead to hypohalous acid accumulation promoting tissue injury. Indeed, myeloperoxidase has garnered significant attention for hypochlorous acid mediated oxidative modifications involved in the development of vascular inflammatory disorders such as atherosclerosis.44 But unlike myeloperoxidase, which requires targeting to vessel wall, peroxidasin is omnipresent at the site of pathology within vascular basement membranes, and therefore primed to generate deleterious oxidants and participate in disease pathogenesis.21,43,44 Collectively, these results establish that peroxidasin forms collagen IV sulfilimine cross-links, a post-translational modification critical for basement membrane integrity and tissue biogenesis, and draw attention to peroxidasin as an oxidant generator embedded within basement membranes readily capable of participating in disease pathogenesis.
Methods
Chemicals
Phloroglucinol, methimazole, potassium iodide, and tetramethylbenzidine were >99% pure, while β-aminopropionitrile, putrescine, and 3-1,2,4-aminotriazole were >98%, >97%, and ~95% pure respectively. All chemicals were obtained from Sigma Chemical Co. (St. Louis, MO).
Collagen IV NC1 Hexamer Isolation
PFHR-9 cells were homogenized in 1% deoxycholate with sonication and the insoluble material isolated after centrifugation at 20,000×g for 15 minutes. The pellet was then extracted with 1M NaCl (or 2M urea in some experiments) + 50 mM Tris-Cl, pH 7.5, and 10 mM Tris-Cl, pH 7.5 and digested in 50 mM Tris-Cl pH 7.5 + 5 mM CaCl2 + 5 mM benzamidine + 25 mM 6-aminocaproic acid + 0.4 mM PMSF + 0.1 mg/mL bacterial collagenase (Worthington; Lakewood, NJ). Collagenase solubilized material was dialyzed against 50 mM Tris-Cl, pH 7.5. NC1 hexamers were purified using anion-exchange chromatography (DE52 Cellulose or Q Sepharose) followed by gel filtration chromatography.
In Vitro Basement Membrane Reactions
PFHR-9 cells treated with potassium iodide (1–10 mM) to eliminate NC1 hexamer cross-links were used for basement membrane isolation. To test halide dependency, halide free conditions were established by washing extensively (at least 5 times) with 10 mM sodium-phosphate pH 7.4. To try to extract or inactivate endogenous basement membrane peroxidase activity, the matrix preparation was extracted twice with 2M guanidine-Cl, 50 mM Tris-Cl pH 7.5, 10 mM EDTA-Na pH 8 followed by extensive washing with 1X PBS. Basement membrane was resuspended in the desired buffer with or without cofactors and inhibitors to examine in vitro NC1 cross-linking under various conditions. Basement membranes were collagenase solubilized to delineate collagen IV NC1 sulfilimine cross-link formation with SDS-PAGE and Coomassie blue staining.
Azide Labeling and Click-Chemistry Biotinylation of Labeled Proteins
PFHR-9 membrane was isolated, washed extensively and resuspended in 1X PBS. Azide (0 to 10 mM) and 1 mM H2O2 were added and allowed to react for 1 hour at 37°C. The matrix was pelleted, washed extensively with 1X PBS, and solubilized with 1X PBS + 2% SDS. Solubilized proteins were reacted with 100 μM TBTA (Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine; Anaspec; Fremont, CA), 1 mM TCEP (Tris(2-carboxyethyl)phosphine hydrochloride; ThermoFisher Pierce; Rockford, IL), 1 mM cupric sulfate, and 100 μM biotin alkyne (PEG4 carboxamide-propargyl biotin; Life Technologies; Grand Island, NY) for 1 hour at 37°C. Click chemistry reactions were quenched with 1 mM AZT (3′-azido-3′-deoxythymidine; Sigma). For avidin-HRP detection, samples were electrophoresed under reducing conditions, transferred to nitrocellulose membranes, and probed with streptavidin-HRP according to manufacturer instructions (ThermoFisher Pierce). To isolate biotinylated proteins, click reaction products were precipitated with 2 volumes of cold acetone to remove reactants, washed with 70% acetone, and then re-solubilized in 1X PBS + 2% SDS. Biotinylated proteins were captured with streptavidin-agarose beads (GE Life Sciences; Piscataway, NJ) and released with boiling for 15 minutes in SDS-PAGE sample buffer containing 50 mM DTT.
Purification of Recombinant Human Peroxidasin
HEK293 cells stably transfected with the human peroxidasin coding sequence27 were grown to confluency and the media was changed to serum free DMEM/F12 + 5 μM hematin + 5 mM sodium butyrate. After 48–60 hours, media was harvested, protease inhibitors were added (0.5 mM PMSF, 1 μg/mL leupeptin, 1 μg/mL pepstatin, and 10 mM EDTA-Na), and proteins were precipitated with 40% ammonium sulfate (226 g/L). Precipitated protein was resuspended at ~1/50 of the original media volume in 0.3M sucrose + 0.1M NaCl + 20 mM Tris-Cl pH 8.5, dialyzed against the same buffer, and chromatographed on a Mono-Q anion exchange column (GE Life Sciences). Enzymatically active fractions were pooled, precipitated to ~1/500 the original media volume of 50 mM NaCl + 10 mM sodium phosphate, pH 7.4 + 3 mM hexadecyltrimethylammonium chloride, and dialyzed against the same buffer. The dialyzed protein was further purified using ultracentrifugation on a 5–20% sucrose gradient. Active fractions were pooled and concentrated to a final concentration of 0.25 – 0.5 mg/mL of purified human peroxidasin.
HEK293 Cell Overlay on Uncross-Linked Collagen IV Networks
PFHR-9 cells were grown in the presence of 50 μM phloroglucinol to produce non-cross-linked collagen IV. Basement membrane was isolated on plate using a modification of a previously published protocol.45 To inactivate endogenous cross-linking activity, the basement membrane was treated with 4M guanidine-Cl + 50 mM Tris-Cl pH 7.5 for 15 minutes and then washed 5 times with 1X PBS. In the first set of experiments, HEK cells stably transfected with human peroxidasin were compared to wildtype HEK293 cells. In follow-up experiments, HEK293T cells were transiently transfected with human peroxidasin coding sequence,27 mouse myeloperoxidase cDNA (Origene, Rockville, MD), mouse lactoperoxidase cDNA (Origene), or empty vector (pCDNA-V5-His-TOPO without insert) using Lipofectamine LTX per manufacturer’s instructions (Life Technologies). In both sets of experiments, HEK293(T) cells were plated on PFHR-9 basement membrane in the presence of 5 μM hematin and 5 mM sodium butyrate. Plates were incubated for 24–48 hours and collagen IV analyzed for NC1 cross-link formation.
Preparation of HOCl and HOBr Solutions
Standard techniques were used to prepare HOCl and HOBr. Further details are provided in Supplementary Methods.
Measurement of Hypohalous Acid Production by Peroxidases
Hypohalous acids were trapped as stable taurine haloamines which oxidize tetramethylbenzidine to yield a colorimetric measure of hypohalous acid concentration and production.17 Further details are outlined in Supplementary Methods.
Mass Spectrometry and Identification of Sulfilimine Cross-Linked Peptides
We used a modification of previously described methods.4 Details are provided in Supplementary Methods.
Drosophila Biochemistry and Genetics
Drosophila collagen IV NC1 hexamer was essentially purified as described for PFHR-9 cells. Standard genetic techniques detailed in Supplementary Methods were utilized.
Statistical Analysis
Statistical analysis was conducted using GraphPad Prism version 5.04 (GraphPad Software, San Diego, CA). Comparisons between two groups utilized two tailed unpaired t-tests, while multiple group comparisons were conducted using ANOVA followed by Tukey’s post-hoc comparisons between specific groups.
Supplementary Material
Acknowledgments
This work was supported by the NIH (RO1 DK18381, DK18381-38S1, and 2PO1 DK065123 to B.G.H.), MDIBL Salisbury Cove Research Fund and FH Epstein Fellowship (to B.G.H), Vanderbilt Division of Nephrology Faculty Development Fund (to R.V.), and a Vanderbilt Physician Scientist Development Award (to G.B.). We acknowledge Dr. Andrew Chisholm (UCSD, San Diego, CA) and Dr. Andrea Page-McCaw (Vanderbilt University, Nashville, TN) for fruitful discussions during the writing of this manuscript. We thank Dr. Miklos Geizst (Semmelweis University, Budapest, Hungary) for the human peroxidasin coding sequence and Dr. Lynn Cooley (Yale University, New Haven, CT) for the Viking GFP protein trap Drosophila line. Parvin Todd, Neonila Danylevych, and Dr. Christo Venkov provided technical assistance.
Footnotes
Author Contributions.
G.B. conducted, designed, and analyzed data from the PFHR-9 cell culture experiments, purified collagen IV NC1 hexamers from Drosophila, and conducted western blotting experiments on Drosophila mutants. C.F.C. conducted mechanistic experiments involving hypohalous acids and peroxidasin. R.M.V. conducted mass spectrometry and analysis. L.I.F. prepared Drosophila materials and C.K.-C. performed Drosophila genetics and confocal microscopy. I.A.E-T. performed overlay experiments involving peroxidasin and other peroxidases. M.R. isolated collagen IV NC1 hexamers and sulfilimine cross-linked peptides for further analysis. J.-S.K. isolated human peroxidasin expressing HEK293 stable cell lines and V.P. established the PFHR-9 cell culture system for these studies. L.I.F. generated Drosophila mutant larvae, antibodies, and protein reagents. L.I.F., J.H.F., and B.G.H. designed the study and wrote the paper along with G.B. All authors discussed the results and commented on the manuscript.
Competing Financial Interests.
The authors have no competing financial interests.
Supplementary information is available online at http://www.nature.com/naturechemicalbiology.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Yurchenco PD. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol. 2011;3:1–27. doi: 10.1101/cshperspect.a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poschl E, et al. Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development. 2004;131:1619–28. doi: 10.1242/dev.01037. [DOI] [PubMed] [Google Scholar]
- 3.Khoshnoodi J, Pedchenko V, Hudson BG. Mammalian collagen IV. Microsc Res Tech. 2008;71:357–70. doi: 10.1002/jemt.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanacore R, et al. A sulfilimine bond identified in collagen IV. Science. 2009;325:1230–4. doi: 10.1126/science.1176811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedchenko V, et al. Molecular architecture of the Goodpasture autoantigen in anti-GBM nephritis. N Engl J Med. 2010;363:343–54. doi: 10.1056/NEJMoa0910500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fessler LI, Fessler JH. Identification of the carboxyl peptides of mouse procollagen IV and its implications for the assembly and structure of basement membrane procollagen. J Biol Chem. 1982;257:9804–10. [PubMed] [Google Scholar]
- 7.Nelson RE, et al. Peroxidasin: a novel enzyme-matrix protein of Drosophila development. EMBO J. 1994;13:3438–47. doi: 10.1002/j.1460-2075.1994.tb06649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adeniyi-Jones SK, Karnovsky ML. Oxidative decarboxylation of free and peptide-linked amino acids in phagocytizing guinea pig granulocytes. The Journal of clinical investigation. 1981;68:365–73. doi: 10.1172/JCI110264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagasaka A, Hidaka H. Effect of antithyroid agents 6-propyl-2-thiouracil and 1-mehtyl-2-mercaptoimidazole on human thyroid iodine peroxidase. The Journal of clinical endocrinology and metabolism. 1976;43:152–8. doi: 10.1210/jcem-43-1-152. [DOI] [PubMed] [Google Scholar]
- 10.Alexander NM. Iodide peroxidase in rat thyroid and salivary glands and its inhibition by antithyroid compounds. The Journal of biological chemistry. 1959;234:1530–3. [PubMed] [Google Scholar]
- 11.Weiss SJ, Klein R, Slivka A, Wei M. Chlorination of taurine by human neutrophils. Evidence for hypochlorous acid generation. The Journal of clinical investigation. 1982;70:598–607. doi: 10.1172/JCI110652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang SS, Trackman PC, Kagan HM. Reaction of aortic lysyl oxidase with beta-aminopropionitrile. The Journal of biological chemistry. 1983;258:4331–8. [PubMed] [Google Scholar]
- 13.Candi E, et al. Biochemical, structural, and transglutaminase substrate properties of human loricrin, the major epidermal cornified cell envelope protein. The Journal of biological chemistry. 1995;270:26382–90. doi: 10.1074/jbc.270.44.26382. [DOI] [PubMed] [Google Scholar]
- 14.Ortiz de Montellano PR, David SK, Ator MA, Tew D. Mechanism-based inactivation of horseradish peroxidase by sodium azide. Formation of meso-azidoprotoporphyrin IX. Biochemistry. 1988;27:5470–6. doi: 10.1021/bi00415a013. [DOI] [PubMed] [Google Scholar]
- 15.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–9. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 16.Obinger C. Chemistry and biology of human peroxidases. Archives of biochemistry and biophysics. 2006;445:197–8. doi: 10.1016/j.abb.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 17.Dypbukt JM, et al. A sensitive and selective assay for chloramine production by myeloperoxidase. Free Radic Biol Med. 2005;39:1468–77. doi: 10.1016/j.freeradbiomed.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci U S A. 2001;98:15050–5. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gotenstein JR, et al. The C. elegans peroxidasin PXN-2 is essential for embryonic morphogenesis and inhibits adult axon regeneration. Development. 2010;137:3603–13. doi: 10.1242/dev.049189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta MC, Graham PL, Kramer JM. Characterization of alpha1(IV) collagen mutations in Caenorhabditis elegans and the effects of alpha1 and alpha2(IV) mutations on type IV collagen distribution. J Cell Biol. 1997;137:1185–96. doi: 10.1083/jcb.137.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng G, Salerno JC, Cao Z, Pagano PJ, Lambeth JD. Identification and characterization of VPO1, a new animal heme-containing peroxidase. Free Radic Biol Med. 2008;45:1682–94. doi: 10.1016/j.freeradbiomed.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coupry I, et al. Ophthalmological features associated with COL4A1 mutations. Arch Ophthalmol. 2010;128:483–9. doi: 10.1001/archophthalmol.2010.42. [DOI] [PubMed] [Google Scholar]
- 23.Favor J, et al. Type IV procollagen missense mutations associated with defects of the eye, vascular stability, the brain, kidney function and embryonic or postnatal viability in the mouse, Mus musculus: an extension of the Col4a1 allelic series and the identification of the first two Col4a2 mutant alleles. Genetics. 2007;175:725–36. doi: 10.1534/genetics.106.064733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gould DB, Marchant JK, Savinova OV, Smith RS, John SW. Col4a1 mutation causes endoplasmic reticulum stress and genetically modifiable ocular dysgenesis. Hum Mol Genet. 2007;16:798–807. doi: 10.1093/hmg/ddm024. [DOI] [PubMed] [Google Scholar]
- 25.Labelle-Dumais C, et al. COL4A1 mutations cause ocular dysgenesis, neuronal localization defects, and myopathy in mice and Walker-Warburg syndrome in humans. PLoS Genet. 2011;7:e1002062. doi: 10.1371/journal.pgen.1002062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Agtmael T, et al. Dominant mutations of Col4a1 result in basement membrane defects which lead to anterior segment dysgenesis and glomerulopathy. Hum Mol Genet. 2005;14:3161–8. doi: 10.1093/hmg/ddi348. [DOI] [PubMed] [Google Scholar]
- 27.Peterfi Z, et al. Peroxidasin is secreted and incorporated into the extracellular matrix of myofibroblasts and fibrotic kidney. Am J Pathol. 2009;175:725–35. doi: 10.2353/ajpath.2009.080693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beal JL, Foster SB, Ashby MT. Hypochlorous acid reacts with the N-terminal methionines of proteins to give dehydromethionine, a potential biomarker for neutrophil-induced oxidative stress. Biochemistry. 2009;48:11142–8. doi: 10.1021/bi901343d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peskin AV, Turner R, Maghzal GJ, Winterbourn CC, Kettle AJ. Oxidation of methionine to dehydromethionine by reactive halogen species generated by neutrophils. Biochemistry. 2009;48:10175–82. doi: 10.1021/bi901266w. [DOI] [PubMed] [Google Scholar]
- 30.Armesto XL, Canle M, Fernandez MI, Garcia MV, Santaballa JA. First steps in the oxidation of sulfur-containing amino acids by hypohalogenation: Very fast generation of intermediate sulfenyl halides and halosulfonium cations. Tetrahedron. 2000;56:1103–1109. [Google Scholar]
- 31.Lavine TF. The formation, resolution, and optical properties of the diastereoisomeric sulfoxides derived from L-methionine. The Journal of biological chemistry. 1947;169:477–91. [PubMed] [Google Scholar]
- 32.Huwiler M, Burgi U, Kohler H. Mechanism of enzymatic and non-enzymatic tyrosine iodination. Inhibition by excess hydrogen peroxide and/or iodide. European journal of biochemistry/FEBS. 1985;147:469–76. doi: 10.1111/j.0014-2956.1985.00469.x. [DOI] [PubMed] [Google Scholar]
- 33.Blair-Johnson M, Fiedler T, Fenna R. Human myeloperoxidase: structure of a cyanide complex and its interaction with bromide and thiocyanate substrates at 1.9 A resolution. Biochemistry. 2001;40:13990–7. doi: 10.1021/bi0111808. [DOI] [PubMed] [Google Scholar]
- 34.Andrews PC, Krinsky NI. A kinetic analysis of the interaction of human myeloperoxidase with hydrogen peroxide, chloride ions, and protons. The Journal of biological chemistry. 1982;257:13240–5. [PubMed] [Google Scholar]
- 35.Taurog A, Dorris ML. Myeloperoxidase-catalyzed iodination and coupling. Archives of biochemistry and biophysics. 1992;296:239–46. doi: 10.1016/0003-9861(92)90568-h. [DOI] [PubMed] [Google Scholar]
- 36.Garver LS, Xi Z, Dimopoulos G. Immunoglobulin superfamily members play an important role in the mosquito immune system. Dev Comp Immunol. 2008;32:519–31. doi: 10.1016/j.dci.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zamocky M, Jakopitsch C, Furtmuller PG, Dunand C, Obinger C. The peroxidase-cyclooxygenase superfamily: Reconstructed evolution of critical enzymes of the innate immune system. Proteins. 2008;72:589–605. doi: 10.1002/prot.21950. [DOI] [PubMed] [Google Scholar]
- 38.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–20. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 39.Touyz RM, Briones AM. Reactive oxygen species and vascular biology: implications in human hypertension. Hypertens Res. 2011;34:5–14. doi: 10.1038/hr.2010.201. [DOI] [PubMed] [Google Scholar]
- 40.Yokoyama M. Oxidant stress and atherosclerosis. Current opinion in pharmacology. 2004;4:110–5. doi: 10.1016/j.coph.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 41.Bai YP, et al. Role of VPO1, a newly identified heme-containing peroxidase, in ox-LDL induced endothelial cell apoptosis. Free radical biology & medicine. 2011;51:1492–500. doi: 10.1016/j.freeradbiomed.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi R, et al. Involvement of vascular peroxidase 1 in angiotensin II-induced vascular smooth muscle cell proliferation. Cardiovasc Res. 2011;91:27–36. doi: 10.1093/cvr/cvr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brandes RP. Vascular peroxidase 1/peroxidasin: a complex protein with a simple function? Cardiovascular research. 2011;91:1–2. doi: 10.1093/cvr/cvr120. [DOI] [PubMed] [Google Scholar]
- 44.Lau D, Baldus S. Myeloperoxidase and its contributory role in inflammatory vascular disease. Pharmacology & therapeutics. 2006;111:16–26. doi: 10.1016/j.pharmthera.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 45.Kramer RH, Bensch KG, Davison PM, Karasek MA. Basal lamina formation by cultured microvascular endothelial cells. The Journal of Cell Biology. 1984;99:692–8. doi: 10.1083/jcb.99.2.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weber S, Dolz R, Timpl R, Fessler JH, Engel J. Reductive cleavage and reformation of the interchain and intrachain disulfide bonds in the globular hexameric domain NC1 involved in network assembly of basement membrane collagen (type IV) European journal of biochemistry/FEBS. 1988;175:229–36. doi: 10.1111/j.1432-1033.1988.tb14188.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.