Summary
The global regulator, Spx, is under proteolytic control exerted by the adaptor YjbH and ATP-dependent protease ClpXP in Bacillus subtilis. While YjbH is observed to bind the Spx C-terminus, YjbH shows little affinity for ClpXP, indicating adaptor activity that does not operate by tethering. Chimeric proteins derived from B. subtilis AbrB and the Spx C-terminus showed that a 28 residue C-terminal section of Spx (AbrB28), but not the last 12 or 16 residues (AbrB12, AbrB16), was required for YjbH interaction and for ClpXP proteolysis, although the rate of AbrB28 proteolysis was not affected by YjbH addition. The result suggested that the YjbH-targeted 28 residue segment of the Spx C-terminus bears a ClpXP-recognition element(s) that is hidden in the intact Spx protein. Residue substitutions in the conserved helix α6 of the C-terminal region generated Spx substrates that were degraded by ClpXP at accelerated rates compared to wild type Spx, and showed reduced dependency on the YjbH activity. The residue substitutions also weakened the interaction between Spx and YjbH. The results suggest a model in which YjbH, through interaction with residues of α6 helix, exposes the C-terminus of Spx for recognition and proteolysis by ClpXP.
Keywords: YjbH, Spx, ClpXP, proteolysis, Bacillus subtilis
Introduction
Proteolysis is an essential process of cellular homeostasis and survival. Protease-catalyzed removal of damaged proteins ensures that toxic, unfolded polypeptides do not accumulate; a process of proteome quality control (Mogk et al., 2011, Moliere & Turgay, 2013). Furthermore, regulatory proteins are expressed according to different growth phases and in response to environmental conditions or stresses, but are often removed proteolytically when no longer needed or when their continued presence could prove detrimental (Gottesman, 2003, Gur et al., 2011, Moliere & Turgay, 2013). To maintain proteome homeostasis, unwanted or damaged proteins are degraded by ATP-dependent proteolytic machines, commonly referred to as AAA+ proteases (ATPase associated with diverse cellular activities) that are composed of unfoldase and peptidase components (Sauer & Baker, 2011). AAA+ proteases are divided into two broad groups: those containing unfoldase and peptidase on separate polypeptides (ClpAP, ClpCP, ClpEP, ClpXP and HslUV) and those possessing both activities on one single polypeptide (LonA, LonB and FtsH) (Sauer & Baker, 2011).
In Bacillus subtilis, there are seven different AAA+ proteases (ClpCP, ClpEP, ClpXP, HslUV, LonA, LonB, and FtsH). These AAA+ proteases recognize a broad range of protein substrates with high degree of specificity. In most cases, there are recognition motifs (degradation tags or degrons) present in the protein substrates that are usually located at the N- or C- terminus, but some reside internally (Schrader et al., 2009). Studies in E. coli ClpX identified five ClpX recognition motifs: three at the N-terminus and two at the C-terminus of the substrate proteins (Flynn et al., 2003). The recognition between the proteases and substrates is through direct interaction or indirect/adaptor-mediated recognition. One of the well-characterized direct recognition motifs recognized by ClpX is the SsrA tag, composed of 11 amino acids (AANDENYALAA) (Gottesman et al., 1998). Proteolysis of SsrA-tagged proteins is accelerated by interaction with SspB, an adaptor that specifically interacts with the appended SsrA peptide and with the N-terminal domain of ClpX, in effect, offering the substrate to the ClpXP proteolytic machine (Flynn et al., 2003). Indirect/adaptor-mediated substrate recognition modulates the protein substrate specificities and enhances proteolysis usually, but not always (Hengge, 2009), by tethering and delivering the substrates to the proteases (Battesti & Gottesman, 2013). In B. subtilis, there are four adaptors identified so far: three adaptors (MecA, McsB, and YpbH) function with ClpCP and one adaptor (YjbH) with ClpX (Kirstein et al., 2007, Larsson et al., 2007, Persuh et al., 2002, Schlothauer et al., 2003).
Previous studies reported that YjbH serves as an adaptor for accelerating Spx proteolysis by ClpXP (Garg et al., 2009, Larsson et al., 2007). A member of the ArsC family of proteins, Spx is a global transcriptional regulator that plays an important role in thiol homeostasis and competence through activation and repression of gene transcription. Exposure to toxic oxidants induces an elevation in Spx protein concentration, which activates the transcription of genes involved in alleviating oxidative stress (Zuber, 2004). Recent studies have provided evidence that Spx activates the transcription of genes whose products function in prevention of protein aggregation resulting from redox dysfunction (Runde et al., 2014). The level and activity of Spx is controlled by three different mechanisms: transcriptional, post-translational (proteolysis) and redox (via a CXXC redox disulfide center) controls. All of these controls ensure the Spx is upregulated when needed but is maintained in low levels during unperturbed growth. Deletion of the yjbH gene results in elevated Spx concentrations (Larsson et al., 2007), and YjbH protein accelerates Spx proteolysis in a reaction containing purified ClpXP (Chan et al., 2012, Garg et al., 2009). Unlike the adaptor, SspB, YjbH in affinity pull-down reactions, shows no detectable affinity for the protease ClpXP (Chan et al., 2012).
A crystal structure of the B. subtilis Spx protein in complex with the C-terminal domain of α subunit (α-CTD) of RNA polymerase provided insights into Spx interaction with RNA polymerase from their interaction interface (Newberry et al., 2005). The last 13 amino acids of Spx (119–131) are missing from the crystal structure probably due to structural disorder. Previous studies showed that the C-terminus of Spx is important for adaptor YjbH binding and ClpXP proteolysis (Chan et al., 2012, Nakano et al., 2003a). Changing the last two amino acids to aspartic acid in Spx (SpxDD) resulted in resistance to proteolysis by ClpXP (Nakano et al., 2003a). Also, deletion of the last 12 amino acids from Spx significantly decreased its interaction with YjbH (Chan et al., 2012) and resulted in Spx protein stability in vivo (Lin and Zuber, 2012). At present, the mechanism of YjbH-mediated, ClpXP-catalyzed Spx proteolysis is not known, however.
In the study presented herein, we identified a conserved motif (IRRFL, α6 helix) at the Spx C-terminus through sequence alignments of 21 Spx orthologs (Fig. 1, S1). We obtained Spx mutants containing residue substitutions in the conserved C-terminal α6 helix and measured their proteolysis rates in the presence and absence of YjbH and examined YjbH interaction with the mutant Spx derivatives. We found that the amino acid substitutions significantly decreased their interaction with YjbH, but elevated the rate of ClpXP-catalyzed proteolysis in the absence of YjbH protein. These results indicated that the conserved residues of the Spx C-terminus are not only required for YjbH interaction and ClpXP recognition, but also contribute to the stabilization of Spx structural architecture that must be acted upon by YjbH to render the C-terminus accessible to ClpXP-catalyzed proteolysis.
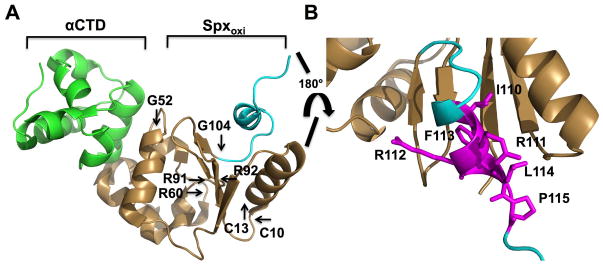
Fig. 1. X-ray structure of B. subtilis Spx in complex with RNA polymerase α C-terminal domain (panel A and B, derived from PDB ID 1Z3E) (Newberry et al., 2005) and sequence alignment of orthologous Spx C-terminal regions.
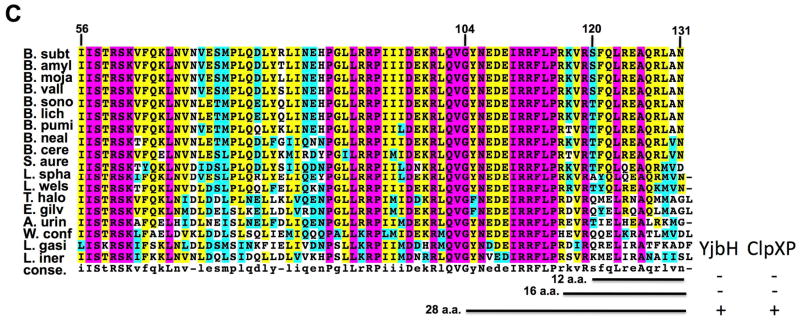
A. Ribbon representation of X-ray structure of B. subtilis Spx [1–103 amino acid (a.a.), gold and C-terminal region, 104–119 a.a., cyan] in complex with αCTD of RNA polymerase (green). Residues of the redox center (C10, C13), in RNA polymerase interaction (G52, R91), promoter recognition (R60, R92), and the start of 28-residue segment of the C-terminal region (G104) are highlighted.
B. C-terminal region of Spx. Residue side chains within the α6 helix (IRRFL) and adjacent proline (115) are highlighted in magenta.
C. Sequence alignment of Spx from 18 organisms (homologous to B. subitlis Spx 56–131 a.a.). Indicated by black horizontal lines are 12 residue (12 a.a., 119–131) and 28 residue (28 a.a.,104–131) tails from C-terminal region of Spx. The conserved residues are boxed, with completely conserved residues in magenta, identical residues in yellow, similar residues in cyan, and less conserved residues in white. B. subt, Bacillus subtilis; B. amyl, Bacillus amyloliquefaciens; B. mojia, Bacillus mojavensis; B. vall, Bacillus vallismortis; B. sono, Bacillus sonorensis; B. lich, Bacillus licheniformis; B. pumi, Bacillus pumilus; B. neal, Bacillus nealsonii; B. cere, Bacillus cereus; S. aure, Staphylococcus aureus; L. spha, Lysinibacillus sphaericus; L. wels, Listeria welshimer; T. halo, Tetragenococcus halophilus; E. gilv, Enterococcus gilvus; A. urin, Aerococcus urinae; W. conf, Weissella confusa; L. gasi, Leuconostoc gasicomiatum; L. iner, Lactobacillus iners. YjbH binding or ClpXP proteolysis of AbrB derivatives bearing the indicated C-terminal segments is indicated with + or − (see data of Fig. 2)
Results
Sequence alignment of Spx orthologs reveals a conserved motif at the C-terminus
The crystal structure of oxidized Spx (1–118) in complex with the α-CTD revealed their interaction at the molecular level. However, the last 13 amino acid residues (119–131) were not resolved, likely due to structural disorder of the Spx C-terminal region (Newberry et al., 2005). In previous studies involving structural, genetic and biochemical analyses of B. subtilis Spx, some residues were identified to be important for RNA polymerase interaction (SpxG52, R91), target promoter DNA binding and positioning of the RNAP σA subunit on the core promoter element of the trxB gene (SpxR60, R92) (Fig. 1A) (Lin et al., 2013, Nakano et al., 2010, Nakano et al., 2000, Newberry et al., 2005). A conserved CXXC motif was also identified as a redox sensing center (Fig. 1A) (Nakano et al., 2005). Recently, the C-terminal tail of Spx was shown to be important for binding to the proteolytic adaptor, YjbH, and for ClpXP protease recognition. More specifically, a deletion of 12 residues from the Spx C-terminus (SpxΔC) significantly decreased the affinity of Spx for YjbH interaction (Chan et al., 2012), and Spx resisted ClpXP proteolysis when its last two amino acids were replaced with aspartic acids (SpxDD) (Nakano et al., 2003a). To investigate which structural features of the Spx C-terminal tail might function in proteolytic control, an alignment of Spx sequences from 21 organisms was inspected, uncovering a conserved motif (IRRFLP) at their C-termini (Fig. 1C, S1). This motif contains the α6 helix and is the last structural element discernable in the crystal structure of the Spx polypeptide (Fig. 1B).
Twenty-eight amino acids of Spx C-terminus are sufficient for YjbH binding and ClpXP-catalyzed proteolysis, but not YjbH-enhanced proteolysis
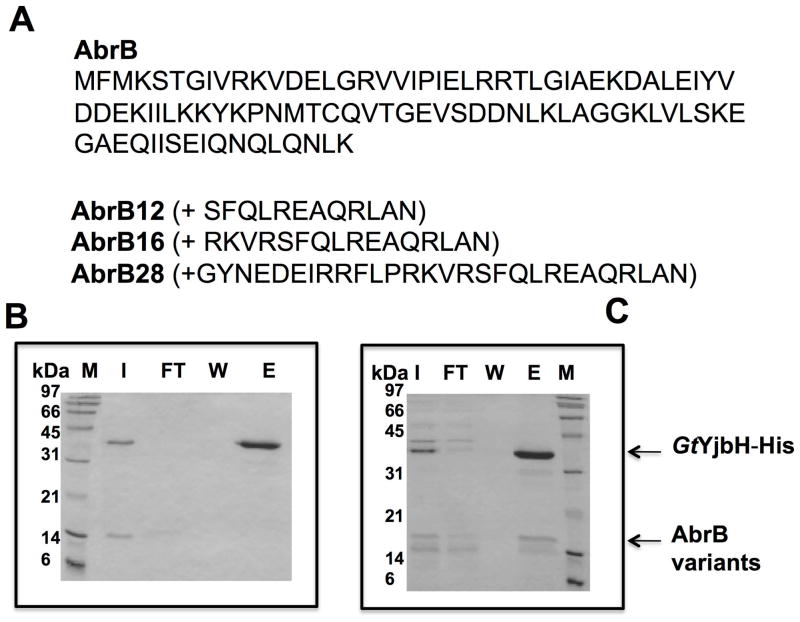
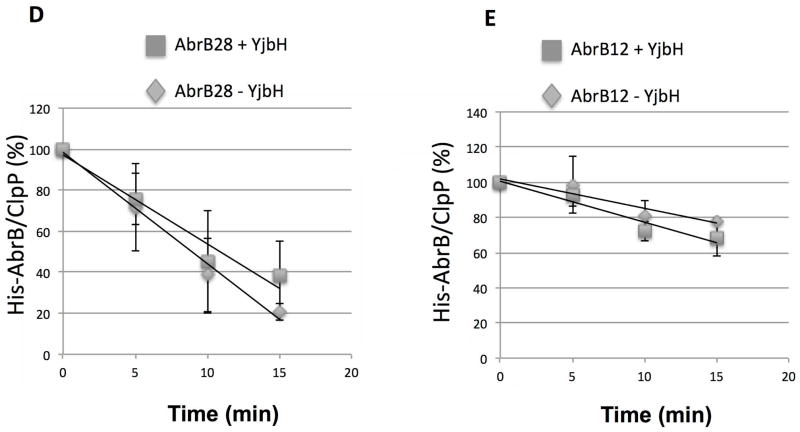
To understand the requirements for Spx interaction with YjbH and ClpXP-catalyzed Spx degradation, we created chimeric proteins that were appended with the C-terminal residues of Spx. AbrB, a protein of similar size to Spx, was selected for chimera construction. The 28 amino acid C-terminal tail feature, containing α6, emerges at position G104 from the four-stranded β sheet of the Spx redox domain consisting of the N- and C-terminal regions of the protein (Newberry et al., 2005) (Fig. 1). Also, the deletion of 12 amino acids from Spx’s C-terminus significantly decreased Spx-YjbH interaction (Chan et al., 2012), and resulted in resistance to ClpXP-catalyzed proteolysis. Therefore, 12, 16, and 28 residue (from G104 to the C-terminal end) segments of the Spx C-terminus were appended to the AbrB (Strauch, 1993) C-terminal end to create AbrB12, AbrB16, and AbrB28, respectively. We then determined if the additions would now render AbrB susceptible to YjbH binding and YjbH-enhanced proteolysis by ClpXP (Fig. 2). The AbrB chimeric proteins are N-terminal hexaHis labeled and purified by Ni-chelate chromatography. Untagged AbrB chimeric proteins were obtained through TEV (tobacco etch virus) protease cleavage followed by Ni-column removal of protease. Geobacillus thermodenitrificans YjbH (GtYjbH) was used in this study and we have shown that GtYjbH complements Bacillus subitlis YjbH (BsYjbH) in vivo and, unlike B. subtilis YjbH, GtYjbH is soluble when expressed in E. coli (Chan et al., 2012). In vitro pull-down reactions with GtYjbH-His6 using a Ni-NTA column showed AbrB28, but not AbrB12 or AbrB16 (Fig. 2A), bound to column-immobilized GtYjbH (Fig. 2B and C, Supplemental Fig. S2). We also tested if the chimeric proteins could be degraded by ClpXP in the presence and absence of GtYjbH. All AbrB variants were degraded by ClpXP but AbrB28 underwent proteolysis at a faster rate than either AbrB12 or AbrB16 (Fig. 2D and E, Fig. S3). The calculated time point at which 50% of substrate was degraded (T50, from data of Fig. 2D and E) of AbrB28 in a ClpXP proteolysis reaction was 8.9 min, whereas the T50 of AbrB12 was 31 min. However, GtYjbH did not significantly accelerate the proteolysis of AbrB12, AbrB28, (Fig. 2D and E) or AbrB16 (data not shown), despite the fact that AbrB28 could bind GtYjbH. The T50 of AbrB28 in a ClpXP proteolysis reaction with added YjbH was 10.9 min. The result demonstrated that addition of the last 28 residues of Spx to a heterologous protein was sufficient to convert the protein to a ClpXP substrate, which is degraded without the need for YjbH activity. The result suggested that, while the C-terminus was required for the recognition of the Spx substrate by ClpX and YjbH, another structural feature of Spx might stabilize the C-terminal end of the intact Spx protein, thus requiring the involvement of YjbH for accelerated proteolysis by ClpXP.
Fig. 2. The 28 residues from Spx C-terminal region are sufficient for YjbH interaction and ClpXP proteolysis.
A. Chimeric protein sequences of AbrB12, AbrB16, and AbrB28 were constructed by fusing the coding sequence of abrB with the last 12, 16, and 28 codons of spx, respectively.
B and C. In vitro Ni affinity pull-down experiments to detect interaction between GtYjbH-His6 and AbrB12 or AbrB28. Binding reactions contained 2.5 μM of each protein, and were incubated at room temperature for 10 min. M, marker; FT, flow-through; W, wash; E, elution (Experimental Procedures for details). Binding data for GtYjbH and AbrB16 in supplemental Fig. S2.
D and E. In vitro proteolysis assay of AbrB28 (D) and AbrB12 (E) in the presence and absence of GtYjbH-His6. AbrB12 or AbrB28 (8 μM), ClpX (3 μM), and ClpP (8 μM) with an ATP-generating system (creatine kinase) were incubated at 37 °C for the times (min) indicated (left). The intensities of ClpP protein in each lane were used as internal controls (Zhang & Zuber, 2007). Plots of time course experiments showing AbrB28 and AbrB12 decay in proteolysis reactions containing ClpXP with (2.6 μM) and without YjbH. Data are from 3–4 replicate reactions with error bars showing standard deviation. Trend-lines were generated with Excel and used to calculate time point at which 50% of substrate was degraded (T50).
Mutations altering the α6 helix of Spx C-terminus affect YjbH binding
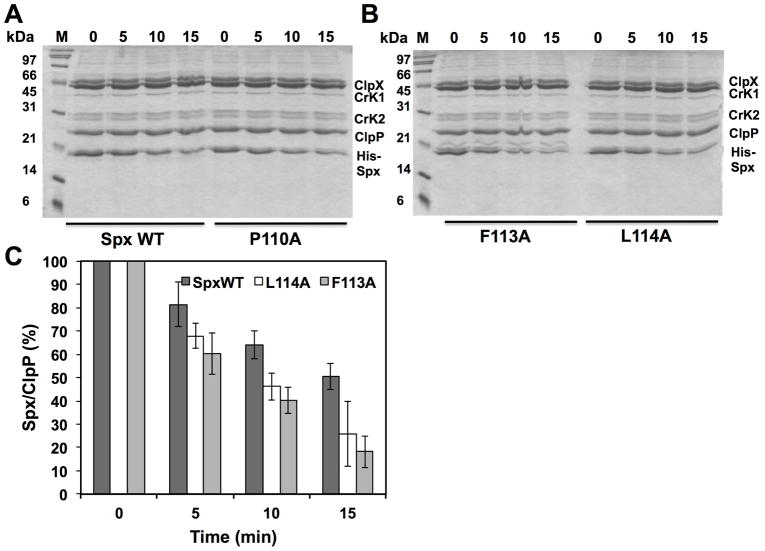
To investigate the importance of the Spx C-terminus for YjbH and ClpXP recognition, we generated alanine residue substitutions of the conserved amino acids in the α6 helix, which was omitted from AbrB16, and purified the resulting mutant proteins. It was observed that Spx mutant proteins (I110A, F113A, L114A, P115A) with N-terminal His6 tag were insoluble when purified from E. coli. An on-column renaturation procedure was used to purify the Spx mutants from the denatured proteins, which yielded pure Spx as a single band on an SDS-polyacrylamide gel (i.e., SpxF113A shown in Fig. S4). The functionality of the renatured proteins was then tested through in vitro transcription of trxB (thioredoxin reductase) promoter DNA (Lin et al., 2013), utilization of which is stimulated by Spx (Nakano et al., 2005). In vitro transcription showed that His-Spx mutants (R111A, R111A/R112A, I110A, F113A, L114A, P115A) are active (Fig. S5), but showed reduced activity compared to the wild-type parent Spx.
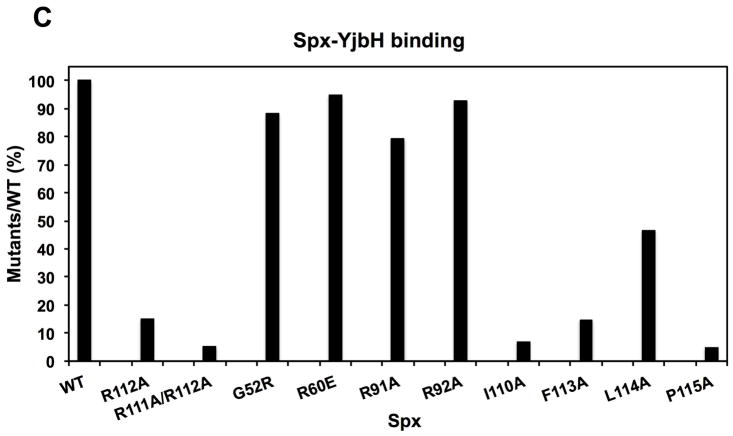
Each C-terminus mutant protein, along with mutants SpxG52R, R60E, R91A, and R92A, were tested for the ability to interact with GtYjbH. In vitro pull down experiments using His-tagged Spx variants identified mutant proteins with significantly reduced affinity for YjbH (Fig. 3A and B). Quantitative analyses showed that the YjbH binding of R112A and F113A decreased by 80% compared to the WT; R111A/R112A, I110A, and P115A decreased by 90%; L114A decreased by 50% (Fig 3C). However, other mutants bearing residue substitutions outside of the C-terminal region, including G52R, R60E, R91A, and R92A, did not show a reduction of YjbH-binding ability (Fig 3C). A control experiment was conducted with His6-Spx that had undergone the denaturation/renaturation procedure used to recover soluble mutant proteins. The denaturant-treated His6-Spx after renaturation bound GtYjbH as well as the untreated His6-Spx protein in a Ni-column pull-down experiment (Fig. S6), indicating that denaturation/renaturation does not effect affinity of His6-Spx for GtYjbH.
Fig. 3. Residue substitutions within the Spx α6 helix significantly affect YjbH interaction with Spx.
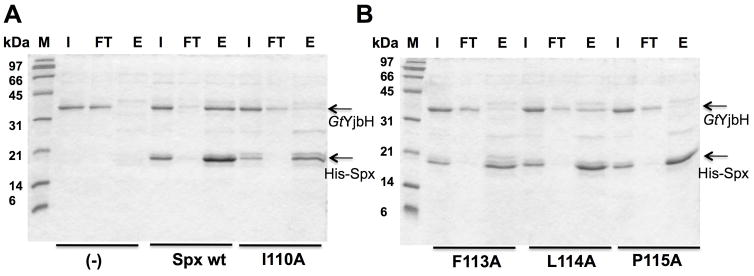
A and B. SDS- PAGE shows in vitro Ni affinity pull-down experiments to detect interaction between GtYjbH and His6-Spx mutants (I110A, F113A, L114A, P115A). M, marker; FT, flow-through; E, elution.
C. A bar graph shows the level of interaction between Spx mutants and GtYjbH. Values are based on band intensity on the gel lanes into which the elution fractions were applied, and is the ratio of Spx:GtYjbH band intensities. The wild-type Spx:GtYjbH band intensity being 100%.
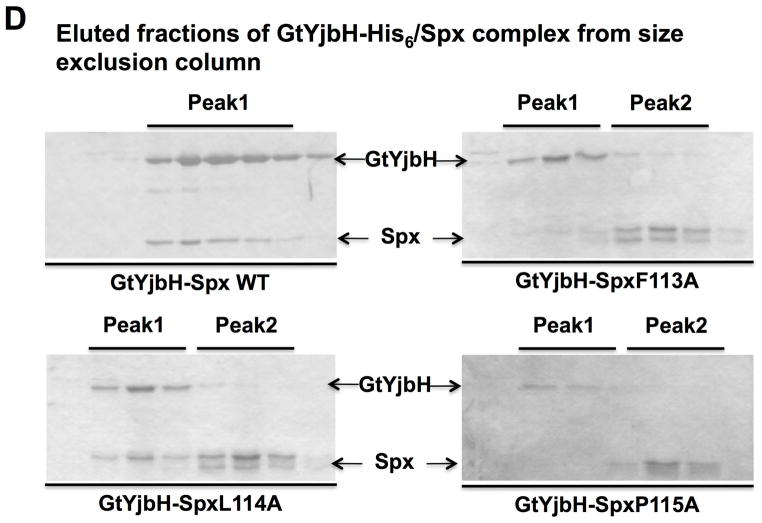
D. SDS-PAGE shows the analyses of peak elution profiles from GtYjbH-His6/Spx (WT, F113A, L114A, and P115A) complex purification using size exclusion column chromatography (Bio-gel P100, BioRad). Using protein standard in gel filtration, 43 kDa (Ovalbumin) eluted in 190 min and 13.7 kDa (Ribonuclease A) in 330 min; Peak1 eluted in 200 min (GtYjbH-His/Spx, 50 kDa) and Peak 2 in 340 min (Spx, 15.5 kDa).
Confirmation of weak YjbH interaction with mutant Spx proteins was obtained by size exclusion chromatography (Fig. 3D), which showed that wild type Spx and GtYjbH co-eluted from a Bio-Gel P-100 column as a single peak of λ280 nm absorbance, while mutant proteins SpxF113A and SpxP115A did not form a complex with GtYjbH (SDS-polyacrylamide gel profile of proteins in two λ280 nm absorbance peaks shown in Fig. 3D). Some interaction between GtYjbH and SpxL114A was observed, but most of the mutant protein eluted off the column after GtYjbH. The results indicated that residues at the C-terminus within the α6 helix were not only essential for structural folding when Spx protein is expressed heterologously, but also for YjbH interaction.
Spx mutants, F113A and L114A, showed enhanced rates of proteolysis by ClpXP in the absence of YjbH in vitro
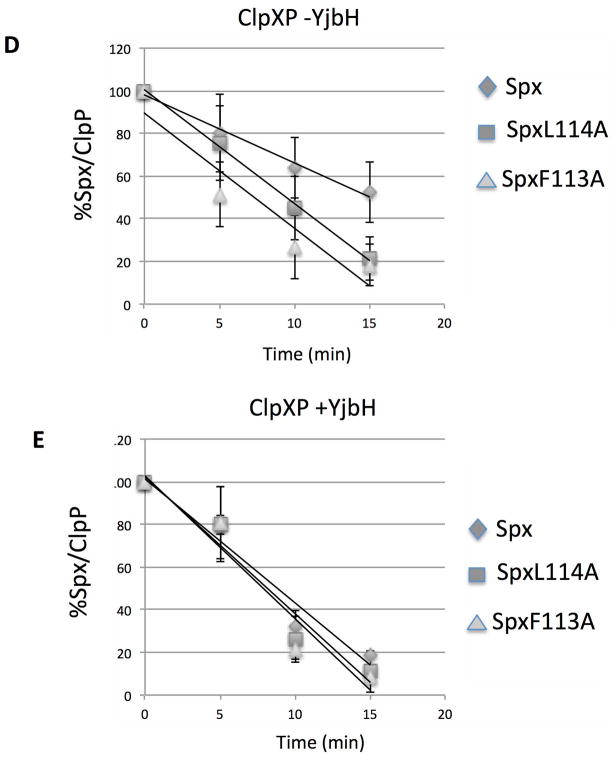
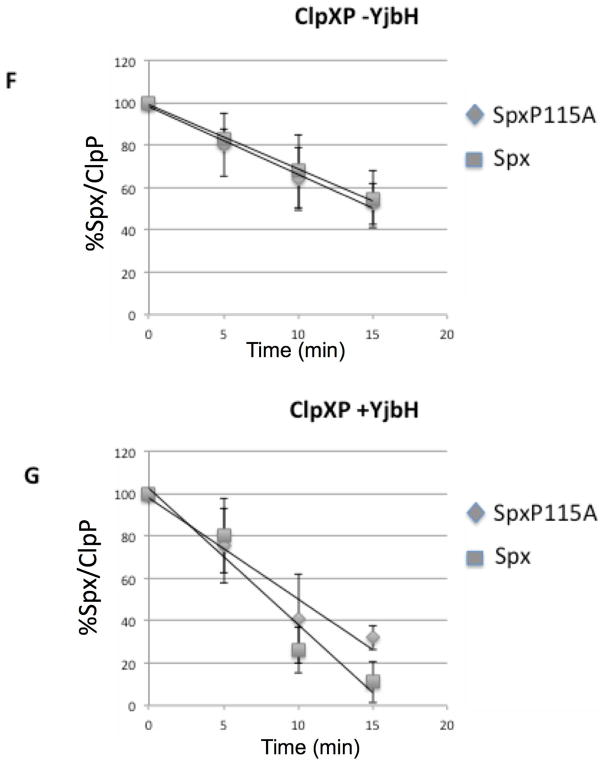
Since the residue substitutions in the Spx C-terminal end affect Spx structurally and the targeted residues were also observed to be part of the YjbH-recognition site, we reasoned that mutationally altering the C-terminal end might result in substrate behavior similar to that observed for AbrB28, in which proteolysis would be less dependent on the YjbH adaptor. In other words, the Spx residue substitutions might change the structure of the C-terminus, rendering it more accessible for ClpXP interaction. Therefore, we tested the mutant Spx proteins as substrates for ClpXP-catalyzed proteolysis in reactions without GtYjbH and with added GtYjbH. Because our in vitro transcription results indicated that all the renatured mutants are active in vitro and therefore had undergone proper overall folding to generate active Spx protein (Fig. S5), we could test the localized C-terminal effects of the residue substitutions (F113A, L114A, P115A) on Spx substrate behavior using in vitro proteolysis assays conducted in the absence of GtYjbH (Fig. 4A, B, and C). The data provided evidence that the mutant SpxF113A and L114A proteins were degraded at faster rates than the wild-type Spx substrate (SpxWT) in proteolysis reactions containing ClpXP (Fig. 4C), whereas SpxP115A was degraded at a rate equal to that of SpxWT. Curve-fitting of substrate decay profiles and calculated T50 of substrate proteins confirmed these findings (Fig. 4D–G). In the absence of GtYjbH, SpxF113A had a T50 of 7.3 min.(±1.8), SpxL114A had a T50 of 9 min. (±1.7), and WT Spx had a T50 of 15.1 min. (±3.2) in ClpXP proteolysis reactions (Fig. 4D). When GtYjbH was added to the reactions, the T50 of wild type Spx was reduced to 8.1 min. (±1.2), but the T50 of SpxF113A and L114A was unchanged; 7.9 (±0.6) and 8.8 (±0.5) min, respectively (Fig. 4E). The SpxP115A mutant had a T50 of 17.4 (±5.5) min in the absence of GtYjbH (Fig. 4F) and showed similar sensitivity to GtYjbH addition, compared to wild-type Spx, with a T50 in ClpXP/GtYjbH reactions of 10.2 (±2.1) min (Fig. 4G). The enhanced ClpXP-catalyzed proteolysis of Spx α6 mutant proteins (F113A and L114A) showed less dependence on YjbH compared to the wild-type Spx parent.
Fig. 4. Spx mutants (F113A and L114A) are degraded faster compared to Spx WT in the absence of YjbH in vitro.
A and B. In vitro proteolysis assay of Spx variants (WT, F113A, L114A, and P115A) in the absence of GtYjbH-His6. Spx variants (8 μM), ClpX (3 μM), and ClpP (8 μM) with an ATP-generating system (creatine kinase) were incubated at 37 °C for the times (min) indicated.
C. A plot of Spx (WT, F113A, and L114A) band intensities against time of reaction in different experiments is shown in the bar graph. The intensities of ClpP protein in each lane were used as internal standards. The Spx/ClpP ratio in 0-min time point is defined as 100%, and the experiments were performed three times to obtain standard deviation.
D–G. Line graphs showing the kinetics of Spx wild-type and mutant protein degradation by ClpXP-catalyzed proteolysis in the absence (D, F) and presence (E, G) of GtYjbH (1 μM). Error bars based on standard deviation, which along with trend lines and substrate T50 (see text) were determined using Excel.
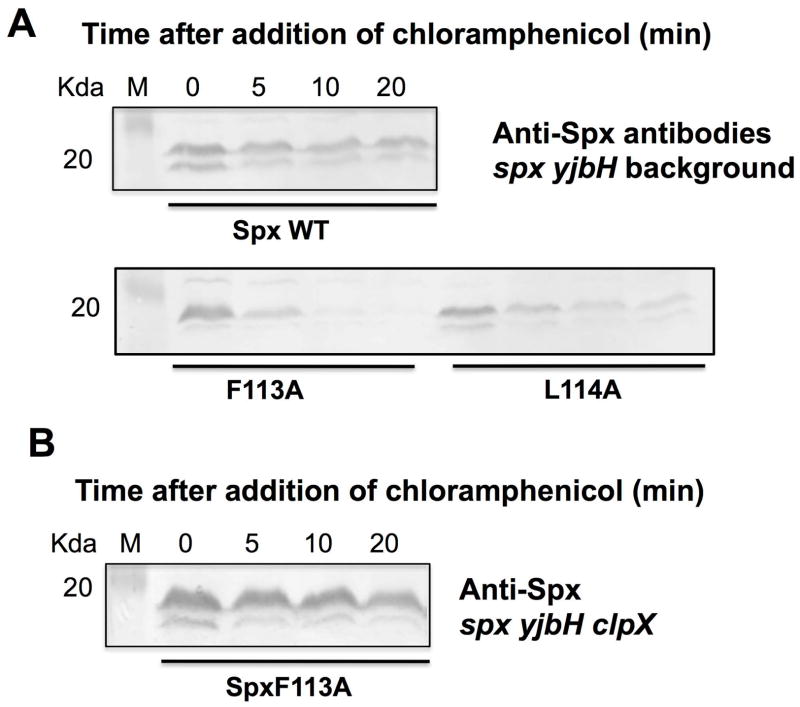
Spx mutants F113A and L114A are less stable in vivo than Spx WT in a yjbH mutant
To determine if the C-terminus mutants are proteolyzed at elevated rates in vivo, the stability of the mutant Spx proteins was tested in strains bearing a deletion of the yjbH gene. The spx mutant alleles were expressed from an IPTG-inducible promoter in the amyE locus of an spx::neo yjbH::tet strain. Cells were grown to mid- exponential stage at 37°C, and 1 mM IPTG induction was applied for 1 hr. The culture was then treated with 0.1 μg/μl chloramphenicol and samples were collected at 0, 5, 10, 20 min time points. Since there is no YjbH present in the cells, Spx turnover would most likely depend on ClpXP. The same experiments were performed in an spx yjbH+ background, but Spx was barely detectable due to the presence of YjbH in the cells even before the addition of chloramphenicol (data not shown).
We then compared the rates of proteolysis of Spx WT and mutant proteins by western blot analysis of cleared cell extracts. The western blot results showed that the F113A and L114A mutant proteins were degraded at elevated rates compared to that of Spx WT (Fig 5A, Supplemental Fig. S7A). SpxF113A degradation resulted in a reduction of protein to 10% by the 10 min time point. However, Spx WT was present at higher levels and maintained a 70% level, which indicated that after the first initial degradation, the rate of proteolysis was reduced (Fig 5A, Supplemental Fig. S7A). The experiments suggested the F113A and L114A substitutions changed the Spx C-terminus structure and exposing the Spx protein to ClpXP-catalyzed proteolysis in vivo. To confirm that the proteolysis was ClpX dependent, a disrupted clpX allele was generated within the yjbH spx Physpac-spxF113A strain and the IPTG/chloramphenicol protocol was performed on cultures of the resultant clpX mutant derivative. Western blot analysis confirmed that the instability of the spxF113A product is dependent on ClpX, as the SpxF113A protein is stable in a clpX mutant (Fig. 5B, Supplemental Fig. S7B). A clpX mutant strain producing the SpxL114A protein showed a severe growth defect, reminiscent of a clpX spx+ strain, and was not studied further.
Fig. 5. Spx mutants (F113A, and L114A) are degraded faster compared to Spx WT in the absence of YjbH in vivo.
A. Western blot analysis of Spx level using anti-Spx antiserum in cells bearing the Physpank- spx (wt, F113A, or P114A) in yjbH spx null mutant background. Cells were grown to exponential phase, induced with 1 mM IPTG for 1 hr, and followed by chloramphenicol treatment. Cell samples were collected at the time points indicated, lysed, and analyzed by western blotting using anti-Spx serum.
B. Spx mutant (F113A) is not degraded in the absence of YjbH and ClpX in vivo. Western blot analysis of Spx level using anti-Spx antiserum in cells bearing the Physpank- spx (F113A) in yjbH spx clpX null mutant background. Cells were grown to exponential phase, induced with 1 mM IPTG for 1 hr, and followed by chloramphenicol treatment. Cell samples were collected at the time points indicated, lysed, and analyzed by western blotting using anti-Spx antiserum.
Recent studies showed that the clpX gene is subject to positive transcription control exerted by Spx (Rochat et al., 2012). We wondered if the differences in the wild-type and mutant Spx stability might be due to altered concentrations of ClpX (Fig S8). In these experiments, the effects of the mutant proteins on ClpX concentration were also examined in strains bearing the rpoAY263C allele, which codes for an RNA polymerase α subunit that is unable to interact with Spx. This was included to determine if any change in ClpX concentration was due to Spx-RNA polymerase interaction. As shown in the western blot of Fig. S8, there is no detectable change in ClpX protein concentration in the strains expressing spx or spx mutant alleles, in either rpoA+ or rpoAY263C genetic backgrounds.
To summarize, the spxF113A and spxL114A mutations resulted in elevated instability due to ClpXP proteolysis (shown in the case of spxF113A) compared to the wild-type parent protein, a result in accordance with the in vitro proteolysis experiments.
Discussion
The results presented herein confirm the role of the C-terminal region of Spx in proteolytic control exerted by YjbH and ClpXP. Additionally, evidence is presented that there exists a site(s) of ClpX recognition within the 28 amino acid C-terminal segment of Spx that is concealed within the protein’s structure. This site(s) is rendered accessible by translocating the 28 residue C-terminal tail of Spx to the C-terminus of another protein, as in the case of the chimera AbrB28 where a ClpX-targeted site is exposed, accounting for the accelerated proteolysis of AbrB28. A ClpX-recognition site might also be exposed through the action of YjbH on intact Spx. We hypothesize that in intact Spx, ClpX-recognition determinants are hidden due to the interactions of the C-terminal helix α6 with the core regions of the Spx structure. This primary structure of the α6 motif is conserved in Spx orthologs, even those in species that do not possess a yjbH gene, suggestive of function beyond proteolytic control. This interaction is likely an important structural determinant in Spx, since the substitutions generated in the α6 motif resulted in protein insolubility (Fig. S4) and reduced Spx activity in vitro (Fig. S5) and in vivo (A. Lin and P. Z., unpublished). Further support of the role played by the C-terminal segment in Spx activity is derived from the phenotype of the original cxs (clpX suppressor) loss-of-function mutations in the spx gene (Nakano et al., 2001). Two of the suppressor mutations conferred substitutions at residues 114 (L->P) and 119 (R->H), both positions in the 28 amino acid C-terminal segment (with L114 part of α6). The side chain of F113 protrudes inward towards the redox domain of Spx (Fig. 1B), suggestive of an interaction to stabilize the C-terminal tail of the protein. Together, these findings and observations strongly suggest that the C-terminal segment of Spx is an important structural component that must be dissociated from the protein’s core to serve as a ClpX-targeting signal. This targeting signal for ClpXP recognition is accompanied by amino acid residues of the extreme C-terminus, which if changed from AN to DD results in a ClpXP resistant form of Spx. Previous work (Larsson et al., 2007) demonstrated that 15 amino acids of the Spx C-terminus appended to GFP (green fluorescent protein) conferred instability that was partially dependent on ClpX.
The SpxP115A mutant, with a substitution of a residue immediately C-terminal to α6, did not show accelerated proteolysis in the ClpXP reaction, and showed a rate of degradation in the presence of GtYjbH similar to the wild type substrate Spx despite reduced affinity for GtYjbH. The proline at position 115 plays a role in YjbH interaction (Fig. 3) yet a substitution of the proline for an alanine had minimal effect on YjbH-mediated proteolysis. In previous work (Chan et al., 2012) very low concentrations of GtYjbH (40x less on a molar basis compared to ClpX and Spx) could efficiently mediate proteolytic turnover of Spx. Future experiments of SpxP115A mutant will test a range of GtYjbH concentrations required for its degradation by ClpXP.
Other Clp protease substrates are known to target multiple sites within the polypeptide chain of substrate proteins. Adaptor and substrate modules within the RepA protein have been shown to function cooperatively in recognition and unfolding that constitutes the reaction pathway of ClpAP-catalyzed proteolysis (Hoskins & Wickner, 2006). Spx might possess a hidden adaptor module in its C-terminal tail that is exposed in AbrB28, in mutant Spx derivatives with residue substitutions that disrupt C-terminal interactions with core Spx protein, or by the action of YjbH. The putative ClpX recognition module of Spx might include the 8 amino acid interval from G104 to R112, as residue changes in F113, L114, and P115 do not inhibit ClpXP proteolysis. This region alone is insufficient for ClpXP-catalyzed Spx proteolysis, however, as the extreme C-terminal sequence is susceptible to mutational changes that block ClpXP-catalyzed proteolysis of Spx (Nakano et al., 2003a).
The region within the C-terminal tail of Spx required for YjbH interaction includes residues of α helix 6 and residues towards the C-terminus, as previous work showed that deleting the C-terminal 12 amino acids weakened YjbH interaction (Chan et al., 2012). The residues of α6 helix are particularly important since they are necessary for YjbH interaction, and are the sites of substitutions that appear to loosen protein structure, rendering the Spx polypeptide chain more accessible for ClpXP interaction and less dependent on YjbH for proteolysis. We postulate that YjbH activity involves interaction with α6 helix residues to disrupt local Spx tertiary structure to promote ClpXP engagement with its substrate.
Previously published data indicated that YjbH does not interact with ClpXP protease (Chan et al., 2012). Furthermore, the simple attachment of the YjbH-binding segment of Spx to another protein does not create an efficient YjbH substrate for proteolysis, which is behavior unlike that of the SspB adaptor protein of E. coli ClpXP (Chien et al., 2007). YjbH was proposed to alter Spx conformation so as to facilitate ClpXP engagement with the Spx substrate (Larsson et al., 2007), a model that is supported by evidence reported herein. This model would require that YjbH interaction with Spx result in a disruption of Spx structure to expose the putative ClpX recognition module, a task not usually associated with the known adaptor proteins that tether substrates to their cognate protease. YjbH mode of action resembles that of RssB, a protein that functions to accelerate the ClpXP-catalyzed degradation of RpoS, the stress induced RNA polymerase sigma subunit of enteric bacteria such as E. coli and Salmonella (Hengge, 2009). Like YjbH, RssB is believed to unfold its substrate, in this case RpoS, to reveal a ClpX interaction surface. RssB does not establish an interaction with ClpX except when in the ternary complex that also includes substrate RpoS protein (Studemann et al., 2003, Zhou et al., 2001). Hence, the term “Proteolytic targeting factor” was proposed (Hengge, 2009) to describe proteolysis-enhancing factors such as RssB, which unlike the adaptors, do not serve as tethers for delivering substrate while in contact with the protease. Others continue to use the term adaptor when referring to RssB, and the term “anti-adaptor” for small Ira peptides that inhibit RssB (Battesti et al., 2013).
Unlike YjbH and RssB, the N-end rule adaptor ClpS operates efficiently when the substrate’s N-terminus is unstructured, which facilitates adaptor interaction (Erbse et al., 2006). SspB and SspBα possess binding specificity towards substrates with SsrA and SsrA-like peptides (Chien et al., 2007), but are not known to function directly in substrate unfolding. Accessory proteins and protein complexes whose activities are linked to proteolytic machines have been uncovered and shown to unfold substrates or to disassemble protein aggregates. The PAN network of ATPases participate in substrate protein unfolding for Archaeal proteasome-catalyzed degradation (Benaroudj & Goldberg, 2000). The GtYjbH preparations used in the study do possess low-level ATPase activity, but it is not affected by Spx interaction, nor is ATP required for Spx interaction (C. M. C. and P. Z., unpublished). YjbH is annotated as a member of a family of protein disulfide isomerases, and most orthologs bear a CXXC motif within their N-terminal region. However, these features seem not to be important for YjbH adaptor-like activity (Engman et al., 2012). Ongoing structural analysis of YjbH-Spx interaction may shed further light on the mechanism of YjbH-enhanced Spx proteolysis.
Experimental procedures
Bacterial strains, plasmids, and chemicals
B. subtilis strains and recombinant plasmids derived from pPROEX-1, pET23a, pDR111 (Britton et al., 2002), and pDG646 (Guerout-Fleury et al., 1995) are listed in Table S1. B. subtilis used in this study are all JH642 derivatives. E. coli strains DH5α and BL21(DE3)pLysS were used for general cloning and protein production operations. Medium components, including LB medium and Difco sporulation medium (DSM) (Nakano et al., 1988, Nicholson & Setlow, 1990), were from Difco. All restriction/modifying enzymes were from New England Biolabs. Oligonucleotide primers (Table S2) were from Invitrogen. The Ni-nitrilotriacetic acid (NTA) resin from 5 PRIME (PerfectPro Ni-NTA agarose) and PCR/plasmid purification kits were from Qiagen (Germany). High-Q anion exchange columns and Bio-Gel P-100 were from Bio-Rad. All analytical grade chemicals were from Sigma-Aldrich unless otherwise stated.
Construction of plasmids
Plasmids containing spx mutant alleles were constructed through QuickChange procedure using Pfu Turbo DNA Polymerase (Stratagene), and plasmid pSN17 (Spx wild type allele) as a template. pEC-57(SpxI110A) was constructed with primers EC79 and EC80; pEC-56(SpxF113A) with EC81 and EC82; pEC-58(SpxL114A) with EC83 and EC84; pEC-59(SpxP115A) with EC85 and EC86. The amplified DNA inserts were digested with NdeI and BamHI and inserted into NdeI-BamHI digested pPROEX-1.
pEC-53(SpxWT), pEC-61(SpxF113A), and pEC-62(SpxL114A) were constructed with primers EC78 and EC64 for PCR using KOD polymerase (Novagen). Using pSN17, pEC-56, and pEC-58 as templates, respectively. The amplified DNA inserts were cleaved with HindIII and NheI and inserted into HindIII-NheI digested pDR111 (Britton et al., 2002).
pEC-27(AbrB12) and pEC-30(ArbB16) were constructed with primer pairs EC35-EC36 and EC35-EC40, respectively, using B. subtilis JH642 genomic DNAs as template. The DNAs were digested with NdeI and NotI and inserted into NdeI-NotI digested pPROEX-1. pEC-31(AbrB28) was constructed with two step PCR: the DNA insert1 was amplified with primers EC35 and EC43 and the DNA insert2 with EC41 and EC42, both using B. subtilis JH642 genomic DNAs as a template. The DNA insert1 and insert2 were denatured under 95 °C for 3 min, slowly annealed under 65 °C for 2 min, and incubated at 37 °C for 2 min. The annealed DNA duplex was used as templates for PCR using primers EC35 and EC41. The amplified DNA inserts were cleaved with NdeI and NotI and inserted into NdeI-NotI digested pPROEX-1.
pEC-66 (166–800 bps clpX fragment) was constructed with primers EC90 and EC91 using B. subtilis JH642 genomic DNAs as template. The DNAs were digested with XbaI inserted into XbaI digested pDG646.
Construction of strains
pEC-53, pEC-61, and pEC-62 were used to transform strain ORB7852 (JH642, spx::neo, yjbH::tet), and transformants were selected on DSM agar containing neomycin 5 μg/ml, tetracycline 12.5 μg/ml, and spectinomycin 75 μg/ml. To confirm the integration of spx variants at the amyE locus, clones were streaked on an LB-starch plate and grown for 16 hr at 37 °C. The resulting amylase-negative strains were ORB8729(WT), ORB8731(F113A), ORB8732(L114A) (amyE:: Physpank-spx variants, spx::neo, yjbH::tet).
pEC-66 was used to transform strain ORB8731 (JH642, spx::neo, yjbH::tet, amyE:: Physpank-spx(F113A)), and transformants were selected on DSM agar containing neomycin 5 μg/ml, tetracycline 12.5 μg/ml, spectinomycin 75 μg/ml, and 1 μg/ml erythromycin/25 μg/ml lincomycin. To confirm the deletion of clpX, clones were expressed to test the expression of clpX in vivo using anti-ClpX antibodies. The resulting ClpX-negative strain was ORB8968 (amyE:: Physpank-spx(F113A), spx::neo, yjbH::tet, clpX::erm).
In vivo Spx stability assay
ORB8729(WT), ORB8731(F113A), ORB8732(L114A), and ORB8968(F113A in clpX) were used to determine in vivo Spx protein level. The strains were grown in culture until OD600 ~0.4–0.5 in DSM containing antibiotics as described above. A final concentration of 1 mM IPTG was added to the cells to induced expression of Spx variants. After 1 hr of IPTG induction, final concentration of 0.1 μg/μl chloramphenicol was added to the cells. Samples of 2 ml culture were collected at time points indicated in the figures. Cell samples were lysed by the bead-beating method (Lin et al., 2012). The cell pellets were suspended in lysis buffer (30 mM Tris-HCl, pH 8.0, 1 mM EDTA) mixed with 0.1 mm disruption glass beads for bacteria (RPI Corp.), disrupted by vortexing for 5 min, and transferred onto ice for 5 min, followed by 2X repeated vortex disruption. The supernatant of each sample was collected after centrifugation (16100 g for 10 min) and the protein concentration was measured with Coomassie protein assay reagent (Pierce). Twenty μg of the total protein for each sample was loaded onto an SDS-PAGE gel, and the Western blot was undertaken using anti-Spx antibodies.
In vitro proteolysis reaction
In vitro proteolysis was carried out as previously reported (Chan et al., 2012). Briefly, reactions of 60 μl reaction containing 50 mM HEPES-KOH (pH 7.6), 50 mM KCl, 10 mM magnesium acetate, 5 mM DTT, 5 mM ATP, 5 mM creatine phosphate, 0.05 U creatine kinase ml−1 (Sigma), and ClpX, ClpP, Spx or AbrB variants and GtYjbH (concentrations are indicated in figure legends) were assembled and incubated at 37 °C. Levels of Spx or AbrB variants after proteolysis were determined as ratios of Spx or AbrB variants to ClpP band intensities, since ClpP concentrations in each set of reactions were equal. The Spx or AbrB variants:ClpP value in a reaction mixture at 0 min time point was defined as 100%. Excel curve-fitting function was employed to establish trend-lines of substrate decay and to calculate time point at which 50% of substrate was degraded (T50).
In vitro affinity interaction assays
Different combinations of proteins (indicated in figure legends) Spx or His6–Spx, GtYjbH or GtYjbH-His, AbrB12, and AbrB28 (all ~2.5 μM) were incubated in buffer A (100 mM Tris/HCl, pH 8.5, 50 mM NaCl) for 10 min at room temperature. The protein mixtures were then applied to a 50 μl Ni2+-NTA affinity agarose column (5 PRIME) pre-equilibrated with buffer A. The column was washed with 10 column volumes (CVs) of buffer W (100 mM Tris/HCl, pH 8.5, 200 mM NaCl, 30 mM imidazole). The bound protein was eluted three times with 1 CV of Buffer E (20 mM Tris/HCl, pH 7.6, 200 mM NaCl, 250 mM imidazole). The input, flow-through, wash and eluted samples were resolved on a 15 % SDS-PAGE gel, followed by ‘Blue silver’ colloidal Coomassie G- 250 protein staining as described previously (Chan et al., 2012).
Protein purification
His6-Spx(R112A and R111A/R112A), His6-AbrB12, His6-AbrB16, and His6-AbrB28 were obtained as follows: the pEC-27(His6-AbrB12), pEC30(His6-AbrB16), and pEC-31(His6- AbrB28) transformed E. coli BL21(DE3)pLysS cells were grown at 37 °C in LB broth containing 50 mg ampicillin ml−1 and 5 mg chloramphenicol ml−1 to OD600 0.4–0.5, and expression was induced with 0.5 mM IPTG for 3 hr at 30 °C. The expressed His6-Spx(R112A and R111A/R112A), His6-AbrB12, His6-AbrB16, and His6-AbrB28 cultures were lysed by passage through French press and the proteins were purified using the same procedure as GtYjbH-His6 (Chan et al., 2012). The untagged His6-Spx(R112A and R111A/R112A), AbrB12, AbrB16, and AbrB28 were purified by removing their hexahistidine tags using γ-TEV (tobacco etch virus) protease. Purification procedures of ClpX, ClpP, GtYjbH-His6, His6-Spx, and Spx were the same as previous studies (Chan et al., 2012, Liu et al., 1999, Nakano et al., 2005, Nakano et al., 2003b, Nakano et al., 2002)..
For His6-Spx mutants (I110A, F113A, L114A, and P115A), the plasmids (pEC-57, pEC-56, pEC-58, and pEC-59, respectively) were transformed to E. coli BL21(DE3)pLysS cells. The expression procedure is the same as AbrB12, AbrB16, and AbrB28 described above. Cells from a 1 L culture were suspended in 15 ml Lysis Buffer (20 mM NaH2PO4, 200 mM NaCl, 10 mM Imidazole). Half tablet of EDTA-free protease inhibitor cocktail (Roche Applied Science) was added per 1L suspended cells. The cells were subjected to freeze-thaw cycles and disrupted by French press at 1,000 psi. The lysate was cleared by centrifugation at 15000 rpm (Sorvall, rotor SL-50T) for 30 min. Since the His6-Spx mutants were not soluble, the supernatants were discarded and the pellets were dissolved in Buffer D (6M GuHCl (Guanidine hydrochloride), 10 mM Tris/HCl, pH 8.0, 100 mM NaH2PO4) for protein denaturation. The dissolved pellet was incubated in 5 ml Ni-NTA resin in a column and gently rotated for 1 hr at 4 °C. After the mixture was passed through the column, the His6-tagged denatured proteins were on-column refolded through a four-step process: washed the column with 50 ml 4 M GuHCl, 10 mM Tris/HCl, pH 8.0, 100 mM NaH2PO4, with 50 ml 3 M GuHCl, 10 mM Tris/HCl, pH 8.0, 100 mM NaH2PO4, with 50 ml 1 M GuHCl, 10 mM Tris/HCl, pH 8.0, 100 mM NaH2PO4, and finally with 50 ml Lysis Buffer. Then, the column was washed with 200 ml Buffer DW (20 mM NaH2PO4, 200 mM NaCl, 20 mM Imidazole). The bound protein was eluted in Buffer DE (20 mM NaH2PO4, 200 mM NaCl, 250 mM Imidazole). Eluted fractions that contain His6-Spx variants were resolved on a 15% SDS- PAGE gel visualized by Coomassie blue staining. The pure Spx proteins were concentrated, buffer-exchanged to Buffer S (20 mM Tris/HCl, pH 7.6, 200 mM NaCl, 10 % glycerol), and stored at 80 °C. Protein concentration was measured using Bio-Rad protein assay, and BSA was used to generate the standard curve.
For size exclusion column analysis of GtYjbH-His6-Spx interaction, 1.2 ml protein sample of GtYjbH-His6-Spx mixture (~30 μM each, 1:1 ratio) was incubated at room temperature for 10 min. After filtration, protein mixture was applied to a size exclusion column for separation (Bio-Gel P-100, Bio-Rad) with Buffer G (20 mM Tris/HCl, pH 7.6, 200 mM NaCl). Eluted peak fractions were analyzed by SDS-PAGE.
In vitro transcription
In vitro transcription was carried as previously published (Lin et al., 2012). A linear trxB promoter DNA template was generated by PCR with oligonucleotides oDYR07-32 and oDYR07-52 that would direct the synthesis of a 66-nucleotide (nt) transcript. In the reaction, trxB linear template (10 nM) and RNA Polymerase with σA (25 nM, 1:1 ratio) were incubated without or with 75 nM Spx variants. Reaction time was 10 min and reactions were carried out same as previously described, analyzed with an 6% polyacrylamide-urea gel and a Typhoon Trio variable imager (GE Healthcare).
Supplementary Material
Acknowledgments
The authors thank Dr. S. M. Barendt for technical assistance and members of the Zuber lab group and Dr. Michiko M. Nakano for helpful discussions. Research support from National Institutes of Health grant GM045898 to P. Z. and NRSA fellowship F32GM099163 to C. M.C.
References
- Battesti A, Gottesman S. Roles of adaptor proteins in regulation of bacterial proteolysis. Curr Opin Microbiol. 2013;16:140–147. doi: 10.1016/j.mib.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Hoskins JR, Tong S, Milanesio P, Mann JM, Kravats A, et al. Anti-adaptors provide multiple modes for regulation of the RssB adaptor protein. Genes Dev. 2013;27:2722–2735. doi: 10.1101/gad.229617.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaroudj N, Goldberg AL. PAN, the proteasome-activating nucleotidase from archaebacteria, is a protein-unfolding molecular chaperone. Nat Cell Biol. 2000;2:833–839. doi: 10.1038/35041081. [DOI] [PubMed] [Google Scholar]
- Britton RA, Eichenberger P, Gonzalez-Pastor JE, Fawcett P, Monson R, Losick R, Grossman AD. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J Bacteriol. 2002;184:4881–4890. doi: 10.1128/JB.184.17.4881-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CM, Garg S, Lin AA, Zuber P. Geobacillus thermodenitrificans YjbH recognizes the C-terminal end of Bacillus subtilis Spx to accelerate Spx proteolysis by ClpXP. Microbiology. 2012;158:1268–1278. doi: 10.1099/mic.0.057661-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien P, Grant RA, Sauer RT, Baker TA. Structure and substrate specificity of an SspB ortholog: design implications for AAA+ adaptors. Structure. 2007;15:1296–1305. doi: 10.1016/j.str.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Engman J, Rogstam A, Frees D, Ingmer H, von Wachenfeldt C. The YjbH adaptor protein enhances proteolysis of the transcriptional regulator Spx in Staphylococcus aureus. J Bacteriol. 2012;194:1186–94. doi: 10.1128/JB.06414-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbse A, Schmidt R, Bornemann T, Schneider-Mergener J, Mogk A, Zahn R, et al. ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature. 2006;439:753–756. doi: 10.1038/nature04412. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Garg SK, Kommineni S, Henslee L, Zhang Y, Zuber P. The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx. J Bacteriol. 2009;191:1268–1277. doi: 10.1128/JB.01289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- Gur E, Biran D, Ron EZ. Regulated proteolysis in Gram-negative bacteria--how and when? Nat Rev Microbiol. 2011;9:839–848. doi: 10.1038/nrmicro2669. [DOI] [PubMed] [Google Scholar]
- Hengge R. Proteolysis of sigmaS (RpoS) and the general stress response in Escherichia coli. Res Microbiol. 2009;160:667–676. doi: 10.1016/j.resmic.2009.08.014. [DOI] [PubMed] [Google Scholar]
- Hoskins JR, Wickner S. Two peptide sequences can function cooperatively to facilitate binding and unfolding by ClpA and degradation by ClpAP. Proc Natl Acad Sci U S A. 2006;103:909–914. doi: 10.1073/pnas.0509154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein J, Dougan DA, Gerth U, Hecker M, Turgay K. The tyrosine kinase McsB is a regulated adaptor protein for ClpCP. EMBO J. 2007;26:2061–2070. doi: 10.1038/sj.emboj.7601655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson JT, Rogstam A, von Wachenfeldt C. YjbH is a novel negative effector of the disulphide stress regulator, Spx, in Bacillus subtilis. Mol Microbiol. 2007;66:669–684. doi: 10.1111/j.1365-2958.2007.05949.x. [DOI] [PubMed] [Google Scholar]
- Lin AA, Walthers D, Zuber P. Residue substitutions near the redox center of Bacillus subtilis Spx affect RNA polymerase interaction, redox control and Spx-DNA contact at a conserved cis-acting element. J Bacteriol. 2013;195:3967–78. doi: 10.1128/JB.00645-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cosby WM, Zuber P. Role of Lon and ClpX in the post-translational regulation of a sigma subunit of RNA polymerase required for cellular differentiation of Bacillus subtilis. Mol Microbiol. 1999;33:415–428. doi: 10.1046/j.1365-2958.1999.01489.x. [DOI] [PubMed] [Google Scholar]
- Mogk A, Huber D, Bukau B. Integrating protein homeostasis strategies in prokaryotes. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004366. pii: a004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moliere N, Turgay K. General and Regulatory Proteolysis in Bacillus subtilis. Subcell Biochem. 2013;66:73–103. doi: 10.1007/978-94-007-5940-4_4. [DOI] [PubMed] [Google Scholar]
- Nakano MM, Hajarizadeh F, Zhu Y, Zuber P. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol Microbiol. 2001;42:383–394. doi: 10.1046/j.1365-2958.2001.02639.x. [DOI] [PubMed] [Google Scholar]
- Nakano MM, Lin A, Zuber CS, Newberry KJ, Brennan RG, Zuber P. Promoter Recognition by a Complex of Spx and the C-Terminal Domain of the RNA Polymerase α Subunit. PLoS ONE. 2010;5:e8664. doi: 10.1371/journal.pone.0008664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MM, Marahiel MA, Zuber P. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J Bacteriol. 1988;170:5662–5668. doi: 10.1128/jb.170.12.5662-5668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MM, Zhu Y, Liu J, Reyes DY, Yoshikawa H, Zuber P. Mutations conferring amino acid residue substitutions in the carboxy-terminal domain of RNA polymerase a can suppress clpX and clpP with respect to developmentally regulated transcription in Bacillus subtilis. Mol Microbiol. 2000;37:869–884. doi: 10.1046/j.1365-2958.2000.02052.x. [DOI] [PubMed] [Google Scholar]
- Nakano S, Erwin KN, Ralle M, Zuber P. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol Microbiol. 2005;55:498–510. doi: 10.1111/j.1365-2958.2004.04395.x. [DOI] [PubMed] [Google Scholar]
- Nakano S, Kuster-Schock E, Grossman AD, Zuber P. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc Natl Acad Sci U S A. 2003a;100:13603–13608. doi: 10.1073/pnas.2235180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Nakano MM, Zhang Y, Leelakriangsak M, Zuber P. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc Natl Acad Sci USA. 2003b;100:4233–4238. doi: 10.1073/pnas.0637648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Zheng G, Nakano MM, Zuber P. Multiple pathways of Spx (YjbD) proteolysis in Bacillus subtilis. J Bacteriol. 2002;184:3664–3670. doi: 10.1128/JB.184.13.3664-3670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberry KJ, Nakano S, Zuber P, Brennan RG. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc Natl Acad Sci USA. 2005;102:15839–15844. doi: 10.1073/pnas.0506592102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson W, Setlow P. Sporulation and Germination. In: Harwood CR, Cutting S, editors. Molecular Biological Methods for Bacillus. New York: John Wiley; 1990. pp. 391–450. [Google Scholar]
- Persuh M, Mandic-Mulec I, Dubnau D. A MecA paralog, YpbH, binds ClpC, affecting both competence and sporulation. J Bacteriol. 2002;184:2310–2313. doi: 10.1128/JB.184.8.2310-2313.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runde S, Moliere N, Heinz A, Maisonneuve E, Janczikowski A, Elsholz AK, et al. The role of thiol oxidative stress response in heat-induced protein aggregate formation during thermotolerance in Bacillus subtilis. Mol Microbiol. 2014;91:1036–1052. doi: 10.1111/mmi.12521. [DOI] [PubMed] [Google Scholar]
- Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- Schlothauer T, Mogk A, Dougan DA, Bukau B, Turgay K. MecA, an adaptor protein necessary for ClpC chaperone activity. Pro Natl Acad Sci U S A. 2003;100:2306–2311. doi: 10.1073/pnas.0535717100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader EK, Harstad KG, Matouschek A. Targeting proteins for degradation. Nature chemical biology. 2009;5:815–822. doi: 10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch MA. AbrB, a transition state regulator. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and Other Gram-Positive Bacteria: Physiology, Biochemistry, and Molecular Biology. Washington D. C: American Society for Microbiology; 1993. pp. 757–764. [Google Scholar]
- Studemann A, Noirclerc-Savoye M, Klauck E, Becker G, Schneider D, Hengge R. Sequential recognition of two distinct sites in sigma(S) by the proteolytic targeting factor RssB and ClpX. EMBO J. 2003;22:4111–4120. doi: 10.1093/emboj/cdg411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zuber P. Requirement of the zinc-binding domain of ClpX for Spx proteolysis in Bacillus subtilis and effects of disulfide stress on ClpXP activity. J Bacteriol. 2007;189:7669–7680. doi: 10.1128/JB.00745-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Gottesman S, Hoskins JR, Maurizi MR, Wickner S. The RssB response regulator directly targets sigma(S) for degradation by ClpXP. Genes Dev. 2001;15:627–637. doi: 10.1101/gad.864401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J Bacteriol. 2004;186:1911–1918. doi: 10.1128/JB.186.7.1911-1918.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.