Abstract
Co-exposure to cigarette smoke and ethanol generates malondialdehyde and acetaldehyde, which can subsequently lead to the formation of aldehyde-adducted proteins. We have previously shown that exposure of bronchial epithelial cells to malondialdehyde-acetaldehyde (MAA) adducted protein increases protein kinase C (PKC) activity and proinflammatory cytokine release. A specific ligand to scavenger receptor A (SRA), fucoidan, blocks this effect. We hypothesized that MAA-adducted protein binds to bronchial epithelial cells via SRA. Human bronchial epithelial cells (BEAS-2B) were exposed to MAA-adducted protein (either bovine serum albumin [BSA-MAA] or surfactant protein D [SPD-MAA]) and SRA examined using confocal microscopy, fluorescent activated cell sorting (FACS), and immunoprecipitation. Differentiated mouse tracheal epithelial cells (MTEC) cultured by air-liquid interface were assayed for MAA-stimulated PKC activity and keratinocyte-derived chemokine (KC) release. Specific cell surface membrane dye co-localized with upregulated SRA after exposure to MAA for 3–7 min and subsided by 20 min. Likewise, MAA-adducted protein co-localized to SRA from 3–7 min with a subsequent internalization of MAA by 10 min. These results were confirmed using FACS analysis and revealed a reduced mean fluorescence of SRA after 3 min. Furthermore, increased amounts of MAA-adducted protein could be detected by Western blot in immunoprecipitated SRA samples after 3 min treatment with MAA. MAA stimulated PKCε-mediated KC release in wild type, but not SRA knockout mice. These data demonstrate that aldehyde-adducted proteins in the lungs rapidly bind to SRA and internalize this receptor prior to the MAA-adducted protein stimulation of PKC-dependent inflammatory cytokine release in airway epithelium.
Keywords: lung, airway epithelial, scavenger receptor, malondialdehyde, acetaldehyde
Introduction
It is well known that excessive alcohol consumption and tobacco use are independently associated with health-related adverse effects (Falk, Yi, & Hiller-Sturmhöfel, 2006). Cigarette smoking has deleterious effects on a person's health when compared to non-smokers. This includes an increased risk of cardiovascular disease, lung cancers, and chronic obstructive pulmonary disease. The adverse effects from cigarette smoking are the leading cause of death in the United States, accounting for 1 in 5 deaths every year (Centers for Disease Control and Prevention, 2012). Excessive alcohol use also has numerous deleterious effects, including increased risk of pancreatitis, acute liver disease and cirrhosis, pneumonia, and ARDS (Falk et al., 2006; Moss, Bucher, Moore, Moore, & Parsons, 1996). These adverse effects are synergistic as smokers are more likely to use alcohol and have an alcohol-use disorder (Falk et al., 2006).
Cigarette smoke and alcohol metabolism both produce reactive aldehydes, which will readily adduct enamines on the amino terminus of protein residues (McCaskill et al., 2011; Tuma et al., 2001). Cigarette smoke contains a high content of acetaldehyde (AA) from the pyrolysis of tobacco. Tobacco smoke also contains a significant number of by-products that will produce reactive oxygen species. These reactive oxygen species will produce malondialdehyde (MDA) through lipid peroxidation. Alcohol metabolism by alcohol dehydrogenase produces AA. Alcohol will also induce oxidative stress, leading to lipid peroxidation and subsequent MDA formation. Together, AA and MDA react through a Schiff base intermediary to form a stable hybrid product, the malondialdehyde-acetaldehyde (MAA) adduct, that will readily adduct to proteins (Tuma et al., 2001).
In the lung, metabolic pathways exist to produce AA and MDA when exposed to alcohol. Cigarette smoke, as well as the metabolites of alcohol, contains both of the necessary aldehydes and reactive oxygen species, adding further AA and MDA to the lung milieu. Thus, MAA-adducted proteins likely form in the lungs of smokers who drink alcohol. In fact, we have shown in a mouse model that MAA is only formed in the lungs of mice co-exposed to both cigarette smoke and alcohol (McCaskill et al., 2011). MAA will readily adduct a number of proteins. MAA adducts have been shown to induce a significant inflammatory response (Hill et al., 1998; Wyatt, Kharbanda, Tuma, & Sisson, 2001). In the lung, we have shown that surfactant proteins are potential targets for MAA adduction (McCaskill et al., 2011).
MAA-adducted proteins have been shown to be potent inflammatory mediators (Hill et al., 1998; Tuma, 2002; Wyatt et al., 2001). Furthermore, MAA-adducted protein stimulates the release of chemokine, IL-8, in an in vitro bronchial epithelial model (Wyatt et al., 2001). This stimulated cytokine release is blocked by PKC inhibitors, implicating PKC in MAA-adducted protein stimulated IL-8 release (Wyatt et al., 2001). Similarly, purified SPD-MAA can induce an IL-8 response when nasally administered to mice that is significantly different from either non-adducted SPD or saline alone (Wyatt et al., 2012). In addition, MAA-adducted protein stimulation of PKC-mediated IL-8 release can be down-regulated by pre-incubation of epithelial cells with fucoidan, a known scavenger receptor A (SRA) ligand (Wyatt et al., 2001). This indicates that SRA is a possible candidate receptor for MAA-adducted proteins, as scavenger receptors are known to readily bind aldehyde species (Duryee et al., 2005; Horiuchi, Murakami, Takata, & Morino, 1986).
Scavenger receptors are a widely varying class of pattern recognition receptors that were initially described by Goldstein et al. in the handling of low-density lipoproteins (Goldstein, Ho, Basu, & Brown, 1979). The family of receptors has continued to expand and is characterized by their ligand, either modified LDL or polyanionic ligand. Adduction of proteins by aldehydes changes their charge in a way that makes them ideal for SRA binding (Duryee et al., 2005). Initially, SRA was found on macrophages and dendritic cells, but SRA can also be found on endothelium and epithelium (Duryee et al., 2005; Limmon et al., 2008; Plüddemann, Neyen, & Gordon, 2007). The cellular response to different ligands binding SRA can stimulate PKC and MAPK (Plüddemann et al., 2007). It has been further shown that numerous kinases, including tyrosine kinase and PKC, can be activated by SRA (Coller & Paulnock, 2001; Hsu, Chiu, Wen, Chen, & Hua, 2001). We have shown that SPD-MAA stimulates PKCε activity (Coller & Paulnock, 2001; Hsu et al., 2001).
Scavenger receptors bind self and non-self pattern associated molecular proteins (PAMPs) to traffic them into the cell. These receptors' role and function is well defined in macrophages and other antigen-presenting cells (Nicoletti et al., 1999). Previously, Duryee et al. described binding of aldehyde-modified proteins, including MAA, in the liver sinusoidal endothelium (Duryee et al., 2005). Scavenger receptors, specifically type A, are also found on bronchial epithelial cells, another cell of the innate immune system (Limmon et al., 2008). Because we previously found that MAA adduct-induced activation of PKC and subsequent release of IL-8 can be blocked by pre-incubation with fucoidan, a known SRA ligand (Wyatt et al., 2001), we therefore hypothesized that SPD-MAA binds to bronchial epithelium via SRA.
Materials and Methods
Cell Lines and Culture
Two types of airway cells were utilized to evaluate MAA and SRA binding and internalization: the human bronchial epithelial cell line, BEAS-2B, and mouse tracheal epithelial cells (MTEC). Both cell types responded similarly to MAA-adducted proteins. BEAS-2Bs were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and maintained in LHC-9/RPMI (1:1 mixture) growth media. MTEC from C57BL/6 wild type and SRA knockout mice on C57BL/6 background (Jackson Laboratory, Bar Harbor, ME) were isolated and cultured on air-liquid interface (ALI). All experimental animal procedures were conducted according to the NIH guidelines for the use of rodents, and the University of Nebraska Medical Center Institutional Animal Care and Use Committee approved all procedures. Once cultured onto ALI, cells were maintained in Ham's F12:DMEM media (1:1) supplemented with 10% FBS, 250 μg/ml amphotericin B, and 10,000 units/mL penicillin/streptomycin, 200 mM glutamine, 40 mg/mL gentamycin, 50 μL of each of the following: insulin, transferrin, epidermal growth factor, and cholera toxin; 200 μL of bovine pituitary extract and 500 μL of retinoic acid, and allowed to differentiate for a period of 2 weeks. Culture media was switched to serum-free Ham's F-12:DMEM + supplements media for 24 h prior to treatment.
Antibodies and Cell Membrane Stain
Rabbit anti-scavenger receptor AI/II and rabbit anti-scavenger receptor BI & BII antibody were purchased from Abcam (Cambridge, MA). For confocal microscopy and fluorescence-activated cell sorting (FACS) analysis, the secondary antibody used was AlexaFluor 488 goat anti-rabbit IgG (Molecular Probes, Invitrogen, Grand Island, NY). The secondary antibody used during Western blotting was horseradish peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, Inc., Grand Island, NY). Cell membranes were stained using 1,1′-Dioctadecyl-6.6′-Di(4-Sulfoophenyl)-3,3,3′,3′-Tetramethylindocarbocyanine [SP-DilC18(3) (Invitrogen)].
Malondialdehyde-Acetaldehyde Synthesis
Bovine serum albumin adducted to MAA (BSA-MAA) and human surfactant protein D adducted to MAA (SPD-MAA) was prepared as previously reported (Xu et al., 1997). Briefly, approximately 1–1.5 mg/mL BSA or SPD was incubated with 1.0 mM acetaldehyde and 1.0 mM MDA in phosphate buffer (0.1 mM). The pH was brought to 7.4, and maintained at 37 °C for 2–4 h. At the end of incubation, the reaction mixture was exhaustively dialyzed against phosphate buffer for 24 h at 4 °C. As previously reported (Wyatt et al., 2012), identical cell responses were observed when using either BSA-MAA or SPD-MAA.
Confocal Microscopy
BEAS-2Bs were cultured onto microscope cover slips to approximately 80–90% confluence. Cells were then incubated with SP-DiIC18(3) at 37 °C for 5 min. Cells were then placed on ice for an additional 15 min or longer. During the incubation on ice, cells were treated with (100 μg/mL) BSA-MAA for 3, 5, 10, 15, 20, and 60 min. Cells were then fixed with 3.7% paraformaldehyde in PBS for 10 min, rinsed once in PBS and then blocked with 1% BSA in PBS for 30 min at room temperature. Cells were incubated with rabbit anti-scavenger receptor A (1:1000 dilution) for 1 h at room temperature or overnight at 4 °C. Cells were then rinsed 5 times for 5 min periods in PBS and incubated with Alexa goat anti-rabbit 488 (1:200) for 1 h at room temperature in a humidified chamber. Cover slips were then rinsed in PBS and mounted onto glass slides using 510 Prolong Gold with DAPI (Invitrogen). The prepared slides were then viewed and analyzed using a Zeiss confocal laser-scanning microscope. All confocal laser-scanning microscope images were obtained at the University of Nebraska Research Core Facility utilizing a Zeiss 510 Meta Confocal Laser Scanning Microscope equipped with four lasers. Zeiss software was used to analyze co-localization of the MAA-adducted protein and scavenger receptor.
Fluorescent Activated Cell Sorting (FACS)
BEAS-2B were plated in 60-mm dishes to 80–90% confluence. While in the adhered state, they were incubated with BSA-MAA for 3, 10, 20, 30, and 60 min. Cells were then rinsed briefly with sterile PBS at 4 °C. Cells were then lifted using a rubber cell lifter. Cells were then placed in 1% paraformaldehyde and pelleted by centrifugation at 250 × g for 5 min at 4 °C. The supernatant was discarded and the pellet was re-suspended and fixed in 3.7% paraformaldehyde for 10 min. The cells were centrifuged and re-suspended in 1 mL PBS with rabbit anti-scavenger receptor A (1:1000). Cells were allowed to incubate for 4 h on ice. After incubating with the primary antibody, 2 mL PBS was added to each tube, the cells were washed 3 times by centrifugation, and re-suspended in PBS. The cells were then re-suspended in 1 mL PBS with Alexa goat anti-rabbit (1:500) and incubated for 1 h on ice. Cells were washed 3 times in PBS. The cells were analyzed within 12 h of being fixed. All FACS analysis was carried out on the FACSCalibur flow cytometer (BD Biosciences, San Jose, CA) at the Cell Analysis Facility at the University of Nebraska Medical Center. Data were reported as percent decrease in mean fluorescence intensity as compared to the control group and analyzed for statistical significance using Prism 5 graph pad software.
Immunoprecipitation
Confluent BEAS-2B were treated with 100 μg/mL BSA-MAA for 1–10 min, washed, and snap-frozen in liquid nitrogen to halt responses. Cells were then thawed and homogenized in 500 μL RIPA buffer (1 M HEPES, 5 M NaCl, 20% Triton X-100, 10% SDS, and 1% BSA) using a 25-gauge needle. Cell homogenates were then incubated for 1 h at room temperature with 500-μL HMEK buffer (30 mM HEPES, pH 7.5, 5 mM MgSO4, 0.5 mM EDTA, and 50 mM KCl) containing 0.5 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC). The crosslinking reaction was halted by boiling for 10 min in 5-μL β-mercaptoethanol. After chilling samples to 4 °C, protein concentrations were standardized to 1 mg/mL in HMEK buffer. Proteins were incubated with 0.33 μg/mL rabbit anti-SRA (Abcam) in TBS (10 mM Tris-HCl, pH 7.5, and 0.15 M NaCl) with 1% NP-40 overnight at 4 °C. SRA was immunoprecipitated with 20 μL Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Dallas, TX) overnight at 4 °C, followed by centrifugation at 250 × g for 5 min at 4 °C. Supernatants were discarded and the precipitate was washed 3 times in TBS+NP-40 containing Tween. Samples were then boiled in SDS-2ME and resolved by PAGE. Transferred proteins were resolved by Western blot. Blots were first probed with anti-MAA antibodies, rinsed, re-blocked, and then re-probed using anti-SRA antibodies. SRA was probed to efficiency of IP and Western blot sample loading.
Western Blot Assay
Protein concentration was determined by Nano Drop spectrophotometer from Thermo Fisher Scientific (Waltham, MA) using the method of Bradford (Bradford, 1976). Approximately 30 μg/mL of protein was loaded into each lane, alongside a positive control. The sample was run on a preformed 20% SDS-PAGE gel (Mini-PROTEAN TGX, Bio-Rad, Hercules, CA) at 180 V for 25 min. The protein was then transferred to nitrocellulose paper using a semi-dry transfer cell (Bio-Rad) at 25 V for 20 min. The nitrocellulose paper was then rinsed with TBS/Tween (50 mM Tris base, 100 mM NaCl, 0.05% Tween-20) for 5 min prior to blocking with 3% dried milk for 1 h at room temperature. The nitrocellulose paper was then incubated with rabbit anti-scavenger receptor A (1:1000) overnight at 4 °C. Transfers were rinsed three times for 5 min each rinse with TBS/Tween. The paper was then incubated with goat anti-rabbit HRP (1:5000) for 1 h at room temperature. Finally, the secondary antibody was detected using the Supersignal West Pico Chemiluminescent substrate kit (Thermo Scientific) and developed on X-ray film. Blots were then stripped and re-probed with rabbit anti-MAA adduct (1:2000) followed by the same secondary as above.
Mouse KC ELISA
To quantitate MAA-stimulated KC release, we utilized differentiated and polarized mouse tracheal epithelial cells grown on air-liquid interface (MTEC ALI). MTEC ALI cultures were treated with either media (Hams F-12:DMEM), SPD, SPD-MAA, fucoidan, or fetuin (all at 100 μg/mL) on the apical surface for 24 h. These supernatants were collected and the basolateral compartments analyzed for KC release. C57Bl/6 KC from MTEC ALI was determined utilizing the R&D Systems DuoSet ELISA kit following the manufacturer's recommended protocol (R&D Systems, USA).
Mouse PKC ε Activity
To determine MAA-stimulated protein kinase C epsilon (PKCε) activity, differentiated and polarized MTEC ALI cultures were treated with either media (Hams F-12:DMEM), SPD, SPD-MAA, fucoidan, or fetuin (all at 100 μg/mL) on the apical surface for 2 h, washed, and cells flash-frozen in liquid nitrogen. PKCε activity was determined in fractionates from cells by direct isoform-specific substrate peptide phosphorylation assays as previously described (Wyatt et al., 2010).
Statistics
All quantitative experiments were performed independently a minimum of three separate times. All data were analyzed using GraphPad Prism (version 4.00 for Windows, GraphPad Software, San Diego CA) and represented as mean ± standard error. Data were analyzed for statistical significance using one-way ANOVA followed by Bonferroni post hoc testing between each condition group. Significance was accepted at the 95% confidence interval.
Results
MAA–adducted protein transiently enhances SRA surface expression in intact bronchial epithelial cells
Human bronchial epithelial cells (BEAS-2B) were treated with 100 μg/mL SPD-MAA for up to 20 min. After treatment with SPD-MAA, cells were probed with anti-SRA antibody (green) to evaluate membrane surface expression of the receptor. In these experiments, cell membranes were also stained with lipophilic carbocyanine derivative SP-DiIC18(3) (red), which readily dissolves into the lipophilic cell surface membrane, conjugating with thiol-containing peptides and proteins. Cell nuclei are shown by DAPI stain (blue). In response to SPD-MAA, surface expression of SRA initially increased at 3–7 min and then decreased with no SRA observed at 20 min (Fig. 1). The co-localization of the SP-DiIC18(3) and SRA (yellow) demonstrated that MAA-adducted protein induced a rapid membrane translocation of SRA, followed by the subsequent internalization of SRA away from the surface membrane (Fig. 1). As a control, no changes in SRA expression were observed when cells were treated with non-adducted SPD (data not shown). A decrease in SRA and membrane co-localization after 7–10 min suggests that SRA is rapidly internalized in response to MAA-adducted protein.
Figure 1.
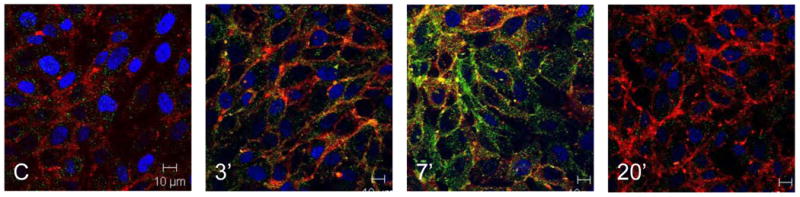
Localization of scavenger receptor A (SRA) with surface membrane in BEAS-2B treated with SPD-MAA. Bronchial epithelial cells (BEAS-2B) were treated with SP-DiIC18(3) to delineate the cell membrane (red). Staining with anti-SRA antibody (green) shows little surface localization of SRA in control (C) unstimulated cells. After 3–7 min of treatment with 100 μg/mL SPD-MAA, SRA co-localizes with cell surface membrane (yellow). Co-localization of membrane and SRA then decreases and is minimally present by 20 min.
MAA-adducted protein co-localizes to SRA in bronchial epithelial cells
To determine if MAA-adducted protein binds directly to SRA in bronchial epithelial cells, BEAS-2B were treated with 100 μg/mL BSA-MAA for 3, 5, 7, and 10 min prior to fixation in 3.7% paraformaldehyde. Cell monolayers were then stained with antibodies against MAA adduct (red) and SRA (green) to determine the localization of these proteins over time. No co-localization (yellow) was observed in the absence of MAA-adducted protein, and little SRA was expressed on the surface of the cells under control conditions (Fig. 2). BSA-MAA appeared to be localized at the surface of the cell membrane by 3 min treatment with a small, but increased amount of co-localized SRA surface expression. However, by 5 min BSA-MAA treatment, the maximum co-localization of BSA-MAA and SRA was observed (Fig. 2). This enhanced co-localization in the cell surface and cytoplasm continued through 7 min BSA-MAA treatment. Internalization of the co-localized BSA-MAA and SRA was observed by decreased fluorescence intensity at 10 min BSA-MAA treatment. Identical patterns of binding and co-localization were observed for both BSA-MAA and SPD-MAA. There was no change in fluorescence at any time point when cells were probed with anti-scavenger receptor B (SRB; data not shown). These data suggest that MAA-adducted protein may be binding to the surface-membrane upregulated SRA as the two proteins temporally co-localize.
Figure 2.
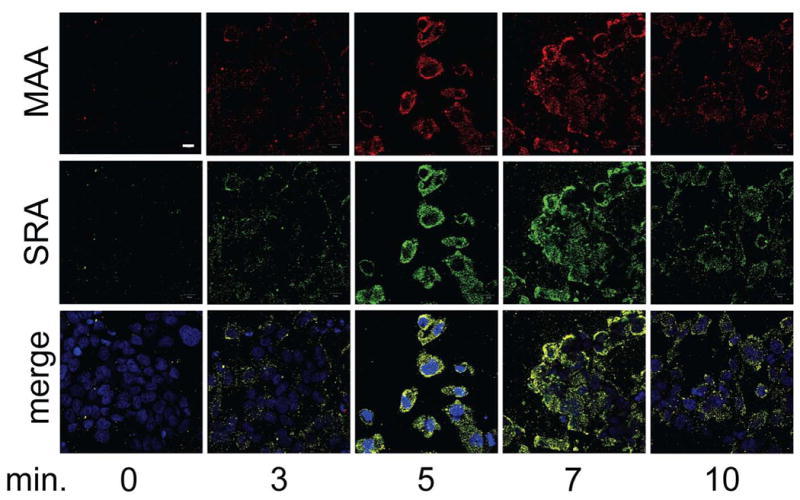
Co-localization of BSA-MAA and SRA in BEAS-2B treated with BSA-MAA. BEAS-2B were treated with 100 μg/mL BSA-MAA for up to 10 min and stained with anti-MAA (red), anti-SRA (green), and DAPI (blue). Maximal co-localization (yellow merge) was transient and observed between 5–7 min MAA stimulation, with a decreased co-localization observed by 10 min. Differential interference contrast (DIC) and DAPI stain were used to identify a consistent number of cells per field (not shown).
MAA–adducted protein modulates SRA content in membrane of bronchial epithelial cells
BEAS-2Bs were treated with 100 μg/mL BSA-MAA or PBS control for 3, 10, and 30 min, stained with anti-SRA antibody, and analyzed by FACS. There was an initial increase, followed by a gradual decrease in surface expression of SRA as evidenced by the FACS overlay of each time point's peak detection (Fig. 3A, MAA). PBS-treated controls produced no shift in peak fluorescence over time as detected by FACS (Fig. 3B, PBS). The mean fluorescence intensity data show that the fluorescent expression of SRA at the cell surface is significantly enhanced at 3 min BSA-MAA treatment, but reaches a significant decrease by 30 min (Fig. 3C). BSA-MAA auto-fluorescence was not detected by FACS. Non-adducted BSA had no effect on SRA expression. Collectively, these data suggest that SRA is upregulated in response to BSA-MAA, resulting in a transient increase in the amount of membrane SRA and that this transient upregulation of SRA in response to MAA exposure may lead to functional binding of the two proteins.
Figure 3.
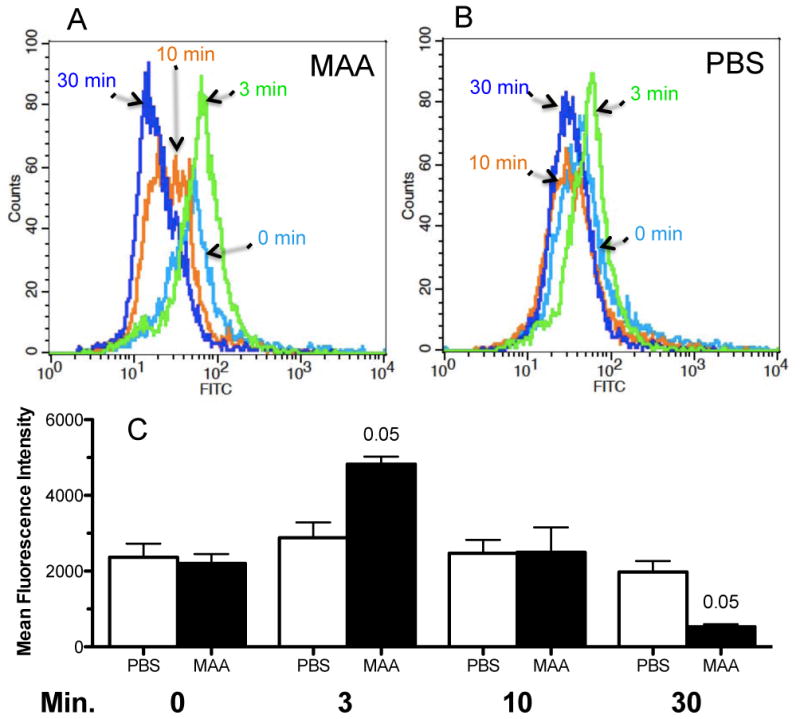
Surface expression of SRA by fluorescence-activated cell sorting (FACS). BEAS-2Bs were treated with 100 μg/mL BSA-MAA or PBS control for 3, 10, and 30 min. (A) FACS analysis shows SRA surface expression decreases with time after BSA-MAA treatment. (B) In contrast, no significant change in SRA surface expression with time is observed after PBS treatment of cells. (C) Average mean fluorescence intensity of three separate experiments treated with either PBS or BSA-MAA from 0–30 min. BSA-MAA treatment resulted in a significant (p < 0.05) increase at 3 min and a significant (p < 0.05) decrease at 30 min in SRA surface expression.
Immunoprecipitated SRA binds MAA-adducted protein
Because the FACS data suggest that BSA-MAA binds to the cell through SRA, as the receptor surface expression initially increases and then decreases in MAA-treated cells by a larger degree when compared to untreated cells, we performed immunoprecipitation assays to determine if SRA binds MAA-adducted protein as a ligand. While a pronounced amount of SRA appears to be detected by immunoprecipitation after 3 min BSA-MAA treatment (Fig. 4), no significant differences in the total average amount of SRA in whole cell homogenates were detected (n = 3 separate experiments). However, a significantly (p < 0.02) increased band of MAA-adducted protein was detected at 3 min after BSA-MAA treatment of the cells in the SRA immunoprecipitations. These data demonstrate that MAA-adducted protein is co-immunoprecipitated with SRA in bronchial epithelium exposed to MAA.
Figure 4.
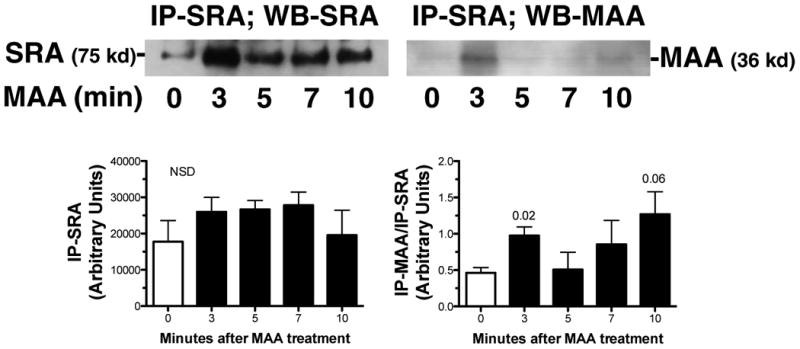
Immunoprecipitation of SRA in bronchial epithelial cell membrane fractions after treatment with BSA-MAA. BEAS-2B were exposed to 100 μg/mL BSA-MAA for up to 10 min and cell membrane fractions were immunoprecipitated with anti-SRA. Immunoprecipitated proteins were resolved by PAGE and probed with anti-SRA and anti-MAA antibodies by Western blot. An increased amount of membrane SRA was immunoprecipitated after 3 min treatment with BSA-MAA. When the same blot is probed with anti-MAA antibodies, MAA-adducted protein is also immunoprecipitated from the membrane fraction by anti-SRA at the 3 min time point.
Functional requirement of SRA in MAA-stimulated mouse tracheal epithelial cells
MAA-adducted proteins have been shown to stimulate inflammatory cytokine release through a PKC-mediated pathway (Wyatt et al., 2001). We have previously shown that PKCε is involved in the production of IL-8, and this could be inhibited in PKCε-dominant negative cells or with PKCε inhibitors (Wyatt et al., 2010). To identify the contribution of differentiated bronchial epithelium to SRA-mediated MAA-stimulated KC release, we utilized differentiated and polarized mouse tracheal epithelial cells grown on an air-liquid interface (MTEC ALI). MTEC ALI cultures were treated with varying media on the apical surface and supernatants were analyzed for KC production from the basolateral surface after 24 h. SPD-MAA stimulated KC production when compared to media (p < 0.01) and fucoidan greatly stimulated KC release (Fig. 5A). However, fucoidan caused a reduced, but significant (p < 0.01) stimulation of KC in MTEC ALI cells derived from SRA KO mice (Fig. 5A). As a negative control for the action of fucoidan, 100 μg/mL fetuin had no effect on KC release from wild type or SRA KO MTEC ALI. Similar to KC release, 100 μg/mL SPD-MAA for 2 h stimulated PKCε in wild type MTEC, but not SRA KO MTEC (Fig. 5B). Fucoidan treatment resulted in significant PKCε activation at 2 h in both wild type and SRA KO MTEC. To control for the presence of normal activatable PKC ε in SRA KO mice, SRA KO MTEC were stimulated with 10 μM DCP-LA, a direct activator of PKCε. DCP-LA significantly stimulated PKCε by 30 min in SRA KO MTEC (data not shown). Likewise, no binding of SPD-MAA (100 μg/mL) was observed on the surface of SRA KO MTEC at any time examined up to 30 min (Fig. 6). Similar to BEAS-2B, wild type MTEC ALI demonstrated an optimum SPD-MAA surface binding (red) to cell monolayers by 5 min treatment with the adducted protein (Fig. 6). A consistent number of cells were contained in each field as determined by DAPI stain (blue). These data confirm that SRA is involved in MAA-adducted protein-stimulated, PKC ε-mediated cytokine production in mouse airway epithelial cells. These data also suggest that fucoidan can stimulate KC release through an alternative mechanism in the absence of SRA.
Figure 5.
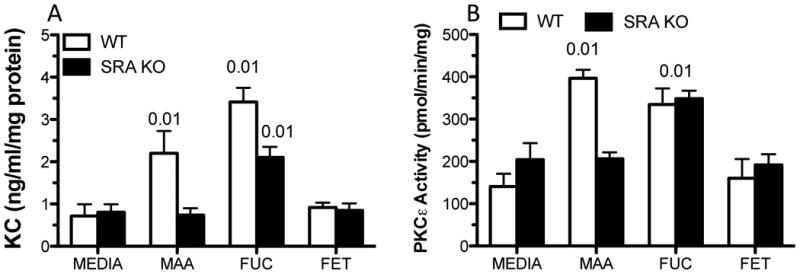
SPD-MAA stimulation of KC in mouse tracheal epithelial cells (MTEC). Wild type (WT) and SRA KO mice were sacrificed and MTECs cultured on air-liquid interface (ALI). Each well was treated with 100 μg/mL SPD-MAA, fucoidan (FUC), or Fetuin (FET) on the apical surface. The basolateral media was analyzed for KC release at 24 h (A). Both SPD-MAA and the SRA ligand, fucoidan, stimulated KC release from WT MTEC. MAA-stimulated release was blocked in differentiated epithelial cells from SRA KO mice. (B) SPD-MAA significantly (p < 0.01 vs. media control) activated PKCε at 2 h in WT, but not SRA KO MTEC. Fucoidan significantly (p < 0.01) stimulated KC and PKCε in both WT and SRA KO MTEC.
Figure 6.
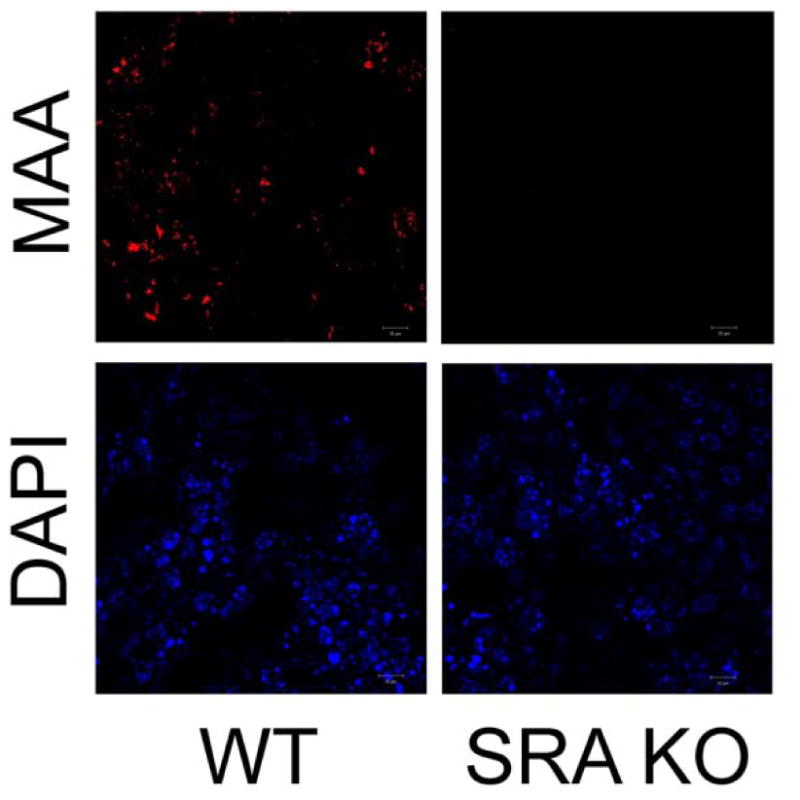
Localization of SPD-MAA binding in wild type and SRA KO mouse tracheal epithelial cells (MTEC) treated with SPD-MAA. BEAS-2B were treated with 100 μg/mL SPD-MAA for up to 10 min and stained with anti-MAA (red) and DAPI (blue). Maximal surface localization (red) was transient from 1–7 min. Representative images shown at 5 min MAA stimulation reveal no MAA binding to SRA KO MTEC. DAPI staining of nuclei was used to control for an equivalent number of cells per field.
Discussion
Because lung surfactant proteins are likely targets for MAA adduction, we hypothesized that such MAA-adducted proteins stimulate the inflammatory response in airway epithelial cells through binding to scavenger receptor A. To address this hypothesis, we measured the expression and localization of SRA under treatment conditions with MAA-adducted protein. In addition, we examined the inflammatory response to MAA-adducted proteins in SRA knockout mice. Consistent with the known function of scavenger receptors to clear endogenous toxins, our findings demonstrate that MAA-adducted protein binds to lung epithelial cells via scavenger receptor A, and that SRA knockout mice have a significantly lower ability to produce MAA-stimulated keratinocyte-derived cytokine (KC), an IL-8 analogue.
Binding of SPD-MAA to SRA was supported by FACS analysis. SPD-MAA treatment demonstrated an eventual decrease in SRA surface expression at 30 min compared to PBS control. It is well known that scavenger receptors, when bound by ligand, will stimulate the receptor and form endosomes to be internalized (Fong & Le, 1999). As SRA is contained in intracellular clathrin-coated pits, up-regulation occurs upon stimulation of SRA protein receptor. Therefore, our FACS data reveal that fluorescence is increased at 3 min when receptor is surface expressed to bind ligand, followed by an internalized decrease beginning at 10 min and clearly pronounced at 30 min.
Western blot analysis from SRA immunoprecipitates further demonstrated that SRA was bound by SPD-MAA and rapidly internalized. By 3 min MAA stimulation, there was a significant increase in SRA expression compared to baseline expression. This would suggest that stimulation of scavenger receptor leads to surface expression up-regulation from coated pits. It has been suggested that the SRA-ligand complex is internalized to the endosome where the ligand is quickly degraded and the scavenger receptor is recycled (Fong, Fong, & Cooper, 1990). We observed a significant rise in SRA expression at 3 min with a general decrease in expression after 5–10 min in the membrane fraction. Subsequent PKC activation did not occur until after 2 h of MAA treatment. This likely represents that the receptor-ligand complex remains in the endosome for some time prior to recycling, as it has been shown that the late endosome provides the most degradation of proteins after 30 min, and up to 60 min (Tjelle, Brech, Juvet, Griffiths, & Berg, 1996).
Binding of SPD-MAA to SRA was further demonstrated by confocal microscopy. BEAS-2Bs exposed to SPD-MAA at varying time points show co-localization of SRA, stained cellular membrane, and MAA-adducted protein to be greatest between 3 and 10 min. The greatest amount of SRA/membrane co-localization was at 7 min, and was eliminated by 20 min. This would be consistent with the rate of internalization of known SRA ligands, including acetylated-LDL. The half-life of acetylated-LDL was previously shown to be less than 4 min in the macrophage (Fong et al., 1990). Oxidized-LDL has been shown to internalize at a rate slower than acetylated-LDL (Fong & Le, 1999). Our data show that the internalization of MAA-adducted protein occurs between 5–10 min, placing the half-life between acetylated and oxidized LDL. We further demonstrated that in bronchial epithelium, fucoidan and acetylated-LDL internalization of SRA was most significant between 10 and 20 min (data not shown). Of interest, it was also noted that there appeared to be clumping of cellular membrane co-localization with SRA that coincided with the time point that showed maximal co-localization. This would be consistent with previous studies demonstrating that upon stimulation, the receptor/ligand complex is internalized in coated pits, likely early endosomes.
We have previously described the inflammatory response produced by SPD-MAA (McCaskill et al., 2011), as it produces a significant amount of KC in mouse lung. Here, we showed that mouse tracheal epithelial cells (MTEC) cultured in air-liquid interface showed a significant KC stimulation in response to SPD-MAA, suggesting that the airway epithelium is a principal source for MAA-stimulated KC release. Again, fucoidan showed a persistent production of KC in the SRA KO when compared to wild type MTEC. In the SRA knockout mouse (SRA KO), the increased KC production when exposed to SPD-MAA is eliminated. The KC production from fucoidan, however, was not eliminated, but reduced. This is most likely explained by the alternative binding of fucoidan to another scavenger receptor in the absence of SRA. As scavenger receptors are known to be somewhat promiscuous, fucoidan has an affinity for and can bind to scavenger receptor class A, B, and C (Platt & Gordon, 1998). In addition, it has previously been described that the inflammatory response from fucoidan was dependent on CD14, and not SRA, but that SRA is the targeted receptor for fucoidan (Kim, Ordija, & Freeman, 2003). However, our observation that fucoidan, but not MAA, stimulates KC in the SRA KO MTEC suggests that MAA-adducted proteins might be more specific ligands for SRA than fucoidan. Indeed, we observed no changes in membrane localization for SRBI/II when airway epithelial cells were treated with MAA-adducted protein.
Unlike previously reported fucoidan-stimulated KC production in the SRA KO macrophage, MTEC show a somewhat decreased fucoidan-stimulated KC release in the SRA KO. This would suggest that in the airway epithelium, SRA functions, in part, as a receptor involved in the phlogistic response. It has previously been shown that in the macrophage, SRA is not involved in direct stimulation of an inflammatory response, but rather helps to negatively regulate the inflammatory response (Cotena, Gordon, & Platt, 2004; Yi et al., 2009). This is of significant importance in that the pulmonary epithelial cell is a first responder to the direct environment. In addition to the barrier mechanism and mucociliary escalator, early activation of the immune response would allow rapid clearance of a microbial agent or environmental irritant before it could produce a systemic response. Scavenger receptors likely have a dual role in the inflammatory response depending on which compartment or cell they are located on, much the same way that the SRA ligand influences the cellular response.
Conclusions
SPD-MAA formation is an important immune stimulator in a mouse model of elevated lung reactive aldehyde load such as that seen during cigarette smoke and alcohol exposure. SPD-MAA will readily bind SRA on bronchial epithelium. We have demonstrated SPD-MAA binding to SRA through FACS, Western blot, and confocal microscopy. Previously, it was shown that SPD-MAA was a potent stimulator of mouse KC. By knocking out the SRA receptor, we can potentially reduce production of airway epithelial-derived KC, and likely inflammation under conditions of cigarette smoke and alcohol co-exposure.
Acknowledgments
This material is the result of work supported with resources and the use of facilities at the Omaha VA Medical Center, Omaha, NE (Department of Veterans Affairs [VA Merit Review] to TAW). This work was supported by NIH-NIAAA (R01AA017993-S1) to TAW, NIH-NIAAA (R01AA017993) to TAW and NIH-NIAAA (R37AA008769) to JHS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Adult cigarette smoking in the United States: Current estimates. 2012 http://www.cdc.gov/tobacco/data_statistics/fact_sheets/adult_data/cig_smoking/
- Coller SP, Paulnock DM. Signaling pathways initiated in macrophages after engagement of type A scavenger receptors. Journal of Leukocyte Biology. 2001;70:142–148. [PubMed] [Google Scholar]
- Cotena A, Gordon S, Platt N. The class A macrophage scavenger receptor attenuates CXC chemokine production and the early infiltration of neutrophils in sterile peritonitis. Journal of Immunology. 2004;173:6427–6432. doi: 10.4049/jimmunol.173.10.6427. [DOI] [PubMed] [Google Scholar]
- Duryee MJ, Freeman TL, Willis MS, Hunter CD, Hamilton BC, 3rd, Suzuki H, et al. Scavenger receptors on sinusoidal liver endothelial cells are involved in the uptake of aldehyde-modified proteins. Molecular Pharmacology. 2005;68:1423–1430. doi: 10.1124/mol.105.016121. [DOI] [PubMed] [Google Scholar]
- Falk DE, Yi HY, Hiller-Sturmhöfel S. An epidemiologic analysis of co-occurring alcohol and tobacco use and disorders: findings from the National Epidemiologic Survey on Alcohol and Related Conditions. Alcohol Research & Health. 2006;29:162–171. [PMC free article] [PubMed] [Google Scholar]
- Fong LG, Fong TA, Cooper AD. Inhibition of mouse macrophage degradation of acetyl-low density lipoprotein by interferon-gamma. The Journal of Biological Chemistry. 1990;265:11751–11760. [PubMed] [Google Scholar]
- Fong LG, Le D. The processing of ligands by the class A scavenger receptor is dependent on signal information located in the cytoplasmic domain. The Journal of Biological Chemistry. 1999;274:36808–36816. doi: 10.1074/jbc.274.51.36808. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, Ho YK, Basu SK, Brown MS. Binding site on macrophages that mediates uptake and degradation of acetylated low density lipoprotein, producing massive cholesterol deposition. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:333–337. doi: 10.1073/pnas.76.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill GE, Miller JA, Baxter BT, Klassen LW, Duryee MJ, Tuma DJ, et al. Association of malondialdehyde-acetaldehyde (MAA) adducted proteins with atherosclerotic-induced vascular inflammatory injury. Atherosclerosis. 1998;141:107–116. doi: 10.1016/s0021-9150(98)00153-1. [DOI] [PubMed] [Google Scholar]
- Horiuchi S, Murakami M, Takata K, Morino Y. Scavenger receptor for aldehyde-modified proteins. The Journal of Biological Chemistry. 1986;261:4962–4966. [PubMed] [Google Scholar]
- Hsu HY, Chiu SL, Wen MH, Chen KY, Hua KF. Ligands of macrophage scavenger receptor induce cytokine expression via differential modulation of protein kinase signaling pathways. The Journal of Biological Chemistry. 2001;276:28719–28730. doi: 10.1074/jbc.M011117200. [DOI] [PubMed] [Google Scholar]
- Kim WS, Ordija CM, Freeman MW. Activation of signaling pathways by putative scavenger receptor class A (SR-A) ligands requires CD14 but not SR-A. Biochemical and Biophysical Research Communications. 2003;310:542–549. doi: 10.1016/j.bbrc.2003.09.049. [DOI] [PubMed] [Google Scholar]
- Limmon GV, Arredouani M, McCann KL, Corn Minor RA, Kobzik L, Imani F. Scavenger receptor class-A is a novel cell surface receptor for double-stranded RNA. FASEB Journal. 2008;22:159–167. doi: 10.1096/fj.07-8348com. [DOI] [PubMed] [Google Scholar]
- McCaskill ML, Kharbanda KK, Tuma DJ, Reynolds JD, DeVasure JM, Sisson JH, et al. Hybrid malondialdehyde and acetaldehyde protein adducts form in the lungs of mice exposed to alcohol and cigarette smoke. Alcoholism: Clinical and Experimental Research. 2011;35:1106–1113. doi: 10.1111/j.1530-0277.2011.01443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss M, Bucher B, Moore FA, Moore EE, Parsons PE. The role of chronic alcohol abuse in the development of acute respiratory distress syndrome in adults. JAMA. 1996;275:50–54. [PubMed] [Google Scholar]
- Nicoletti A, Caligiuri G, Törnberg I, Kodama T, Stemme S, Hansson GK. The macrophage scavenger receptor type A directs modified proteins to antigen presentation. European Journal of Immunology. 1999;29:512–521. doi: 10.1002/(SICI)1521-4141(199902)29:02<512::AID-IMMU512>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Platt N, Gordon S. Scavenger receptors: diverse activities and promiscuous binding of polyanionic ligands. Chemistry & Biology. 1998;5:R193–203. doi: 10.1016/s1074-5521(98)90156-9. [DOI] [PubMed] [Google Scholar]
- Plüddemann A, Neyen C, Gordon S. Macrophage scavenger receptors and host-derived ligands. Methods. 2007;43:207–217. doi: 10.1016/j.ymeth.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Tjelle TE, Brech A, Juvet LK, Griffiths G, Berg T. Isolation and characterization of early endosomes, late endosomes and terminal lysosomes: their role in protein degradation. Journal of Cell Science. 1996;109(Pt 12):2905–2914. doi: 10.1242/jcs.109.12.2905. [DOI] [PubMed] [Google Scholar]
- Tuma DJ. Role of malondialdehyde-acetaldehyde adducts in liver injury. Free Radical Biology & Medicine. 2002;32:303–308. doi: 10.1016/s0891-5849(01)00742-0. [DOI] [PubMed] [Google Scholar]
- Tuma DJ, Kearley ML, Thiele GM, Worrall S, Haver A, Klassen LW, et al. Elucidation of reaction scheme describing malondialdehyde-acetaldehyde-protein adduct formation. Chemical Research in Toxicology. 2001;14:822–832. doi: 10.1021/tx000222a. [DOI] [PubMed] [Google Scholar]
- Wyatt TA, Kharbanda KK, McCaskill ML, Tuma DJ, Yanov D, DeVasure J, et al. Malondialdehyde-acetaldehyde-adducted protein inhalation causes lung injury. Alcohol. 2012;46:51–59. doi: 10.1016/j.alcohol.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt TA, Kharbanda KK, Tuma DJ, Sisson JH. Malondialdehyde-acetaldehyde-adducted bovine serum albumin activates protein kinase C and stimulates interleukin-8 release in bovine bronchial epithelial cells. Alcohol. 2001;25:159–166. doi: 10.1016/s0741-8329(01)00177-x. [DOI] [PubMed] [Google Scholar]
- Wyatt TA, Slager RE, Heires AJ, DeVasure JM, Vonessen SG, Poole JA, et al. Sequential activation of protein kinase C isoforms by organic dust is mediated by tumor necrosis factor. American Journal of Respiratory Cell and Molecular Biology. 2010;42:706–715. doi: 10.1165/rcmb.2009-0065OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Thiele GM, Kearley ML, Haugen MD, Klassen LW, Sorrell MF, et al. Epitope characterization of malondialdehyde-acetaldehyde adducts using an enzyme-linked immunosorbent assay. Chemical Research in Toxicology. 1997;10:978–986. doi: 10.1021/tx970069t. [DOI] [PubMed] [Google Scholar]
- Yi H, Yu X, Gao P, Wang Y, Baek SH, Chen X, et al. Pattern recognition scavenger receptor SRA/CD204 down-regulates Toll-like receptor 4 signaling-dependent CD8 T-cell activation. Blood. 2009;113:5819–5828. doi: 10.1182/blood-2008-11-190033. [DOI] [PMC free article] [PubMed] [Google Scholar]


