Abstract
BACKGROUND AND PURPOSE
Recent data have indicated that α3β4* neuronal nicotinic (n) ACh receptors may play a role in morphine dependence. Here we investigated if nACh receptors modulate morphine physical withdrawal.
EXPERIMENTAL APPROACHES
To assess the role of α3β4* nACh receptors in morphine withdrawal, we used a genetic correlation approach using publically available datasets within the GeneNetwork web resource, genetic knockout and pharmacological tools. Male and female European-American (n = 2772) and African-American (n = 1309) subjects from the Study of Addiction: Genetics and Environment dataset were assessed for possible associations of polymorphisms in the 15q25 gene cluster and opioid dependence.
KEY RESULTS
BXD recombinant mouse lines demonstrated an increased expression of α3, β4 and α5 nACh receptor mRNA in the forebrain and midbrain, which significantly correlated with increased defecation in mice undergoing morphine withdrawal. Mice overexpressing the gene cluster CHRNA5/A3/B4 exhibited increased somatic signs of withdrawal. Furthermore, α5 and β4 nACh receptor knockout mice expressed decreased somatic withdrawal signs compared with their wild-type counterparts. Moreover, selective α3β4* nACh receptor antagonists, α-conotoxin AuIB and AT-1001, attenuated somatic signs of morphine withdrawal in a dose-related manner. In addition, two human datasets revealed a protective role for variants in the CHRNA3 gene, which codes for the α3 nACh receptor subunit, in opioid dependence and withdrawal. In contrast, we found that the α4β2* nACh receptor subtype is not involved in morphine somatic withdrawal signs.
CONCLUSION AND IMPLICATIONS
Overall, our findings suggest an important role for the α3β4* nACh receptor subtype in morphine physical dependence.
Introduction
Opiate addiction from long-term use of prescription analgesics and illicit substances remains a major public health problem not only in the United States but around the world (Manchikanti et al., 2012). An important component of opiate dependence is withdrawal, an aversive syndrome that occurs upon cessation of drug use and constitutes a powerful motivator for addicted individuals to continue to use drugs, even in the face of adverse consequences. The withdrawal syndrome is characterized in opioid-dependent humans as extremely unpleasant and manifests as tachycardia, hypertension, sweating, diarrhoea, vomiting, irritability, anxiety, shakes and insomnia. Current treatment for opiate withdrawal, which includes maintenance therapy with replacement opiates, methadone or buprenorphine, suffers from serious limitations. These limitations consist of dependence liability and withdrawal upon abrupt cessation (Kuhlman et al., 1998; Dyer et al., 1999). Therefore, the development of novel non-opioid pharmacotherapies for opiate withdrawal is critical.
A growing body of evidence supports the argument that neuronal nicotinic receptors (nACh receptors) may play an important role in opiate withdrawal. Recent rodent studies demonstrated that the administration of nicotinic antagonists attenuates signs of morphine withdrawal. For example, the non-selective nicotinic antagonists, mecamylamine and bis (2, 2, 6, 6-tetramethyl-4-piperidinyl) sebacate (BTMPS), were found to attenuate both spontaneous and naloxone-precipitated somatic signs in rats undergoing morphine withdrawal (Taraschenko et al., 2005; Hall et al., 2011). One particular nACh receptor subtype that has been indicated in decreasing somatic signs in precipitated morphine withdrawal is the α3β4* nACh receptor subtype (where * denotes the possible inclusion of additional subunits, see Alexander et al., 2013). However, these antagonists, including dextromethorphan, bupropion and 18-methoxycoronaridine (18-MC), have poor or partial selectivity for the α3β4* nACh receptor subtype and produced variable levels of decrease in the somatic manifestations of naloxone-precipitated morphine withdrawal in rats (Rho and Glick, 1998; Panchal et al., 2005; Taraschenko et al., 2005). Altogether, these results imply a role for the α3β4* nACh receptor subtype in morphine withdrawal. However, the conclusions remain limited because the selectivity issues of drugs used.
The α3β4* nACh receptor subtype is an interesting candidate since the 15q25 gene cluster, which contains the CHRNA5/CHRNA3/CHRNB4 genes, coding for the α5, α3 and β4 nACh receptor subunits, respectively, has emerged as a candidate region contributing to risk of heavy smoking, nicotine dependence and smoking-related diseases in humans (Bierut, 2009). Rodent studies have also confirmed an important role for α5, α3 and β4 nACh receptor subunits in nicotine withdrawal and aversion (Wada et al., 1990; Salas et al., 2004a,b; 2009; Jackson et al., 2010; Frahm et al., 2011). The α5 nACh receptor subunit can co-assemble with the α3β4* nACh receptor subtype to form functional receptors in the peripheral ganglia, as well as centrally, in the medial habenula (MHb) and interpeduncular nucleus (IPN; Wada et al., 1990; Zoli et al., 1995; Quick et al., 1999; Whiteaker et al., 2002). These brain regions were recently reported to be involved in nicotine withdrawal and intake (Salas et al., 2009; Fowler and Kenny, 2012).
This attractive hypothesis prompted us to investigate whether: (i) the α3β4* nACh receptor subtype plays a role in morphine physical dependence in mice; and (ii) the possible association of polymorphisms in human CHRNA5/CHRNA3/CHRNB4 genes with morphine dependence and withdrawal exists. To test the hypothesis that α3β4* receptors are involved in morphine somatic withdrawal, we used genetic correlation analyses across the BXD recombinant inbred (RI) mouse panel, selective α3β4* nACh receptor antagonists and finally transgenic mice with gene deletions or overexpression to elucidate the contribution of the α5, α3 and β4 nACh receptor subunits to morphine physical withdrawal. Lastly, human genetic association analyses were conducted using the Study of Addiction: Genetics and Environment (SAGE) European-American and African-American datasets to determine if variants in the 15q25 gene cluster are associated with opioid dependence.
Methods
All studies involving animals are reported in accordance with the ARRIVE guidelines for reporting experiments involving animals (Kilkenny et al., 2010; McGrath et al., 2010). In addition, all receptor and drug nomenclature conforms to British Journal of Pharmacology's Concise Guide to Pharmacology (Alexander et al., 2013).
Animals
Male adult C57BL/6J mice were purchased from Jackson Laboratories. Mice null for the α5 (Jackson Laboratories, Bar Harbor, ME, USA), α4 and β2 (Institut Pasteur, Paris, France) nACh receptor subunits and their wild-type (WT) littermates were bred in an animal care facility at Virginia Commonwealth University (Richmond, VA). β4 nACh receptor knockout (KO) mice were generated and bred at Baylor College of Medicine (Houston, TX, USA) as described previously (Xu et al., 1999). α5, α4, β2 and β4 mice were backcrossed at least 8 to 12 generations to C57BL/6J mice (Jackson Laboratories). Mutant/transgenic and WT littermates were obtained from crossing heterozygous mice. Transgenic mice overexpressing the human cluster CHRNA5/A3/B4 (TgCHRNA5/A3/B4) (Gallego et al., 2012) were obtained at Barcelona Biomedical Research Park (PRBB, Barcelona, Spain) by crossing transgenic mice with hybrid B6/SJL-F1 female mice (F1–F5). The non-transgenic littermates obtained from crosses of TgCHRNA3/A5/B4 mice and B6/SJL-F1J females served as controls. These transgenic mice bear increased [125I]-epibatidine + 5I-A-85380 binding sites in brain regions where α3β4* is endogenously expressed and are hypersensitive to high doses of nicotine (Gallego et al., 2012). Animals were 8–12 weeks of age at the start of the experiments and were group-housed (three to five per cage) under a 12 h light/dark cycle in a 21°C humidity-controlled AAALAC-approved animal care facility with ad libitum access to food and water. Experiments were performed during the light cycle and were approved by the Institutional Animal Care and Use Committee of Virginia Commonwealth University, Baylor College of Medicine, and the PRBB ethical committee.
General chronic morphine administration and precipitated withdrawal protocol
Mice were injected with saline or morphine s.c. over the course of 8 days as follows: days 1 and 2, mice received 25 mg·kg−1 morphine 2× a day. Days 3 and 4, mice received 50 mg·kg−1 morphine 2× a day. Days 5 and 6, mice received 80 mg·kg−1 morphine 2× a day. Days 7 and 8, mice received 100 mg·kg−1 2× a day. On Day 9, mice were injected with 100 mg·kg−1 morphine in the morning. Two hours after morphine injection, mice were injected with naloxone (1 mg·kg−1, s.c.) to precipitate somatic signs. Somatic signs were observed for 30 min immediately following naloxone injection.
Mice were individually placed in Plexiglass cages and were observed and scored for the manifestation of different withdrawal signs including the total number of jumps, wet dog shakes, paw tremors, backing, ptosis and diarrhoea. Results were reported as the average of the total signs per group. All testing was conducted in a blind manner.
Antagonist administration for morphine-precipitated withdrawal protocol
For groups receiving the antagonist AuIB via i.c.v., mice underwent the chronic morphine administration protocol as described in the previous section. However, on the evening of day 8, approximately 2 h after the last morphine injection, mice were anaesthetized with sodium pentobarbital (45 mg·kg−1 i.p.), and a scalp incision was made to expose the bregma. During anesthesia, mice were evaluated for pain with toe pinch prior to the procedure and checked every 3–5 min. Unilateral injection sites were prepared using a 26-gauge needle with a sleeve of polyurethane tubing to control the depth of the needle at a site 2 mm rostral and 2 mm lateral to the bregma at a depth of 2 mm. Animals were sutured, leaving the injection site accessible with gentle displacement of the scalp to enable an injection volume of 5 μL. Mice were allowed to recover overnight. On day 9, mice were injected with 100 mg·kg−1 morphine in the morning. Two hours after morphine injection, mice were injected i.c.v. with either vehicle or AuIB (1.75 or 3.5 pmol). Five minutes after AuIB or vehicle treatment, mice were injected with naloxone (1 mg·kg−1, s.c.) to precipitate somatic signs. Somatic signs were observed for 30 min immediately following naloxone injection.
At the end of the study, a subset of animals were injected i.c.v. with 5 μL of cresyl violet dye 10 min before a lethal overdose of 65 mg·kg−1 sodium pentobarbital and perfused to confirm drug diffusion from the injection site to the lateral ventricle. Brain slices were collected, and mice were observed to have blue cresyl violet dye in both lateral ventricles.
For groups treated with AT-1001, on day 9, 2 h after morphine injection, mice were injected i.p. with vehicle or AT-1001 (1 or 3 mg·kg−1) and, 15 min later, with naloxone. Somatic signs were observed for 30 min immediately following naloxone injection as described above.
To assess the role of α4β2* nACh receptors in morphine withdrawal, in a separate group, mice received the same morphine protocol as described in the previous section and 2 h after morphine injection on withdrawal day, mice were injected s.c. with vehicle or DHβE (3 mg·kg−1). Five minutes after DHβE or vehicle treatment, mice were injected with naloxone (1 mg·kg−1, s.c.) to precipitate somatic signs. Somatic signs were observed following naloxone injection as described above.
A similar protocol of morphine-precipitated withdrawal was followed with studies using α5, α4, β2 and β4 nACh receptor KO mice and their WT counterparts. Control groups treated with chronic saline were also assessed. Each treatment group contained n = 6–8 mice.
A slightly different administration protocol was used to investigate morphine dependence in the TgCHRNA5/A3/B4 model due to the different genetic background used (hybrid C57/SJL strain). Since WT mice showed a ceiling effect with the previously described protocol (data not shown), we reduced the dosage of morphine as follows: day 1, mice received 20 mg·kg−1 morphine 2× a day. Day 2, mice received 30 mg·kg−1 morphine 2× a day. Day 3, mice received 40 mg·kg−1 morphine 2× day, days 4 and 5, mice received 50 mg·kg−1 morphine 2× a day. On the morning of day 6, mice received 50 mg·kg−1 morphine and approximately 2 h after the last morphine injection, mice were injected with naloxone (1 mg·kg−1, s.c.) to precipitate somatic signs. This protocol allowed the detection of increased morphine withdrawal sensitivity in transgenic mice, avoiding the ceiling effect of high doses. Somatic signs were observed following naloxone injection as described above.
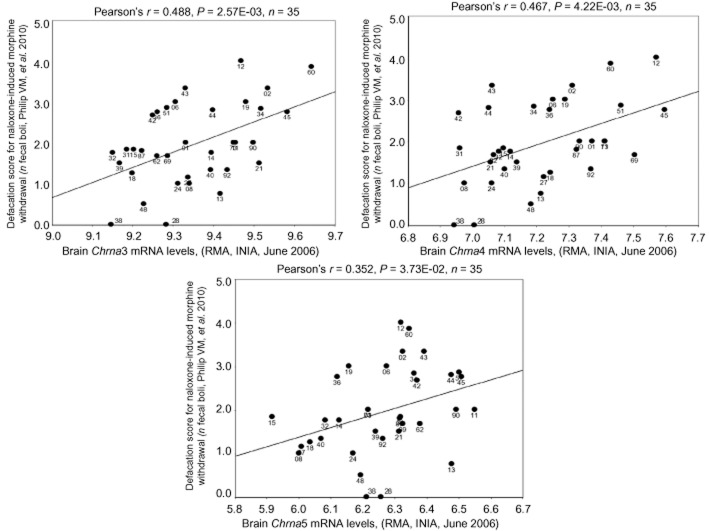
α3, α5 and β4 nicotinic subunit gene expression correlates with morphine withdrawal somatic signs in the BXD inbred mouse panel
Previous mouse genetic studies have phenotyped a number of morphine-induced behavioural responses across the BXD RI mouse panel (Schadt et al., 2003). Analysing phenotype data along with gene expression data by means of genetic mapping and/or genetic correlations allows for the detection of putative candidate genes whose variation in expression might be responsible for the variation in behaviour observed across the BXD panel (Schadt et al., 2003). We took a genetic correlation approach using publically available datasets within the GeneNetwork web resource. As described in Philip et al. (2010), BXD mice were given an acute i.p. injection of 50 mg·kg−1 morphine and 3 h later, withdrawal was precipitated with 30 mg·kg−1 of naloxone. While this mode of acute morphine exposure is not similar to our repeated morphine administration protocol, it is well established that a single injection or relatively short-term chronic infusion of morphine and other drugs can elicit withdrawal-like signs (‘acute dependence’) in rodents. This high dose of naloxone is routinely used to induce acute precipitated morphine withdrawal in mice. The numerous similarities in the behavioural and neurobiological mechanisms mediating acute dependence and those mediating withdrawal following relatively long-term drug exposure has led to the suggestion that the effects elicited in acute dependence paradigms are valid indices of drug withdrawal and have similar predictive outcomes (Harris and Gewirtz, 2005). Withdrawal signs scored were number of jumps, faecal boli, urine puddles, locomotion, vertical activity, horizontal activity and somatic signs such as wet dog shakes, ptosis, salivation, abnormal posture and abdominal contractions. We correlated those morphine withdrawal phenotypes with basal mRNA levels of Chrna3, Chrna5 and Chrnb4 in whole brain [forebrain and midbrain dataset: INIA Brain mRNA M430 (Jun06) RMA] and many regions (hypothalamus, hippocampus, VTA, nucleus accumbens, prefrontral cortex, cerebellum, amygdala and neocortex). All comparisons used the genetic Pearson product moment correlations within GeneNetwork.
Opioid dependence human genetic analysis
Male and female European-American (n = 2772) and African-American (n = 1309) subjects from the SAGE dataset were used to assess the possible association of polymorphisms in the 15q25 gene cluster and opioid dependence. Genetic data (genotypes and phenotypes) were downloaded from dbGaP under the protocol of genetic study of nicotine dependence by X. Chen. The SAGE study is comprised of three independent studies: the Collaborative Genetic Study of Nicotine Dependence (COGEND), the Collaborative Study on the Genetics of Alcoholism (COGA) and the Family Study of Cocaine Dependence (FSCD). Genotype and phenotype data from each of the three studies were used in the analysis. European-American and African-American data were treated as two different datasets. The phenotypes tested were case/control status for opioid withdrawal and Diagnostic and Statistical Manual of Mental Disorders-Fourth Edition (DSM-IV) opioid dependence. To conduct the analysis, single nucleotide polymorphisms (SNPs) in the 15q25 gene cluster were imputed in both datasets using IMPUTE2 (Howie et al., 2009; 2011; 2012) with the 1000 Genomes Phase 1 integrated variant set (March 2012 release) as a reference panel. A total of 363 SNPs were imputed in the region containing the α3, α5 and β4 nACh receptor genes, though some markers were non-polymorphic in either dataset. Of these, 293 SNPs were shared between datasets and were used for subsequent analysis. To increase the power of our analysis, a meta-analysis was conducted using results from both datasets (n = 4081).
Statistical analysis
For all animal data, statistical analyses were performed using StatView® (SAS, Cary, NC, USA). Data were analysed with one-way anova with treatment as the between subject factor or two-way anova with treatment and genotype as between subject factors for the nicotinic KO and TgCHRNA5/A3/B4 mice withdrawal studies. P values of less than 0.05 were considered significant. Significant results were further analysed using the Neuman–Keuls post hoc test.
Statistical analysis for genetic association studies was performed using the PLINK software (Purcell et al., 2007). Opioid withdrawal and DSM-IV opioid dependence were treated as dichotomized variables and were analysed using logistic regression. Age, sex, study (COGA, COGEND or FSCD), and principal components to control for population stratification within the sample were used as covariates. The GWAMA program (Magi and Morris, 2010) was used for a random effects meta-analysis of the results from both datasets. Cochrane's Q statistic P values were calculated to measure between-study heterogeneity. Correction for multiple testing was carried out on all tests jointly for the meta-analysis using single nucleotide polymorphism spectral decomposition (Nyholt, 2004), a correction for multiple testing based on the linkage disequilibrium (LD) structure of the markers tested. Based on the LD structure of the 293 markers assessed, the corrected P-value threshold was P < 0.0006.
Drugs
DHβE was purchased from RBI (Natick, MA, USA). The α3β4*nACh receptor-selective antagonist α-conotoxin AuIB (AuIB) was purified from the venom of the ‘court cone’, Conus aulicus, blocks the α3β4* nACh receptor subtype with >100-fold higher potency than other nicotinic receptor combinations, such as α3β2* and α4β4* (Luo et al., 1998). AT-1001, a high affinity α3β4* nACh receptor antagonist (Zaveri et al., 2010; Toll et al., 2012) was provided by Astraea Therapeutics (Mountain View, CA, USA). Morphine sulfate and naloxone were obtained from the National Institute on Drug Abuse (Baltimore, MD, USA). AT-1001 was dissolved in 20% DMSO: emulphor : saline solution in a 1:1:18 ratio, and the drug was injected i.p. The rest of the compounds were dissolved in physiological saline (0.9% sodium chloride) and administered to each animal by s.c. or i.c.v. injection. The doses for AuIB (1.75 and 3.5 pmol) were calculated based on the functional IC50 at α3β4 nACh receptors (Luo et al., 1998). The doses of AT-1001 were based on the recent in vivo studies with nicotine (Toll et al., 2012).
Results
Neuronal Chrna3, Chrna4 and Chrna5 mRNA expression significantly correlates with defecation following naloxone-precipitated morphine withdrawal
As an initial screen to identify whether α3, α5 and β4 nicotinic receptor subunits might influence morphine withdrawal phenotypes, we utilized a genetic correlation analysis across the BXD inbred mouse panel using publically available datasets within the GeneNetwork web resource. We found significant correlations between mRNA levels of Chrna3, Chrna5 and Chrnb4, in different brain regions (such as prefrontal cortex, ventral tegmental area and cerebellum), which were associated with various withdrawal signs (see Supporting Information Table S1). As an example, we illustrated, in Figure 1, the significant positive correlations between Chrna3, Chrna5 and Chrnb4 and the magnitude of defecation (number of fecal boli) in male mice during naloxone-precipitated morphine withdrawal. These correlations suggest that higher mRNA expression of α3, α5, and β4 subunits within the forebrain and/or midbrain is associated with a larger withdrawal response to morphine, as measured by defecation severity. Based on these results, we considered the possibility that changes in the levels of α3, α5 and β4 subunits may affect morphine withdrawal in mice.
Figure 1.
Correlation of α3, α5 or β4 with somatic signs across BXD strains treated with morphine. Using a publically available BXD inbred mouse panel dataset, significant positive correlations between (A) Chrna3, (B) Chrna5 and (C) Chrnb4 mRNA in the forebrain and midbrain and the magnitude of defecation in males (number of fecal boli) during naloxone-precipitated morphine withdrawal were demonstrated.
Morphine withdrawal modulation in transgenic TgCHRNA5/A3/B4 mice and β4 and α5 nACh receptor KO mice
Mice overexpressing the CHRNA5/A3/B4 cluster exhibited increased number of somatic signs compared with their WT littermates in morphine somatic withdrawal, resulting in a significant main effect of genotype [F(1,27) = 5.74, P < 0.05] and treatment [F(1,27) = 32.20, P < 0.0001]. Moreover, post hoc analysis demonstrated significant differences between TgCHRNA5/A3/B4-treated and WT-treated mice (P < 0.05; Figure 2A). The increased signs in TgCHRNA5/A3/B4-treated mice were mainly attributed to the manifestation of higher jumping behaviour, upon naloxone-precipitated withdrawal, as revealed by a significant genotype × treatment interaction [F(1,27) = 6.21, P < 0.05], followed by a post hoc test ( Table 1). Furthermore, a higher percentage of TgCHRNA5/A3/B4 mice presented diarrhoea compared with their WT counterparts (Table 1).
Figure 2.
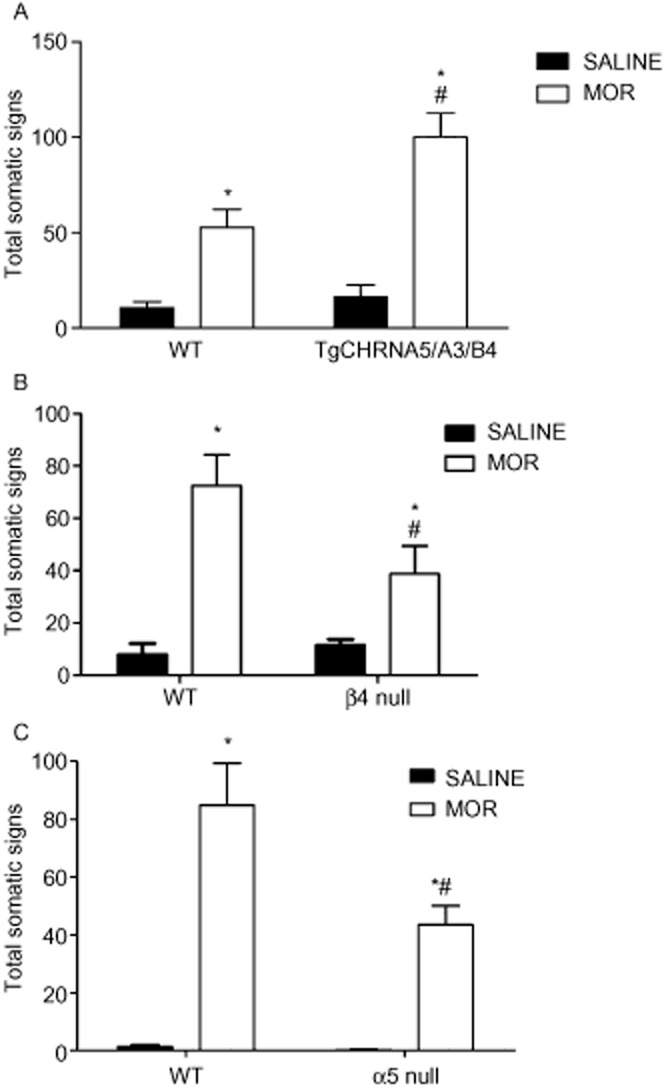
Overexpression of the genomic cluster CHRNA5/A3/B4 in mice increases somatic signs associated with morphine withdrawal. TgCHRNA5/A3/B4 morphine-dependent mice demonstrate a significant increase in withdrawal somatic signs compared with their WT counterparts. Data are expressed as mean ± SEM of n = 6–9 mice per group. *P < 0.05 versus saline control; #P < 0.05 versus WT-treated mice. (B) Conversely, β4 nACh receptor KO morphine-dependent mice expressed significant decreases in naloxone-precipitated somatic withdrawal signs compared with their WT counterparts. Data are expressed as mean ± SEM. of n = 6–8 mice per group. *P < 0.05 versus saline control; #P < 0.05 versus WT-treated mice. (C) Furthermore, α5 nACh receptor KO morphine-dependent mice demonstrated a significant decrease in withdrawal somatic signs compared with their WT counterparts. Data are expressed as mean ± SEM. of n = 6–8 mice per group. *P < 0.05 versus saline control; #P < 0.05 versus WT-treated mice. MOR, morphine.
Table 1.
Somatic signs of morphine withdrawal in mice overexpressing the genomic cluster CHRNA5/A3/B4
| Individual somatic signs | WT SAL | TgCHRNA5/A3/B4 SAL | WT MOR | TgCHRNA5/A3/B4 MOR |
|---|---|---|---|---|
| Paw tremors | 2.4 ± 0.9 | 7.3 ± 4.6 | 13.8 ± 4.9 | 25.4 ± 6.9 |
| Head shakes | 0.4 ± 0.4 | 1.4 ± 0.5 | 1.7 ± 0.4 | 2.5 ± 0.4 |
| Backing | 0 ± 0 | 0 ± 0 | 8.6 ± 2.9 | 4.4 ± 1.7 |
| Ptosis | 0.6 ± 0.6 | 0 ± 0 | 4.8 ± 1.0 | 4.0 ± 0.9 |
| Jumping | 0 ± 0 | 0.3 ± 0.2 | 8.3 ± 4.5* | 42.8 ± 9.0*# |
| Body tremor | 0.9 ± 0.9 | 0.6 ± 0.4 | 2.3 ± 1.0 | 3.8 ± 0.8 |
| Occurrence of diarrhoea (% of mice) | 0 | 0 | 33 | 88 |
Mice were made dependent on morphine for 5 days. On day 6, mice were administered naloxone (1 mg·kg−1, s.c.) before observing individual physical signs and % occurrence of diarrhoea. Data are expressed as mean ± SEM of n = 6–9 mice per group.
Denotes P < 0.05 versus saline control.
P < 0.05 versus WT-treated mice.
MOR, morphine.
Conversely, the increase in total somatic signs observed in β4 nACh receptor WT mice was significantly reduced in β4 nACh receptor KO mice, revealing a significant main effect of treatment [F(1,22) = 181.9, P < 0.0001] and genotype [F(1,22) = 19.77, P < 0.001] (Figure 2B). This was due to a decrease in head shakes resulting in a genotype × treatment interaction [F(1,22) = 30.00, P < 0.0001] and body tremors resulting in a genotype × treatment interaction [F(1,22) = 12.48, P < 0.001] upon naloxone-precipitated withdrawal, but not jumps in the β4 nACh receptor KO mice compared with their WT counterparts (Table 2). In contrast, all individual signs were attenuated in morphine-dependent α5 nACh receptor KO mice (Table 3). Indeed, the increase in total somatic signs observed in α5 nACh receptor WT mice was significantly reduced in the α5 KO group (Figure 2C), demonstrating a significant main effect of treatment [F(1,18) = 41.47, P < 0.001] and genotype [F(1,18) = 4.67, P < 0.05] in α5 nACh receptor KO and WT. In addition, two-way anova revealed the difference was due to the decrease in signs of paw tremors [F(1,18) = 10.77, P < 0.004], head shakes [F(1,18) = 14.94, P < 0.001] and jumping [F(1,18) = 60.43, P < 0.0001].
Table 2.
Knockout of β4 gene reduced somatic signs in morphine-treated mice
| Individual somatic signs | β4 WT SAL | β4 KO SAL | β4 WT MOR | β4 KO MOR |
|---|---|---|---|---|
| Paw tremors | 4.5 ± 0.8 | 7.2 ± 0.5 | 22.8 ± 5.1* | 11.9 ± 2.5# |
| Head shakes | 2.8 ± 0.8 | 4.3 ± 0.5 | 25.5 ± 6.1* | 8.9 ± 2.0# |
| Backing | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Ptosis | 0 ± 0 | 0 ± 0 | 2.6 ± 0.7 | 1.3 ± 0.7 |
| Jumping | 0.7 ± 0.3 | 0 ± 0 | 15.6 ± 7.2* | 14 ± 5.8* |
| Body tremor | 0 ± 0 | 0 ± 0 | 6.9 ± 1.0* | 2.7 ± 0.6*# |
| Occurrence of diarrhoea (% of mice) | 0 | 0 | 50 | 0 |
Mice were made dependent on morphine for 8 days. On day 9, mice were administered naloxone (2 mg·kg−1) before observing individual physical signs expressed as mean ± SEM and % occurrence of diarrhoea.
n = 8 mice per group.
P < 0.05 from saline group.
P < 0.05 from morphine WT group.
MOR, morphine; SAL, saline.
Table 3.
Knockout of α5 gene reduced somatic signs in morphine-treated mice
| Individual somatic signs | α5 WT SAL | α5 KO SAL | α5 WT MOR | α5 KO MOR |
|---|---|---|---|---|
| Paw tremors | 1.2 ± 0.4 | 0.3 ± 0.2 | 33 ± 13.2* | 12.5 ± 3.0# |
| Head shakes | 0.3 ± 0.2 | 0 ± 0 | 1.8 ± 0.7* | 2.3 ± 0.7*# |
| Backing | 0 ± 0 | 0 ± 0 | 2.5 ± 0.9 | 1.2 ± 0.3 |
| Ptosis | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Jumping | 0 ± 0 | 0.2 ± 0.2 | 44.5 ± 7.7* | 26.0 ± 4.88*# |
| Body tremor | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| Occurrence of diarrhoea (% of mice) | 0 | 0 | 100 | 50 |
Mice were made dependent on morphine for 8 days. On day 9, mice were administered naloxone (2 mg·kg−1) before observing individual physical signs expressed as mean ± SEM and % occurrence of diarrhoea.
*Denotes difference from vehicle group. n = 8 mice per group.
*P < 0.05 from control group.
P < 0.05 from Morphine WT group.
MOR, morphine; SAL, saline.
α3β4* nACh receptor antagonists block physical signs of morphine withdrawal
We further studied the role of α3β4* nACh receptor inhibition in morphine withdrawal by using two selective α3β4* antagonists in morphine-dependent mice. Male mice chronically treated with saline or morphine received i.c.v. injections of either vehicle or the α3β4* antagonist, AuIB (1.75 or 3.5 pmol per mouse) before naloxone (2 mg·kg−1, s.c.) challenge. As expected, naloxone precipitated a significant increase in total somatic signs in morphine-treated vehicle-injected mice [F(4,18) = 3.35, P < 0.05] while AuIB significantly reduced naloxone-precipitated somatic signs [F(4,28) = 3.058, P < 0.05)] (Figure 3A). All individual signs were attenuated by AuIB treatment (Table 4). Similarly, AT-1001, another selective α3β4* antagonist active after systemic administration, given at 1 and 3 mg·kg−1 blocked naloxone-precipitated somatic signs [F(4,27) = 112.361, P < 0.0001] in morphine-dependent mice (Figure 3B). The highest doses of AuIB (3.5 pmol) and AT-1001 (3 mg·kg−1) had no significant effect in saline-naloxone or morphine-saline-treated mice.
Figure 3.
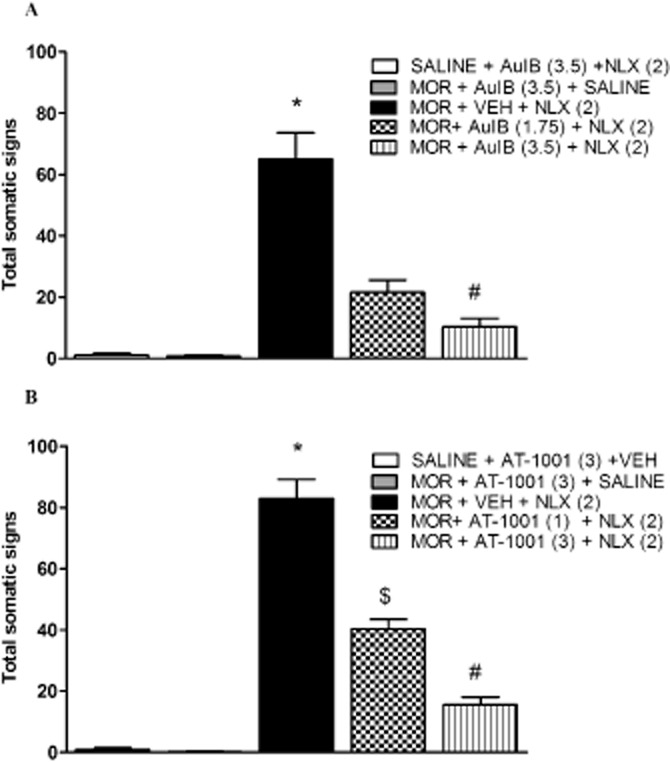
AuIB and AT-1001 reduce physical morphine withdrawal signs in C57Bl/6 mice. Morphine-dependent mice pretreated with (A) 1.75 or 3.5 pmol AuIB displayed a decrease in somatic withdrawal signs. *P < 0.05 versus all treatment groups; #P < 0.05 versus morphine (MOR) + AuIB (1.75 pmol) + naloxone (NLX) (2) group. (B) Similarly, 1 and 3 mg·kg−1 AT-1001 reduced the expression of somatic withdrawal signs. *P < 0.05 versus all treatment groups; #P < 0.05 versus MOR+ AT-1001 (1) + NLX (2) group. The total withdrawal signs measure consists of paw tremors, head shakes, backing, ptsosis, diarrhoea, jumping and other miscellaneous signs. Data are expressed as mean ± SEM. of n = 6–8 mice per group. VEH, vehicle.
Table 4.
AuIB treatment dose-dependently attenuated all individual signs in morphine somatic withdrawal
| Individual somatic signs | SAL + AuIB (3.5) + NLX (2) | MOR + AuIB (3.5) + SAL | MOR + SAL + NLX (2) | MOR + AuIB (1.75) + NLX (2) | MOR + AuIB (3.5) + NLX (2) |
|---|---|---|---|---|---|
| Paw tremors | 0 ± 0 | 0 ± 0 | 40.4 ± 5.7* | 12.0 ± 2.0# | 3.0 ± 1.1# |
| Head shakes | 1 ± 1.0 | 1.0 ± 0.5 | 4.1 ± 1.0 | 1.1 ± 0.5# | 1.6 ± 1.0# |
| Backing | 0 ± 0 | 0 ± 0 | 1.1 ± 0.4 | 0 ± 0 | 0 ± 0 |
| Ptosis | 0 ± 0 | 0 ± 0 | 1.0 ± 0.3 | 0.3 ± 0.3 | 1.0 ± 1.0 |
| Writhing | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0.1 ± 0.1 | 0.4 ± 0.4 |
| Jumping | 0 ± 0 | 0 ± 0 | 18.0 ± 3.0* | 7.7 ± 3.0# | 4.2 ± 3# |
| Body tremor | 0 ± 0 | 0 ± 0 | 1.0 ± 0.3 | 0.4 ± 0.2 | 0.4 ± 0.2 |
| Occurrence of diarrhoea (% of mice) | 0 | 0 | 100 | 43 | 14 |
Mice were made dependent on morphine for 8 days. On day 9, mice were administered naloxone (2 mg·kg−1) before observing individual physical signs expressed as mean ± SEM and % occurrence of diarrhoea.
n = 6 mice per group.
P < 0.05 from control group. Dose of AuIB is shown in pmol.
P < 0.05 from morphine + VEH + NLX (2) group.
MOR, morphine; SAL, saline; NLX, naloxone.
Role of the α4β2* nACh receptor subtype in morphine physical dependence
As a control, we examined the role of α4β2* nACh receptor inhibition in morphine withdrawal. Male mice chronically treated with saline or morphine were treated with vehicle or the β2* selective nACh receptor antagonist DHβE (3 mg·kg−1, s.c.) 5 min before naloxone (2 mg·kg−1, s.c.) challenge. Mice treated with morphine showed significant naloxone-precipitated somatic signs [F(3,20) = 25.79, P < 0.0001]. DHβE failed to significantly alter the intensity of morphine withdrawal (Figure 4A). DHβE at the dose tested (3 mg·kg−1, s.c.) had no significant effect in saline-naloxone or morphine-saline-treated mice. Furthermore, morphine withdrawal signs were not significantly altered in the α4 (Figure 4B) [F(1,22) = 0.24, P > 0.5] or the β2 (Figure 4C) [F(1,31) = 2.47, P > 0.1] nACh receptor KO mice compared with their respective WT controls. There was no significant increase in control, saline-treated, WT or KO mice challenged with naloxone. In addition, there was no significant difference in individual somatic signs between genotypes.
Figure 4.
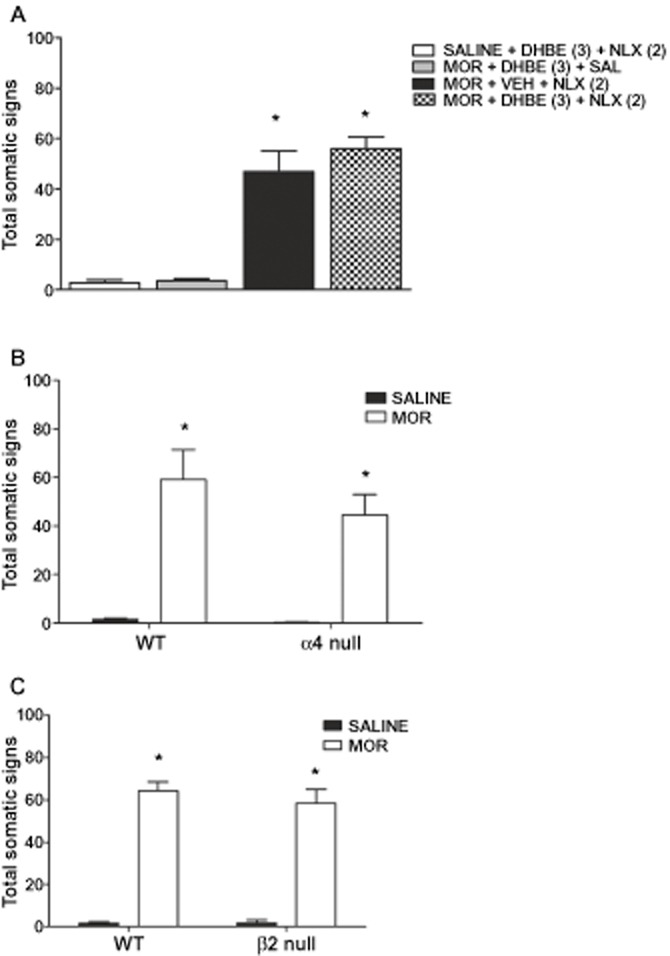
Role of the α4β2* subtype in morphine withdrawal somatic signs. Morphine-dependent C57Bl/6 mice treated with (A) vehicle or DHβE (2 mg·kg−1, s.c.) and challenged with naloxone (NLX) did not alter somatic withdrawal signs nor did (B) β2nACh receptor KO or (C) α4 nACh receptor KO compared with their respective WT littermates. *P < 0.05 versus saline and drug control. MOR, morphine; VEH, vehicle.
Variants in the 15q25 gene cluster are associated with opioid withdrawal and DSM-IV opioid dependence
Prompted by the results from animal studies, we performed association analyses using the SAGE European-American and African-American datasets to determine if variants in the 15q25 gene cluster are associated with opioid dependence based on case/control status of opioid withdrawal and DSM-IV opioid dependence. Results shown in Table 5 represent the top three most significant associations detected of the 293 SNPs analysed. The variants rs112712252 and rs190825809, both located in introns in the CHRNA3 gene, were significantly associated with a protective effect against opioid withdrawal and DSM-IV opioid dependence in both European and African-American populations. The variant rs116932868, also located in an intron in CHRNA3, was also associated with a protective effect for both phenotypes measured in European-Americans, but only for DSM-IV opioid dependence in African-Americans, though the significance in opioid withdrawal was marginal (P = 0.06). Meta-analysis also revealed a significant protective effect for these three variants in both phenotypes. All three variants survived correction for multiple testing for both phenotypes.
Table 5.
Variants in the 15q25 gene cluster are associated with opioid dependence
| SAGE EA, n = 2772 | SAGE AA, n = 1309 | Combined, n = 4081 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Phenotype | SNP | Allele | OR | P | OR | P | OR | P | Q_P |
| Withdrawal | rs112712252 | G | 0.31 | 7.3E−04 | 0.31 | 0.01 | 0.31 | 3.1E−05* | 0.99 |
| rs190825809 | C | 0.33 | 0.001 | 0.32 | 0.02 | 0.33 | 9.6E−05* | 0.98 | |
| rs116932868 | G | 0.31 | 0.001 | 0.33 | 0.06 | 0.32 | 2.0E−04* | 0.92 | |
| rs16969968 | A | 1.15 | 0.22 | 1.49 | 0.32 | 1.17 | 0.14 | 0.54 | |
| DSM-IV | rs112712252 | G | 0.29 | 1.8E−04 | 0.27 | 0.006 | 0.28 | 3.5E−06* | 0.91 |
| rs190825809 | C | 0.28 | 3.0E−04 | 0.28 | 0.01 | 0.29 | 1.1E−05* | 0.91 | |
| rs116932868 | G | 0.30 | 3.3E−04 | 0.28 | 0.03 | 0.28 | 2.4E−05* | 0.99 | |
| rs16969968 | A | 1.25 | 0.04 | 1.68 | 0.22 | 1.28 | 0.02 | 0.50 | |
Association analysis conducted in the SAGE European American (EA) and African-American (AA) datasets shows that variants in the 15q25 gene cluster are significantly associated with a protective effect against opioid dependence. Uncorrected P values are shown for individual datasets. Cochran's Q statistic P values (Q_P) show no significant heterogeneity between datasets. Significant results are bold and underlined.
Denotes results that survived the single nucleotide polymorphism spectral decomposition correction for multiple testing threshold (P < 0.0006).
Although the α5 SNP rs16969968 was not found to be among the top three most significant associations in this study, we also report the results of this variant in Table 5 due to its significant role in nicotine dependence, and to support previous findings showing an association with this variant in opioid dependence (Erlich et al., 2010; Sherva et al., 2010). The rs16969968 marker was significantly associated with risk for DSM-IV opioid dependence in the European-American population, and in the meta-analysis, though this P-value did not survive correction for multiple testing. The variant was not associated with opioid withdrawal in this study.
A LD block of the top three most significant SNPs and rs16969968 is shown in Figure 5. While rs16969968 is not in LD with the other three SNPs, rs112712252, rs190825809 and rs116932868 are in strong LD in the European-American sample (Figure 5A), African-American sample (Figure 5B) and the combined sample (Figure 5C), suggesting that these SNPs are strongly correlated to opioid dependence.
Figure 5.
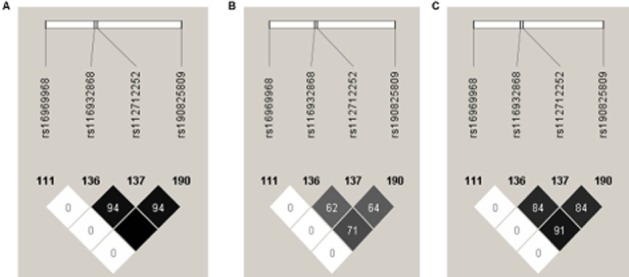
Linkage disequilibrium (LD) blocks containing r2 values for (A) the SAGE European-American dataset, (B) the SAGE African-American dataset and (C) both SAGE datasets combined. The darker the shading and/or higher numbers represent a stronger correlation between two markers.
Discussion
The goal of the present study was to elucidate the role of α3β4* nACh receptors in a mouse model of physical morphine withdrawal using multiple approaches: mouse genetics, preclinical pharmacology and human genetic association studies. Our BXD RI mouse panel data demonstrated that higher mRNA expression of α3, α5 and β4 nACh receptor subunits within the forebrain and/or midbrain is associated with higher intensity of one of the morphine withdrawal signs. This notion that α5, α3 and β4 nACh receptor subunits are involved in morphine dependence is confirmed in mice bearing extra copies of the cluster CHRNA5/A3/B4, thus mice with increased expression of α5, α3 and β4 nACh receptor subunits exhibit enhanced somatic signs of withdrawal. In agreement with this, mouse genetic KO and pharmacological studies demonstrated that the blockade of α3β4* nACh receptors reduced the expression and development of the physical signs associated with morphine withdrawal. Furthermore, the α5 nACh receptor subunit, which can co-assemble with the α3β4* nACh receptor subtype, seems to be partially mediating the somatic signs of morphine withdrawal. Lastly, association analyses in two human datasets showed that variants of the CHRNA3 gene are associated with opioid dependence and withdrawal.
Our genetic correlation analyses of morphine withdrawal phenotype data and gene expression data across the BXD RI panel demonstrate that mRNA levels of α3, α5 and β4 nACh receptor subunits in many brain regions are associated with higher magnitude of morphine somatic signs. In particular, these subunits within the forebrain, midbrain and other brain regions are associated with higher magnitude of defecation in male mice during naloxone-precipitated morphine withdrawal. Overall, the BXD RI data suggest that higher mRNA expression of α3, α5 and β4 subunits in the brain is associated with a larger withdrawal response to morphine. Moreover, mice overexpressing the cluster CHRNA5/A3/B4 with increased α3, α5 and β4 expression demonstrated enhanced morphine physical withdrawal compared with their WT counterparts.
Indeed, the reduction in morphine withdrawal signs in the α5 nACh receptor KO mice and the increase in somatic signs in the TgCHRNA5/A3/B4 mouse model suggest that the α3α5β4* nACh receptor subtype is involved in the somatic symptoms of morphine abstinence in the mouse. Interestingly, the α5 nACh receptor subunit co-assembles with α3β4* nACh receptor subtypes to form functional receptors in the periphery (ganglia) and CNS (MHb and IPN; Wada et al., 1990; Zoli et al., 1995; Quick et al., 1999; Whiteaker et al., 2002). In addition, TgCHRNA5/A3/B4 exhibit increased activation of the medial habenula upon nicotine administration (Gallego et al., 2012). Our current data does not allow us to discern whether α3β4* nACh receptors that incorporate the α5 subunit are responsible for the morphine withdrawal increase detected in TgCHRNA5/A3/B4. In nicotine somatic withdrawal signs, neuronal α3β4* nACh receptor subtypes mediate somatic signs of withdrawal independently of α5 nACh receptor subunits (Jackson et al., 2013).
Conversely, the α3β4* selective antagonist, AuIB, given centrally, blocked the expression of morphine physical signs in a dose-related manner. We believe that the doses used (1.75 or 3.5 pmol per mouse) in our current study selectively blocked the α3β4* nACh receptor subtype. Our highest dose of 3.5 pmol would yield, based on diffusion studies from an estimated tissue concentration of ≈ 0.7 μM, a value similar to the IC50 value (0.75 μM) obtained with rat α3β4 nACh receptors expressed in Xenopus oocytes (Luo et al., 1998) and the IC50 value (2.2 μM) that inhibits nicotine-induced hippocampal noradrenaline secretion (Fu et al., 1999). AuIB is 100-fold more potent at α3β4* nACh receptors compared with other heteromeric nicotinic receptor combinations and 10-fold more potent at α3β4* than at the α7 homomeric nACh receptor subtype (Luo et al., 1998). The role of α3β4* nACh receptors in morphine withdrawal is further supported by the experiments with another selective antagonist, AT-1001 (Toll et al., 2012). The pharmacological results were supported by the data obtained in β4 nACh receptor KO mice, demonstrating a significant reduction of total somatic signs compared with their WT littermates, and by the correlation results with brain α3, α5 and β4 nACh receptor subunit mRNA levels in the BXD RI mice. Our findings complement previous findings with non-selective nicotinic antagonists, mecamylamine and BTMPS (Hall et al., 2011; Taraschenko et al., 2005), and with 18-MC, a moderately selective α3β4* nACh receptor antagonist, which have been shown previously to reduce morphine physical dependence signs in rats (Taraschenko et al., 2005). The importance of the α3β4* nACh receptor subtype in morphine physical dependence is highlighted by the observation that α4β2* nACh receptors, the major heteromeric subtype expressed in the CNS, do not participate in morphine somatic withdrawal signs. Neither the selective β2* nACh receptor antagonist, DHβE, nor the β2 and α4 nACh receptor KO mice demonstrated significant alterations in somatic signs of withdrawal.
While the brain regions that mediate the blocking effect of α3β4* antagonists on morphine physical dependence were not investigated in our experiments, the limited brain distribution of α3β4* nACh receptors suggests the MHb-IPN pathway as a possible site. In line with this suggestion, local administration of the α3β4* nACh receptor antagonist, 18-MC into the MHb and IPN significantly reduces somatic morphine withdrawal signs in rats (Panchal et al., 2005). However, it is possible that other brain regions such as the ventral tegmental area, hippocampus and cortex, where α3, α5 and β4 nACh receptor subunits are also found, may be involved in morphine withdrawal (Millar and Gotti, 2009). Interestingly, Neugebauer et al. (2013) recently reported that knocking down the β4 subunits in the MHb did not reduce the number of jumps during somatic morphine withdrawal. These findings are similar to our data with the β4 KO mice, which did not differ in the number of jumps compared with their WT counterparts. In addition, our findings with BXD mice gene correlations suggest this possibility. Interestingly, our pharmacological and genetic approaches indicate that the α3β4* nACh receptor subtype mostly mediates naloxone-precipitated jumping behaviour and diarrhoea somatic signs. These two manifestations of morphine physical dependence are principally mediated by the locus coeruleus (LC; Maldonado et al., 1992), a hindbrain region where α3β4* nACh receptors are also expressed (Lena et al., 1999). Therefore, we cannot exclude their possible involvement in morphine dependence since direct infusion of 18-MC into the LC is able to attenuate most of the somatic signs associated with withdrawal (Panchal et al., 2005).
Polymorphisms in the CHRNA5/CHRNA3/CHRNB4 gene cluster were recently associated with increased risk and severity of opioid dependence (Erlich et al., 2010; Sherva et al., 2010), further supporting a role for nACh receptors in mediating aspects of morphine withdrawal. In agreement, the results of our human genetic association study implicate a protective role for the rs112712252, rs190825809 and rs116932868 variants, located within the CHRNA3 gene in opioid withdrawal and DSM-IV criteria. These results support findings from the current animal study, where α3β4* nACh receptor antagonists significantly reduced physical morphine withdrawal signs in the mouse. To our knowledge, this is the first known study to identify significant variation in the CHRNA3 gene implicated in opioid dependence phenotypes. Because the markers are in LD (i.e. the markers are strongly correlated), it is difficult to identify the causal variant in this case and we cannot rule out the possibility that these markers serve as proxies for a causal variant that was not identified in this study. Nonetheless, our findings identify protective variants in the CHRNA3 gene in opioid dependence and withdrawal. Our assessment of the rs16969968 variant, located in CHRNA5, did not survive correction for multiple testing, but for the DSM-IV phenotype in the combined sample, produced a similar odds ratio and P value similar to that observed in the Sherva et al. (2010) study, which also used DSM-IV opioid dependence as a phenotypic measure. In both studies, rs16969968 was associated with risk for opioid dependence. Overall, the human results identify alleles with reduced risk for opioid dependence in the 15q25 gene cluster. Future studies should involve haplotype analyses across the genetic region to identify the most significant marker combinations that contribute to opioid dependence phenotypes.
In summary, our findings suggest that neuronal α3β4* nACh receptors are a potential target for treating physical morphine dependence, and may be involved in mechanisms of physical drug withdrawal in general.
Acknowledgments
The authors would like to thank Tie Han for his technical assistance with the withdrawal studies. This research was supported by National Institute on Drug Abuse (grant DA032246) to M. I. D., National Institute on Alcohol Abuse and Alcoholism (grants AA017828 and AA016667) to M. F. M., National Institute of Mental Health (grant MH-020030) to K. J. J., National Institute of General Medical Sciences (GM48677 and GM103801) and NIDA (grant DA017173) and NCI (grant U19CA148127), Area 2 to M. D. B., Catalan Agency for Administration of University and Research (AGAUR2009SGR1313) and Spanish Ministry of Education and Sciences (SAF2010-16427).
The SAGE samples (PI: Laura J. Bierut) were GWAS datasets sponsored by the National Human Genome Research Institute. Funding support for the SAGE Study was provided through the NIH Genes, Environment and Health Initiative (GEI) (U01HG004422). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH [GEI (U01HG004438)], the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse and the NIH contract ‘High throughput genotyping for studying the genetic contributions to human disease’ (HHSN268200782096C). The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/about.html through dbGaP accession number phs000092.v1.p.
Glossary
- AuIB
α-conotoxin AuIB
- KO
knockout
- nACh receptors
nicotinic ACh receptors
- WT
wild-type
Authors' contributions
Participated in research design: Muldoon, Perez, Maldonado, Dierssen, De Biasi, Jackson, Chen and Damaj. Conducted experiments: Muldoon, Jackson, Perez, Molas, Harenza, Anwar and Rais. Contributed new reagents or analytic tools: McIntosh and Zaveri. Performed data analysis: Muldoon, Perez, Molas, Jackson, Chen, De Biasi and Damaj. Wrote or contributed to the writing of the manuscript: Muldoon, Jackson, Perez, Molas, Harenza, Rais, Anwar, Zaveri, Dierssen, Maskos, McIntosh, Miles, Chen, De Biasi and Damaj.
Conflict of interest
All of the authors declare no conflict of interest.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
http://dx.doi.org/10.1111/bph.12741
Table S1 Correlation of α3, α5 or β4 with somatic signs across BXD strains treated with morphine in various brain regions. Using a publically available BXD inbred mouse panel dataset (Philip et al., 2010), we assessed significant correlations between Chrna3, Chrna5 and Chrnb4 mRNA levels in several brain regions and the scores of various morphine withdrawal signs. Locomotion is defined by the number beam breaks. Horizontal is defined by the horizontal distance traveled in the open field test.
References
- Alexander SPH, Benson HE, Faccenda E, Pawson AJ, Sharman JL, Spedding M, et al. The Concise Guide to PHARMACOLOGY 2013/14: Ligand-gated ion channels. Br J Pharmacol. 2013;170:1582–1606. doi: 10.1111/bph.12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ. Nicotine dependence and genetic variation in the nicotinic receptors. Drug Alcohol Depend. 2009;104:S64–S69. doi: 10.1016/j.drugalcdep.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer KR, Foster DJ, White JM, Somogyi AA, Menelaou A, Bochner F. Steady-state pharmacokinetics and pharmacodynamics in methadone maintenance patients: comparison of those who do and do not experience withdrawal and concentration-effect relationships. Clin Pharmacol Ther. 1999;65:685–694. doi: 10.1016/S0009-9236(99)90090-5. [DOI] [PubMed] [Google Scholar]
- Erlich PM, Hoffman SN, Rukstalis M, Han JJ, Chu X, Kao WH, et al. Nicotinic acetylcholine receptor genes on chromosome 15q25.1 are associated with nicotine and opioid dependence severity. Hum Genet. 2010;128:491–499. doi: 10.1007/s00439-010-0876-6. [DOI] [PubMed] [Google Scholar]
- Fowler CD, Kenny PJ. Habenular signaling in nicotine reinforcement. Neuropsychopharmacology. 2012;37:306–307. doi: 10.1038/npp.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frahm S, Slimak MA, Ferrarese L, Santos-Torres J, Antolin-Fontes B, Auer S, et al. Aversion to nicotine is regulated by the balanced activity of β4 and α5 nicotinic receptor subunits in the medial habenula. Neuron. 2011;12:522–535. doi: 10.1016/j.neuron.2011.04.013. [DOI] [PubMed] [Google Scholar]
- Fu Y, Matta SG, McIntosh JM, Sharp BM. Inhibition of nicotine-induced hippocampal norepinephrine release in rats by alpha-conotoxins MII and AuIB microinjected into the locus coeruleus. Neurosci Lett. 1999;266:113. doi: 10.1016/s0304-3940(99)00293-1. [DOI] [PubMed] [Google Scholar]
- Gallego X, Molas S, Amador-Arjona A, Marks MJ, Robles N, Murtra P, et al. Overexpression of the CHRNA5/A3/B4 genomic cluster in mice increases the sensitivity to nicotine and modifies its reinforcing effects. Amino Acids. 2012;43:897–909. doi: 10.1007/s00726-011-1149-y. [DOI] [PubMed] [Google Scholar]
- Hall BJ, Pearson LS, Terry AV, Jr, Buccafusco JJ. The use-dependent, nicotinic antagonist BTMPS reduces the adverse consequences of morphine self-administration in rats in an abstinence model of drug seeking. Neuropharmacology. 2011;61:798–806. doi: 10.1016/j.neuropharm.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AC, Gewirtz JC. Acute opioid dependence: characterizing the early adaptations underlying drug withdrawal. Psychopharmacology (Berl) 2005;178:353–366. doi: 10.1007/s00213-005-2155-0. [DOI] [PubMed] [Google Scholar]
- Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson KJ, Marks MJ, Vann RE, Chen X, Gamage TF, Warner JA, et al. The role of α5 nicotinic acetylcholine receptors in the behavioral and pharmacological effects of nicotine in mice. J Pharmacol Exp Ther. 2010;334:137–146. doi: 10.1124/jpet.110.165738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson KJ, Sanjakdar SS, Muldoon PP, McIntosh JM, Damaj MI. The α3β4* nicotinic acetylcholine receptor subtype mediates nicotine reward and physical nicotine withdrawal signs independently of the α5 subunit in the mouse. Neuropharmacology. 2013;70:228–235. doi: 10.1016/j.neuropharm.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG. NC3Rs Reporting Guidelines Working Group. Br J Pharmacol. 2010;160:1577–1579. doi: 10.1111/j.1476-5381.2010.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman JJ, Jr, Levine B, Johnson RE, Fudala PJ, Cone EJ. Relationship of plasma buprenorphine and norbuprenorphine to withdrawal symptoms during dose induction, maintenance and withdrawal from sublingual buprenorphine. Addiction. 1998;93:549–559. doi: 10.1046/j.1360-0443.1998.93454910.x. [DOI] [PubMed] [Google Scholar]
- Lena C, de Kerchove D'Exaerde A, Cordero-Erausquin M, Le Novere N, del Mar Arroyo-Jimenez M, Changeux JP. Diversity and distribution of nicotinic acetylcholine receptors in the locus ceruleus neurons. Proc Natl Acad Sci U S A. 1999;96:12126–12131. doi: 10.1073/pnas.96.21.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Kulak JM, Cartier GE, Jacobsen RB, Yoshikami D, Olivera BM, et al. α-Conotoxin AuIB selectively blocks α3β4 nicotinic acetylcholine receptors and nicotine-evoked norepinephrine release. J Neurosci. 1998;18:8571–8579. doi: 10.1523/JNEUROSCI.18-21-08571.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Drummond G, McLachlan E, Kilkenny C, Wainwright C. Guidelines for reporting experiments involving animals: the ARRIVE guidelines. Br J Pharmacol. 2010;160:1573–1576. doi: 10.1111/j.1476-5381.2010.00873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado R, Stinus L, Gold LH, Koob GF. Role of different brain structures in the expression of the physical morphine withdrawal syndrome. J Pharmacol Exp Ther. 1992;261:669–677. [PubMed] [Google Scholar]
- Manchikanti L, Helm S, 2nd, Fellows B, Janata JW, Pampati V, Grider JS, et al. Opioid epidemic in the United States. Pain Physician. 2012;15(3 Suppl):ES9–ES38. [PubMed] [Google Scholar]
- Millar NS, Gotti C. Diversity of vertebrate nicotinic acetylcholine receptors. Neuropharmacology. 2009;56:237–246. doi: 10.1016/j.neuropharm.2008.07.041. [DOI] [PubMed] [Google Scholar]
- Neugebauer NM, Einstein EB, Lopez MB, McClure-Begley TD, Mineur YS, Picciotto MR. Morphine dependence and withdrawal induced changes in cholinergic signaling. Pharmacol Biochem Behav. 2013;109:77–83. doi: 10.1016/j.pbb.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal V, Taraschenko OD, Maisonneuve IM, Slick GD. Attenuation of morphine withdrawal signs by intracerebral administration of 18-methoxycoronaridine. Eur J Pharmacol. 2005;525:98–104. doi: 10.1016/j.ejphar.2005.09.060. [DOI] [PubMed] [Google Scholar]
- Philip VM, Duvvuru S, Gomero SB, Ansah TA, Blaha CD, Cook MN, et al. High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav. 2010;9:129–159. doi: 10.1111/j.1601-183X.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick MW, Ceballos RM, Kasten M, McIntosh MJ, Lester R. α3β4 subunit-containing nicotinic receptors dominate function in rat medial habenula neurons. Neuropharmacology. 1999;38:769–783. doi: 10.1016/s0028-3908(99)00024-6. [DOI] [PubMed] [Google Scholar]
- Rho B, Glick SD. Effects of 18-methoxycoronaridine on acute signs of morphine withdrawal in rats. Neuroreport. 1998;9:1283–1285. doi: 10.1097/00001756-199805110-00004. [DOI] [PubMed] [Google Scholar]
- Salas R, Cook KD, Bassetto L, De Biasi M. The alpha3 and beta4 nicotinic acetylcholine receptor subunits are necessary for nicotine-induced seizures and hypolocomotion in mice. Neuropharmacology. 2004a;47:401–407. doi: 10.1016/j.neuropharm.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Salas R, Pieri F, De Biasi M. Decreased signs of nicotine withdrawal in mice null for the β4 nicotinic acetylcholine receptor subunit. J Neurosci. 2004b;24:10035–10039. doi: 10.1523/JNEUROSCI.1939-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas R, Sturm R, Boulter J, De Biasi M. Nicotinic receptors in the habenulo-interpeduncular system are necessary for nicotine withdrawal in mice. J Neurosci. 2009;29:3014–3018. doi: 10.1523/JNEUROSCI.4934-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Sherva R, Kranzler HR, Yu Y, Logue MW, Poling J, Arias AJ, et al. Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology. 2010;35:1921–1931. doi: 10.1038/npp.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraschenko OD, Panchal V, Maisonneuve IM, Glick SD. Is antagonism of alpha3beta4 nicotinic receptors a strategy to reduce morphine dependence? Eur J Pharmacol. 2005;513:207–218. doi: 10.1016/j.ejphar.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Toll L, Zaveri NT, Polgar WE, Jiang F, Khroyan TV, Zhou W, et al. AT-1001: a high affinity and selective α3β4 nicotinic acetylcholine receptor antagonist blocks nicotine self-administration in rats. Neuropsychopharmacology. 2012;37:1367–1376. doi: 10.1038/npp.2011.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada E, McKinnon D, Heinemann S, Patrick J, Swanson LW. The distribution of mRNA encoded by a new member of the neuronal nicotinic acetylcholine receptor gene family (alpha 5) in the rat central nervous system. Brain Res. 1990;526:45–53. doi: 10.1016/0006-8993(90)90248-a. [DOI] [PubMed] [Google Scholar]
- Whiteaker P, Peterson CG, Xu W, McIntosh JM, Paylor R, Beaudet AL. Involvement of the alpha3 subunit in central nicotinic receptor populations. J Neurosci. 2002;22:2522–2529. doi: 10.1523/JNEUROSCI.22-07-02522.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Orr-Urtreger A, Nigro F, Gelber S, Sutcliffe CB, Armstrong D, et al. Multiorgan autonomic dysfunction in mice lacking the beta2 and the beta4 subunits of neuronal nicotinic acetylcholine receptors. J Neurosci. 1999;19:9298–9305. doi: 10.1523/JNEUROSCI.19-21-09298.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaveri N, Jiang F, Olsen C, Polgar W, Toll L. Novel α3β4 nicotinic acetylcholine receptor-selective ligands. Discovery, structure-activity studies, and pharmacological evaluation. J Med Chem. 2010;53:8187–8191. doi: 10.1021/jm1006148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoli M, Le Novère N, Hill JA, Changeux JP. Developmental regulation of nicotinic Ach receptor mRNAs in the rat central and peripheral nervous system. J Neurosci. 1995;3:1912–1939. doi: 10.1523/JNEUROSCI.15-03-01912.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Correlation of α3, α5 or β4 with somatic signs across BXD strains treated with morphine in various brain regions. Using a publically available BXD inbred mouse panel dataset (Philip et al., 2010), we assessed significant correlations between Chrna3, Chrna5 and Chrnb4 mRNA levels in several brain regions and the scores of various morphine withdrawal signs. Locomotion is defined by the number beam breaks. Horizontal is defined by the horizontal distance traveled in the open field test.