Abstract
During past decades the major impact of DNA damage on cancer as ‘disease of the genes’ has become abundantly apparent. In addition to cancer recent years have also uncovered a very strong association of DNA damage with many features of (premature) aging. The notion that DNA repair systems not only protect against cancer but equally against too fast aging has become evident from a systematic, integral analysis of a variety of mouse mutants carrying defects in e.g. transcription-coupled repair with or without an additional impairment of global genome nucleotide excision repair and the corresponding segmental premature aging syndromes in man. A striking correlation between the degree of the DNA repair deficiency and the acceleration of specific progeroid symptoms has been discovered for those repair systems that primarily protect from the cytotoxic and cytostatic effects of DNA damage. These observations are explained from the perspective of nucleotide excision repair mouse mutant and human syndromes. However, similar principles likely apply to other DNA repair pathways including interstrand crosslink repair and double strand break repair and genome maintenance systems in general, supporting the notion that DNA damage constitutes an important intermediate in the process of aging.
DNA damage and its consequences
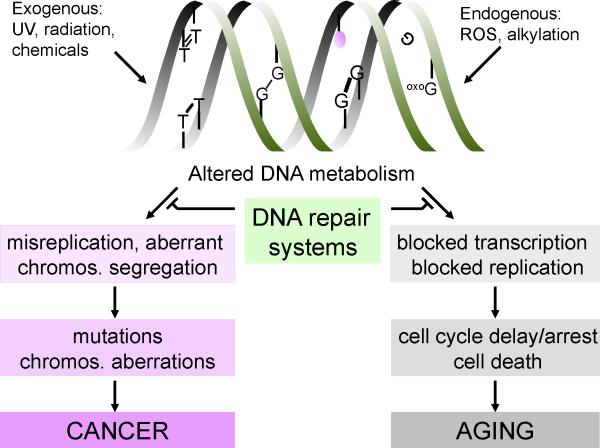
Our genome lies under continuous attack of environmental and endogenous agents and suffers from intrinsic chemical instability and aberrant products of DNA metabolism. For example, UV light gives rise to cyclobutane pyrimidine dimers (CPDs) and pyrimidine-(6,4)-pyrimidone photoproducts (6-4PPs) with mild and strong helix-distorting properties, respectively. Ionizing radiation can cause both different kinds of single and double-strand breaks in DNA and numerous types of oxidative lesions. Chemotherapeutics used in cancer therapy, and other environmental chemical agents, present for example in polluted air and tobacco smoke, induce a plethora of DNA lesions, including intra- and inter-strand cross-links and mono-adducts. In addition, endogenous agents cause a wide variety of DNA lesions. Metabolic processes including oxidative respiration lead to the formation of reactive oxygen species (ROS) and many other reactive compounds, which damage proteins, lipids and DNA. Although ROS participate in beneficial physiological processes as growth factor signal transduction [1] and elimination of foreign pathogens, they also cause wreckage to the cell's interior, including a broad spectrum of oxidative DNA lesions, such as 8-oxo-2’-deoxyguanosine (8-oxodG), thymine glycols, cyclopurines, as well as different types of single and double-strand breaks (SSBs, DSBs resp.) [2]. In addition to reactive oxygen and nitrogen species and lipid peroxidation products, also endogenous alkylating agents, estrogen and cholesterol metabolites and reactive carbonyl species may damage biomolecules within the cell, including DNA. Finally, lesions in DNA can also form without a direct damaging agent. For example, spontaneous hydrolysis or modifications of nucleotides occurs in cells, which leaves non-informative a-basic sites or altered, miscoding nucleotides (See figure 1, top part)[3]. Trapped topoisomerase intermediates may end up as protein-DNA crosslinks. In total estimates of the number of lesions that occur under normal conditions in the genome may range from 104 to 105 damages per mammalian cell, per day [3].
Figure 1.
Different sources of DNA damage and the cellular consequences
Lesions in DNA have immediate effects on cell function as well as long-term consequences (see Figure 1). For instance, DNA lesions that are misinterpreted by the replication machinery may induce permanent changes in the genetic information. These mutations, as well as other changes in DNA (rearrangements, deletions, insertions, loss of heterozygosity and numerical chromosomal aberrations) caused by DSBs or missegregation can -on the long-term- give rise to cancer or when occurring in the germ line inborn diseases. Alternatively, DNA injury may acutely interfere with transcription in all cells and with replication in proliferating cells [4-5], causing dysfunctioning and -depending on the damage load, cell type and stage of differentiation- transient or permanent cell cycle arrest or (programmed) cell death [6]. The active process of intended cell death called apoptosis may eliminate cells at risk of malignant transformation. Also cellular senescence, i.e. limited proliferative potential followed by irreversible growth arrest, can neutralize potentially malignant cells [7]. Different cell types exhibit different responses to DNA damage [8]. For instance, pluripotent embryonic stem cells lack the p53-dependent G1/S checkpoint and show increased apoptosis after treatment with several types of DNA-damaging agents compared to e.g. differentiated keratinocytes [9] [10]. On the other hand, postmitotic cells such as neurons or more differentiated somatic stem cells such as bulge stem cells of the hairshaft may prefer to stay alive albeit with DNA damage thereby avoiding the need for cell replacement as long as risk for cancer is acceptable [11]. Importantly, cellular dysfunction or depletion of proliferative capacity of cells by senescence or apoptosis can contribute to aging [12-13]. These processes lead to compromised tissue homeostasis, most likely through diminished self-renewal and/or altered tissue structure [14]. For example, cells that are lost via apoptosis might be replaced by progenitors and in time this may exhaust the regenerative capacity of a tissue. The above DNA damage dilemma, the choice between cell survival (with the risk of mutagenesis) and cell death or senescence, represents the trade-off between cancer and aging, posed by damage to the DNA.
Repair mechanisms
To counteract the deleterious effects of DNA damage, the cell is equipped with a wide variety of genome-caretaking mechanisms [15-16] (Figure 2). One important component is a transient cell cycle arrest, which can provide an extended time window for repair [17], carried out by a set of complementary DNA repair systems each specialized in a subclass of DNA lesions (reviewed by [15-16]). However, some types of DNA damage escape detection by repair and persist in DNA. A clear example is the class of UV-induced CPDs, which in rodents are overlooked in large parts of the genome. Such lesions may be bypassed during replication by a dedicated replication machinery at the risk of mutation induction. This process is called trans-lesion synthesis and involves an array of specialized polymerases each capable of synthesizing DNA opposite a specific class of damage in the template at a relatively low error rate [18-20].
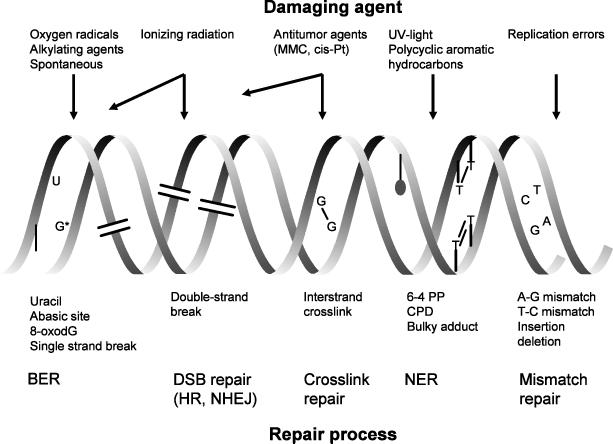
Figure 2. DNA lesions and repair mechanisms.
Top of the figure: examples of common DNA-damaging agents. Middle part: corresponding (classes of) lesions in DNA. Lower part: various repair pathways for these types of lesions.
Nucleotide excision repair discussed in more detail below, acts by a ‘cut-and-patch’ damage excision mechanism, in which damage recognition, opening of the DNA helix around the lesion, dual incision of the damage-containing strand at some distance from the lesion, gap-filling replication and final ligation of the newly synthesized piece to the pre-existing strand are the main successive steps (reviewed by [15, 21-22]). This repair mechanism can remove numerous types of helix-distorting and bulky lesions.
Base excision repair (BER) is another multi-step ‘cut-and-patch’ type damage excision mechanism that involves the concerted interplay of different proteins, targeting more subtle types of DNA injury. A battery of DNA glycosylases with partly overlapping lesion specificity recognizes and removes the altered base from the DNA, followed by incision of the damaged strand at the resulting abasic site to generate a single strand break. Subsequent steps of DNA endprocessing, short or long patch repair synthesis using the intact complementary strand as template and ligation complete the repair reaction. The different glycosylases allow BER to remove a wide variety of nucleotide modifications that only subtly alter DNA structure (reviewed by [23-24]). As BER removes base adducts from ROS, methylation, deamination, and hydroxylation, it is considered as the main guardian against DNA lesions caused by endogenous cellular metabolism.
Mismatch repair is a ‘cut-and-patch’-like excision mechanism that does not repair damaged DNA, rather it corrects replication mistakes: single base-base mismatches and small insertion/deletion loops caused by erroneous base incorporation, and slippage of DNA polymerases during replication or recombination (reviewed by [25]). Mismatch detection and removal closely follows the replication machinery and involves resynthesis of the new DNA strand from the mismatch. As such this system prevents accumulation of these mutagenic lesions and is mainly important in the prevention of cancer. This is illustrated by the fact that mutations in mismatch repair genes cause hereditary non-polyposis colorectal cancer or HNPCC [26].
DSBs are deleterious lesions that arise from ionizing radiation, free radicals, chemicals, and inhibitors of topoisomerases or are formed during replication of SSBs. Cells are equipped with at least two major DSB repair systems: homologous recombination and non-homologous end joining (NHEJ) (reviewed by [27-28]). The first uses the identical sequence of the sister chromatid, present after DNA replication as a template to repair the DSB in an error-free manner. This involves intricate DNA gymnastics to find and properly align the broken ends to the intact sequence of the sister chromatid and accurate copy in the missing sequence followed by properly disentangling the intertwined DNA strands. In contrast, NHEJ simply ligates two DNA ends together without the use of minor sequence homology and operates predominantly but not exclusively in G1 and beginning of S, when there is no identical sister chromatid. Since this process sticks both ends together it is more error-prone: it may make mistakes by loss or addition of sequences or by combining the wrong ends.
Cross-link repair is capable of removing highly toxic inter-strand cross-links, involving the numerous components of the Fanconi Anemia-BRCA pathway in, what seems to become very complex series of DNA transactions. The reaction mechanism is still poorly understood, but recently significant progress has been made using a Xenopus-based in vitro system that is capable of repairing a site-directed interstrand cross link in a plasmid sequence [29-30]. Cross-link repair seems to be mainly restricted to the S-phase of the cell cycle and to involve a series of still poorly characterized steps. First, the DNA replication machinery is arrested by the crosslink, perhaps from both sides. Subsequently, incisions are made on both sides of the crosslink in one of the strands. The lesion is unhooked and replicational bypass (which may not be error-free at the site of the unhooked lesion in the template) occurs using one or more translesion polymerases, after which the other incised strand is repaired using the newly synthesized intact strand as template for HR repair. NER may eliminate the unhooked lesion in the non-incised parental strand and copy in the missing information using the repaired complementary strand as template. Crosslink repair depends on unique repair proteins and in addition on many factors borrowed from other repair processes (NER, homologous recombination, mismatch repair and trans-lesion synthesis) [31-32] Although it is believed that the main pathway of interstrand crosslink repair occurs in S-phase, recently clear evidence has emerged for a G1 pathway as well (recent review [33]).
Nucleotide excision repair
One of the most versatile DNA repair mechanisms is nucleotide excision repair (NER). This repair system can remove a remarkably wide variety of structurally unrelated helix-distorting lesions mostly of exogenous but also of endogenous origin. For example, NER is responsible for the removal of 6-4PPs (and in humans albeit slowly CPDs) induced by ubiquitous UV light. NER can also remove bulky chemical adducts (e.g. induced by tobacco smoke), intra-strand cross-links and several forms of oxidative damage such as cyclopurines induced by ROS [15, 34].
NER is composed of two sub-pathways that differ in the way lesions are recognized: global genome NER and transcription-coupled NER (Figure 3). In global genome NER, the XPC/hHR23 complex is the primary lesion detector, that probes the entire genome for helix-distorting injuries, such as UV-induced 6-4PPs. As a consequence CPDs, that only mildly disturb the helix, are inefficiently repaired. These require the assistance of the XPE/UV-DDB complex for (slow) repair. In contrast, blockage of elongating RNA polymerase II by CPDs or other distorting damage in the template strand triggers transcription-coupled NER to quickly restore the vital process of transcription [35]. This repair system requires the CSB, CSA and many other proteins [22]. In both global genome NER and transcription-coupled NER, the unique lesion recognition step is followed by a common core repair reaction, initiated by recruitment of the ten-subunit, multi-functional transcription factor TFIIH. TFIIH contains the XPB and XPD helicases that unwind ~30bp of DNA around the lesion using their ATPase-dependent helicase activity [36]. XPA then demarcates the damage and together with the single-strand-binding protein complex RPA stabilizes the NER intermediate. Next, two endonucleases ERCC1/XPF and XPG cleave respectively 5’ and 3’ of the lesion at the transition of single to double stranded DNA in the damaged strand [37]. Finally, the 24-32 base ssDNA gap is filled by the replication machinery and the resulting nick is sealed by ligase I or ligase III [38]. In vivo, the NER complex assembles rapidly at the site of damage and eliminates it in a reaction that takes about 4 minutes [39]. Since global genome NER repairs lesions in the entire genome it is important for preventing mutations and consequently cancer. Transcription-coupled NER repairs only distorting lesions that actually block transcription. This repair process may be part of a broader transcription-coupled repair (TCR) system, that includes also repair of non-helix-distorting, transcription-stalling damage that after initial steps common with transcription-coupled NER (involving CSB, CSA, TFIIH and XPG) may proceed with the most appropriate repair pathway for the specific damage, such as BER [40]. Since TCR deals only with specific lesions that occur in the transcribed strand, it is a minor fraction of the total. Hence, TCR is not critical for preventing mutations but is essential for enabling unhindered transcription, which is crucial for cellular survival.
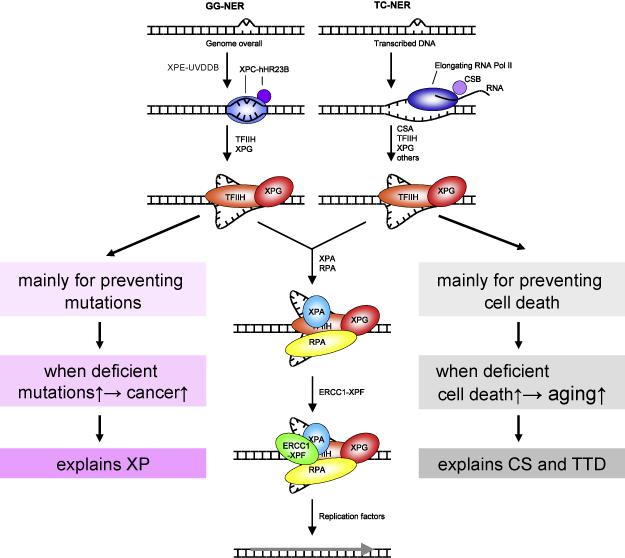
Figure 3. Mechanism of nucleotide excision repair.
Nucleotide excision repair (NER) is comprised of two subpathways, global genome NER (GG-NER) and transcription-coupled NER (TC-NER). In GG-NER, the XPC/hHR23 protein complex recognizes the helix-distorting lesion. For some less distorting lesions, overlooked by XPC/hHR23, the XPE/UV-DDB complex assists in damage detection (not indicated). When RNA polymerase II is stalled by a distorting lesion in the template strand, TC-NER is initiated by the CSB protein, later follwed by the CSA gene product. In both GG-NER and TC-NER the lesion recognition step is followed by recruitment of TFIIH. The XPB and XPD helicases from the 10-subunit TFIIH complex unwind the DNA around the lesion. The initial open complex is stabilized by XPG, XPA , which verifies the lesion and RPA which binds the opposite intact single-stranded DNA. The structure-specific endonucleases XPG and ERCC1/XPF cleave 3’ and 5’ of the lesion, respectively. The resulting 24-32 nucleotide fragment, containing the lesion, is removed and the remaining single-strand gap filled by the replication machinery and the resulting nick is sealed by ligase I or ligase III.
Repair-related disorders
A variety of rare (autosomal, mostly recessive) disorders illustrate the importance of DNA repair mechanisms and other genome caretaking processes. Many DNA repair syndromes show as one of the main characteristics an elevated cancer risk highlighting the importance of DNA repair in preventing mutations and consequently cancer. As mentioned, hereditary non-polyposis colorectal cancer, inherited in a dominant manner is caused by mutations in mismatch repair genes, in which tumour cells have lost the wild-type allele and as a consequence accumulate mutations at a dramatically elevated rate [26]. On the other hand autosomal, recessive adenomatous polyposis can be caused by biallelic mutations in MYH, a glycosylase involved in BER [41-42]. Ataxia telangiectasia, ataxia telangiectasia-like disorder and Nijmegen breakage syndrome, Bloom's syndrome are DSB-repair related disorders and all display cancer predisposition and in several cases aspects of accelerated aging as well as hypersensitivity to ionizing radiation and chromosomal instability [43]. Mutations in one of a large set of genes involved in cross-link repair underlie the cancer-prone syndrome Fanconi's anemia [32]. Inherited defects in NER cause three distinct rare, autosomal recessive disorders: xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy (reviewed by [44-45]).
Xeroderma pigmentosum
The diagnostic features of xeroderma pigmentosum (XP) are a dry scaly skin (xeroderma), abnormal pigmentation in sun-exposed skin areas (pigmentosum), striking photosensitivity, and a >1000-fold increased risk of developing UV-induced skin cancer (mainly squamous cell carcinomas and basal cell carcinomas). However, these symptoms are heterogeneous in occurrence as well as in severity among different XP patients. In addition to skin cancer predisposition, patients have a 10-20 fold increased risk of developing several types of internal cancers before the age of 20 [45].
Complementation studies based on fusion of fibroblasts from different patients have shown the involvement of at least 7 NER genes: XPA through XPG. XP-C and XP-E are specifically deficient in the global genome NER pathway [46-49] leaving the transcription-coupled NER intact. This is probably why, except for the high cancer risk, XP-C and XP-E patients generally show mild XP features. The other five complementation groups display defects in both global genome and transcription-coupled (NE)R to varying degrees. Many mutations in XP genes do not cause a complete inactivation of proteins and thus provide the cell with residual repair capacity, explaining at least in part the milder forms of XP (reviewed in [45]). Many XP patients develop almost normally but die of cancer, which reduces their average life span by approximately 30 years [50]. About 20-30% of XP patients show progressive neurological degeneration with a variable course. Features observed in these patients include diminished or absent deep tendon reflexes, high frequency hearing loss, abnormal gait, swallowing difficulties and mental retardation [51]. A minority of patients exhibit microcephaly and growth retardation [45, 52-54]. The neurological symptoms in XP are caused primarily by neuronal degeneration [55-56]. The diagnosis of XP is based on reduced levels of DNA repair synthesis after UV-irradiation of skin fibroblasts.
Cockayne syndrome and XP/CS
As with XP, patients with Cockayne syndrome (CS) display photohypersensitivity of the skin. However, CS is not associated with sun-induced pigmentation abnormalities and increased skin cancer risk probably due to fully functional global genome NER and increased apoptosis of damaged, premutagenic cells because of impaired TC-(NE)R. CS patients display severe additional features not observed in XP, including a characteristic facies with progressive features of deep sunken eyes, prominent ears and wizened appearance (birdlike face), short stature, kyphosis, arrested sexual development, caries, thin hair, cachexia and osteoporosis. Neurological symptoms include microcephaly, increased deep tendon reflexes, progressive sensorineural deafness, progressive visual loss with pigmentary retinal degeneration and involvement of the lens and cornea, progressive ataxia, delayed psychomotor development, and mental retardation [51]. Brain imaging shows absence of myelin, cerebral atrophy and prominent calcification of among others the basal ganglia [51]. The cerebral atrophy is due to degeneration of neurons, but the most striking neuropathological feature is abnormal myelination [55-56]. Many of these features resemble aging and classify CS as a segmental progeroid disease (reviewed by [45, 57]). The mean reported age of death, mainly caused by respiratory infections, resulting from a poor overall condition, is 12.5 years.
Cerebro-oculo-facio-skeletal syndrome (COFS) can be regarded as a very severe form of CS [58]. Symptoms include reduced birth weight, early microcephaly with subsequent brain atrophy, reduced white matter, patchy grey matter, hypotonia, deep-set eyes and cataract. In addition, movement is markedly decreased and joint contractures are common. Similar to XP, CS is a heterogeneous disease. In order to diagnose CS, cellular studies must show impaired UV survival, proficient global genome NER, but defective transcription-coupled NER, measured as lack of RNA synthesis recovery, i.e. re-start of transcription after a temporary UV-induced arrest. Defects in two genes, CSA and CSB, can cause CS [57]. However, a correlation between complementation group and severity of CS has not been observed. Only few COFS patients have been reported but the genes involved are CSB, XPG, XPD and ERCC1 [59] [60] [61]. In addition, a subset of mutations in XPB, XPD and XPG can lead to the combined phenotype of XP and CS including predisposition to cancer [45]. Patients with the XP/CS complex have the skin and eye phenotype as observed in XP in addition to the somatic and neurological abnormalities of CS including short stature, immature sexual development and retinal degeneration [51, 62]. The reported patients show progressive neurological degeneration.
Trichothiodystrophy and XP/TTD
A third inherited NER-deficient photosensitive disorder is trichothiodystrophy (TTD). TTD involves a broad and varying spectrum of symptoms and as a result the nomenclature can be confusing. Over the years, many syndromes, such as the syndrome of Pollitt, Tay's syndrome, Sabinas syndrome, ONMR syndrome (onychotrichodysplasia with neutropenia and mental retardation), and Amish brittle hair disease have been described that retrospectively belong to the spectrum of TTD [63-65]. PIBIDS is an acronym that is often used for the combination of symptoms displayed by TTD patients: photosensitivity, ichthyosis (scaly skin), brittle hair and nails, impaired intelligence, decreased fertility, and short stature [45, 64]. TTD encompasses PIBIDS, IBI(D)S and BI(D)S as well as SIBIDS (osteosclerosis or skeletal abnormality and IBIDS) [45, 66]. The hallmark of TTD is brittle hair with it's characteristic ‘tiger tail’ pattern (when viewed with polarised microscopy) that is dry, sparse and easily broken, as a result of the reduced amount of cysteine-rich (sulphur-rich) matrix proteins in the hair shaft [67].
A recent comprehensive review describes features of 112 patients that were included based on the presence of at least two of four abnormalities: (1) brittle hair and/or hair shaft abnormalities; (2) tiger tail banding; (3) decreased sulphur or cysteine content of hair; and (4) a DNA repair abnormality [65]. The spectrum of clinical features varied from mild disease with only hair involvement to severe disease with profound developmental defects, recurrent infections and a high mortality at a young age. The median age of these patients was 6 years (ranging from 12 weeks to 47 years of age). DNA repair abnormalities or gene defects were reported in 41 patients; 32 carried mutations in XPD, two in XPB and two in TTDA. Five showed cellular UV hypersensitivity, but no specific gene defect was determined. In addition, 6 patients had a mutation in the newly discovered TTDN1 gene, which does not correlate with a NER defect. The majority of patients exhibited hair, skin and nail abnormalities; most frequently ichthyosis (65%) and photosensitivity (42%). 86% of patients presented with developmental delay or intellectual impairment. A notably sociable or outgoing behaviour, which is also a feature in CS, was observed in 17% of patients. Further neurological abnormalities included microcephaly (50%), abnormal gait/ataxia (26%), sensorineural hearing loss (~20%). In 23% of patients neuro-imaging revealed abnormalities including dysmyelination, cerebellar atrophy, and dilated ventricles, which are similar to features found in CS. Growth abnormalities (81%) including short stature (73%) as well as facial dysmorphism (66%) and an aged or “bird-like” appearance (8%) were recorded. Ocular abnormalities were noted most frequently cataracts (29%). Infections were common (46%) and most reported deaths were related to infections. In total 38% patients showed radiographic bone abnormalities including (axial) osteosclerosis, delayed bone age, (distal) osteopenia, and kyphosis. Haematologic abnormalities as anaemia and neutropenia, sexual abnormalities like hypogonadism, cryptorchidism or delayed pubertal development, and cardiac defects including cardiomyopathy, pulmonic stenosis and ventricular septal defect were reported in respectively 21%, 14% and 7% of patients. Several of these features are shared with CS including the remarkable absence of (skin)cancer. Like CS, TTD can be categorized as a segmental premature aging syndrome.
TTD results from phenotype-specific mutations in the XPB and XPD helicase genes and TTDA, which codes for the smallest TFIIH subunit p8, or the recently discovered TTDN1 gene, with an unknown function [68-71]. As mentioned above most TTD patients have a defect in XPD. Some patients with a combined form of XP/TTD have been described [72], so far only with mutations in XPD. Intriguingly, XPD and to a lesser extent XPB mutations can thus cause a spectrum of diseases: XP, XP with additional neurological features, COFS, TTD and the combinations XP/TTD and XP/CS. The paradox of one gene causing these very different clinical phenotypes is discussed further in the next paragraph.
TFIIH mutations and disease outcome
Mutations in XP-B and XP-D can cause XP, XP/CS as well as PIBIDS [73]. How can symptoms as neurodysmyelination, early cessation of development and brittle hair originate from a repair defect, particularly in view of the notion that these features are not observed in e.g. completely NER-deficient XPA patients [73]?. The first clue came from the discovery that the repair proteins XPB and XPD are in fact subunits of TFIIH, a protein complex with an additional other function: transcription initiation [74]. Both XPB and XPD are DNA helicases unwinding DNA in the 3’-5’ and 5’-3’ direction respectively [75-76]. The TFIIH complex opens DNA around the lesion in DNA repair as well as around the promoter in initiation of basal transcription [22, 77-81]. The role of XPD in these two processes is, however, quite different. Whereas DNA repair requires the helicase activity of XPD, merely the structural presence of (even enzymatically inactive) XPD is sufficient for transcription initiation to ensure stability of the complex [55, 82-84]. In contrast, the XPB helicase activity is essential for transcription initiation [36, 55]. As a consequence many mutations in XPD result in severe repair defects while they have little effect on transcription and are thus viable [55]. Indeed, numerous different viable mutations have been detected in the XPD gene whereas only few mutations in XPB are tolerated [85-90]. Still, genotype-phenotype relationships have turned out to be exceedingly complicated in view of the multiple functions of the TFIIH complex. TFIIH consist of 10 subunits of which the XPB helicase, p62, p52, p44, p34 and p8 form a tight ‘core’ complex [71] (see also review by Egly and Coin in this article of DNA Repair). The XPD helicase interacts with p44 and thus serves as a bridge between the core and the ternary cyclin-activating-kinase (CAK) complex, consisting of CDK7, MAT1 and cyclinH [71, 91-93], which also occurs in a free form in the cell. Mutations in the XPD protein can work out differently for all of the many functions of TFIIH: initiation of RNA-pol I transcription, RNA-pol II transcription, some forms of transcription regulation, global genome NER, transcription-coupled NER and likely general TCR, that involves repair of helix-distorting and non-distorting transcription-blocking lesions. Possibly, TFIIH has implications for cell-cycle regulation as well [74, 94-99]. The multiple engagements of TFIIH provide an explanation for the extraordinary clinical heterogeneity and pleiotropic effects of inherited mutations in TFIIH subunits.
For instance, mutations causing UV-sensitive TTD are exclusively found in components of the TFIIH complex, which uniquely is implicated in basal transcription initiation. Consequently, a link between defective basal transcription and the TTD-specific features of brittle hair has been made [66, 73, 100-104]. TTD hair shafts show transverse fractures, a ‘tiger-tail’ pattern (Fig. 3) and severely damaged or absent cuticle, caused by a strong reduction in the class of ultra-high sulphur-rich matrix proteins. These proteins are made up for 30% or more of cysteine residues involved in disulfide cross-linking of keratin filaments [45]. Similarly, the cutaneous symptoms, such as acanthosis and hyperkeratosis, can be explained by reduced transcription of skin-specific, differentiation-related genes such as SPRR2, a member of the small proline-rich protein (SPRR) family expressed in the dermis [64]. SPRR2 encodes a structural component of the cornified envelope and is expressed in the final stage of terminal differentiation [64]. The TTD-specific features of hair, skin and nails can be due to XPD mutations, which alter the stability of the entire complex. When during terminal differentiation the de novo synthesis of the complex does not compensate anymore for the reduced half life, keratinocytes may run out of TFIIH before the completion of terminal differentiation. As a consequence the final genes, encoding proteins that provide strength to the hair, skin and nails by crosslinking the keratin filaments, are not sufficiently transcribed and hair and nails are brittle and skin scaly. A comparable reasoning can easily explain the observation that many TTD patients with an XPD mutation have reduced levels of β-globin in erythrocytes without a defect in a haemoglobin gene [55, 105], also due to diminished transcription of this gene at the end of the erythrocyte differentiation.
Studies of the crystal structure of the XPD protein have recently given some insight in the genotype-phenotype correlation of XPD mutations which could not be understood solely by their position along the gene as adjacent mutations can cause different diseases [106-107] [108-109] (see also article by Fuss and Tainer in this issue of DNA Repair). The XPD protein is folded into four domains, two helicase domains (HD1 and HD2), the 4FeS domain involved in DNA- and ATP-binding and the Arch domain, named by its arch-shaped conformation [106]. XP mutations are located along the ATP-binding edge of HD1 and the DNA-binding channel of HD1 and impair helicase activity essential for NER [106]. XP/CS mutations not only impair helicase activity, but also have an effect on the functional flexibility of the HD1 and HD2 domains which likely affects protein-protein interactions within the TFIIH complex as well as with other critical protein partners including XPG [106]. TTD mutations may affect helicase activity, but map to sites in all four domains expected to cause framework defects impacting TFIIH integrity [106]. These mutations are thought to result in destabilization of the interaction with p44 or the XPD structure itself and therefore TFIIH [108].
TTD mouse model
To unravel the clinical intricacies associated with TFIIH mutations several mouse models have been generated, carrying precisely the same point mutations in Xpd and Xpb as encountered in patients. Mice carrying an XPD point mutation (Arg722→Trp (R722W)) found in several TTD patients, recapitulate the human disorder to a remarkable extent and display partial defects in transcription and repair [103, 110]. In addition, it was observed that TTD mice develop premature aging features [100]. In fact, this initial observation of premature aging features was the first hint of a connection between TTD and premature aging, which was not noted before with patients. After initial normal development TTD mice acquire an ‘aged’ appearance: they show early hair greying, cachexia and kyphosis. In addition, average life span ranged from less than 12 months to somewhat over 1,5 year compared to more than 2 years for wild type littermates. Closer examination revealed patchy depigmentation of the skin which was observed earlier and more frequently than in wild type littermates and hyperplasia of sebaceous glands as observed in human aging. TTD females displayed ovarian dysfunction and appeared to lose fertility early. At 6 months of age, TTD mice showed mild normochrome anemia and an enlarged spleen. In addition, significantly reduced levels of the branced-chain amino acids (valine, leucine, and isoleucine) were found indicative of starvation, not caused by aberrant food uptake or malabsorption.
A systematic extensive analysis of the overall phenotype of TTD mice in a pure C57Bl/6 background [111] revealed three types of pathology: (1) pathology consistent with accelerated aging, (2) trichothiodystrophy-specific pathology and (3) pathology consistent with dietary restriction. The first type includes osteoporosis of the femur, kyphosis, abnormalities in liver, kidney and lymphoid tissue, aortic sarcopenia and reduced hypodermal fat. For example, the increased levels of lipofuscin pigmentation in TTD livers can be related to oxidative damage. The second type of pathology harbours anorectal prolapse, which in addition to a general conditional decline is a common cause of death in TTD mice. Furthermore, typical skin lesions as acanthosis, hyperkeratosis and sebaceous hyperplasia are present in this group. Notably, the pathology in the last type of pathology that among others includes reduced hypodermal fat, is not caused by a difference in food uptake between TTD and C57Bl/6 control mice. This dietary restriction pathology is in concordance with the observed reduced body weight in TTD mice.
Other mouse models showing premature aging
In many progeroid disorders the underlying defect is now known to be a disturbance in DNA metabolism or genome maintenance [112]. Mouse models are powerful tools in the analysis of the effect of defective genome maintenance on aging. Today, a large number of mouse mutants with defective genome maintenance that also display symptoms of accelerated aging are available [112]. Besides TTD mice, transcription-coupled repair-deficient CSB mice show several features of accelerated aging. In both TTD and CSB mice, the features of premature aging are severely enhanced by crossing them to XPA mutant mice that have a total NER defect but no overt features of premature aging. For example, TTD/XPA mice develop dramatically runted growth and extreme cachexia resulting in a severely shortened life span on average of only 3-5 weeks [100]. In addition, they show disturbed gait and spinal kyphosis. This suggests that a complete NER defect, in combination with a defect in transcription-coupled repair that renders the transcription machinery very sensitive to lesions in transcribed genes, is responsible for the markedly accelerated aging of the double mutant mice [13]. In addition, Ku80 mice with a defect in NHEJ exhibit an early onset of aging characteristics as among others a shortened life span, cancer, reduced weight, osteopenia, and alopecia. In Ercc1 and Xpf mutant mice, with a defect in NER and inter-strand cross-link repair, liver and kidney abnormalities reminiscent of accelerated aging are present in addition to reduced life span, body weight and progressive neuronal dysfunction [98]. In case of the Ercc1/Xpf mice later the corresponding human XFE syndrome was identified with many prominent features of premature aging, noted before in the mouse, strengthening the validity of mouse models for understanding the human syndromes (see also article by Gregg et al. in this issue of DNA Repair). In fact, as discussed below in the vast majority of human progeroid disorders the underlying defect is a problem in DNA metabolism which strongly supports the hypothesis that DNA damage is an important factor in the aging process.
Aging and genome maintenance
Aging is commonly defined as progressive loss of function accompanied by decreasing fertility and increasing mortality with advancing age [113]. Time-dependent accumulation of damage is generally accepted to be the cause of aging [1, 13, 113]. Already in 1957, Harman postulated the ‘free radical theory’ of aging speculating that endogenous radicals were generated in cells and resulted in a pattern of cumulative damage [1]. However, endogenously produced ROS do not only cause damage to lipids, proteins and DNA, but also function as intracellular signalling molecules [1]. So ideally there should be a balance: reducing detrimental effects of ROS while keeping oxidative metabolism and the role of ROS as intracellular signalling molecules intact. Defence mechanisms against the deleterious effects of ROS include anti-oxidant scavenging, damage removal, and repair. In addition, it has been claimed that MMR, BER, NER and DSB repair become less efficient with age leading to accumulation of mutations [114]. As discussed above, inborn defects in proteins involved in DNA repair also give rise to symptoms that resemble segmental accelerated aging [13]. As ROS are believed to result in at least 70 different types of DNA lesions [15] and the various DNA repair systems are highly specific, it is not surprising that defects in various DNA repair proteins occasionally result in segmental and not full reconstitution of aging. Further evidence supporting that premature aging is caused by defects in genome maintenance comes from additional progeroid disorders as Werner syndrome, Bloom syndrome and Rothmund-Thomson syndrome. These syndromes are all triggered by mutations in Rec-Q like DNA helicases. Rec-Q like helicases are involved in DNA repair and replication and thereby safeguard genome stability. Mutations in the genes coding for these helicases lead to accelerated aging features such as early alopecia (loss of hair), osteoporosis, malignancies, atherosclerosis, diabetes, cataracts, telangiectasia, skin atrophy and greying of hair [115]. Ataxia telangiectasia (AT) is caused by Ataxia Telangiectasia mutated (ATM) protein, which is a DNA damage sensing signalling protein kinase. Symptoms of AT include skin atrophy/sclerosis, telangiectasia, immuno-deficiencies, malignancies (mainly lymphomas), greying of hair and neurodegeneration ([13] and references therein). Mutations in the LMNA gene, which encodes for type A nuclear lamins are responsible for Hutchinson-Gilford progeria syndrome (HGPS) and atypical Werner syndrome. LMNA is a structural component of the nuclear envelope and is involved in regulating mitotic signalling pathways. Mutations in LMNA cause nuclear fragility, rendering the nucleus sensitive to mechanical stress, reduced mitotic stability, shortened telomere length and diminished DNA repair [116-117]. HGPS is known as a progeria of childhood with features including atrophy of subcutaneous fat, alopecia, short stature, premature atherosclerosis and a panel of musculoskeletal abnormalities. In conclusion, genome maintenance is not only important in preventing cancer, but has a role in preventing premature aging as well.
Acknowledgements
The authors’ work is supported by the Dutch Science Organization, Medical Sciences, the Netherlands Genomics Initiative, the European Commission (Integrated Project on DNA Repair and LifeSpan, EULSHG-CT-2007-036894), the National Institutes of Health (1PO1AG17242-02), the Dutch Cancer Society, and the International Agency for Research on Cancer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408(6809):239–47. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 2.Cadet J, et al. Oxidative damage to DNA: formation, measurement and biochemical features. Mutat Res. 2003;531(1-2):5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362(6422):709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 4.Lehmann AR, Kirk-Bell S, Mayne L. Abnormal kinetics of DNA synthesis in ultraviolet light-irradiated cells from patients with Cockayne's syndrome. Cancer Res. 1979;39(10):4237–41. [PubMed] [Google Scholar]
- 5.Mayne LV, Lehmann AR. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne's syndrome and xeroderma pigmentosum. Cancer Res. 1982;42(4):1473–8. [PubMed] [Google Scholar]
- 6.Bernstein C, et al. DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis. Mutat Res. 2002;511(2):145–78. doi: 10.1016/s1383-5742(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 7.Campisi J. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 2001;11(11):S27–31. doi: 10.1016/s0962-8924(01)02151-1. [DOI] [PubMed] [Google Scholar]
- 8.de Waard H. Genome caretaking and differentiation, in Cell Biology and Genetics. Erasmus University Rotterdam; Rotterdam: 2004. p. 141. [Google Scholar]
- 9.Aladjem MI, et al. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr Biol. 1998;8(3):145–55. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- 10.de Waard H, et al. Cell-type-specific consequences of nucleotide excision repair deficiencies: Embryonic stem cells versus fibroblasts. DNA Repair (Amst) 2008;7(10):1659–69. doi: 10.1016/j.dnarep.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Sotiropoulou PA, et al. Bcl-2 and accelerated DNA repair mediates resistance of hair follicle bulge stem cells to DNA-damage-induced cell death. Nat Cell Biol. 2010;12(6):572–82. doi: 10.1038/ncb2059. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell JR, Hoeijmakers JH, Niedernhofer LJ. Divide and conquer: nucleotide excision repair battles cancer and ageing. Curr Opin Cell Biol. 2003;15(2):232–40. doi: 10.1016/s0955-0674(03)00018-8. [DOI] [PubMed] [Google Scholar]
- 13.Hasty P, et al. Aging and genome maintenance: lessons from the mouse? Science. 2003;299(5611):1355–9. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- 14.Chen JH, Hales CN, Ozanne SE. DNA damage, cellular senescence and organismal ageing: causal or correlative? Nucleic Acids Res. 2007;35(22):7417–28. doi: 10.1093/nar/gkm681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411(6835):366–74. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 16.Friedberg EC. DNA damage and repair. Nature. 2003;421(6921):436–40. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 17.Bartek J, Lukas J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol. 2001;13(6):738–47. doi: 10.1016/s0955-0674(00)00280-5. [DOI] [PubMed] [Google Scholar]
- 18.Baynton K, Fuchs RP. Lesions in DNA: hurdles for polymerases. Trends Biochem Sci. 2000;25(2):74–9. doi: 10.1016/s0968-0004(99)01524-8. [DOI] [PubMed] [Google Scholar]
- 19.Friedberg EC, Wagner R, Radman M. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science. 2002;296(5573):1627–30. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- 20.McGowan CH. Running into problems: how cells cope with replicating damaged DNA. Mutat Res. 2003;532(1-2):75–84. doi: 10.1016/j.mrfmmm.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Aboussekhra A, et al. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80(6):859–68. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 22.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13(7):768–85. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 23.Fortini P, et al. The base excision repair: mechanisms and its relevance for cancer susceptibility. Biochimie. 2003;85(11):1053–71. doi: 10.1016/j.biochi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Krokan HE, et al. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000;476(1-2):73–7. doi: 10.1016/s0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- 25.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7(5):335–46. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 26.Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21(6):1174–9. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 27.Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22(37):5792–812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- 28.van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2001;2(3):196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- 29.Knipscheer P, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326(5960):1698–701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raschle M, et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134(6):969–80. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho TV, Scharer OD. Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ Mol Mutagen. 2010;51(6):552–66. doi: 10.1002/em.20573. [DOI] [PubMed] [Google Scholar]
- 32.Moldovan GL, D'Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–49. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muniandy PA, et al. DNA interstrand crosslink repair in mammalian cells: step by step. Crit Rev Biochem Mol Biol. 2010;45(1):23–49. doi: 10.3109/10409230903501819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith JR, Pereira-Smith OM. Replicative senescence: implications for in vivo aging and tumor suppression. Science. 1996;273(5271):63–7. doi: 10.1126/science.273.5271.63. [DOI] [PubMed] [Google Scholar]
- 35.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51(2):241–9. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 36.Coin F, Oksenych V, Egly JM. Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol Cell. 2007;26(2):245–56. doi: 10.1016/j.molcel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Staresincic L, et al. Coordination of dual incision and repair synthesis in human nucleotide excision repair. Embo J. 2009;28(8):1111–20. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moser J, et al. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol Cell. 2007;27(2):311–23. doi: 10.1016/j.molcel.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 39.Hoogstraten D, et al. Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol Cell. 2002;10(5):1163–74. doi: 10.1016/s1097-2765(02)00709-8. [DOI] [PubMed] [Google Scholar]
- 40.Banerjee D, et al. Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J Biol Chem. 2011;286(8):6006–16. doi: 10.1074/jbc.M110.198796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al-Tassan N, et al. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat Genet. 2002;30(2):227–32. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 42.Cheadle JP, Sampson JR. Exposing the MYtH about base excision repair and human inherited disease. Hum Mol Genet. 2003;12:R159–65. doi: 10.1093/hmg/ddg259. Spec No 2. [DOI] [PubMed] [Google Scholar]
- 43.Thompson LH, Schild D. Recombinational DNA repair and human disease. Mutat Res. 2002;509(1-2):49–78. doi: 10.1016/s0027-5107(02)00224-5. [DOI] [PubMed] [Google Scholar]
- 44.de Boer J, Hoeijmakers JH. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000;21(3):453–60. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- 45.Bootsma D, et al. Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. McGraw-Hill; New York: 2001. [Google Scholar]
- 46.Hwang BJ, et al. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci U S A. 1999;96(2):424–8. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang JY, et al. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol Cell. 2000;5(4):737–44. doi: 10.1016/s1097-2765(00)80252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venema J, et al. Xeroderma pigmentosum complementation group C cells remove pyrimidine dimers selectively from the transcribed strand of active genes. Mol Cell Biol. 1991;11(8):4128–34. doi: 10.1128/mcb.11.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venema J, et al. The residual repair capacity of xeroderma pigmentosum complementation group C fibroblasts is highly specific for transcriptionally active DNA. Nucleic Acids Res. 1990;18(3):443–8. doi: 10.1093/nar/18.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kraemer KH, Lee MM, Scotto J. DNA repair protects against cutaneous and internal neoplasia: evidence from xeroderma pigmentosum. Carcinogenesis. 1984;5(4):511–4. doi: 10.1093/carcin/5.4.511. [DOI] [PubMed] [Google Scholar]
- 51.Kraemer KH, et al. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship. Neuroscience. 2007;145(4):1388–96. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kraemer KH, Lee MM, Scotto J. Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch Dermatol. 1987;123(2):241–50. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- 53.Itoh M, et al. Neurodegeneration in hereditary nucleotide repair disorders. Brain Dev. 1999;21(5):326–33. doi: 10.1016/s0387-7604(99)00033-9. [DOI] [PubMed] [Google Scholar]
- 54.Brooks PJ. DNA repair in neural cells: basic science and clinical implications. Mutat Res. 2002;509(1-2):93–108. doi: 10.1016/s0027-5107(02)00222-1. [DOI] [PubMed] [Google Scholar]
- 55.Lehmann AR. The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev. 2001;15(1):15–23. doi: 10.1101/gad.859501. [DOI] [PubMed] [Google Scholar]
- 56.Brooks PJ, Cheng TF, Cooper L. Do all of the neurologic diseases in patients with DNA repair gene mutations result from the accumulation of DNA damage? DNA Repair (Amst) 2008;7(6):834–48. doi: 10.1016/j.dnarep.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nance MA, Berry SA. Cockayne syndrome: review of 140 cases. Am J Med Genet. 1992;42(1):68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- 58.Graham JM, Jr., et al. Cerebro-oculo-facio-skeletal syndrome with a nucleotide excision-repair defect and a mutated XPD gene, with prenatal diagnosis in a triplet pregnancy. Am J Hum Genet. 2001;69(2):291–300. doi: 10.1086/321295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laugel V, et al. Cerebro-oculo-facio-skeletal syndrome: three additional cases with CSB mutations, new diagnostic criteria and an approach to investigation. J Med Genet. 2008;45(9):564–71. doi: 10.1136/jmg.2007.057141. [DOI] [PubMed] [Google Scholar]
- 60.Laugel V, et al. Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum Mutat. 2010;31(2):113–26. doi: 10.1002/humu.21154. [DOI] [PubMed] [Google Scholar]
- 61.Jaspers NG, et al. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am J Hum Genet. 2007;80(3):457–66. doi: 10.1086/512486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lindenbaum Y, et al. Xeroderma pigmentosum/cockayne syndrome complex: first neuropathological study and review of eight other cases. Eur J Paediatr Neurol. 2001;5(6):225–42. doi: 10.1053/ejpn.2001.0523. [DOI] [PubMed] [Google Scholar]
- 63.McCuaig C, et al. Trichothiodystrophy associated with photosensitivity, gonadal failure, and striking osteosclerosis. J Am Acad Dermatol. 1993;28(5 Pt 2):820–6. doi: 10.1016/0190-9622(93)70109-7. [DOI] [PubMed] [Google Scholar]
- 64.Itin PH, Sarasin A, Pittelkow MR. Trichothiodystrophy: update on the sulfur-deficient brittle hair syndromes. J Am Acad Dermatol. 2001;44(6):891–920. doi: 10.1067/mjd.2001.114294. quiz 921-4. [DOI] [PubMed] [Google Scholar]
- 65.Faghri S, et al. Trichothiodystrophy: a systematic review of 112 published cases characterises a wide spectrum of clinical manifestations. J Med Genet. 2008;45(10):609–21. doi: 10.1136/jmg.2008.058743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bergmann E, Egly JM. Trichothiodystrophy, a transcription syndrome. Trends Genet. 2001;17(5):279–86. doi: 10.1016/s0168-9525(01)02280-6. [DOI] [PubMed] [Google Scholar]
- 67.Price VH, et al. Trichothiodystrophy: sulfur-deficient brittle hair as a marker for a neuroectodermal symptom complex. Arch Dermatol. 1980;116(12):1375–84. doi: 10.1001/archderm.116.12.1375. [DOI] [PubMed] [Google Scholar]
- 68.Weeda G, et al. A mutation in the XPB/ERCC3 DNA repair transcription gene, associated with trichothiodystrophy. Am J Hum Genet. 1997;60(2):320–9. [PMC free article] [PubMed] [Google Scholar]
- 69.Stefanini M, et al. Xeroderma pigmentosum (complementation group D) mutation is present in patients affected by trichothiodystrophy with photosensitivity. Hum Genet. 1986;74(2):107–12. doi: 10.1007/BF00282072. [DOI] [PubMed] [Google Scholar]
- 70.Nakabayashi K, et al. Identification of C7orf11 (TTDN1) gene mutations and genetic heterogeneity in nonphotosensitive trichothiodystrophy. Am J Hum Genet. 2005;76(3):510–6. doi: 10.1086/428141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Giglia-Mari G, et al. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat Genet. 2004;36(7):714–9. doi: 10.1038/ng1387. [DOI] [PubMed] [Google Scholar]
- 72.Broughton BC, et al. Two individuals with features of both xeroderma pigmentosum and trichothiodystrophy highlight the complexity of the clinical outcomes of mutations in the XPD gene. Hum Mol Genet. 2001;10(22):2539–47. doi: 10.1093/hmg/10.22.2539. [DOI] [PubMed] [Google Scholar]
- 73.Bootsma D, Hoeijmakers JH. DNA repair. Engagement with transcription. Nature. 1993;363(6425):114–5. doi: 10.1038/363114a0. [DOI] [PubMed] [Google Scholar]
- 74.Schaeffer L, et al. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260(5104):58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- 75.Schaeffer L, et al. The ERCC2/DNA repair protein is associated with the class II BTF2/TFIIH transcription factor. Embo J. 1994;13(10):2388–92. doi: 10.1002/j.1460-2075.1994.tb06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weber CA, et al. Molecular cloning and biological characterization of a human gene, ERCC2, that corrects the nucleotide excision repair defect in CHO UV5 cells. Mol Cell Biol. 1988;8(3):1137–46. doi: 10.1128/mcb.8.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holstege FC, van der Vliet PC, Timmers HT. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. Embo J. 1996;15(7):1666–77. [PMC free article] [PubMed] [Google Scholar]
- 78.Tirode F, et al. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell. 1999;3(1):87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 79.Evans E, et al. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. Embo J. 1997;16(21):6559–73. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Araujo SJ, et al. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 2000;14(3):349–59. [PMC free article] [PubMed] [Google Scholar]
- 81.Sugasawa K, et al. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001;15(5):507–21. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Winkler GS, et al. TFIIH with inactive XPD helicase functions in transcription initiation but is defective in DNA repair. J Biol Chem. 2000;275(6):4258–66. doi: 10.1074/jbc.275.6.4258. [DOI] [PubMed] [Google Scholar]
- 83.Feaver WJ, et al. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell. 1993;75(7):1379–87. doi: 10.1016/0092-8674(93)90624-y. [DOI] [PubMed] [Google Scholar]
- 84.Sung P, et al. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. Embo J. 1988;7(10):3263–9. doi: 10.1002/j.1460-2075.1988.tb03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Broughton BC, et al. Mutations in the xeroderma pigmentosum group D DNA repair/transcription gene in patients with trichothiodystrophy. Nat Genet. 1994;7(2):189–94. doi: 10.1038/ng0694-189. [DOI] [PubMed] [Google Scholar]
- 86.Takayama K, et al. Defects in the DNA repair and transcription gene ERCC2 in the cancer-prone disorder xeroderma pigmentosum group D. Cancer Res. 1995;55(23):5656–63. [PubMed] [Google Scholar]
- 87.Takayama K, et al. Defects in the DNA repair and transcription gene ERCC2(XPD) in trichothiodystrophy. Am J Hum Genet. 1996;58(2):263–70. [PMC free article] [PubMed] [Google Scholar]
- 88.Kobayashi T, et al. Mutations in the XPD gene leading to xeroderma pigmentosum symptoms. Hum Mutat. 1997;9(4):322–31. doi: 10.1002/(SICI)1098-1004(1997)9:4<322::AID-HUMU4>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 89.Botta E, et al. Analysis of mutations in the XPD gene in Italian patients with trichothiodystrophy: site of mutation correlates with repair deficiency, but gene dosage appears to determine clinical severity. Am J Hum Genet. 1998;63(4):1036–48. doi: 10.1086/302063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Taylor EM, et al. Xeroderma pigmentosum and trichothiodystrophy are associated with different mutations in the XPD (ERCC2) repair/transcription gene. Proc Natl Acad Sci U S A. 1997;94(16):8658–63. doi: 10.1073/pnas.94.16.8658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Drapkin R, et al. Human cyclin-dependent kinase-activating kinase exists in three distinct complexes. Proc Natl Acad Sci U S A. 1996;93(13):6488–93. doi: 10.1073/pnas.93.13.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reardon JT, et al. Isolation and characterization of two human transcription factor IIH (TFIIH)-related complexes: ERCC2/CAK and TFIIH. Proc Natl Acad Sci U S A. 1996;93(13):6482–7. doi: 10.1073/pnas.93.13.6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Egly JM. The 14th Datta Lecture. TFIIH: from transcription to clinic. FEBS Lett. 2001;498(2-3):124–8. doi: 10.1016/s0014-5793(01)02458-9. [DOI] [PubMed] [Google Scholar]
- 94.Harper JW, Elledge SJ. The role of Cdk7 in CAK function, a retro-retrospective. Genes Dev. 1998;12(3):285–9. doi: 10.1101/gad.12.3.285. [DOI] [PubMed] [Google Scholar]
- 95.Keriel A, et al. XPD mutations prevent TFIIH-dependent transactivation by nuclear receptors and phosphorylation of RARalpha. Cell. 2002;109(1):125–35. doi: 10.1016/s0092-8674(02)00692-x. [DOI] [PubMed] [Google Scholar]
- 96.Le Page F, et al. Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell. 2000;101(2):159–71. doi: 10.1016/s0092-8674(00)80827-2. [DOI] [PubMed] [Google Scholar]
- 97.Chen J, et al. Xpd/Ercc2 regulates CAK activity and mitotic progression. Nature. 2003;424(6945):228–32. doi: 10.1038/nature01746. [DOI] [PubMed] [Google Scholar]
- 98.Hoeijmakers JH. From xeroderma pigmentosum to the biological clock contributions of Dirk Bootsma to human genetics. Mutat Res. 2001;485(1):43–59. doi: 10.1016/s0921-8777(00)00079-3. [DOI] [PubMed] [Google Scholar]
- 99.Iben S, et al. TFIIH plays an essential role in RNA polymerase I transcription. Cell. 2002;109(3):297–306. doi: 10.1016/s0092-8674(02)00729-8. [DOI] [PubMed] [Google Scholar]
- 100.de Boer J, et al. Premature aging in mice deficient in DNA repair and transcription. Science. 2002;296(5571):1276–9. doi: 10.1126/science.1070174. [DOI] [PubMed] [Google Scholar]
- 101.Dubaele S, et al. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol Cell. 2003;11(6):1635–46. doi: 10.1016/s1097-2765(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 102.Vermeulen W, et al. Clinical heterogeneity within xeroderma pigmentosum associated with mutations in the DNA repair and transcription gene ERCC3. Am J Hum Genet. 1994;54(2):191–200. [PMC free article] [PubMed] [Google Scholar]
- 103.de Boer J, et al. A mouse model for the basal transcription/DNA repair syndrome trichothiodystrophy. Mol Cell. 1998;1(7):981–90. doi: 10.1016/s1097-2765(00)80098-2. [DOI] [PubMed] [Google Scholar]
- 104.Botta E, et al. Reduced level of the repair/transcription factor TFIIH in trichothiodystrophy. Hum Mol Genet. 2002;11(23):2919–28. doi: 10.1093/hmg/11.23.2919. [DOI] [PubMed] [Google Scholar]
- 105.Viprakasit V, et al. Mutations in the general transcription factor TFIIH result in beta-thalassaemia in individuals with trichothiodystrophy. Hum Mol Genet. 2001;10(24):2797–802. doi: 10.1093/hmg/10.24.2797. [DOI] [PubMed] [Google Scholar]
- 106.Fan L, et al. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133(5):789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wijnhoven SW, et al. Accelerated aging pathology in ad libitum fed Xpd(TTD) mice is accompanied by features suggestive of caloric restriction. DNA Repair (Amst) 2005;4(11):1314–24. doi: 10.1016/j.dnarep.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 108.Liu H, et al. Structure of the DNA repair helicase XPD. Cell. 2008;133(5):801–12. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wolski SC, et al. Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol. 2008;6(6):e149. doi: 10.1371/journal.pbio.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.de Boer J, et al. Mouse model for the DNA repair/basal transcription disorder trichothiodystrophy reveals cancer predisposition. Cancer Res. 1999;59(14):3489–94. [PubMed] [Google Scholar]
- 111.Wijnhoven SW, et al. Accelerated aging pathology in ad libitum fed Xpd(TTD) mice is accompanied by features suggestive of caloric restriction. DNA Repair (Amst) 2005 doi: 10.1016/j.dnarep.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 112.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361(15):1475–85. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 113.Kirkwood TB, Austad SN. Why do we age? Nature. 2000;408(6809):233–8. doi: 10.1038/35041682. [DOI] [PubMed] [Google Scholar]
- 114.Gorbunova V, et al. Changes in DNA repair during aging. Nucleic Acids Res. 200735(22):7466–74. doi: 10.1093/nar/gkm756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brosh RM, Jr., Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007;35(22):7527–44. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Martin GM, Oshima J. Lessons from human progeroid syndromes. Nature. 2000;408(6809):263–6. doi: 10.1038/35041705. [DOI] [PubMed] [Google Scholar]
- 117.Mounkes LC, Stewart CL. Aging and nuclear organization: lamins and progeria. Curr Opin Cell Biol. 2004;16(3):322–7. doi: 10.1016/j.ceb.2004.03.009. [DOI] [PubMed] [Google Scholar]