Significance
Nonalcoholic fatty liver disease (NAFLD) is a common chronic hepatic disease affecting up to 25% of subjects in the developed world, and represents a progressive illness eventually leading to liver fibrosis and cirrhosis. This study reports an extensive set of human liver lipid droplet (LD)-associated proteins and an array of proteins differentially expressed in human NAFLD. We also uncover 17β-hydroxysteroid dehydrogenase-13, a newly identified LD-associated protein, as a pathogenic molecule in the development of NAFLD. The present study provides a potential link between LD proteins and the pathogenesis of hepatic steatosis.
Keywords: lipogenesis, SCDR9, HSDI7β13
Abstract
Nonalcoholic fatty liver disease (NAFLD) is characterized by a massive accumulation of lipid droplets (LDs). The aim of this study was to determine the function of 17β-hydroxysteroid dehydrogenase-13 (17β-HSD13), one of our newly identified LD-associated proteins in human subjects with normal liver histology and simple steatosis, in NAFLD development. LDs were isolated from 21 human liver biopsies, including 9 cases with normal liver histology (group 1) and 12 cases with simple steatosis (group 2). A complete set of LD-associated proteins from three liver samples of group 1 or group 2 were determined by 2D LC-MS/MS. By comparing the LD-associated protein profiles between subjects with or without NAFLD, 54 up-regulated and 35 down-regulated LD-associated proteins were found in NAFLD patients. Among them, 17β-HSD13 represents a previously unidentified LD-associated protein with a significant up-regulation in NAFLD. Because the 17β-HSD family plays an important role in lipid metabolism, 17β-HSD13 was selected for validating the proteomic findings and exploring its role in the pathogenesis of NAFLD. Increased hepatic 17β-HSD13 and its LD surface location were confirmed in db/db (diabetic) and high-fat diet-fed mice. Adenovirus-mediated hepatic overexpression of human 17β-HSD13 induced a fatty liver phenotype in C57BL/6 mice, with a significant increase in mature sterol regulatory element-binding protein 1 and fatty acid synthase levels. The present study reports an extensive set of human liver LD proteins and an array of proteins differentially expressed in human NAFLD. We also identified 17β-HSD13 as a pathogenic protein in the development of NAFLD.
Nonalcoholic fatty liver disease (NAFLD) has become a major health concern worldwide (1). NAFLD comprises a morphological spectrum of liver lesions ranging from simple triglyceride (TG) accumulation in hepatocytes (simple steatosis) to inflammatory and hepatocellular ballooning injury (nonalcoholic steatohepatitis), eventually leading to fibrosis and cirrhosis (2). NAFLD is a well-documented risk factor for hyperlipidemia (3), type 2 diabetes (4), and hepatocarcinoma (5). Histologically, NAFLD is characterized by a significant accumulation of lipid droplets (LDs) (1), subcellular organelles consisting of a neutral lipid core covered by a monolayer of phospholipids and many associated proteins (6–9). LDs provide a temporary storage site for accumulated TGs resulting from either enhanced de novo synthesis or increased exogenous uptake of fatty acids in hepatocytes. Stored TGs in LDs are mobilized for the assembly of very low-density lipoproteins or used for peroxisomal and mitochondrial β-oxidation under tight control from multiple nuclear transcription factors (10).
An LD is now recognized as an organelle involved in lipid storage and metabolism, membrane traffic, and signal transduction (11). Increased accumulation of LDs is associated with many metabolic diseases, including obesity, hepatosteatosis, and atherosclerosis. In addition to a monolayer of phospholipids, LDs are also covered by many proteins (12), which have been considered to play an important role in the dynamic regulation of the size and lipid contents of LDs (13).
The hallmark feature of the pathogenesis of NAFLD is the accumulation of TGs in the liver, which results in large hepatic LDs (14). Although a number of studies have provided useful information about the protein composition of LDs in many different species, ranging from bacteria to mammals (12), there has yet to be a report detailing the proteome of human liver LDs. In the present study, we performed a proteomic study of human liver LDs purified from human subjects with or without simple steatosis. We also compared protein profiles of LDs of human livers with or without simple fatty liver. Among many differentially expressed proteins, 17β-hydroxysteroid dehydrogenase-13 (17β-HSD13) represents a previously unidentified human liver LD-associated protein, which is significantly up-regulated in the hepatic LDs of patients with fatty liver and enhances hepatic lipogenesis in normal mouse liver and cultured human hepatocytes.
Results
Histological and Morphological Analyses of the Livers of Human Subjects With or Without Simple Steatosis.
Histological and morphological examination showed typical hepatocellular steatosis and ballooning, without inflammation and fibrosis in patients with simple steatosis (SI Appendix, Fig. S1 A–C). Electron microscopic examination demonstrated the presence of LDs of various sizes in the cytoplasm of hepatocytes of patients with simple steatosis (SI Appendix, Fig. S1D). Clinical features of all patients are summarized in SI Appendix, Table S1.
Isolation and Characterization of LDs from Human Liver.
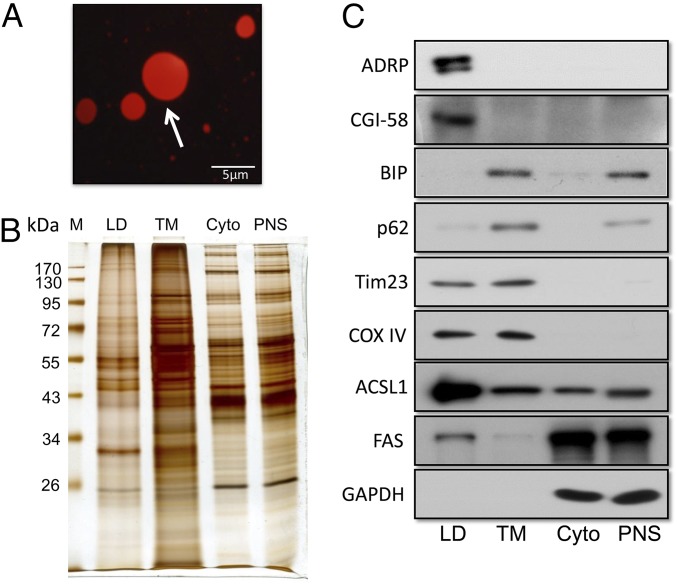
Human liver LDs were purified using a modification of a method described previously (15, 16). The quality of the preparation was first examined by fluorescence microscopy using Nile red staining (Fig. 1A). LDs stained with Nile red showed spherical structures of various sizes. Consistent with a previous report, the sizes of most LDs ranged between 1 and 3 μm, which is smaller than fat cell LDs and bigger than LDs from skeletal muscle (17). To assess the quality of the isolated LDs, equal amounts of proteins from LD, total membrane, cytosol (Cyto), and postnuclear supernatant (PNS) fractions were separated by SDS/PAGE. Silver staining revealed that the LD protein profile was quite different from the profiles of the other three fractions (Fig. 1B). To assess the purity of the isolated LDs, subcellular organelle and compartment-specific markers were tested in the four fractions, which included three LD-specific markers [adipose differentiation-related protein (ADRP), acyl-CoA synthetase long-chain family member 1 (ACSL1), and comparative gene identification-58 (CGI-58)], one cytoplasmic marker (GAPDH), two endoplasmic reticulum (ER) markers (p62 and BIP), and two mitochondrial markers (Tim23 and COX IV) (Fig. 1C). As shown in Fig. 1C, we found that all three LD-specific marker proteins were highly enriched in the LD fraction. The mitochondrial membrane proteins Tim23 and COX IV were also identified in the LD fraction. These findings indicate that the liver LD preparations were of high purity and free of contaminations by other organelles, except for mitochondria. The presence of mitochondrial proteins in the LD fraction was consistent with previous reports that LDs interact with mitochondria, mediated by SNAP 23 (18).
Fig. 1.
Purification and validation of LDs using immunoblot analysis. (A) Isolated human liver LDs were stained with Nile red and imagined by fluorescence microscope. Arrow points to a lipid droplet. (B) Silver-stained gel was used to compare the protein profiles of different subcellular fractions. (C) Proteins separated by SDS/PAGE were transferred to a PVDF membrane and immune blotted with indicated antibodies. The primary antibodies of ADRP, ACSL1, and CGI58 were used to confirm the enrichment of LDs. BIP (ER-specific marker), Tim23, and COX IV (mitochondrion-specific markers) were used to represent the indicated organelles. GAPDH was used to exclude contamination of cytosol. PNS, postnuclear supernatant; TM, total membrane.
Analysis of Liver LD Proteome of Human Subjects Without NAFLD.
To obtain a complete set of LD-associated proteins, three liver LD preparations isolated from three human subjects without NAFLD were separated using gel electrophoresis and silver staining, showing very similar protein patterns (SI Appendix, Fig. S2A). LD proteins were then analyzed by 2D LC-MS/MS. A total of 596 proteins were identified in the first preparation, 555 in the second preparation, and 501 in the third preparation (SI Appendix, Tables S2–S4). Collectively, a total of 713 proteins were found in three preparations (SI Appendix, Table S5).
To enhance the validity of the data, only proteins with two or more unique peptides and present in all three preparations were considered to be putative human liver LD proteins (SI Appendix, Fig. S2B). Overall, we identified 148 proteins, which we consider to be valid constituents of the human liver LD proteome (SI Appendix, Table S6). Among these proteins, 66 (44.6%) have been identified in LDs previously, 26 (17.6%) of which have previously been reported as possible liver LD proteins in human hepatoma cell lines and mouse livers (19, 20). The remaining 82 (55.4%) proteins are, to our knowledge, first described here in LDs.
The 148 LD-associated proteins were categorized into 10 groups based on their functional characteristics and subcellular locations. As shown in SI Appendix, Fig. S2C, the most abundant proteins were involved in lipid metabolism (23.6%, 35 proteins) (SI Appendix, Table S6). Besides fatty acid synthase and ligase, several sterol and alcohol dehydrogenases were also identified. Furthermore, apolipoprotein A-I and B-100 were detected in liver LDs, which is consistent with previous reports (17, 21). These data are in agreement with the notion that the liver is the center for sterol metabolism and alcohol detoxification, and further suggest that LDs may be involved in these processes. Another major group was cytoskeleton proteins (12.8%, 19 proteins) that were reported to be related to LD movement (22). Proteins involved in membrane trafficking also made up a notable part of all identified LD proteins (14 proteins, 9.5%). These molecules play important roles in LD fusion, fission, and interaction with other organelles (23, 24). Mitochondrial and ER proteins were also found in the LD fraction, constituting 15 (10.1%) and 13 (8.8%) proteins, respectively, indicating tight LD–mitochondrial and LD–ER interactions, as reported previously in different tissues and cell lines (23, 25, 26). Seven cell-signaling proteins (4.7%), 5 chaperones (3.4%), 5 protein translation- and degradation-related proteins (3.4%), and 31 other proteins (20.9%) were also identified in LDs, indicating that LD may have other functions as an active organelle.
Comparative LD Proteomics Between Human Subjects With or Without Simple Steatosis.
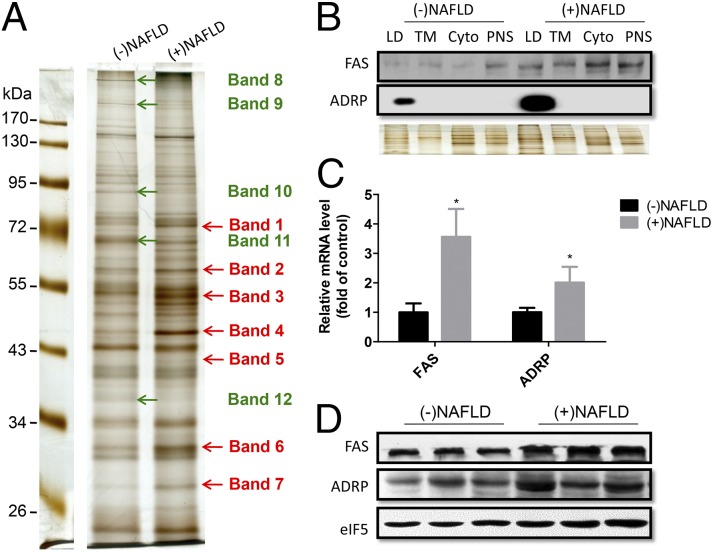
Equal amounts of protein samples pooled either from three patients with simple steatosis or three human subjects with histologically normal liver were separated by SDS/PAGE and processed using silver staining to exhibit protein profiles (Fig. 2A). Seven bands (bands 1–7) with significantly higher intensity in simple steatosis and five bands (bands 8–12) with increased intensity in normal liver were excised and processed for proteomic analysis. Of the 89 proteins isolated, 54 proteins were found to be up-regulated in NAFLD, and 35 proteins were shown to be down-regulated (SI Appendix, Table S7). Three perilipin family proteins, including perilipin, ADRP, and Tip 47 were significantly up-regulated in patients with simple steatosis, consistent with the importance of the perilipin family in the pathogenesis of fatty liver (27). Interestingly, a group of cytochrome P450 family proteins, such as cyp2E1, cyp4A11, and cyp2C9, was also found to be associated with the LDs and up-regulated in fatty liver, perhaps suggesting that these cytochrome P450 enzymes are involved in the development of NAFLD. Many mitochondrial proteins were found to be down-regulated in the LDs of patients with simple steatosis, possibly suggesting that mitochondrial dysfunction is associated with the pathogenesis of fatty liver (SI Appendix, Table S7). To confirm these findings, several proteins were selected and determined with respect to differences in their expression between patients with simple steatosis and those without simple steatosis. As shown in Fig. 2B, increased ADRP and fatty acid synthase (FAS) expression was confirmed in the LD fractions of patients with simple steatosis, although FAS was also expressed in other fractions (Fig. 2B). In addition, ADRP and FAS expression was found to be up-regulated in the fatty livers at both the mRNA level and the protein level (Fig. 2 C and D).
Fig. 2.
Identification of LD-associated proteins which show altered expression in NAFLD livers versus control livers. (A) Equal protein loads of pooled LD fractions isolated from human livers with or without NAFLD were separated by SDS/PAGE and silver stained. The arrows indicate the bands selected, excised from the gel, and processed for 2D LC-MS/MS analyses. (B) Western blot analysis confirms up-regulation of ADRP and FAS in LD fraction of NAFLD shown in SI Appendix, Table S7. Silver staining served as an even loading control. (C) Real-time PCR analysis reveals that ADRP and FAS mRNA levels were significantly up-regulated. n = 7, *P < 0.05 vs. controls. β-Actin was used as an internal control. (D) Western blot assay of ADRP and FAS protein expression levels using whole-tissue lysates of control and fatty livers. eIF5 was used as an internal control.
We then used the STRING 9.0 program to examine the association between the identified proteins and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database to define potential functional interactions among control and fatty liver LD proteins (SI Appendix, Fig. S3A). Among the 35 down-regulated proteins in fatty livers, we did not detect significant enrichment in any KEGG pathway. However, for the 54 LD proteins up-regulated in fatty liver, there was an enrichment in 4 KEGG pathways with P values <0.01 (i.e., retinol metabolism, metabolism of xenobiotics by cytochrome P450, drug metabolism, and linoleic acid metabolism) (SI Appendix, Table S8). In the retinol metabolism pathway, 11 enzymes involved were up-regulated in the LDs of fatty livers (SI Appendix, Fig. S3B).
Up-Regulation of 17β-HSD13 in the LD Fraction of Fatty Liver.
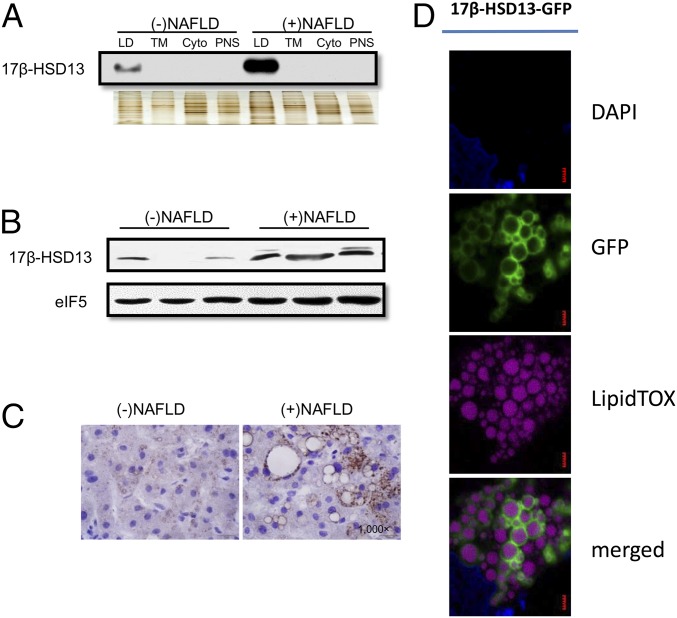
Among the many newly identified LD-associated proteins, 17β-HSD13 was found to be localized in the LD fraction of human fatty liver. Because our mass spectra results showed 17β-HSD13 to be one of the most abundant proteins in human liver LDs and the hydroxysteroid (17β) dehydrogenase family plays an important role in lipid metabolism, 17β-HSD13 was selected for further exploration of its role in the pathogenesis of NAFLD. 17β-HSD13 was highly abundant in mouse livers of both genders (SI Appendix, Fig. S4A). To confirm its association with LDs, the LD fractions from human subjects with normal and fatty liver were loaded with three other subcellular fractions with equal amount of proteins and separated by SDS/PAGE. As shown in Fig. 3A, 17β-HSD13 was exclusively present in the LD fractions and its expression markedly up-regulated in the LD fraction of fatty liver. To further confirm this finding, Western blot and immunohistochemistry assays were used to determine the expression level of 17β-HSD13 and its subcellular localization in the livers of patients with or without fatty liver. 17β-HSD13 protein expression was significantly up-regulated in fatty livers (Fig. 3B). It was mainly localized to the surface of LDs (Fig. 3C). This finding was further supported by an immunofluorescence analysis in which Huh7 cells were transfected with the 17β-HSD13–GFP plasmid. As shown in Fig. 3D, 17β-HSD13 was exclusively localized to the surfaces of LDs in Huh7 cells, supporting the conclusion that 17β-HSD13 is an LD-associated protein in human liver.
Fig. 3.
LD surface localization of 17β-HSD13 in human liver and cultured hepatocytes. (A) Western blot analysis showing that 17β-HSD13 was markedly up-regulated in LD fraction of NAFLD. Silver staining served as an even loading control. (B) Immunoblot assay using whole liver lysates demonstrating that 17β-HSD13 was significantly up-regulated in fatty livers. (C) Immunostaining of 17β-HSD13 showing that 17β-HSD13 was localized at the surface of LDs in human livers. Note: More intense staining of 17β-HSD13 in fatty liver than control liver. (D) LD surface localization of GFP-tagged 17β-HSD13 (17β-HSD13–GFP) in Huh7 cells. Green: 17β-HSD13-GFP; Mauve: LipidTOX Deep Red; Blue: DAPI. (Scale bar, 1 µm.)
Up-Regulation of 17β-HSD13 in Steatotic Liver of db/db Mice and Mice Fed with a High-Fat Diet.
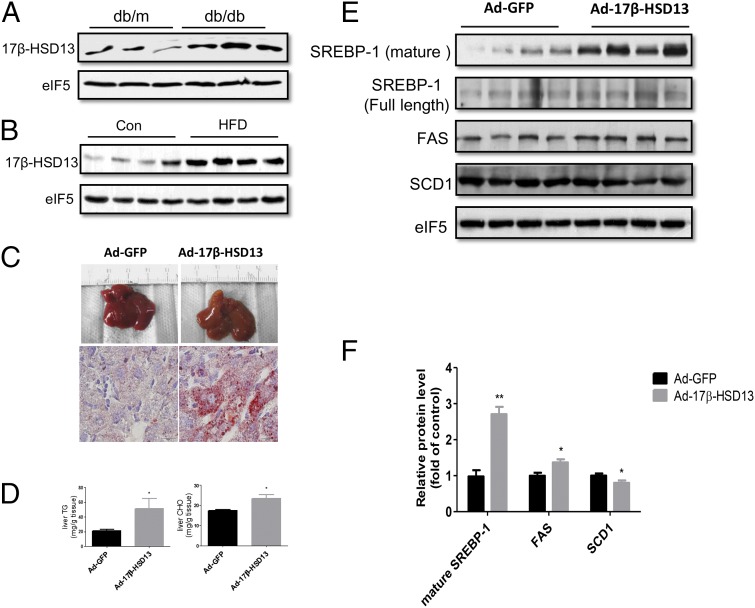
As the first step in characterizing the role of 17β-HSD13 in the pathogenesis of NAFLD, we examined its expression levels in db/db (diabetic) mice, a murine model of fatty liver and diabetes. Compared with db/m mice without fatty liver, db/db mice exhibited massive lipid accumulation in the liver as shown in our previous reports (28, 29). 17β-HSD13 expression was significantly up-regulated in the liver of db/db mice (Fig. 4A). Similarly, 17β-HSD13 expression was significantly increased in the livers of mice fed with a high-fat diet (HFD) for 3 mo (Fig. 4B). These findings suggest that 17β-HSD13 may play an important role in the pathogenesis of fatty liver in mice as well.
Fig. 4.
17β-HSD13 enhanced lipogenesis in mouse liver. (A and B) db/db mice and mice fed with a HFD, two murine fatty liver models, were used to examine 17β-HSD13 expression levels. (A) Western blot analysis of 17β-HSD13 protein expression levels in db/m and db/db mice. (B) Immunoblot analysis of 17β-HSD13 protein expression levels in normal diet and HFD-fed mice. (C) Hepatic overexpression of 17β-HSD13 via tail-vein administration of adenoviruses encoding 17β-HSD13 for 4 d resulted in a fatty liver phenotype as assessed by morphological examination (Upper) and Oil red O staining (Lower, magnification: 1,000×). Adenovirus expressing GFP (Ad-GFP) was used as a control. (D) Liver TG and CHO levels were tested 4 d after the adenovirus injection. Hepatic overexpression significantly increased liver TG and CHO contents. *P < 0.05 vs. Ad-GFP, n = 5. (E) Western blot analysis of protein involved in lipogenesis, including SREBP-1, FAS, and SCD1. (F) Quantitative analysis of 17β-HSD13 and the protein expression levels shown in C. *P < 0.05 and **P < 0.01 vs. Ad-GFP, n = 5.
17β-HSD13 Promotes Lipogenesis in Mouse Livers and Cultured Hepatocytes.
To determine the role of 17β-HSD13 in the pathogenesis of NAFLD, 17β-HSD13 was overexpressed in the livers of normal C57BL/6 mice via an adenovirus-based approach (SI Appendix, Fig. S4 B and C). As shown in Fig. 4C, hepatic overexpression of 17β-HSD13 for 4 d markedly induced neutral lipid accumulation as assessed by morphological examination and Oil red O staining. Enhanced lipogenesis was further confirmed by the finding that hepatic TG contents were significantly increased in the livers with 17β-HSD13 overexpression (Fig. 4D). However, hepatic overexpression of 17β-HSD13 had little effect on plasma TG and cholesterol (CHO) levels (SI Appendix, Fig. S4D). To elucidate the underlying mechanisms, we measured expression levels of genes involved in lipogenesis by Western blot and found that mature sterol regulatory element-binding protein 1 (SREBP-1) levels were significantly increased in mouse livers with 17β-HSD13 overexpression (Fig. 4 E and F). Consistently, expression of FAS, a target gene of SREBP-1, was also significantly up-regulated following 17β-HSD13 overexpression using adenovirus infection. However, SCD1 expression level was reduced (Fig. 4 E and F). The genes involved in fatty acid β-oxidation showed no change or a slight decrease in expression levels.
To further confirm the lipogenic effect of 17β-HSD13 in the liver, HepG2 cells were transfected with pEGFP-N1 plasmid or 17β-HSD13–EGFP fusion protein. As shown in SI Appendix, Fig. S5A, overexpression of 17β-HSD13 significantly increased the number and size of LDs. Similar changes were observed in Huh7 cells, another human hepatocyte cell line (SI Appendix, Fig. S5 B and C), suggesting that lipogenesis was markedly enhanced.
Discussion
NAFLD is becoming a major chronic liver disorder that affects at least 10% of the general population worldwide (30). Although NAFLD is considered a complicated disease resulting from the interaction of environmental factors and a susceptible polygenic background, the underlying mechanisms remain largely uncharacterized. To our knowledge, the present study has, for the first time, established a database of the LD-associated proteins of normal human livers using a proteomic approach. By comparing protein expression profiles, 89 differentially displayed proteins were identified in the LDs of NAFLD patients. Among these proteins, 17β-HSD13 was found to be a most abundant novel LD-associated protein, which is significantly up-regulated in NAFLD livers. 17β-HSD13 expression was also markedly increased in the livers of db/db mice and HFD-fed mice. Overexpression of 17β-HSD13 in the livers of C57BL/6 mice and cultured hepatocyte lines resulted in significantly increased lipogenesis and an increase in the number and size of LDs, respectively. Enhanced lipogenesis in the liver was found to be associated with increased activity of SREBP-1 and its downstream target gene FAS. Collectively, these findings suggest 17β-HSD13 as an LD-associated protein that may play an important role in the pathogenesis of NAFLD.
The present study identified 148 LD-associated proteins from three normal human livers. As expected, the perilipin family of proteins including ADRP, TIP47, and S3-12 were found in the LDs of human livers. Our study provides direct evidence that perilipin is expressed on the surface of LDs of normal human livers, which may play an important role in the pathogenesis of NAFLD. In support of this finding, the expression levels of perilipin, as well as ADRP and TIP47, have been found to be significantly up-regulated in the livers of patients with NAFLD, supporting the possibility that enhanced expression of the perilipin family members contributes to hepatic ectopic lipid accumulation and the increased number of LDs in fatty liver (31–33).
The present study also identified FAS as an LD-associated protein. FAS catalyzes the biosynthesis of saturated fatty acids and has been thought to be a soluble protein in the cytoplasm. The finding of FAS in LD proteome indicates that FAS may be associated with LDs. Increased expression of FAS in hepatic LDs of patients with NAFLD suggests that FAS may act on the surface of LDs to increase fatty acid synthesis and promote the development of NAFLD. In addition, we also found that the retinoid metabolism pathway was enhanced in the livers of patients with NAFLD. More than 90% of total body retinol is stored in liver stellate cells with unknown function. Given the pathogenic role of oxidative stress in NAFLD and the potent antioxidant effect of vitamin A, it is possible that the retinoic acid metabolism pathway may play a hepatoprotective role in the pathogenesis of NFALD (34).
Among many proteins in NAFLD, 17β-HSD13 was identified as a new LD-associated protein (20). It was first cloned from a human liver cDNA library in 2007, and originally named short-chain dehydrogenase/reductase 9 (SCDR9). In the present study, we found that 17β-HSD13 was among one of the most abundantly expressed LD proteins specifically localized on the surface of LDs, and its level was up-regulated in the livers of patients and mice with NAFLD. Overexpression of 17β-HSD13 resulted in an increase in the number and size of LDs, whereas gene silencing of 17β-HSD13 attenuated oleic acid-induced LD formation in cultured hepatocytes (data not shown). Consistently, hepatic overexpression of 17β-HSD13 in C57BL/6 mice significantly increased lipogenesis and TG contents in the livers, leading to a fatty liver phenotype possibly because of enhanced SREBP1 transcription activity. These findings demonstrate that 17β-HSD13 is a novel LD-associated protein, which may play an important role in the pathogenesis of NAFLD.
The family of 17β-HSDs consists of 14 enzymes responsible for reduction or oxidation of sex hormones, fatty acids, and bile acids in vivo (35). These enzymes differ in tissue distribution, subcellular localization, catalytic preference, and have diverse substrate specificities as they also catalyze the conversions of other substrates than steroids, as for example lipids and retinoids (36). To date, the function of 17β-HSD13 remains unknown. However, some of the 17β-HSD family members, including 17β-HSD-4, -7, -10, and -12, have been shown to participate in CHO and fatty acid metabolism, suggesting that 17β-HSD13 might also play a role in lipid metabolic pathways. A recent study reported that hepatic up-regulation of 17β-HSD13 was observed in patients with fatty liver, which is consistent with our findings and further supports a role of this enzyme in the pathogenesis of NAFLD (37).
In conclusion, we identified 148 LD-associated proteins in normal human livers. For 89 differentially expressed proteins, the PAT (perilipin, adipophilin, and TIP47) family members, FAS, and enzymes involved in retinoic acid metabolism were up-regulated in the livers of patients with NAFLD. In addition, 17β-HSD13 was identified as a novel LD-associated protein that may play an important role in the pathogenesis of NAFLD and represents a potential therapeutic target for the treatment of fatty liver.
Materials and Methods
Human Samples.
Clinical information and liver samples from patients were obtained after approval by the Ethical Committee on Human Research of the participating hospitals and with patient consent. NAFLD was diagnosed according to the guidelines of the American Association for the Study of Liver Diseases (38). Because the severity of NAFLD among the patients ranged from relatively benign simple steatosis to progressive nonalcoholic steatohepatitis and fibrosis, the liver histology of patients with NAFLD (steatosis on biopsy and compatible clinical features in the absence of an alcohol intake greater than 20 g/d) was staged according to the Brunt scoring system by a board-certified pathologist at the Department of Pathology at Peking University Health Science Center (39). Patients with other known causes of liver disease, including hepatitis B surface antigen or hepatitis C virus positivity, granulomatous liver disease, hereditary hemochromatosis, and known use of methotrexate, tamoxifen, or corticosteroids were excluded from this analysis. Liver samples were obtained from 21 patients who were admitted to Peking University First Hospital and Peking University People’s Hospital for the diagnosis and treatment of hepatic diseases. All 21 cases were subjected to liver biopsies during surgery. A portion of each liver was removed and some of it used for LD isolation. The remaining parts of the removed liver tissue were fixed in 4% (wt/vol) PFA, embedded in paraffin, or flash-frozen in liquid nitrogen for RNA and protein analyses.
Histopathologic evaluation was performed using H&E staining, Oil red O staining, and Masson staining methods. Only those patients who exhibited at least 5% steatosis, without obvious inflammation, fibrosis [stage 3 (septal/bridging)] or cirrhosis (stage 4 fibrosis) were included in the present study. All 21 cases were evaluated histologically, with 12 cases of simple steatosis and 9 without steatosis being diagnosed.
Purification of LDs.
LDs were purified using a modification of the method used by Liu et al. (15, 16). We collected liver samples from patients with or without simple fatty liver and put them into ice-cold PBS containing 0.2 mM PMSF. Then, all of the tissue samples were transferred to 10 mL buffer A (25 mM tricine, pH 7.6, 250 mM sucrose) plus 0.2 mM PMSF and homogenized with a Dounce type glass-Teflon homogenizer on ice 10 times. The PNS fraction was obtained by centrifugation of the liver homogenate at 3,000 × g for 10 min. Next, 10 mL PNS was loaded into an SW40 Ti tube and 2 mL Buffer B (20 mM Hepes, pH 7.4, 100 mM KCl, and 2 mM MgCl2) was carefully loaded on top. The samples were then centrifuged at 12,628 × g for 20 min at 4 °C. The white band containing LDs at the top of the gradients was collected in 1.5-mL EP tubes and centrifuged at 14,000 × g for 5 min. After carefully removing the underlying solution, LDs were gently resuspended in 200 μL of buffer B. This procedure was repeated four times.
In-Solution Digestion and 2D LC-MS/MS Analysis.
The LD proteomic analysis was carried out based on a previously reported method (40). Briefly, the LD protein pellet was dissolved in 20 μL of freshly prepared 8 M urea, reduced with 10 mM DTT at 56 °C for 1 h, and treated with 40 mM iodoacetamide in the dark for 45 min to block the sulfhydryl groups. Trypsin was added to a ratio of 1:10 (1:20–40 for silver staining) relative to total protein content and the sample was incubated at 37 °C overnight. After 12 h, FA (Formic acid) was added to end the digestion. The sample was dried by speed-vacuum. Additional FA was added to dissolve the sample. The tryptic peptide mixtures were analyzed using a 2D-HPLC system coupled to a linear ion trap mass spectrometer LTQ ion trap (Thermo Fishier Scientific). MS/MS data were analyzed using the SEQUEST algorithm (v2.8) against the National Center for Biotechnology Information Refseq human database that was released on Jan. 3, 2011. The database was reversed and attached to estimate the false-discovery rate. All searches were performed using a precursor mass tolerance of 3 Da calculated using average isotopic masses. The SEQUEST outputs were then analyzed using the commercial software BioWorks (Rev.3.3.1) from Thermo Electron. The filter settings for peptides were as follows: XCorr ≥ 1.5 (+1 ions), 2.0 (+2 ions), 2.5 (+3 ions); Delta CN ≥ 0.08; Sp ≥ 500; Rsp ≤ 5.
Animal Studies.
Eight-week-old male C57BL/6 mice (Jackson Laboratory) were used to determine the tissue distribution of 17β-HSD13 and for hepatic overexpression of 17β-HSD13 via tail-vein injection of adenoviruses expressing 17β-HSD13 with the dose of 1.0 × 109 pfu per mouse. C57BL/6 mice fed on a control or a HFD [Diet #MD45%fat and #MD10%fat (Medicience)] for 3 mo were used to determine 17β-HSD13 expression in fatty liver (41). Eight- to 12-wk-old male insulin-resistant db/db mice on a C57BKS background and age- and sex-matched db/m mice were used to determine 17β-HSD13 expression in the livers. All procedures were approved by the Institutional Animal Care and Use Committee of Peking University Health Science Center.
Measurement of Hepatic and Serum TG and CHO Content.
Total TG and CHO were extracted from mouse liver and quantitated by use of TG and CHO assay kits (42). Serum samples were obtained from mice for TG and CHO measurements.
Immunohistochemistry.
The immunohistochemistry study was performed as previously described (43). For 17β-HSD13 immunohistochemistry, paraffin-embedded sections were deparaffinized, rehydrated, blocked with 3% H2O2 in H2O for 8 min, and incubated with 1% BSA in PBS for 20 min. The sections were then incubated overnight at 4 °C with a rabbit anti-human 17β-HSD13 primary antibody (Abcam, ab122036) with a dilution ratio of 1:200 in 1% BSA. After rinsing three times for 3 min in PBS, HRP-conjugated secondary antibody (goat anti-rabbit IgG, pv-6001; ZSGB-Bio) was applied for 20 min at room temperature. The samples were then treated with a DAB chromogen reagent (pv-6001) until the sections were developed. Hematoxylin was used to stain the nucleus. Images were captured in an Olympus microscope.
Construction of a Human 17β-HSD13-GFP Fusion Protein.
A full-length 17β-HSD13 cDNA was amplified by PCR using total human liver cDNA and the primers 5′-atgaacatcatcctagaaatcc-3′ (with XhoI site) and 5′-tcaatggtgatggtgatgatgtttcattttgattttgt-3′(with KpnI site) and cloned into the pEGFP-N1 plasmid, which was predigested with XhoI and KpnI. The resulting plasmid was named the17β-HSD13-GFP plasmid.
Confocal Microscopy.
Using the Lipofectamine 2000 transfection reagent (Invitrogen), cultivated Huh7 cells were transfected with the 17β-HSD13–GFP plasmid for 24 h before immunofluorescence analysis. Cells were fixed in 4% PFA, permeabilized with 0.1% Triton X-100, and labeled with primary antibodies. Nile red (Sigma Chemical) was then added in a 1/1,000 (vol/vol) ratio and incubated for an additional 15 min before being mounted with coverslips.
Construction of an Adenovirus Containing a Full-Length Human 17β-HSD13.
A recombinant adenovirus driving the expression of human 17β-HSD13 was constructed using a modified form of the method used by Luo et al. (29). A full-length 17β-HSD13 cDNA fragment was amplified by PCR and subcloned into a pAdTrack-CMV shuttle vector. The PmeI-digested vector was used for transformation into AdEasy BJ5183 cells. Correct recombination of the resulting viral vector was confirmed by restriction digestions. Finally, the PacI-digested viral DNA was transfected into human embryonic kidney 293 cells for virus (Ad-17β13) production and amplification.
Transmission Electron Microscopy.
Human liver samples were examined with transmission electron microscopy using ultrathin sectioning. The liver sample was prefixed in 2.5% (wt/vol) glutaraldehyde in PBS (pH 7.4) for 2 d at 4 °C. Dehydration was carried out in an ascending concentration series of ethanol at room temperature. Then samples were embedded in Spurr’s resin. Ultrathin sections were viewed in an electron microscope (Japan, JEM-1230).
Bioinformatics Analysis.
STRING 9.0 (http://string.embl.de/), Gene ontology database (http://geneontology.org/), and KEGG (www.genome.jp/kegg) were used for protein interaction, biological function, and pathway analysis, respectively.
Statistical Analysis.
Quantitative variables are expressed as mean and SD. Comparisons between groups were performed using Pearson χ2, Wilcoxon or Student t tests, as appropriate. Statistical analyses were performed using Prism 5.0.
Supplementary Material
Acknowledgments
We thank Dr. Qing Xu from China Rehabilitation Research Center, Dr. Weihua Zhu from Peking University People’s Hospital, and Dr. Yujian Niu from the Institute of Liver Transplantation of the 301 Hospital for helping to collect liver samples; and T. Guan for his assistance in editing the manuscript. This work was supported by Grants 2012CB517504, 2011CBA009000, 2010CB912500, and 2011ZX09102-011-12 from the Ministry of Science and Technology; Grants 81030003, 81390351, 81121061, 81270932, 61273228, 81270275, and 81200511 from the Natural Science Foundation; and Grant E-0004 from the Robert A. Welch Foundation (to J.-Å.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410741111/-/DCSupplemental.
References
- 1.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: Old questions and new insights. Science. 2011;332(6037):1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qureshi K, Abrams GA. Metabolic liver disease of obesity and role of adipose tissue in the pathogenesis of nonalcoholic fatty liver disease. World J Gastroenterol. 2007;13(26):3540–3553. doi: 10.3748/wjg.v13.i26.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrence HR, et al. Novel and potent 17beta-hydroxysteroid dehydrogenase type 1 inhibitors. J Med Chem. 2005;48(8):2759–2762. doi: 10.1021/jm049045r. [DOI] [PubMed] [Google Scholar]
- 4.Bulla GA, et al. Genome-wide analysis of hepatic gene silencing in hepatoma cell variants. Genomics. 2012;100(3):176–183. doi: 10.1016/j.ygeno.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Chang BH, et al. Protection against fatty liver but normal adipogenesis in mice lacking adipose differentiation-related protein. Mol Cell Biol. 2006;26(3):1063–1076. doi: 10.1128/MCB.26.3.1063-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin S, Parton RG. Lipid droplets: A unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7(5):373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 7.Murphy DJ. The dynamic roles of intracellular lipid droplets: From archaea to mammals. Protoplasma. 2012;249(3):541–585. doi: 10.1007/s00709-011-0329-7. [DOI] [PubMed] [Google Scholar]
- 8.Thiam AR, Farese RV, Jr, Walther TC. The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol. 2013;14(12):775–786. doi: 10.1038/nrm3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farese RV, Jr, Walther TC. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139(5):855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, Robert A, Luu-The V. Cloning and characterization of human form 2 type 7 17beta-hydroxysteroid dehydrogenase, a primarily 3beta-keto reductase and estrogen activating and androgen inactivating enzyme. J Steroid Biochem Mol Biol. 2005;94(1-3):173–179. doi: 10.1016/j.jsbmb.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 11.Beckman M. Cell biology. Great balls of fat. Science. 2006;311(5765):1232–1234. doi: 10.1126/science.311.5765.1232. [DOI] [PubMed] [Google Scholar]
- 12.Yang L, et al. The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J Lipid Res. 2012;53(7):1245–1253. doi: 10.1194/jlr.R024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLauchlan J. Lipid droplets and hepatitis C virus infection. Biochim Biophys Acta. 2009;1791(6):552–559. doi: 10.1016/j.bbalip.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Adams LA, Lindor KD. Nonalcoholic fatty liver disease. Ann Epidemiol. 2007;17(11):863–869. doi: 10.1016/j.annepidem.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 15.Liu P, et al. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem. 2004;279(5):3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- 16.Ding Y, et al. Isolating lipid droplets from multiple species. Nat Protoc. 2013;8(1):43–51. doi: 10.1038/nprot.2012.142. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, et al. Proteome of skeletal muscle lipid droplet reveals association with mitochondria and apolipoprotein a-I. J Proteome Res. 2011;10(10):4757–4768. doi: 10.1021/pr200553c. [DOI] [PubMed] [Google Scholar]
- 18.Jägerström S, et al. Lipid droplets interact with mitochondria using SNAP23. Cell Biol Int. 2009;33(9):934–940. doi: 10.1016/j.cellbi.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Wu CC, Howell KE, Neville MC, Yates JR, 3rd, McManaman JL. Proteomics reveal a link between the endoplasmic reticulum and lipid secretory mechanisms in mammary epithelial cells. Electrophoresis. 2000;21(16):3470–3482. doi: 10.1002/1522-2683(20001001)21:16<3470::AID-ELPS3470>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 20.Fujimoto Y, et al. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim Biophys Acta. 2004;1644(1):47–59. doi: 10.1016/j.bbamcr.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Ohsaki Y, Cheng J, Fujita A, Tokumoto T, Fujimoto T. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol Biol Cell. 2006;17(6):2674–2683. doi: 10.1091/mbc.E05-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welte MA, Gross SP, Postner M, Block SM, Wieschaus EF. Developmental regulation of vesicle transport in Drosophila embryos: Forces and kinetics. Cell. 1998;92(4):547–557. doi: 10.1016/s0092-8674(00)80947-2. [DOI] [PubMed] [Google Scholar]
- 23.Zehmer JK, et al. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 2009;9(4):914–921. doi: 10.1002/pmic.200800584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boström P, et al. SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat Cell Biol. 2007;9(11):1286–1293. doi: 10.1038/ncb1648. [DOI] [PubMed] [Google Scholar]
- 25.Ohsaki Y, Cheng J, Suzuki M, Fujita A, Fujimoto T. Lipid droplets are arrested in the ER membrane by tight binding of lipidated apolipoprotein B-100. J Cell Sci. 2008;121(Pt 14):2415–2422. doi: 10.1242/jcs.025452. [DOI] [PubMed] [Google Scholar]
- 26.Ozeki S, et al. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J Cell Sci. 2005;118(Pt 12):2601–2611. doi: 10.1242/jcs.02401. [DOI] [PubMed] [Google Scholar]
- 27.Wolins NE, Rubin B, Brasaemle DL. TIP47 associates with lipid droplets. J Biol Chem. 2001;276(7):5101–5108. doi: 10.1074/jbc.M006775200. [DOI] [PubMed] [Google Scholar]
- 28.Huxley R. Asia Pacific Cohort Studies Collaboration The impact of modifiable risk factors on mortality from prostate cancer in populations of the Asia-Pacific region. Asian Pac J Cancer Prev. 2007;8(2):199–205. [PubMed] [Google Scholar]
- 29.Luo, et al. A protocol for rapid generation of recombinant adenoviruses using AdEasy system. Nat Protoc. 2007;2(5):1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 30.Nagayoshi Y, et al. Characterization of 17beta-hydroxysteroid dehydrogenase type 4 in human ovarian surface epithelial cells. Mol Hum Reprod. 2005;11(9):615–621. doi: 10.1093/molehr/gah215. [DOI] [PubMed] [Google Scholar]
- 31.Kengne AP, et al. Asia Pacific Cohort Studies Collaboration Systolic blood pressure, diabetes and the risk of cardiovascular diseases in the Asia-Pacific region. J Hypertens. 2007;25(6):1205–1213. doi: 10.1097/HJH.0b013e3280dce59e. [DOI] [PubMed] [Google Scholar]
- 32.Straub BK, Stoeffel P, Heid H, Zimbelmann R, Schirmacher P. Differential pattern of lipid droplet-associated proteins and de novo perilipin expression in hepatocyte steatogenesis. Hepatology. 2008;47(6):1936–1946. doi: 10.1002/hep.22268. [DOI] [PubMed] [Google Scholar]
- 33.Yim J. Science in the Orient. An interview with Jeongbin Yim, President of the Asia-Pacific International Molecular Biology Network, Seoul, South Korea. Interview by Holger Breithaupt and Samuel Caddick. EMBO Rep. 2007;8(7):622–625. doi: 10.1038/sj.embor.7401024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu ZJ, Lee WJ, Zhu BT. Selective insensitivity of ZR-75-1 human breast cancer cells to 2-methoxyestradiol: Evidence for type II 17beta-hydroxysteroid dehydrogenase as the underlying cause. Cancer Res. 2005;65(13):5802–5811. doi: 10.1158/0008-5472.CAN-04-3714. [DOI] [PubMed] [Google Scholar]
- 35.Moeller G, Adamski J. Integrated view on 17beta-hydroxysteroid dehydrogenases. Mol Cell Endocrinol. 2009;301(1–2):7–19. doi: 10.1016/j.mce.2008.10.040. [DOI] [PubMed] [Google Scholar]
- 36.Marchais-Oberwinkler S, et al. 17β-Hydroxysteroid dehydrogenases (17β-HSDs) as therapeutic targets: Protein structures, functions, and recent progress in inhibitor development. J Steroid Biochem Mol Biol. 2011;125(1-2):66–82. doi: 10.1016/j.jsbmb.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 37.Kampf C, et al. The human liver-specific proteome defined by transcriptomics and antibody-based profiling. FASEB J. 2014;28(7):2901–2914. doi: 10.1096/fj.14-250555. [DOI] [PubMed] [Google Scholar]
- 38.Tchédam Ngatcha B, Luu-The V, Labrie F, Poirier D. Androsterone 3alpha-ether-3beta-substituted and androsterone 3beta-substituted derivatives as inhibitors of type 3 17beta-hydroxysteroid dehydrogenase: Chemical synthesis and structure-activity relationship. J Med Chem. 2005;48(16):5257–5268. doi: 10.1021/jm058179h. [DOI] [PubMed] [Google Scholar]
- 39.Brunt EM, Janney CG, Di Bisceglie AM, Neuschwander-Tetri BA, Bacon BR. Nonalcoholic steatohepatitis: A proposal for grading and staging the histological lesions. Am J Gastroenterol. 1999;94(9):2467–2474. doi: 10.1111/j.1572-0241.1999.01377.x. [DOI] [PubMed] [Google Scholar]
- 40.Brozic P, Golob B, Gomboc N, Rizner TL, Gobec S. Cinnamic acids as new inhibitors of 17beta-hydroxysteroid dehydrogenase type 5 (AKR1C3) Mol Cell Endocrinol. 2006;248(1-2):233–235. doi: 10.1016/j.mce.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 41.Farrell GC. New hepatitis C guidelines for the Asia-Pacific region: APASL consensus statements on the diagnosis, management and treatment of hepatitis C virus infection. J Gastroenterol Hepatol. 2007;22(5):607–610. doi: 10.1111/j.1440-1746.2007.04969.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang Q, et al. Abrogation of hepatic ATP-citrate lyase protects against fatty liver and ameliorates hyperglycemia in leptin receptor-deficient mice. Hepatology. 2009;49(4):1166–1175. doi: 10.1002/hep.22774. [DOI] [PubMed] [Google Scholar]
- 43.Wu CC, Yates JR, 3rd, Neville MC, Howell KE. Proteomic analysis of two functional states of the Golgi complex in mammary epithelial cells. Traffic. 2000;1(10):769–782. doi: 10.1034/j.1600-0854.2000.011004.x. [DOI] [PubMed] [Google Scholar]