Significance
Obligate aerobes require survival strategies to persist in temporarily oxygen-deprived environments. In this article, we reveal a previously unidentified survival mechanism for obligately aerobic bacteria. Under oxygen-limiting conditions, the saprophytic actinomycete Mycobacterium smegmatis can rapidly switch between fermentative hydrogen production and hydrogen oxidation coupled to either oxygen or fumarate reduction depending on electron acceptor availability. To our knowledge, these results demonstrate for the first time (i) hydrogen production in an obligate aerobe, (ii) the unambiguous confirmation of fermentation in a mycobacterium and (iii) strong evidence that hydrogen has a role in survival and not just growth.
Abstract
Oxygen availability is a major factor and evolutionary force determining the metabolic strategy of bacteria colonizing an environmental niche. In the soil, conditions can switch rapidly between oxia and anoxia, forcing soil bacteria to remodel their energy metabolism accordingly. Mycobacterium is a dominant genus in the soil, and all its species are obligate aerobes. Here we show that an obligate aerobe, the soil actinomycete Mycobacterium smegmatis, adopts an anaerobe-type strategy by activating fermentative hydrogen production to adapt to hypoxia. This process is controlled by the two-component system DosR-DosS/DosT, an oxygen and redox sensor that is well conserved in mycobacteria. We show that DosR tightly regulates the two [NiFe]-hydrogenases: Hyd3 (MSMEG_3931-3928) and Hyd2 (MSMEG_2719-2718). Using genetic manipulation and high-sensitivity GC, we demonstrate that Hyd3 facilitates the evolution of H2 when oxygen is depleted. Combined activity of Hyd2 and Hyd3 was necessary to maintain an optimal NAD+/NADH ratio and enhanced adaptation to and survival of hypoxia. We demonstrate that fermentatively-produced hydrogen can be recycled when fumarate or oxygen become available, suggesting Mycobacterium smegmatis can switch between fermentation, anaerobic respiration, and aerobic respiration. Hydrogen metabolism enables this obligate aerobe to rapidly meet its energetic needs when switching between microoxic and anoxic conditions and provides a competitive advantage in low oxygen environments.
There are frequent shifts in the biological, chemical, and physical components of soil ecosystems across both space and time. To grow and survive in such heterogeneous environments, microorganisms must quickly adapt to changing environmental conditions. Oxygen availability is a particularly important environmental pressure (1). The best-studied soil bacteria are facultative anaerobes that can use fermentation or anaerobic respiration (e.g., nitrate respiration) to sustain growth; these include copiotrophs of the phyla α-proteobacteria (e.g., Paracoccus pantotrophus) and β-proteobacteria (e.g., Ralstonia eutropha). However, many obligate aerobes also colonize the soil matrices (2), for example, members of the actinobacterial genera Mycobacterium, Streptomyces, and Rhodococcus (2–4). Adopting an oligotrophic lifestyle, these organisms can survive oxygen deprivation in the absence of growth. Their persistence and survival depends on metabolic strategies to generate a proton-motive force and maintain redox homeostasis during hypoxia and anoxia (5, 6). However, there remains an incomplete understanding of the mechanisms such organisms use to achieve this.
Adaptation to transient and prolonged oxygen deprivation has been demonstrated in both saprophytic mycobacteria (e.g., M. smegmatis) and pathogenic species (e.g., M. tuberculosis) (7, 8). M. tuberculosis uses anaerobic electron sinks such as nitrate and possibly fumarate (9, 10). In contrast to the pathogen, M. smegmatis lacks a functional nitrate reductase (10, 11), and it is unclear if one of the putative succinate dehydrogenase/fumarate reductase operons (9, 12) acts as a fumarate reductase. However, it can scavenge oxygen using a cytochrome bd complex that is up-regulated under hypoxia (13, 14) and can enhance its metabolic plasticity using three phylogenetically distinct [NiFe]-hydrogenases for both hydrogen oxidation and possibly hydrogen production (15). It remains unclear whether any Mycobacterium can use fermentation during oxygen limitation. This genus is generally modeled to be strictly respiratory, given their F1Fo-ATP synthase is required for growth even on fermentable carbon sources (16, 17). However, a recent observation of succinate secretion during hypoxia suggests that some mycobacteria may in fact have the capacity to excrete fermentative end products (9, 12). It has also been suggested, but never proven, that mycobacteria dissipate excess reductant by coupling the reoxidation of NAD(P)H to the production of H2 (14, 15, 18, 19).
Adaptation to hypoxia is usually under tight transcriptional regulation. The actinobacterial-specific two-component system DosS/DosT-DosR controls the initial hypoxia-induced response in the majority of mycobacteria. The heme-binding histidine kinases DosS and DosT sense the redox state and oxygen concentration of the cell (20, 21). Both kinases in turn activate DosR, a LuxR-type helix-turn-helix transcriptional activator that binds 18-bp palindromic sequences in promoter regions (22–24). Despite being present in all sequenced mycobacteria except M. leprae (25), this transcriptional regulator has mainly been studied for its role in the pathogenesis of M. tuberculosis. Microarray and computational predictions suggest that DosR activates 48 genes in M. tuberculosis (22, 26), all of which are known to be up-regulated during hypoxia (27). The current literature shows that DosR is involved in maintaining cellular redox balance and viability during the transition from oxic to hypoxic conditions (5, 6), but the molecular basis for this remains to be resolved, and the physiological roles of the majority of the DosR-activated gene products remain to be determined.
Although there have been numerous studies on the adaptation of pathogenic mycobacteria to anoxic environments, there is still little knowledge of the molecular and biochemical mechanisms that enable environmental mycobacteria (the majority of this genera) to adapt to anoxia (14, 28). DosR (MSMEG_5244) appears to have a central role in the hypoxia response of M. smegmatis (29), and its deletion results in rapid loss of cell viability during oxygen limitation (30). Although the regulon has been predicted (25), it has never been experimentally determined. In this work, we determined the DosR regulon of M. smegmatis to gain insight into the global transcriptional response of an obligately aerobic soil bacterium to oxygen deprivation. We show that DosR of M. smegmatis primarily mediates intracellular redox homeostasis by activating hydrogen production. Through coordinating the expression and activity of its three phylogenetically distinct [NiFe]-hydrogenases, Hyd1 (MSMEG_2262-2263), Hyd2 (MSMEG_2719-2720), and Hyd3 (MSMEG_3928-3931), M. smegmatis is able to switch between fermentation and hydrogen oxidation coupled to either oxygen or fumarate reduction depending on electron acceptor availability.
Results
DosR Activates Transcription of 49 Genes in M. smegmatis.
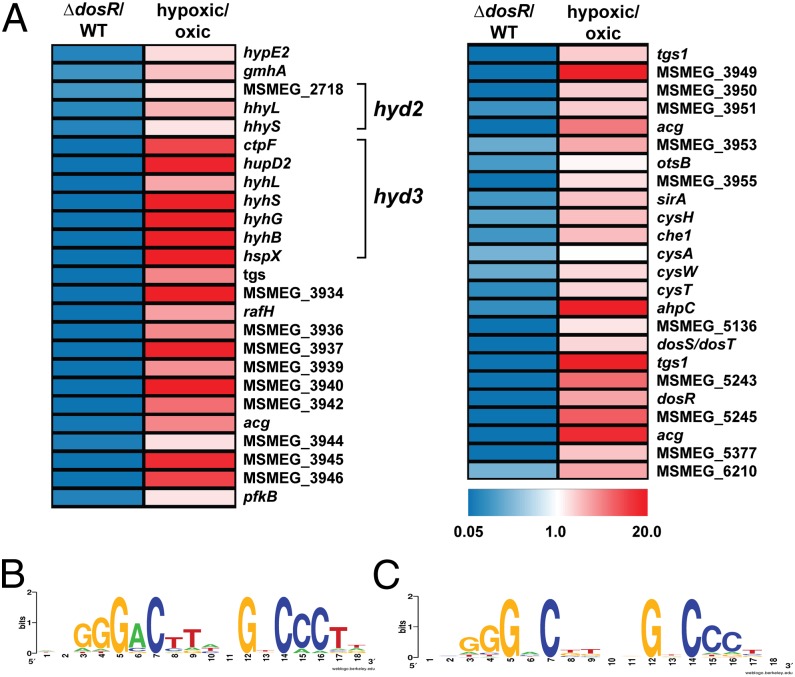
To determine the DosR regulon, the transcriptomes of M. smegmatis mc2155 and the ΔdosR cells were compared by microarray at 10 h following induction of oxygen limitation. We performed quantitative RT-PCR to confirm the quality of the microarray data (Fig. S1). In total, 132 genes showed a significantly lower expression ratio (below 0.5-fold threshold level, P < 0.05) in the ΔdosR strain (Dataset S1). To confirm whether DosR directly regulates these genes, we manually identified palindromic 18-bp sequences similar to the DosR consensus motif of M. tuberculosis encoded upstream of the operons and genes up-regulated in the WT strain. A computational search for candidate DosR sites in the M. smegmatis genome using a custom positional weight matrix identified 355 putative DosR-binding sites across the whole genome. Combining these findings with the microarray results, we predict that the DosR regulon of M. smegmatis comprises 11 operons and 49 genes (Fig. 1A and Table S1). All of these genes were previously observed to be up-regulated at 0.6% oxygen saturation vs. 50% oxygen saturation in continuous culture at a doubling time of 70 h (14) (Table S1).
Fig. 1.
(A) Heat map of the DosR regulon of M. smegmatis. Expression ratios (fold change) are depicted for ∆dosR vs. WT (Left) and WT hypoxia vs. WT oxia (Right) (14). A tabulated DosR regulon is shown in Table S1. (B) WebLogo of 10 DosR binding sites of genes with the lowest expression ratio based on microarray analysis of ∆dosR vs. WT. (C) WebLogo of 32 genes with DosR biding sites that show significantly lower expression in ∆dosR.
The majority of these genes clustered in two regions of the genome: MSMEG_3926-3954 and MSMEG_5241-5246. These regions include the genes encoding for DosR (MSMEG_5244) and a further DosR homolog (MSMEG_3944). In agreement with a previous report (30), MSMEG_3944 was not able to compensate for the loss of DosR. The extent of activation of each promoter appears to depend on the number of DosR binding sites; whereas most DosR-dependent genes are preceded by a single DosR-binding site, there are six strongly activated loci preceded by two putative DosR-binding sites (Table S1). The percentage match to the consensus sequence also influenced extent of activation; consistently, there was greater sequence conservation in the WebLogos of the 10 genes most activated by DosR (Fig. 1B) compared with the entire regulon (Fig. 1C).
Transcription of Two [NiFe]-Hydrogenases Depends on DosR.
Numerous components of the DosR regulon of M. smegmatis are also DosR dependent in pathogenic mycobacteria, including multiple universal stress proteins (30, 31), a ribosome stabilizing-factor (32, 33), and putative energy metabolism genes (e.g., diacylglycerol acyltransferases, nitroreductases) (34, 35) (Tables S1 and S2). The DosR regulon of M. smegmatis also controls the expression of some genes that are not DosR dependent or are absent from the pathogen (Table S2), most notably 10 genes involved in hydrogen metabolism and 6 genes linked to sulfur metabolism (Fig. 1 and Table S1).
The operon MSMEG_3932-3926 (15) is the most strongly activated component of the DosR regulon and is up-regulated by up to 125-fold by the regulator during hypoxia (Fig. 1A and Table S1). The DosR binding site is located 67 bp upstream of the translational start site of MSMEG_3932 (Table S1) and falls well into the promoter region of hyd3 whose operon structure and transcriptional regulation were described previously (15). This seven-gene operon encodes the four subunits and the endopeptidase of the [NiFe]-hydrogenase Hyd3, as well as the apparently unrelated membrane-bound chaperone HspX/Acr (36) and the putative calcium-exporting P-type ATPase CtpF (37). We have previously postulated that Hyd3, as a member of the group 3b [NiFe]-hydrogenases (NADP-linked bidirectional hydrogenase), may dissipate excess reductant as H2 (14, 15). Our data also surprisingly show that Hyd2, a group 5 [NiFe]-hydrogenase (actinobacteria-type uptake hydrogenase), is activated by DosR from a previously unidentified promoter. In contrast to Hyd3, this enzyme acts as a high-affinity, oxygen-dependent uptake hydrogenase during carbon limitation (15, 38). Neighboring operons encoding a putative sulfate ABC importer (CysTWA) (MSMEG_4530-4532) and a sulfite reductase (SirA), phosphoadenosine phosphosulfate reductase, and siroheme chelatase (Che1) (MSMEG_4527-4529) also exhibited a fivefold decrease in expression in ∆dosR. Consistent with the hypoxic regulation of these genes, DosR-binding sites were identified upstream of cysT and sirA (Table S1).
DosR-Regulated Hydrogenase Evolves H2 When Electron Acceptors Are Depleted.
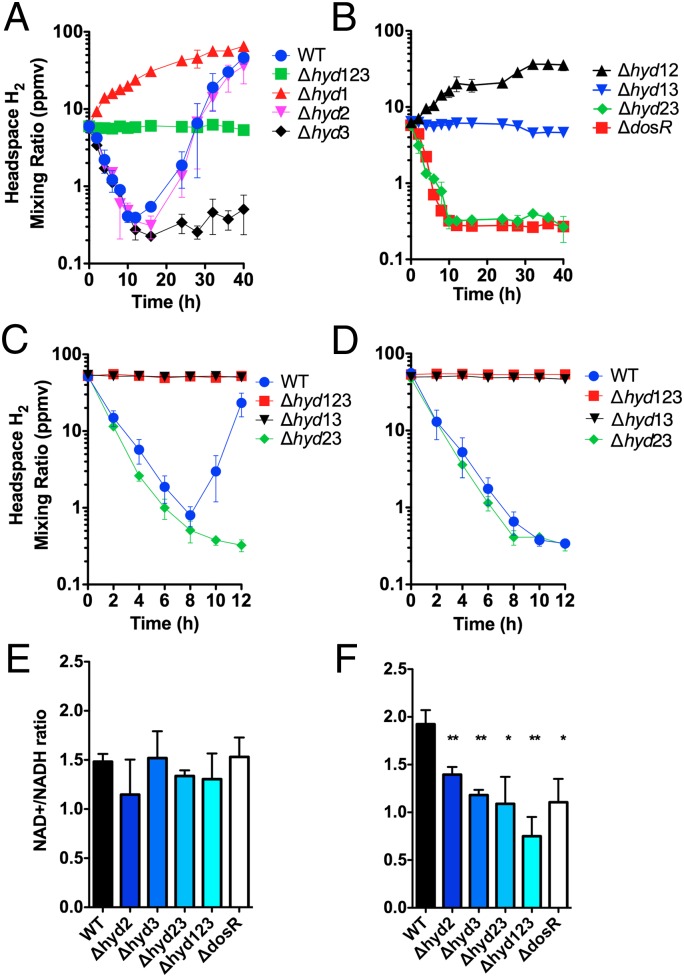
To address the hypothesis that the DosR-regulated hydrogenases may dissipate excess reductant as H2, we used GC to measure the change in the mixing ratios of H2 in the headspace of sealed serum vials (Fig. 2). Activity was compared between WT M. smegmatis mc2155 and strains harboring markerless deletions of its three [NiFe]-hydrogenases (15) (Table S2). Consistent with DosR being inactive in these conditions, the hydrogenase activity of the WT and ΔdosR strains was identical during exponential phase and carbon limitation; net H2 oxidation was observed in both strains due to the combined activity of two uptake hydrogenases (Fig. S2). Net H2 oxidation also was detected following the initial onset of oxygen limitation in the WT strain. The headspace H2 was oxidized from the initial mixing ratio (6.3 ppmv, 0 h) to subatmospheric concentrations (0.39 ppmv, 12 h). H2 oxidation under oxygen limitation is entirely dependent on Hyd1; the activity was maintained in the Δhyd23 strain (Fig. 2B) and eliminated in the Δhyd1 strain (Fig. 2A). We have previously shown that Hyd1 is a rapid-acting, high-affinity group 2a [NiFe]-hydrogenase (38) that is up-regulated during oxygen limitation in a manner independent of DosR (15). H2 oxidation could not be observed at later time points, likely because the remaining O2 is depleted to levels insufficient to sustain respiratory activity.
Fig. 2.
(A and B) H2 oxidation and evolution by oxygen-limited cultures of M. smegmatis mc2155 and derivatives. H2 mixing ratios were measured on entry into stationary phase due to oxygen limitation and at regular intervals over 40 h. (A) WT and triple mutant compared with single hydrogenase KO strains. (B) DosR mutant compared with double hydrogenase KO mutants. (C and D) H2 recycling by cultures of M. smegmatis mc2155 and derivatives. All cultures were limited for O2 over 5 d. They were subsequently spiked with (C) 0.5% O2 or (D) 1 mM sodium fumarate (degassed). H2 mixing ratios were measured on spiking of culture and at regular intervals over 40 h. The initial headspace mixing ratios were adjusted to 55 ppmv. (E and F) Measurement of NAD/NADH ratios in oxygen-limited cultures of M. smegmatis (E) during midexponential phase before oxygen depletion and (F) 5 d following entry into stationary phase due to oxygen limitation. *P < 0.05, **P < 0.01 difference relative to WT bars. In all panels, error bars show SD from three biological replicates.
At ∼24 h following the onset of oxygen limitation, net hydrogen evolution was clearly observed in the WT strain. The H2 mixing ratio increased 100-fold from 0.39 ppmv (12 h) to 46 ppmv (40 h). Consistent with our predictions, analysis of the Δhyd12 (Fig. 2B) and Δhyd3 (Fig. 2A) mutant strains confirms that the group 3b [NiFe]-hydrogenase Hyd3 is solely responsible for hydrogen evolution. This activity was eliminated in the ΔdosR strain, thus confirming that DosR is responsible for activating Hyd3 and hence H2 production. Analysis of the Δhyd1 and Δhyd12 mutant strains suggests H2 evolution occurs from the onset of oxygen limitation; however, the H2 is recycled by Hyd1, and net oxidation prevails if there is sufficient oxygen present. Despite being up-regulated by DosR in this condition, no clear enzymatic activity was observed for the group 5 [NiFe]-hydrogenase Hyd2; we speculate that the enzyme is dependent on other hydrogenases, has a lower-than-observable activity, or behaves bidirectionally in this condition.
Net H2 oxidation was restored in long-term oxygen-limited cultures by injection of trace volumes of O2 into the headspace (Fig. 2C). The WT strain rapidly oxidized the H2 present in the headspace (initial mixing ratio = 51 ppmv; final mixing ratio = 0.80 ppmv). When this electron acceptor was exhausted, net H2 evolution was again observed. Analysis of the Δhyd23 strain confirms that the hydrogen oxidation activity is also Hyd1 dependent. Supplementation of 1 mM fumarate also restored net oxidation of H2 (Fig. 2D). As the fumarate supply was in excess, hydrogen evolution did not resume. These data provide evidence that hydrogen oxidation can be coupled to the reduction of the traditionally anaerobic acceptor fumarate when oxygen is depleted. Succinate production has previously been shown to have an essential role in adaptation of mycobacteria to hypoxia; the multifunctional compound is capable of being oxidized for ATP synthesis, used for anaplerosis, or excreted as a potential fermentative end product (9, 12). Hence, transferring electrons from H2 to fumarate may provide a mechanism to sustain both the proton-motive force and ATP synthesis through anaerobic respiration while maintaining redox balance in the cell.
Hydrogen Cycling Maintains Intracellular Redox Homeostasis.
The ratios of NAD+/NADH were determined to address whether hydrogen production contributes to the reoxidation of coenzymes (Fig. 2 E and F). During the exponential phase, the NAD+/NADH ratios were similar between all strains (Fig. 2E). Following entry into oxygen limitation, there was a significant and reproducible twofold reduction in ratios of the cofactors in the mutants lacking hydrogenases compared with the WT strain. Whereas the WT strain continued to maintain higher concentrations of oxidized to reduced NAD ([NAD+]/[NADH] = 1.92 ± 0.28), these cofactors were at close to equal concentrations in the Δhyd2 (1.39 ± 0.07), Δhyd3 (1.18 ± 0.09), Δhyd123 (0.75 ± 0.29), and ΔdosR (1.10 ± 0.29) strains following 5 d of oxygen limitation (Fig. 2F). These data provide evidence that strains lacking Hyd3 and Hyd2 are impaired in the ability to maintain redox balance and reoxidize cofactors during oxygen limitation.
We propose that Hyd3 regulates redox balance by directly coupling the oxidation of NAD(P)H to evolution of H2. Although group 3b [NiFe]-hydrogenases are widespread in actinobacteria and proteobacteria (38), until now, they have only been characterized in thermophilic archaea. Hyd3 shares >30% sequence identity with the cytosolic hydrogenases of Pyrococcus furiosus (39, 40) and Thermococcus kodakaraensis (41), including motifs predicted to bind a FAD moiety, a [NiFe] center, and a relay of six [FeS] clusters. Like these enzymes, Hyd3 is likely to evolve H2 by directly transferring electrons liberated from NAD(P)H at the diaphorase module (hyhG/hyhB) to the [NiFe] center of the hydrogenase module (hyhL/hyhS) (Fig. S3). It is unclear whether the M. smegmatis enzyme acts as a bifunctional sulfhydrogenase like its archaeal homologs (39). H2S production and media acidification were detected when M. smegmatis became oxygen limited in the presence of 0.1% elemental sulfur. However, this occurred even in the Δhyd123 strain. Instead, we postulate that the DosR-activated gene products of sirA, cysH, che1, and cysTWA are responsible for this process, with the sulfite reductase dissipating excess electrons from ferredoxin as the diffusible gas H2S. A thorough characterization and quantification of this process has yet to be conducted.
Hydrogen Metabolism Is Essential for Adaptation to Oxygen Depletion and Sustains Long-Term Survival in Hypoxic Conditions.
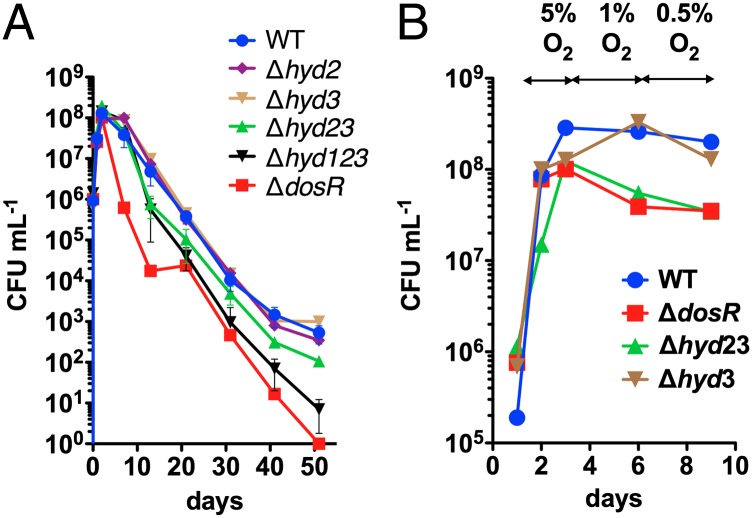
To determine the importance of hydrogen metabolism during oxygen depletion, the survival of the M. smegmatis strains was measured during long-term incubation in sealed serum vials (Fig. 3A). Consistent with previous observations (30), the dosR mutant dramatically lost viability on induction of hypoxia. Complementation experiments have previously demonstrated that this phenotype is primarily, but not completely, due to loss of the ribosome-stabilizing factor RafH (32). Our experiments show that survival also depends on the combined activity of the three hydrogenases from M. smegmatis, all of which are up-regulated under oxygen limitation (14). Although the single mutants behaved similarly to the WT, the long-term survival of the Δhyd23 and Δhyd13 strains was reduced, and a strain lacking all three hydrogenases Δhyd123 showed a two-log difference in survival after 50 d (Fig. 3A). These data provide evidence that both H2 evolution and reoxidation are both important for long-term viability.
Fig. 3.
(A) Long-term survival of oxygen-limited cultures in rubber-stoppered serum vials. Error bars show SD from three biological replicates. (B) Adaptation to gradual decrease in oxygen concentration in gas-mix controlled bioreactors. Percentage O2 represents oxygen concentration in gas mix.
In contrast to the natural soil environment of M. smegmatis, gasses cannot escape and will accumulate in the sealed serum vial model. Therefore, to study the importance of DosR-controlled hydrogenase metabolism during gradual depletion of oxygen in more detail, we used fully controlled bioreactors with a gas-mixing feature to precisely control oxygen concentration. We collected corroborating evidence that the DosR-controlled Hyd2 and Hyd3 are involved in the adaptation phase to decreasing oxygen concentration. In fact, the Δhyd23 KO strain behaved identically to ΔdosR during adaptation to decreasing oxygen concentration in gas mix-controlled bioreactors (Fig. 3B).
Discussion
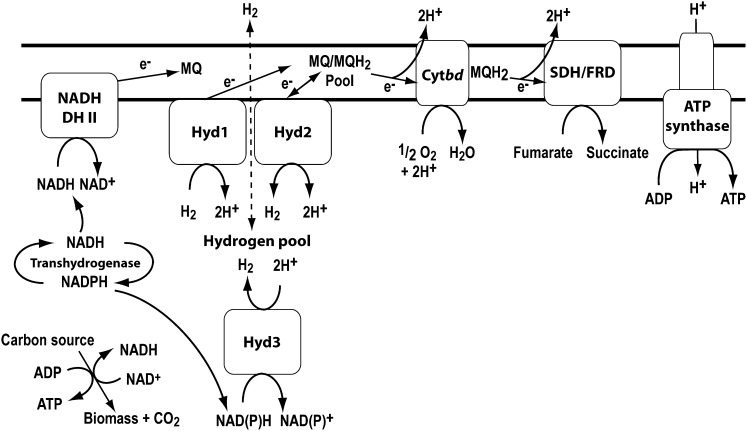
This study provides, to our knowledge, the first unambiguous confirmation that a bacterium that is strictly oxygen dependent for growth can adopt fermentative hydrogen production in response to oxygen limitation. Our data collectively show that a group 3b [NiFe]-hydrogenase, Hyd3, is responsible for this process. Transcription of this enzyme only occurs during oxygen limitation (15), and microarray analysis demonstrates that genes encoding its four subunits are the most strongly activated components of the DosR regulon. Loss of Hyd3 activity affects the NAD+/NADH ratios of the cell and, in combination with Hyd2, impairs long-term adaptation and survival during oxygen limitation. Based on phylogenetic analysis and sequence alignments, it is probable that the enzyme directly couples NAD(P)H oxidation to H2 evolution. This mechanism may also aid in recycling excess NADH, given transhydrogenases can interconvert electrons between NADH and NADPH; one such enzyme (MSMEG_2748) is known to be up-regulated during oxygen limitation in M. smegmatis (Fig. 4) (14).
Fig. 4.
Summary of the respiratory and fermentative processes taking place in cells of M. smegmatis exposed to hypoxia and anoxia. NADH DH II, NADH dehydrogenase type II; Hyd, hydrogenase; Cytbd, cytochrome bd oxidase; SDH, succinate dehydrogenase; FRD, fumarate reductase.
Hydrogen fermentation has been observed in strictly respiratory organisms, but never in an obligate aerobe. The group 3d [NiFe]-hydrogenase of Ralstonia eutropha, which grows through aerobic and nitrate respiration, transiently reverses directionality to couple NADH reoxidation to H2 generation during anaerobiosis (42, 43). In contrast to this system, the tight regulation and sustained activity of Hyd3 under oxygen limitation in M. smegmatis is consistent with Hyd3 being specifically selected to catalyze the fermentative production of H2 during dormancy. The conservation of Hyd3 homologs across other actinobacteria, e.g., mycobacteria, streptomycetes, and rhodococci (38), suggests that hydrogen fermentation may be a widespread, but previously unanticipated, mechanism for long-term survival among obligate aerobic soil bacteria.
The diverse other genes identified to be DosR dependent are likely to enhance the adaptability of M. smegmatis in response to shifting partial pressures of oxygen. Universal stress proteins, the ribosome-associated protein RafH, and the membrane-bound chaperone HspX may contribute to the robustness of the cell in response to stressful conditions. These proteins are widely DosR associated in mycobacteria, and phenotypic studies have demonstrated their importance in mediating the dormancy response. In addition to hydrogen fermentation, it is probable that DosR activates other changes in energy metabolism. For example, triacylglycerol accumulation provides the organism with an energy-rich fuel source for the continuation of dormancy and the transition back into growth when aerobic conditions are restored. In addition, our data are consistent with sulfite reductase (SirA) acting as an alternative terminal electron acceptor in the absence of oxygen in a dosR-dependent manner. The DosR-regulated loci encoding CysTWA, SirA, CysH, and Che1 might serve to import, activate, and reduce sulfate via sulphite to H2S. SirA has been shown to catalyze sulfite reduction in M. smegmatis (44).
Such fermentation reactions clearly provide an elegant solution to the problem of dissipating excess reductant during oxygen limitation, given the end product H2 (and possibly H2S) is a readily diffusible gas. However, it is apparent that M. smegmatis can recycle the hydrogen produced when traces of aerobic or anaerobic electron acceptors are present (Fig. 4). This process is DosR independent and is catalyzed by membrane-associated uptake hydrogenase Hyd1. Hydrogen recycling may be more favorable than hydrogen dissipation due to much greater ATP yields that can be generated through oxidative phosphorylation compared with substrate-level phosphorylation. We postulate that H2 reoxidation provides a mechanism to sustain a proton-motive force while bypassing the need for cofactors associated with catabolism of organic carbon sources, such as NAD. Anaerobic respiration coupling H2 oxidation to fumarate reduction may be a particularly effective way to maintain redox balance while generating a proton gradient, given it results in production of the multifunctional redox valve and potential fermentation product succinate (Fig. 4). A third hydrogenase Hyd2 is also up-regulated in a DosR-dependent manner under hypoxia; although GC studies were unable to resolve its catalytic activity, it is clearly important for maintaining redox balance and viability in this condition.
We conclude that M. smegmatis maintains proton-motive force and intracellular redox homeostasis during hypoxia by rapidly switching between fermentation, anaerobic respiration, and aerobic respiration depending on electron acceptor availability. This ability is likely a competitive advantage in low oxygen environments and could be a widespread strategy among soil actinobacteria harboring these hydrogenases.
Methods
Bacterial Strains and Growth Conditions.
All bacterial strains used in this study are listed in Table S3. M. smegmatis mc2155 (45) and derived mutants (14, 15, 30) were maintained on LB agar plates supplemented with 0.05% (wt/vol) Tyloxapol (Sigma-Aldrich). For broth culture, M. smegmatis was grown in Hartmans de Bont (HdB) minimal medium supplemented with 25 mM glycerol, 0.05% Tyloxapol, and 10 µM NiSO4 (46). All cultures were inoculated to an initial optical density of 0.005 using a hypodermic syringe. Cultures were incubated at 37 °C with agitation (200 rpm) in 30 mL medium in 120-mL rubber-stoppered serum vials. Cultures entered stationary phase upon depletion of headspace oxygen (at OD ∼ 2.2/1.5 × 108 CFU/mL), as indicated by the decolorization of 1.5 µg/mL methylene blue. Optical densities to assess growth were measured at 600 nm (OD600) in a Jenway 6300 spectrometer. Cultures were diluted in 0.85% saline to bring the OD600 below 0.5 when measured in cuvettes of 1-cm light path length. To count colony forming units (CFU/mL), each culture was serially diluted in PBS (pH 7.0) and spotted onto agar plates. All survival studies were performed with biological triplicates, and plates were spotted in technical quadruplicate.
Growth in Bioreactors.
Cells were cultivated in fully controlled parallel bioreactors (DASGIP DASbox; Julich). The culture volume was 200 mL with a headspace of 100 mL and stirring rate was maintained at 100 rpm. N2, CO2, and air (TechAir) were delivered from separate gas tanks, mixed by the DASbox system, and delivered to the bioreactors at a flow rate of 15 sL/h. The gas mixture was adjusted at indicated time points to maintain decreasing concentrations of O2 by replacing air with increasing concentrations of N2. At the indicated time points, 1 mL of cell culture was withdrawn, and dilutions were plated for CFU as described above.
RNA Extraction.
M. smegmatis mc2155 and ΔdosR (MSMEG_5244) cells were grown synchronously in sealed serum vials. At 10 h following oxygen depletion (methylene blue decolorization), the cells were harvested for RNA extraction and microarray analysis. Cultures of 10 mL were mixed with 20 mL cold glycerol saline [3:2 (vol/vol); −20 °C], centrifuged (27,000 × g, 20 min, −20 °C), and resuspended in glycerol saline [1:1 (vol/vol); −20 °C]. Total RNA was extracted using TRIZOL reagent (Invitrogen) according to the manufacturer's instructions. Cell lysis was achieved by three cycles of bead-beating in a Mini-Beadbeater (Biospec) at 5,000 rpm for 30 s. DNA was removed from the RNA preparation by treatment with 2 U RNase-free DNase using the TURBO DNA-free kit (Ambion), according to the manufacturer's instructions. The concentration and purity of the RNA were determined using a NanoDrop ND-1000 spectrophotometer, and its integrity was confirmed on a 1.2% agarose gel.
Microarray Analysis.
Microarray analysis was performed using arrays provided by the Pathogen Functional Genomics Research Center (PFGRC) funded by the National Institute of Allergy and Infectious Diseases. Analysis was performed as previously described (47) based on protocols standard operating procedure M007 and M008 from The Institute of Genomic Research (TIGR) (48).
Quantitative Real-Time PCR.
cDNA was synthesized from 1 µg of RNA for each sample with the SuperScript III Reverse Transcriptase Kit (Invitrogen). After cDNA synthesis, real-time PCR was conducted according to the manufacturer with PlatinumSYBRGreen qPCR SuperMix-UDG with ROX (Invitrogen). Primers (Integrated DNA Technologies) for 10 genes (Table S4) were designed with the publicly available Primer3 software. Primer optimization was performed. The real-time PCR reactions were conducted in ABI Prism7500 (Applied Biosystems), and the results were normalized to the gene sigA (MSMEG_2758) as an endogenous control.
Promoter Analysis.
The 500-bp regions immediately 5′ to the annotated start codons of the DosR-dependent genes identified by microarray were scanned manually for sequences similar to previously reported M. tuberculosis DosR consensus sequence (TSGGGACTWWAGTCCCSA) (22). Similar sequences were observed in 32 putative promoters, which were used to create a custom positional weight matrix (PWN) for a virtual footprint analysis of the M. smegmatis genome using PRODORIC (49). This analysis identified 355 putative DosR-binding sites across the whole genome. WebLogos were created using a publicly available program (50). For comparative genomic analysis, the consensus sequence was also used to predict DosR-regulated genes in other mycobacteria.
GC Measurements.
Cultures of M. smegmatis mc2155 and derivatives were grown in serum vials until they entered stationary phase due to oxygen limitation. Standard H2 gas (1,000 ppmv ± 2%; Messer Schweiz) was added to the headspaces to obtain initial mixing ratios of 6.3 ± 0.7 ppmv H2. The change in mixing ratio was measured as a function of time using gas chromatography. Aliquots of headspace air (250 µL) were extracted using a gas-tight syringe (VICI Precision Sampling) and loaded on to a Trace Analytical Reduced Gas Analyzer (SRI Instruments). Serum vials were continuously agitated (120 rpm on a rotary shaking incubator) to enhance transfer of H2 from the gas phase to the liquid phase. To measure recycling of H2, vials containing cultures that had been oxygen deprived for 5 d were spiked with either 0.5% O2 or 1 mM sodium fumarate that had been bubbled with pure N2 for at least 2 min. The initial mixing ratios of headspace H2 were normalized to 55 ± 5 ppmv H2. All experiments were performed using biological triplicates. To avoid gas buildup, the serum vials used in this study were only sealed with rubber stoppers after autoclaving. The reproducibility of the measurements was assessed before and during each time course by analysis of standard H2 gas (2.0 ppmv ± 5%, 50 ppmv ± 2%, and 1,000 ppmv ± 2% H2; Messer Schweiz).
Measurement of NAD+/NADH Ratios.
One-milliliter samples of M. smegmatis cultures were withdrawn during exponential phase (OD ∼ 0.5) and at 5 d following the induction of stationary phase. Each sample was centrifuged (15,000 × g, 3 min) and resuspended in either 0.3 mL of 0.2 M HCl (for NAD+ extraction) or 0.3 mL of 0.2 M NaOH (for NADH extraction). The cultures were heated (55 °C, 10 min), cooled (0 °C, 5 min), and neutralized with either 0.3 mL of 0.1 M NaOH (for NAD+ extraction) or 0.3 mL of 0.1 M HCl (for NADH extraction). After centrifugation (15,000 × g, 3 min), the supernatants were collected and transferred into cuvettes containing 50 µL of 1 M bicine (pH 8.0), 50 µL of 40 mM EDTA (pH 8.0), 50 µL of 4.2 mM 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT), and 50 µL of 16 mM phenazine ethosulfate. NAD+ and NADH concentrations were measured by addition of 50 µL ethanol and 5 U yeast alcohol dehydrogenase II (51). The rate of reduction of MTT was measured spectrophotometrically at 570 nm and was proportional to the concentration of standards of each cofactor.
Supplementary Material
Acknowledgments
We thank Huw Williams for providing the DosR mutant. This work and M.B. were financially supported by a Marsden grant from the Royal Society of New Zealand and National Institutes of Health (NIH) Grant AI26170. C.G. was supported by an Otago postgraduate scholarship from the University of Otago. G.M.C. was supported by a James Cook fellowship from the Royal Society of New Zealand. W.R.J. was supported by NIH Grants AI26170 and R01AI097548-01A1. C.G.’s travel to the Max Planck Institute for Terrestrial Microbiology was funded by the Sandy Smith Memorial Scholarship, the Maurice & Phyllis Paykel Trust, the Otago School of Medical Sciences, and the Department of Microbiology and Immunology at the University of Otago.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE56083).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407034111/-/DCSupplemental.
References
- 1.Voroney RP. 2007. The soil habitat. Soil Microbiology, Ecology, and Biochemistry, ed Paul EA (Academic Press, Oxford, UK), 3rd Ed, pp 25–49. [Google Scholar]
- 2.Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol. 2006;72(3):1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Keulen G, Alderson J, White J, Sawers RG. The obligate aerobic actinomycete Streptomyces coelicolor A3(2) survives extended periods of anaerobic stress. Environ Microbiol. 2007;9(12):3143–3149. doi: 10.1111/j.1462-2920.2007.01433.x. [DOI] [PubMed] [Google Scholar]
- 4.Falkinham JO., 3rd Nontuberculous mycobacteria in the environment. Clin Chest Med. 2002;23(3):529–551. doi: 10.1016/s0272-5231(02)00014-x. [DOI] [PubMed] [Google Scholar]
- 5.Rao SP, Alonso S, Rand L, Dick T, Pethe K. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2008;105(33):11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leistikow RL, et al. The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J Bacteriol. 2010;192(6):1662–1670. doi: 10.1128/JB.00926-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wayne LG, Lin KY. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect Immun. 1982;37(3):1042–1049. doi: 10.1128/iai.37.3.1042-1049.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dick T, Lee BH, Murugasu-Oei B. Oxygen depletion induced dormancy in Mycobacterium smegmatis. FEMS Microbiol Lett. 1998;163(2):159–164. doi: 10.1111/j.1574-6968.1998.tb13040.x. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe S, et al. Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLoS Pathog. 2011;7(10):e1002287. doi: 10.1371/journal.ppat.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sohaskey CD. Nitrate enhances the survival of Mycobacterium tuberculosis during inhibition of respiration. J Bacteriol. 2008;190(8):2981–2986. doi: 10.1128/JB.01857-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sohaskey CD, Wayne LG. Role of narK2X and narGHJI in hypoxic upregulation of nitrate reduction by Mycobacterium tuberculosis. J Bacteriol. 2003;185(24):7247–7256. doi: 10.1128/JB.185.24.7247-7256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eoh H, Rhee KY. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2013;110(16):6554–6559. doi: 10.1073/pnas.1219375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kana BD, et al. Characterization of the cydAB-encoded cytochrome bd oxidase from Mycobacterium smegmatis. J Bacteriol. 2001;183(24):7076–7086. doi: 10.1128/JB.183.24.7076-7086.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berney M, Cook GM. Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia. PLoS ONE. 2010;5(1):e8614. doi: 10.1371/journal.pone.0008614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berney M, Greening C, Hards K, Collins D, Cook GM. Three different [NiFe] hydrogenases confer metabolic flexibility in the obligate aerobe Mycobacterium smegmatis. Environ Microbiol. 2014;16(1):318–330. doi: 10.1111/1462-2920.12320. [DOI] [PubMed] [Google Scholar]
- 16.Tran SL, Cook GM. The F1Fo-ATP synthase of Mycobacterium smegmatis is essential for growth. J Bacteriol. 2005;187(14):5023–5028. doi: 10.1128/JB.187.14.5023-5028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andries K, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307(5707):223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 18.Boshoff HI, Barry CE., 3rd Tuberculosis - metabolism and respiration in the absence of growth. Nat Rev Microbiol. 2005;3(1):70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- 19.He H, Bretl DJ, Penoske RM, Anderson DM, Zahrt TC. Components of the Rv0081-Rv0088 locus, which encodes a predicted formate hydrogenlyase complex, are coregulated by Rv0081, MprA, and DosR in Mycobacterium tuberculosis. J Bacteriol. 2011;193(19):5105–5118. doi: 10.1128/JB.05562-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar A, Toledo JC, Patel RP, Lancaster JR, Jr, Steyn AJ. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci USA. 2007;104(28):11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sousa EH, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA. DosT and DevS are oxygen-switched kinases in Mycobacterium tuberculosis. Protein Sci. 2007;16(8):1708–1719. doi: 10.1110/ps.072897707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park HD, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48(3):833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Florczyk MA, et al. A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires Rv3133c (dosR or devR) for expression. Infect Immun. 2003;71(9):5332–5343. doi: 10.1128/IAI.71.9.5332-5343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wisedchaisri G, Wu M, Sherman DR, Hol WG. Crystal structures of the response regulator DosR from Mycobacterium tuberculosis suggest a helix rearrangement mechanism for phosphorylation activation. J Mol Biol. 2008;378(1):227–242. doi: 10.1016/j.jmb.2008.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerasimova A, Kazakov AE, Arkin AP, Dubchak I, Gelfand MS. Comparative genomics of the dormancy regulons in mycobacteria. J Bacteriol. 2011;193(14):3446–3452. doi: 10.1128/JB.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voskuil MI, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198(5):705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherman DR, et al. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc Natl Acad Sci USA. 2001;98(13):7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cook GM, et al. Physiology of mycobacteria. Adv Microb Physiol. 2009;55:81–182, 318–319. doi: 10.1016/S0065-2911(09)05502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayuri BG, Bagchi G, Das TK, Tyagi JS. Molecular analysis of the dormancy response in Mycobacterium smegmatis: Expression analysis of genes encoding the DevR-DevS two-component system, Rv3134c and chaperone alpha-crystallin homologues. FEMS Microbiol Lett. 2002;211(2):231–237. doi: 10.1111/j.1574-6968.2002.tb11230.x. [DOI] [PubMed] [Google Scholar]
- 30.O’Toole R, et al. A two-component regulator of universal stress protein expression and adaptation to oxygen starvation in Mycobacterium smegmatis. J Bacteriol. 2003;185(5):1543–1554. doi: 10.1128/JB.185.5.1543-1554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drumm JE, et al. Mycobacterium tuberculosis universal stress protein Rv2623 regulates bacillary growth by ATP-Binding: Requirement for establishing chronic persistent infection. PLoS Pathog. 2009;5(5):e1000460. doi: 10.1371/journal.ppat.1000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trauner A, Lougheed KE, Bennett MH, Hingley-Wilson SM, Williams HD. The dormancy regulator DosR controls ribosome stability in hypoxic mycobacteria. J Biol Chem. 2012;287(28):24053–24063. doi: 10.1074/jbc.M112.364851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar A, et al. Mycobacterium tuberculosis DosR regulon gene Rv0079 encodes a putative, ‘dormancy associated translation inhibitor (DATIN)’. PLoS ONE. 2012;7(6):e38709. doi: 10.1371/journal.pone.0038709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Y, Coates AR. Mycobacterium tuberculosis acg gene is required for growth and virulence in vivo. PLoS ONE. 2011;6(6):e20958. doi: 10.1371/journal.pone.0020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Low KL, et al. Triacylglycerol utilization is required for regrowth of in vitro hypoxic nonreplicating Mycobacterium bovis bacillus Calmette-Guerin. J Bacteriol. 2009;191(16):5037–5043. doi: 10.1128/JB.00530-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu G, et al. Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate. Mol Microbiol. 2006;59(5):1417–1428. doi: 10.1111/j.1365-2958.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- 37.Rosch JW, Sublett J, Gao G, Wang Y-D, Tuomanen EI. Calcium efflux is essential for bacterial survival in the eukaryotic host. Mol Microbiol. 2008;70(2):435–444. doi: 10.1111/j.1365-2958.2008.06425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greening C, Berney M, Hards K, Cook GM, Conrad R. A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. Proc Natl Acad Sci USA. 2014;111(11):4257–4261. doi: 10.1073/pnas.1320586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma K, Schicho RN, Kelly RM, Adams MW. Hydrogenase of the hyperthermophile Pyrococcus furiosus is an elemental sulfur reductase or sulfhydrogenase: Evidence for a sulfur-reducing hydrogenase ancestor. Proc Natl Acad Sci USA. 1993;90(11):5341–5344. doi: 10.1073/pnas.90.11.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma K, Weiss R, Adams MW. Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J Bacteriol. 2000;182(7):1864–1871. doi: 10.1128/jb.182.7.1864-1871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanai T, Ito S, Imanaka T. Characterization of a cytosolic NiFe-hydrogenase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J Bacteriol. 2003;185(5):1705–1711. doi: 10.1128/JB.185.5.1705-1711.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuhn M, Steinbüchel A, Schlegel HG. Hydrogen evolution by strictly aerobic hydrogen bacteria under anaerobic conditions. J Bacteriol. 1984;159(2):633–639. doi: 10.1128/jb.159.2.633-639.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warnecke-Eberz U, Friedrich B. Three nitrate reductase activities in Alcaligenes eutrophus. Arch Microbiol. 1993;159(5):405–409. [Google Scholar]
- 44.Pinto R, Harrison JS, Hsu T, Jacobs WR, Jr, Leyh TS. Sulfite reduction in mycobacteria. J Bacteriol. 2007;189(18):6714–6722. doi: 10.1128/JB.00487-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4(11):1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 46.Berney M, Weimar MR, Heikal A, Cook GM. Regulation of proline metabolism in mycobacteria and its role in carbon metabolism under hypoxia. Mol Microbiol. 2012;84(4):664–681. doi: 10.1111/j.1365-2958.2012.08053.x. [DOI] [PubMed] [Google Scholar]
- 47.Hümpel A, Gebhard S, Cook GM, Berney M. The SigF regulon in Mycobacterium smegmatis reveals roles in adaptation to stationary phase, heat, and oxidative stress. J Bacteriol. 2010;192(10):2491–2502. doi: 10.1128/JB.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hegde P, et al. 2000 A concise guide to cDNA microarray analysis. Biotechniques 29(3):548–550, 552–554, 556. [Google Scholar]
- 49.Münch R, et al. Virtual Footprint and PRODORIC: An integrative framework for regulon prediction in prokaryotes. Bioinformatics. 2005;21(22):4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- 50.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leonardo MR, Dailly Y, Clark DP. Role of NAD in regulating the adhE gene of Escherichia coli. J Bacteriol. 1996;178:6013–6018. doi: 10.1128/jb.178.20.6013-6018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]