Significance
Despite advances in cardiac imaging technologies such as MRI, computed tomography, and ultrasound it has not been possible to image cardiac function in animal models at the cellular level owing to insufficient resolution and excess motion. As a result, our understanding of cardiac biology and function is derived from either whole-organ imaging techniques or from isolated in vitro heart preparations and cellular model systems. We demonstrate a technique for intravital optical microscopy that compensates for motion of the contracting heart and allows for measurement of contractile function at the single-cell level. These approaches will enable studies of complex and dynamic cellular physiology in the beating heart and should be widely useful for studying heart disease in animal models.
Keywords: intravital micoscopy, molecular imaging, cardiovascular imaging, fluorescence, pacing
Abstract
Knowledge of cardiomyocyte biology is limited by the lack of methods to interrogate single-cell physiology in vivo. Here we show that contracting myocytes can indeed be imaged with optical microscopy at high temporal and spatial resolution in the beating murine heart, allowing visualization of individual sarcomeres and measurement of the single cardiomyocyte contractile cycle. Collectively, this has been enabled by efficient tissue stabilization, a prospective real-time cardiac gating approach, an image processing algorithm for motion-artifact-free imaging throughout the cardiac cycle, and a fluorescent membrane staining protocol. Quantification of cardiomyocyte contractile function in vivo opens many possibilities for investigating myocardial disease and therapeutic intervention at the cellular level.
With knowledge of the molecular bases for cardiovascular diseases expanding rapidly, a considerable void exists in our ability to phenotype heart function at the subcellular scale in vivo and in real time. Understanding how the fundamental unit of myocardial function, the cardiomyocyte, responds, adapts, and ultimately fails in response to stress, both individually and in a network of cells contributing to whole-organ function, is central to unraveling mechanisms of disease and designing novel therapies aimed at molecular pathways. The ability to measure single cardiomyocyte contractile function in vivo in the native environment is not possible using existing techniques.
Optical microscopy has potential to assess cardiomyocyte structure and function in rodent models (1). Intravital confocal and two-photon microscopy have been used in combination with fluorescence molecular imaging probes in cancer research, immunology, and the neurosciences to reveal biological processes at the cellular level in living organisms (2). Application of intravital techniques for imaging the beating heart in rodent models has been significantly limited by motion from cardiac contraction and respiration, and most studies as a result have used noncontracting Langendorf heart preparations (3–10) or transplanted heart models (11). These model systems do not allow investigation of cardiomyocyte biology in the native heart under physiologic conditions. A few studies have achieved intravital imaging in orthotopic hearts at relatively modest spatial and temporal resolutions that prevent visualization of subcellular structures. Essential to these techniques are methods of macrostabilization such as affixing the heart with sutures (12), compressing the heart with a coverslip (11, 13), bonding the heart with a mechanical stabilizer (14), or suction-based devices (15, 16). Although tissue stabilization methods alone can facilitate very-low-resolution cardiac imaging (e.g., microvasculature, cellular recruitment, and flow), they do not achieve the necessary temporal or spatial resolution for subcellular imaging of cardiomyocytes throughout the cardiac cycle. To further overcome residual motion artifacts, retrospective gating algorithms for segmented microscopy have been explored (14), but the retrospective nature and poor temporal resolution of the methods limit their application. Prospective gating schemes have also been previously used for imaging zebrafish specimens (17).
Here we describe a microscopy method for subcellular resolution and motion-artifact-free imaging of contracting cardiomyocytes in vivo in the native mouse heart. The platform technique is based on four innovations: (i) an optimized fluorescent staining protocol to label myocyte subcellular structure, (ii) a highly effective tissue stabilizer, (iii) a prospective cardiac gating approach using cardiac pacing, and (iv) an image processing algorithm, prospective sequential segmented microscopy (PSSM), for motion-artifact-free subcellular imaging at any point in the cardiac cycle. These advances combined enable intravital imaging of individual cardiomyocytes with high temporal and spatial resolution and quantification of sarcomere length changes during the single-cell contractile cycle.
Results
Given the array of available transgenic mouse models of disease, we sought an approach that can be readily applied to any mouse. Prior work with two-photon microscopy in explanted hearts has shown that sarcomere length can be quantified using fluorescent staining of cardiomyocyte transverse tubule cell membranes (4, 18). We therefore developed a protocol for staining cardiomyocyte structures in vivo using the water-soluble dye di-2-anepeq (Materials and Methods). The dye effectively labels all cell membranes, including cardiomyocyte outer membranes and transverse tubules, endothelial cells, and circulating blood cells in vivo (Fig. S1).
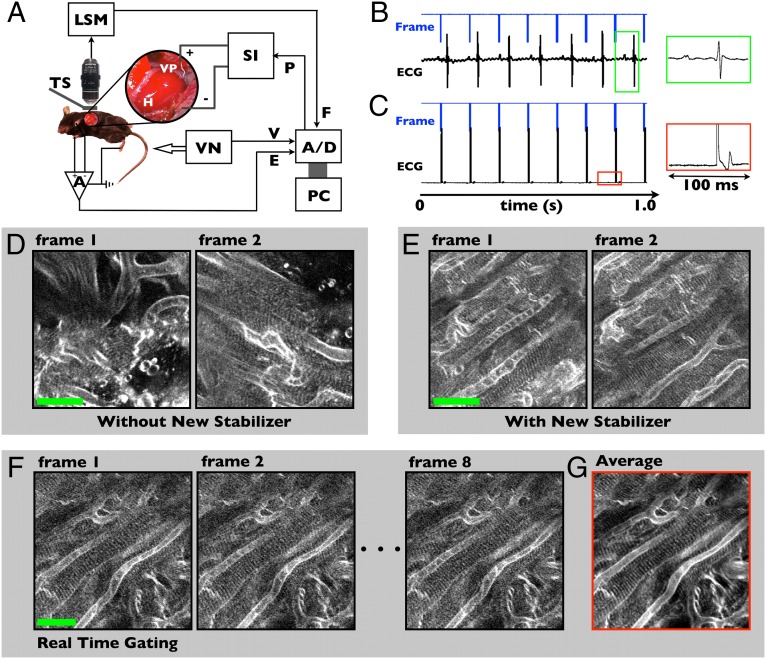
The technique was designed to work with commercial confocal or two-photon microscopes for wide applicability (Fig. 1A and Fig. S2B). Imaging is performed through a left thoracotomy incision providing direct access to the heart, with the animal anesthetized and mechanically ventilated. Cardiac pacing is performed through a small-diameter insulated wire lead, which is sutured into the left ventricular apex (Fig. 1A, Inset, and Fig. S2A). The laser scanning microscope provides a frame signal corresponding with microscope acquisition (8 Hz), which serves as the master timing clock for the system. All timing waveforms are recorded through a data acquisition system interfacing to a personal computer. In retrospective gating mode, the microscope acquisition is not synchronized to the native heart beat (Fig. 1B). In prospective mode, each frame signal is used to trigger a pacing waveform, and each microscope image is then precisely synchronized with the cardiac cycle (Fig. 1C). Typical fast-mode microscope acquisition rates of 7–10 frames per second correspond to physiologic heart rates in the mouse of 400–600 beats per minute.
Fig. 1.
Real-time prospective cardiac gating for intravital microscopy. (A) System schematic. Laser scanning microscope, LSM, acquisition is synchronized to the cardiac cycle using pacing. The microscope frame signal, F, controls the pacemaker drive signal, P, which is delivered by a stimulus isolator, SI, through a ventricular pace wire, VP, secured to the apex of the heart, H, with a suture. Reproducibility of heart motion is ensured with a tissue stabilizer, TS. A, differential amplifier; A/D, analog-to-digital converter; E, electrocardiogram; PC, personal computer; V, ventilation timing signal; VN, mechanical ventilator. (B and C) Timing waveforms demonstrate asynchronous acquisition (B) and synchronization of the acquisition with the cardiac cycle using pacing (C). The insets illustrate ventricular capture with widening of the paced electrocardiogram (ECG) complex (C), compared with the native ECG (B). (D and E) A grid stabilizer (Fig. S2 C and D) provides improved suppression of motion artifact for single-myocyte imaging. Sequential two-photon microscopy frames using a previously demonstrated ring stabilizer (D) have significant frame-to-frame variation, whereas the grid stabilizer allows tracking of individual myocytes (E) (Movie S1). (F and G) Real-time cardiac gating removes myocyte motion from images with high repeatability over seconds. Summing over a series of eight frames (1 s) demonstrates the high degree of stabilization and enables improved signal-to-noise without loss of resolution in the averaged image (G). Movie S2 demonstrates real-time gating. (Scale bars in D–F, 25 µm.)
Excellent tissue stabilization proved critical to enable imaging of contracting cardiomyocytes. Gross cardiac motion in three dimensions must be suppressed to visualize and track the same myocyte at each point in the cardiac cycle. A grid stabilizer temporarily bonded to the pericardium on the surface of the heart (Fig. S2 C and D) provided the desired motion suppression while allowing a field of view encompassing several myocytes. Ungated two-photon images using a previous ring stabilizer (Fig. 1D and Movie S1) demonstrate excess motion during cardiac contraction, with significant frame-to-frame variation (14). The grid stabilizer effectively suppresses motion artifact to allow identification and tracking of individual cardiomyocytes (Fig. 1E and Movie S1).
Using cardiac pacing to perform prospective gating, myocyte contractile motion can be removed from the images (Fig. 1F and Movie S2). Frame-to-frame variation is due only to motion from blood flow, which is not synchronized to the acquisition during pacing. Motion from ventilation can be eliminated either using prospective triggering of the ventilator or by introducing brief pauses (seconds in duration) in the ventilator drive waveform, which was done for simplicity in this work. Excellent frame-to-frame gated image stability (Fig. S3) prevents loss of image resolution from motion artifact and allows the system to approach the resolution of the optics in tissue. Image averaging can be performed to improve signal-to-noise while maintaining subcellular resolution of transverse tubule structure measuring <2 µm (Fig. 1G). Real-time prospective gating also enables advanced imaging modes in vivo, including depth stacks and 3D imaging as well as image mosaicking (Fig. S4). Importantly, each line in the prospective gated image corresponds to a different point in the cardiac cycle and synchronization is highly stable from one frame to the next. The image of the myocardium captured by the microscope in prospective gated mode is essentially frozen from one frame to the next. The structure in the gated image sequence is, however, distorted by the presence of contractile deformation of the tissue surface. To remove spatial distortion present in the images and to visualize contraction of the myocardium, each line of the image must be sampled at all points in time throughout the contractile cycle.
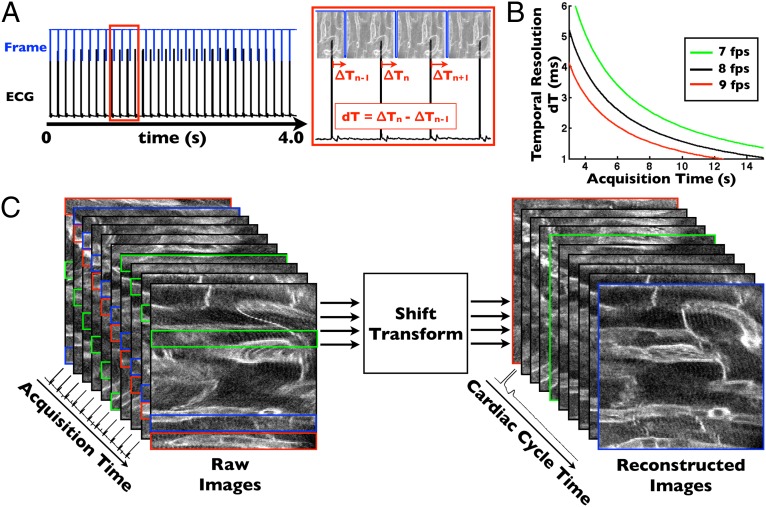
We developed an image acquisition algorithm called PSSM, which uses cardiac pacing to efficiently construct true motion-artifact-free images at every point in the cardiac cycle. Even typical fast confocal and two-photon microscopy modes (8–16 frames per second) suffer the limitation that image acquisition time is relatively long compared with the cardiac cycle in the mouse. Because cardiorespiratory motion is periodic and made highly reproducible on the microscale by the tissue stabilizer, sequential segmented acquisition schemes can be devised for intravital microscopy, where an image is sampled over successive sequential cardiac cycles, analogous to algorithms used in magnetic resonance imaging (19). To implement PSSM, we program a slight frequency shift between the microscope acquisition rate and the pacemaker rate, resulting in sequential phase shifts between acquired images (Fig. 2A). The frequency difference between the microscope and heart rate determines the acquisition time over which an entire cardiac cycle is sampled, with a longer acquisition time providing higher temporal resolution (Fig. 2B). Over a period of seconds, the entire cardiac cycle can be sampled with millisecond time resolution and without loss of image resolution. Image processing is accomplished with a time-shift transform of the acquired image sequence (Fig. 2C, Movie S3, and Materials and Methods). Each image in the PSSM sequence represents a single point in time and the sequence of images spans the cardiac cycle with the specified temporal resolution (Fig. S5). Compared with algorithms using retrospective gating, PSSM is highly efficient, using each line of the acquired image sequence to contribute to the processed images and thereby minimizing overall acquisition time. PSSM enables high-resolution imaging of cardiomyocyte subcellular structure, including visualization of transverse tubule structure, microvasculature, endothelial cells, and cell nuclei (Fig. 3 A–D). Elimination of motion artifact in the images improves image resolution compared with ungated images and enables averaging to increase signal-to-noise (Fig. S6). In vivo image resolution and quality are comparable in the PSSM images to those taken ex vivo (Fig. S1). Individual cardiomyocyte contraction is highly reproducible from beat to beat, as evidenced by the ability to construct images of contracting myocytes with high resolution over multiple heart beats using the PSSM technique (Movies S3 and S4).
Fig. 2.
Prospective sequential segmented microscopy enables motion-artifact-free imaging of the beating heart. (A) Introduction of a precise frequency difference between the microscope acquisition rate and the cardiac pacing rate enables high-resolution temporal sampling of the cardiac cycle over multiple heart beats. Each successive frame is slightly shifted from the prior with respect to the cardiac cycle by an amount dT, which defines the temporal sampling resolution. (B) Temporal sampling resolution is determined by the acquisition time of the gated sequence, which is inversely related to the frequency difference between the microscope frame rate and the paced heart rate. (C) Image processing scheme. Fast raster-scanned image lines are time-shifted to construct an image at a specific point in the cardiac cycle. Each constructed image consists of lines acquired over multiple heart beats. Movie S3 shows raw and processed image sequences.
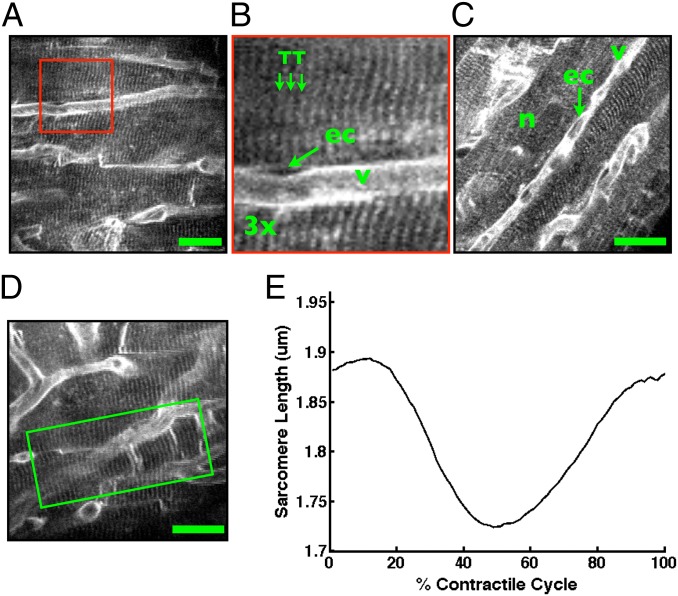
Fig. 3.
Intravital microscopy of cardiomyocyte structure and function in the beating heart. (A–D) Two-photon images reveal subcellular structure in individual cardiomyocytes in the contracting heart. Capillary vessels, v, endothelial cells, ec, cell nuclei, n, and cardiomyocyte transverse tubule structure, TT, are clearly resolved. Image in B is a zoom view of the boxed region in A. Images here are in diastole. Motion-artifact-free images can be formed at every point in the cardiac cycle (Figs. S5 and S6), however, allowing visualization of myocyte contraction. Movies S3 and S4 demonstrate contracting cardiomyocytes in vivo in the beating heart. (E) Measurement of the single-cell contractile cycle from an individual cardiomyocyte (shown in D and Movie S4). Mean sarcomere length is computed over a region of interest (green box in D) at every point in the cardiac cycle by Fourier transform analysis of the sarcomere striations in PSSM images (Fig. S7 and Materials and Methods). (Scale bars in A, C, and D, 20 µm.)
We next measured the single cardiomyocyte contractile cycle in vivo in the beating heart (Fig. 3 D and E). We analyzed PSSM images for individual cardiomyocytes and measured average sarcomere length at each point in the cardiac cycle using a frequency analysis algorithm (Materials and Methods and Fig. S7). The algorithm analyzes the mean sarcomere frequency of a selected region of interest at each point of the cardiac cycle to compute the spatially averaged cardiomyocyte contractile cycle. Manual measurements of sarcomere length made directly from a PSSM image exhibit a distribution of sarcomere lengths within the region of interest (Fig. S8 A and B). A manual average measurement along a single line at the center of the region of interest yields good correspondence with the contractile cycle produced by the algorithm over the entire region of interest (Fig. S8C), with small differences in the curve attributable to the differences in the number and positions of the sarcomeres included in the respective average measurements.
Discussion
We describe an advanced approach for intravital imaging of cardiomyocytes in the beating heart that provides motion-artifact-free images with millisecond time resolution and subcellular spatial resolution at any point in the cardiac cycle. This technology, based on multiple advances, enables visualization and quantitative analysis of single-cell function. The method is broadly applicable and suitable for use with commercial microscopes and any mouse strain. We demonstrate imaging in the mouse heart in vivo, but these methods should scale to larger animal models as well and will also readily apply to the widely used Langendorf ejecting heart preparation (20). The general gating algorithm can also be applied with other imaging modalities.
Compared with previous work, the present results demonstrate a substantial and enabling improvement in both spatial and temporal resolution. Prior approaches using tissue stabilization schemes alone (11, 15) were not capable of imaging with subcellular resolution and did not provide precise gating to account for artifacts introduced with contraction. Without gating, spatial resolution suffers and temporal resolution is very low owing to the constraints of the frame rate of the microscope. Our prior approach introduced retrospective gating schemes (14) but was only capable of providing high-spatial-resolution images in diastole when the heart motion is at a minimum. The prospective gating technique eliminates motion artifact to below the expected optical resolution of the microscope in tissue (Fig. S3), thereby enabling spatial image resolutions comparable to those achieved ex vivo. Furthermore, the temporal resolution of prior retrospective approaches is low (Table S1). Using prospective gating and taking advantage of the high reproducibility of motion ensured by the stabilizer, the current approach overcomes these limitations to enable resolution and quantitative measurement of repeating events during the contractile cycle.
In cardiovascular physiology, the single-cardiomyocyte contractile cycle is a fundamental measure of the state of the cell. Just as the electrocardiogram and pressure–volume loops describe the contractile physiology of the whole heart, measurement of cell length and contractile shortening describe the physiology of the cardiomyocyte. Measurement of single-cell contractile function is challenging owing to the demands of precisely localizing the myocyte in space and time over the cardiac cycle and of imaging sarcomere length changes near the resolution limit of the microscope. As a result, the cardiomyocyte contractile cycle is typically measured from isolated myocytes in a culture dish. This measurement, however, cannot accurately reflect the complex interaction of a cardiomyocyte with its native environment to determine overall heart function. The degree to which single-cardiomyocyte contractile function varies among myocytes in a network of cells in the same heart and how these cells respond individually and collectively to pharmacotherapy in vivo remain important questions in cardiovascular physiology. The techniques described here will be useful to characterize this heterogeneity in cardiomyocyte function and its implications in healthy and diseased hearts.
Frequency analysis methods have been applied previously for the measurement of sarcomere length using microscopy in the excised rodent heart (3, 4) and in isolated cardiac myocytes (21) as well as in skeletal muscle (22). There are multiple challenges in measuring sarcomere spacing in the beating heart with microscopy. These include the changes in cell orientation during contraction, the intrinsic distribution of sarcomere lengths within and between myocytes, and the impact of other nonmyocyte components of the heart tissue on the measurement algorithm (e.g., vessels). We developed our approach (Fig. S7) considering prior work and in analogy to the typical algorithms used in commercially available myocyte imaging systems for in vitro preparations (4, 23, 24). Previous efforts to optimize algorithms for sarcomere length estimation in isolated cardiomyocytes can also be considered for intravital microscopy (21).
Combined with transgenic mouse models of disease and molecularly targeted fluorescent probes, intravital microscopy molecular imaging offers a powerful way to study individual cardiomyocyte physiology and cell-to-cell heterogeneity. Beyond measurement of contractile function, our methods will be useful for imaging and quantitative analysis of other dynamic events in the myocardium and microvasculature, including membrane potential changes and arrhythmia, calcium signaling, leukocyte trafficking, cell–cell communication, and investigation of cellular responses to injury and pharmacologic intervention.
Materials and Methods
Microscope Setup.
The cardiomyocyte imaging system (Fig. 1A and Fig. S2) was developed using a commercially available confocal and multiphoton imaging system (FV1000-MPE; Olympus). A tunable modelocked Ti:sapphire laser provided short-pulse illumination for two-photon imaging (MaiTai DeepSee; Spectra Physics). A low-magnification air objective (XL Fluor 2×/340 NA 0.14; Olympus) was used in confocal mode for alignment whereas a high-N.A. water-immersion objective with near-infrared correction (XLPlan N 25× N.A. 1.05; Olympus) was used for high-magnification confocal and two-photon microscopy of cardiomyocytes. The microscope ran in fast imaging mode with bidirectional scanning at 8–16 frames per second and images were recorded using the standard software interface for the commercial unit running on a personal computer. A frame timing signal generated by the microscope hardware consisting of a pulse train at the acquisition frame rate was acquired using a data acquisition analog-to-digital converter (ADC) card (PCI-6229; National Instruments) and a second personal computer. A single electrocardiogram lead and a timing synchronization pulse train from the animal ventilator (INSPIRA ASV 55-7058; Harvard Apparatus) were also acquired. Custom-designed software was developed in Labview (National Instruments) for recording of timing signals and generation of a pacing waveform. The pacing voltage waveform from the ADC passed through a stimulus isolator (2200; A-M Systems) to produce a pacing current stimulus of 2-ms pulses with amplitude set to reach the ventricular capture threshold, typically 100–300 µA. The ECG was recorded using a low-noise differential amplifier (DP-301; Warner Instruments).
Tissue Stabilizer.
The tissue stabilizer (Fig. S2C) consisted of a machined stainless steel ring measuring 3.5 mm in diameter with 2.2-mm inner diameter and 0.5-mm thickness. An electron microscopy grid (12433-CU; Ted Pella) with 3.0-mm diameter and 230-µm hole diameter was affixed to the ring to construct a grid stabilizer. The stabilizer also has a thin ring of silicone on the upper surface to hold a drop of water on the tissue surface for use with water-immersion objective lenses. The entire unit was soldered to a stainless steel extension arm, which was then mounted on a base plate with translation and rotation capability for precise positioning.
Mice.
A total of 65 C57BL/6 mice were used for the experiments and all procedures were approved by the Institutional Animal Care and Use Committee at the Massachusetts General Hospital. Mice were between 8 and 20 wk of age with weights between 20–30 g. The breakdown of mice for the experiments included 10 mice for testing and optimization of the grid stabilizer, 10 mice for testing the pacing hardware and developing the surgical protocol, 15 mice for developing the staining protocol, and 30 mice for development and validation of the prospective cardiac gating and sequential segmented microscopy acquisition protocols for myocyte imaging.
Tissue Staining.
Multiple membrane-specific fluorescent stains were tested for in vivo labeling of cardiomyocyte structure (di-4-anepps, di-8-anepps, RH237, CellMask, and di-2-anepeq, all available from Invitrogen). Direct topical staining of the surgically exposed heart proved relatively ineffective compared with i.v. labeling. The water-soluble dye di-2-anepeq provided optimal brightness for i.v. injection (Fig. S1). Stock solution of 5 mM concentration was made from 5 mg di-2-anepeq dissolved in 1.83 mL deionized water and stored at ±20 °C. For a single experiment, 60–80 µL of the stock solution was injected i.v. via a catheter in the tail vein 30–60 min before imaging. Assuming distribution in a typical blood volume of 2.0 mL, the estimated average cellular level concentration of the stain was <200 µM. Two-photon excitation of the dye was optimal at 910-nm wavelength and epifluorescence was collected using a band-pass optical filter between 575 and 630 nm. Several animals were observed over 24 h after injection of the stain and demonstrated normal activity with no obvious adverse effects from the stain administration.
Surgical Preparation.
Mice were induced with anesthesia using 3–4% isoflurane mixed with 2 L/min of 100% oxygen and maintenance anesthesia was provided with 1.5–2.5% isoflurane mixed with oxygen. Analgesia was given with 0.1 mg/kg of buprenorphine administered before the surgical procedure. Animals were intubated using a 22-gauge angiocath as an endotracheal tube and mechanically ventilated with a small animal ventilator (INSPIRA ASV 55-7058; Harvard Apparatus), with tidal volume and respiratory rate parameters set by weight-based manufacturer guidelines. Normal saline (0.5–1.0 mL) was given by i.p. injection to prevent dehydration during the surgical procedure. Animal body temperature was maintained at 37 °C using a temperature-controlled heat plate (Olympus) and a surgical drape over the imaging field to prevent heat loss (Fig. S2B). A left thoracotomy was performed in the fourth left intercostal space and the anterior and lateral aspects of the left ventricle were exposed using retractors (Fig. S2A). The parietal pericardium was carefully incised, allowing direct access to the thin visceral pericardium and the myocardial surface. For pacing, the terminal 1-mm length of a small-diameter insulated wire (diameter 50 μm bare/114 μm coated; AM Systems) was stripped away and secured in contact with the ventricular myocardium using a superficial 8.0 nylon suture (Ethicon) (25). The tissue stabilizer was then bonded directly to the epicardial surface of the heart using a thin (<100 µm) layer of cyanoacrylate adhesive (e.g., VetBond). Adhesive was applied only to the metal regions of the grid stabilizer, leaving unobstructed observation windows of myocardium without glue contact for microscopy (Fig. S1D). To enable complete control of ventilation, animals were given a low-dose skeletal muscle paralytic, pancuronium, at 0.1–0.2 mg/kg dosing, by i.v. tail vein injection. At the completion of imaging experiments the mice in these experiments were killed using a CO2 gas chamber in accordance with approved protocol.
Image Acquisition.
Images were streamed to disk during acquisition using the standard Olympus software interface. Timing waveforms were acquired with Labview. With the stabilizer in place, images were acquired first with the low-magnification air objective lens to align the grid stabilizer to the illumination path. Then, using multiphoton mode, images were acquired in ungated fashion to pan and survey for regions of interest (Fig. 1E and Movie S1). Images were acquired at eight frames per second with 256 × 256 pixels, and optical zoom between 2× and 8× was used to increase image resolution at smaller field of view. When an area of interest was identified, prospective gating with cardiac pacing (Fig. 1F and Movie S2) was used to acquire images without myocyte motion and to assess capillary flow in real time. PSSM was then used to analyze single-myocyte contractile function. During PSSM sequences, the ventilator was transiently paused to eliminate residual motion artifact from changes in lung volume. This was similar to a breath-hold maneuver frequently used during mechanical ventilation of humans and was well tolerated by the animals. PSSM sequences with millisecond time resolution require ventilator pauses of only several seconds. Alternatively, prospective triggering of the ventilator can be used if PSSM sequences of long duration are required. Table S1 and Movie S5 compare acquisition modes.
Image Characterization Using Fluorescent Beads.
Fluorescent polystyrene beads (FluoSpheres, 10 µm diameter; Invitrogen) were directly injected into the left ventricle of a mouse (50-μL injection) in vivo. The beads become trapped in the capillaries in the heart and can be imaged as fiducial markers for image stability measurements. Gated image sequences of the same bead (Fig. S3A) positioned at multiple locations in the cardiac cycle were analyzed for the variation of the centroid position of the bead across multiple cardiac cycles. The distance of the bead in each image was computed relative to the average position of the bead in the image sequence, and the SD of this distance across the sequence of images was plotted as centroid variation (Fig. S3B).
PSSM Image Processing and Data Analysis.
Image analysis was performed in ImageJ and Matlab. Presented images are not filtered. The raw PSSM image data were processed as a 3D matrix (row, column, and time) and reconstruction of the cardiac cycle was efficiently performed using a matrix shift applied along the time dimension to each image row (Fig. 2C). The shift size, Si, measured in pixels along the time dimension in the PSSM data matrix, can be represented for each row in the processed matrix as Si = NI * i/NR, where i is the row index, NR is the number of rows in the image, and NI is the number of images in the PSSM sequence. For acquisition frequency, fA, and pacing frequency, fP, the acquisition time, T, for the PSSM sequence is T = 1/(fP − fA) and the number of images in the PSSM sequence, NI = fA/(fP − fA). The temporal resolution, dT, for the PSSM sequence is then dT = 1/(NI *fA) (Fig. 2B). A brief segment of Matlab code used to perform the matrix shift is included in SI Appendix. For contractile function analysis (Fig. 3 D and E and Fig. S7), individual images from the PSSM sequence, each representing a single point in the cardiac cycle, were analyzed using Fourier transformation. Images were first rotated and interpolated to place sarcomere striations perpendicular to the image lines. A region of interest (ROI) was then chosen over the myoycyte. Individual lines within the ROI were then band-pass–filtered for spatial frequencies between 1 and 3 µm using an eighth-order Chebyshev Type II filter. A Blackman–Harris window was applied to the filtered lines to minimize edge effects from the line scans. Frequency content of individual lines was determined using a fast Fourier transform (FFT) and the frequency spectra were then averaged over all lines in the ROI to produce an average frequency spectrum for the ROI. The mean sarcomere length for a specific point in the cardiac cycle was determined as the inverse of the peak frequency of the averaged frequency spectrum for the ROI. Moving average 15-point temporal smoothing was applied to the averaged frequency spectrum in the plotted trace (Fig. 3E). Analogous approaches for sarcomere length measurement have been performed in isolated cardiac myocytes (23, 24), noncontracting Langendorf heart preparations (3, 4), and skeletal muscle in vivo (22). Manual measurements of sarcomere length (Fig. S8) were measured in ImageJ from a PSSM image in diastole by defining sub regions within the predefined ROI used for FFT analysis. The average sarcomere length for each subregion was calculated from the measured distance of five sarcomeres. The manual measurement of the contractile cycle was made from PSSM images at multiple points in the cardiac cycle by taking the average sarcomere length over the same span of sarcomeres located along a single line through the center of the ROI for each image.
Supplementary Material
Acknowledgments
We thank Matthias Nahrendorf for insight on animal experiments. The project was funded by National Institutes of Health (NIH) Contracts HHSN268201000044C and R01EB006432. A.D.A. was funded by NIH Training Grant T32HL094301.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401316111/-/DCSupplemental.
References
- 1.Rubart M. Two-photon microscopy of cells and tissue. Circ Res. 2004;95(12):1154–1166. doi: 10.1161/01.RES.0000150593.30324.42. [DOI] [PubMed] [Google Scholar]
- 2.Pittet MJ, Weissleder R. Intravital imaging. Cell. 2011;147(5):983–991. doi: 10.1016/j.cell.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botcherby EJ, et al. Fast measurement of sarcomere length and cell orientation in Langendorff-perfused hearts using remote focusing microscopy. Circ Res. 2013;113(7):863–870. doi: 10.1161/CIRCRESAHA.113.301704. [DOI] [PubMed] [Google Scholar]
- 4.Bub G, et al. Measurement and analysis of sarcomere length in rat cardiomyocytes in situ and in vitro. Am J Physiol Heart Circ Physiol. 2010;298(5):H1616–H1625. doi: 10.1152/ajpheart.00481.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hama T, Takahashi A, Ichihara A, Takamatsu T. Real time in situ confocal imaging of calcium wave in the perfused whole heart of the rat. Cell Signal. 1998;10(5):331–337. doi: 10.1016/s0898-6568(97)00136-8. [DOI] [PubMed] [Google Scholar]
- 6.Kaneko T, Tanaka H, Oyamada M, Kawata S, Takamatsu T. Three distinct types of Ca(2+) waves in Langendorff-perfused rat heart revealed by real-time confocal microscopy. Circ Res. 2000;86(10):1093–1099. doi: 10.1161/01.res.86.10.1093. [DOI] [PubMed] [Google Scholar]
- 7.Kaneko T, et al. Real-time two-photon microscopy and its application for in situ imaging. Acta Histochem Cytochem. 2001;34:399–403. [Google Scholar]
- 8.Matsumoto-Ida M, Akao M, Takeda T, Kato M, Kita T. Real-time 2-photon imaging of mitochondrial function in perfused rat hearts subjected to ischemia/reperfusion. Circulation. 2006;114(14):1497–1503. doi: 10.1161/CIRCULATIONAHA.106.628834. [DOI] [PubMed] [Google Scholar]
- 9.Minamikawa T, Cody SH, Williams DA. In situ visualization of spontaneous calcium waves within perfused whole rat heart by confocal imaging. Am J Physiol. 1997;272(1 Pt 2):H236–H243. doi: 10.1152/ajpheart.1997.272.1.H236. [DOI] [PubMed] [Google Scholar]
- 10.Rubart M, Wang E, Dunn KW, Field LJ. Two-photon molecular excitation imaging of Ca2+ transients in Langendorff-perfused mouse hearts. Am J Physiol Cell Physiol. 2003;284(6):C1654–C1668. doi: 10.1152/ajpcell.00469.2002. [DOI] [PubMed] [Google Scholar]
- 11.Li W, et al. Intravital 2-photon imaging of leukocyte trafficking in beating heart. J Clin Invest. 2012;122(7):2499–2508. doi: 10.1172/JCI62970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chilian WM, Layne SM. Coronary microvascular responses to reductions in perfusion pressure. Evidence for persistent arteriolar vasomotor tone during coronary hypoperfusion. Circ Res. 1990;66(5):1227–1238. doi: 10.1161/01.res.66.5.1227. [DOI] [PubMed] [Google Scholar]
- 13.Dumont EA, et al. Real-time imaging of apoptotic cell-membrane changes at the single-cell level in the beating murine heart. Nat Med. 2001;7(12):1352–1355. doi: 10.1038/nm1201-1352. [DOI] [PubMed] [Google Scholar]
- 14.Lee S, et al. Real-time in vivo imaging of the beating mouse heart at microscopic resolution. Nat Commun. 2012;3:1054. doi: 10.1038/ncomms2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung K, et al. Endoscopic time-lapse imaging of immune cells in infarcted mouse hearts. Circ Res. 2013;112(6):891–899. doi: 10.1161/CIRCRESAHA.111.300484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vinegoni C, Lee S, Gorbatov R, Weissleder R. Motion compensation using a suctioning stabilizer for intravital microscopy. Intravital. 2012;1(2):115–121. doi: 10.4161/intv.23017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor JM, Girkin JM, Love GD. High-resolution 3D optical microscopy inside the beating zebrafish heart using prospective optical gating. Biomed Opt Express. 2012;3(12):3043–3053. doi: 10.1364/BOE.3.003043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bu G, Adams H, Berbari EJ, Rubart M. Uniform action potential repolarization within the sarcolemma of in situ ventricular cardiomyocytes. Biophys J. 2009;96(6):2532–2546. doi: 10.1016/j.bpj.2008.12.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finn JP, Edelman RR. Black-blood and segmented k-space magnetic resonance angiography. Magn Reson Imaging Clin N Am. 1993;1(2):349–357. [PubMed] [Google Scholar]
- 20.Liao R, Podesser BK, Lim CC. The continuing evolution of the Langendorff and ejecting murine heart: New advances in cardiac phenotyping. Am J Physiol Heart Circ Physiol. 2012;303(2):H156–H167. doi: 10.1152/ajpheart.00333.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson P, Kalda M, Vendelin M. Real-time determination of sarcomere length of a single cardiomyocyte during contraction. Am J Physiol Cell Physiol. 2013;304(6):C519–C531. doi: 10.1152/ajpcell.00032.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Llewellyn ME, Barretto RP, Delp SL, Schnitzer MJ. Minimally invasive high-speed imaging of sarcomere contractile dynamics in mice and humans. Nature. 2008;454(7205):784–788. doi: 10.1038/nature07104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cazorla O, Wu Y, Irving TC, Granzier H. Titin-based modulation of calcium sensitivity of active tension in mouse skinned cardiac myocytes. Circ Res. 2001;88(10):1028–1035. doi: 10.1161/hh1001.090876. [DOI] [PubMed] [Google Scholar]
- 24.Fan D, Wannenburg T, de Tombe PP. Decreased myocyte tension development and calcium responsiveness in rat right ventricular pressure overload. Circulation. 1997;95(9):2312–2317. doi: 10.1161/01.cir.95.9.2312. [DOI] [PubMed] [Google Scholar]
- 25.Berul CI, Aronovitz MJ, Wang PJ, Mendelsohn ME. In vivo cardiac electrophysiology studies in the mouse. Circulation. 1996;94(10):2641–2648. doi: 10.1161/01.cir.94.10.2641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.