Significance
When DNA breakage results in a 3′-PO4 terminus, the end is considered “dirty” because it cannot prime repair synthesis by DNA polymerases or sealing by classic DNA ligases. This paper shows how the noncanonical ligase RtcB can evade the dirty end problem by transferring GMP to the DNA 3′-PO4 to form a DNA3′pp5′G cap. DNA capping blocks 3′ end-resection and healing, while permitting DNA synthesis from the cap guanosine–3′-OH primer by polymerases. Cap-primed synthesis embeds a pyrophosphate linkage in DNA. A 3′ decapping activity inherent in the disease-associated repair enzyme aprataxin suggests that DNA capping is a dynamic process.
Keywords: DNA repair, 3′ exonuclease
Abstract
Many biological scenarios generate “dirty” DNA 3′-PO4 ends that cannot be sealed by classic DNA ligases or extended by DNA polymerases. The noncanonical ligase RtcB can “cap” these ends via a unique chemical mechanism entailing transfer of GMP from a covalent RtcB–GMP intermediate to a DNA 3′-PO4 to form DNA3′pp5′G. Here, we show that capping protects DNA 3′ ends from resection by Escherichia coli exonucleases I and III and from end-healing by T4 polynucleotide 3′ phosphatase. By contrast, the cap is an effective primer for DNA synthesis. E. coli DNA polymerase I and Mycobacterium DinB1 extend the DNAppG primer to form an alkali-labile DNApp(rG)pDNA product. The addition of dNTP depends on pairing of the cap guanine with an opposing cytosine in the template strand. Aprataxin, an enzyme implicated in repair of A5′pp5′DNA ends formed during abortive ligation by classic ligases, is highly effective as a DNA 3′ decapping enzyme, converting DNAppG to DNA3′p and GMP. We conclude that the biochemical impact of DNA capping is to prevent resection and healing of a 3′-PO4 end, while permitting DNA synthesis, at the price of embedding a ribonucleotide and a pyrophosphate linkage in the repaired strand. Aprataxin affords a means to counter the impact of DNA capping.
The synthesis and repair of DNA are driven by enzymatic reactions at 3′ ends. The terminal 3′-OH is the nucleophile that primes 3′–5′ phosphodiester bond formation by DNA polymerases and DNA ligases. To contend with the many situations in biology when incision of DNA generates a 3′-PO4 (a so-called “dirty” end, which cannot be extended by polymerases or sealed by classic ligases), nature has evolved an assortment of end-healing enzymes that remove the 3′-PO4 and/or resect 3′ nucleotides (1). We recently described a different fate for DNA 3′-PO4 ends at the hands of the unconventional ligase RtcB (2, 3), which adds a guanylate “cap” to form DNA3′pp5′G (4). DNA capping by RtcB is highly efficient, whether at the 3′-PO4 ends of ssDNAs or at a nick in duplex DNA (4). DNA 3′ capping has tantalizing implications. For analogy, one need only consider the multifaceted role of the 5′ cap structure, m7GpppN–, in RNA metabolism (5). Here, we address the consequences of DNA 3′ capping for the reactions of exemplary exonucleases, phosphatases, and polymerases at 3′ ends. In addition, we illuminate aprataxin as a DNA 3′ decapping enzyme.
Results and Discussion
DNA Capping Confers Resistance to End Resection by 3′ Exonucleases and 3′ Phosphatase.
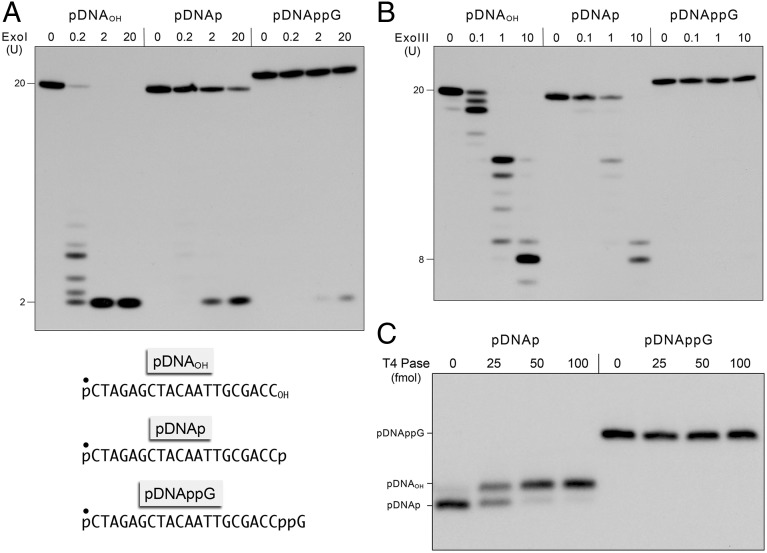
Escherichia coli exonucleases I and III are DNA repair enzymes that hydrolyze 3′-OH DNA ends in a stepwise fashion to liberate 5′ deoxyribonucleoside monophosphate (dNMP) mononucleotide products. ExoI acts processively on ssDNA with a 3′-OH end but has minimal activity at a 3′-PO4 end (6, 7). By contrast, ExoIII distributively resects 3′-OH and 3′-PO4 DNA ends, the latter by virtue of its intrinsic DNA 3′ phosphatase activity (7–9). Here, we gauged the effect of DNA 3′ capping on end resection by these two canonical 3′ exonucleases. In the experiment shown in Fig. 1A, 5′ 32P-labeled 20-mer DNA strands of an identical nucleobase sequence, with 3′-OH (pDNAOH), 3′-PO4 (pDNAp), or 3′ capped (pDNAppG) termini (chemical structures of the 3′ termini are shown in Fig. S1) were reacted with increasing concentrations of ExoI. The 32P-labeled products were resolved by denaturing PAGE. Two units (U) of ExoI sufficed to convert the pDNAOH substrate quantitatively to a dinucleotide product; 0.2 U effected resection of 97% of the input 20-mer to yield a mixture of short oligonucleotides (2-mer to 8-mer). The pDNAp substrate was about two orders of magnitude less sensitive to digestion than pDNAOH (i.e., 2 and 20 U of ExoI converted 43% and 75%, respectively, of the input 20-mer pDNAp to dinucleotide product) (Fig. 1A). The capped DNAppG substrate was even more refractory, such that 2 and 20 U of ExoI converted only 4% and 15%, respectively, of the input capped DNA to dinucleotide product. From these data, we estimate that the 3′ cap confers at least 500-fold resistance to ExoI vs. a 3′-OH and 10-fold resistance vs. a 3′-PO4.
Fig. 1.
DNA capping confers resistance to end resection by 3′ exonucleases and end-healing by polynucleotide 3′ phosphatase. (A) Reaction mixtures (10 μL) containing 67 mM glycine-KOH (pH 9.5); 6.7 mM MgCl2; 10 mM β-mercaptoethanol; 0.2 pmol of 5′ 32P-labeled pDNAOH, pDNAp, or pDNAppG substrates (with the 5′ 32P-label denoted by ●, Lower) and 0, 0.2, 2, or 20 U of E. coli exonuclease I were incubated at 37 °C for 20 min. (B) Reaction mixtures (10 μL) containing 10 mM Bis-Tris⋅propane⋅HCl (pH 7.0); 10 mM MgCl2; 1 mM DTT; 0.2 pmol of 5′ 32P-labeled pDNAOH, pDNAp, or pDNAppG; and 0, 0.1, 1, or 10 U of E. coli exonuclease III were incubated at 37 °C for 20 min. The exonuclease reactions were quenched with 10 μL of 90% (vol/vol) formamide and 50 mM EDTA, and the products were analyzed by urea-PAGE and visualized by autoradiography. The sizes (in nucleotides) of the input substrate and the major products of 3′ resection are indicated on the left. (C) Reaction mixtures (10 μL) containing 100 mM Tris⋅actetate (pH 6.0); 10 mM MgCl2; 2 mM DTT; 0.2 pmol of 5′ 32P-labeled pDNAp or pDNAppG; and 0, 25, 50, or 100 fmol of T4 Pnkp 3′ phosphatase (Pase) as specified were incubated at 22 °C for 20 min. The reactions were quenched with formamide and EDTA, and the products were analyzed by urea-PAGE and visualized by autoradiography. The positions of the pDNAOH, pDNAp, and pDNAppG strands are indicated on the left.
Treatment of pDNAOH with 0.1, 1, and 10 U of ExoIII converted 77%, >99%, and >99%, respectively, of the input substrate to shorter species; consistent with a distributive profile, the sizes of the product clusters shortened with increasing ExoIII, with the predominant product being an 8-mer at 10 U (Fig. 1B). The pDNAp substrate was less sensitive to ExoIII (0.1, 1, and 10 U converted 9%, 55%, and 99%, respectively, of the input substrate to shorter species), although the product size distribution was basically unchanged, reflecting the extra step of 3′-PO4 hydrolysis before the onset of the 3′ phosphodiesterase reaction. The salient finding was that the pDNAppG substrate was refractory to resection, with only 3% of the input capped substrate being shortened by 10 U of ExoIII.
The polynucleotide 3′ phosphatase (Pase) domain of bacteriophage T4 polynucleotide kinase-phosphatase (Pnkp) exemplifies a widely distributed acyl-phosphatase family of 3′ end-healing enzymes involved in nucleic acid repair (10, 11). Reaction of 25, 50, and 100 fmol of T4 Pase with the 20-mer pDNAp elicited progressive conversion of 49%, 90%, and 92%, respectively, of the input substrate to a slower migrating pDNAOH product (Fig. 1C). By contrast, the pDNAppG substrate was completely resistant to end-healing by up to 100 fmol of T4 Pase (Fig. 1C).
These experiments illuminate a protective function for the 3′ cap against DNA exonucleases and 3′ phosphatase, analogous to that of the 5′ RNA cap against 5′ exoribonucleases and phosphomonoesterases.
DNA3′pp5′G Ends Are Extended by DNA Polymerases.
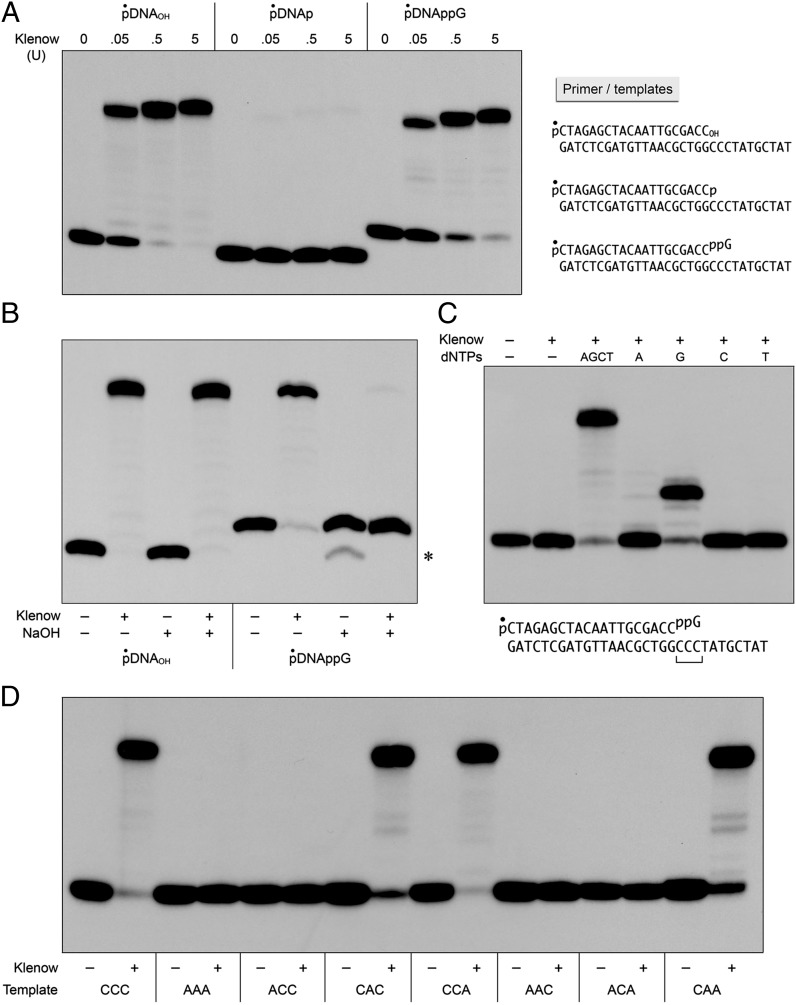
The resistance of capped DNA ends to 3′ processing raises the important question of their fate once formed, especially when the cap is introduced at 3′-PO4 ends at duplex DNA gaps. A key issue is whether the cap guanylate might prime DNA synthesis by DNA repair polymerases. To answer this question, we focused first on a prototypal A family polymerase, the E. coli DNA polymerase I (Pol1) Klenow fragment (specifically a mutant Klenow polymerase devoid of proofreading 3′ exonuclease). We compared the ability of Klenow Pol1 to extend a series of primer-templates formed by annealing the 5′ 32P-labeled 20-mer pDNAOH, pDNAp, or pDNAppG strand to a complementary 31-mer DNA strand to form a 20-bp duplex with an 11-nt single-strand 5′ tail (Fig. 2A). The sequence of the tail immediately flanking the duplex junction consisted of three consecutive deoxycytidines that could potentially pair with the 3′ cap guanylate of the pDNAppG strand (detailed view of the chemical structure of the 3′ capped primer-template is shown in Fig. S2). Reaction of the pDNAOH primer-template with 0.05, 0.5, and 5 U of Klenow Pol1 in the presence of all four dNTPs resulted in the extension of 57%, 96%, and 98%, respectively, of the input primer strand (Fig. 2A). At 0.05 U of Pol1, the predominant outcome was fill-in synthesis to the end of the template strand; increasing Klenow Pol1 to 0.5 and 5 U resulted in the addition of a single extra nontemplated nucleotide. As expected, the pDNAp primer-template was ineffective (Fig. 2A). Less than 1% of the input pDNAp strand was filled in by 5 U of Klenow Pol1. The instructive finding was that the cap guanylate served as an effective primer for Klenow Pol1, with 0.05, 0.5, and 5 U of enzyme catalyzing the extension of 35%, 81%, and 93%, respectively, of the input pDNAppG strand (Fig. 2A).
Fig. 2.
DNAppG primes DNA synthesis by E. coli polymerase I. (A) Cap priming of DNA synthesis. Reaction mixtures (10 μL) containing 10 mM Tris⋅HCl (pH 7.9); 50 mM NaCl; 10 mM MgCl2; 1 mM DTT; 4 mM dNTPs; 0.2 pmol of 5′ 32P-labeled pDNAOH, pDNAp, or pDNAppG primer-templates (Right); and 0, 0.05, 0.5, or 5 U of Klenow Pol1 as specified were incubated at 37 °C for 20 min. (B) Cap-primed DNA synthesis embeds the cap ribonucleotide. Reaction mixtures (10 μL) containing 10 mM Tris⋅HCl (pH 7.9), 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 4 mM dNTPs, 0.2 pmol of 5′ 32P-labeled pDNAOH or pDNAppG primer-template, and either no enzyme (−) or 5 U of Klenow Pol1 (+) were incubated at 37 °C for 20 min. The reaction mixtures were adjusted to 50 mM EDTA and then supplemented with either 1.2 μL of 3 M NaOH (+) or 1.2 μL of 3 M NaCl (−) and incubated at 22 °C overnight, after which the NaOH mixtures were neutralized by adding 1.2 μL of 3 M HCl. The samples were analyzed by urea-PAGE and visualized by autoradiography. (Right) Asterisk denotes that a minority of the input 32P-labeled pDNAppG strand was converted by the alkali treatment to a species that migrated about one nucleotide step faster during PAGE. Because this species was not observed upon alkali treatment of the pDNApp(rG)pDNA product, we speculate that alkali might promote attack of a cap ribose hydroxyl on the cap phosphates to form pDNAp or pDNAOH. (C) dNTP requirement for cap-primed DNA synthesis. Reaction mixtures (10 μL) containing 10 mM Tris⋅HCl (pH 7.9); 50 mM NaCl; 10 mM MgCl2; 1 mM DTT; 0.2 pmol of 5′ 32P-labeled pDNAppG primer-template (Lower); no dNTPs (−), 4 mM dNTPs (AGCT), or 1 mM individual dNTPs (A, G, C, or T); and no enzyme (−) or 5 U of Klenow Pol1 (+) were incubated at 37 °C for 20 min. (D) Template requirement for cap-primed DNA synthesis. Reaction mixtures (10 μL) containing 10 mM Tris⋅HCl (pH 7.9), 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 4 mM dNTPs, 0.2 pmol of 5′ 32P-labeled pDNAppG primer-templates with different trinucleotide template sequences flanking the primer 3′ end (as specified below the lanes), and either no enzyme (−) or 5 U of Klenow Pol1 (+) were incubated at 37 °C for 20 min. The reaction mixtures were supplemented with formamide and EDTA, and the products were analyzed by urea-PAGE and visualized by autoradiography.
Direct proof that the cap guanosine 3′-OH was the primer nucleophile for the first step of dNMP addition by Klenow Pol1 was obtained by treating the products of fill-in synthesis on the pDNAOH and pDNAppG primer-templates with NaOH (0.3 M for 16 h at 22 °C), which will cleave any ribonucleoside embedded in a DNA polynucleotide via base-catalyzed attack of the ribose O2′ on the (rN)pN phosphodiester. Whereas extension of the pDNAOH primer yielded a filled-in strand that was resistant to alkali (i.e., an all-DNA strand), the filled-in product of pDNAppG extension was sensitive to alkali, being converted to a shorter pDNAppGp species (Fig. 2B). We conclude that the cap guanylate, and perforce the cap pyrophosphate linkage, is embedded in the product of DNA synthesis: pDNApp(rG)pDNA.
We conducted similar studies of the utilization of 3′ capped DNA as a primer for Mycobacterium smegmatis DinB1, a 463-aa Y family DNA repair polymerase (Fig. S3). Reaction of the pDNAOH primer-template with 0.1 and 1 pmol of recombinant DinB1 in the presence of all four dNTPs resulted in the extension of 87% and 95%, respectively, of the input primer strand (Fig. S3A). At 0.1 pmol of DinB, the predominant product had undergone three steps of dNMP addition; increasing DinB1 to 1 and 10 pmol resulted in fill-in to the end of the template strand. The pDNAp primer-template was unreactive, with less than 0.4% of the input pDNAp strand being extended by 10 pmol of DinB1 (Fig. S3A). The 3′ cap was an effective primer, with 0.1, 1, and 10 pmol of DinB1 catalyzing the extension of 79%, 91% and 93%, respectively, of the input pDNAppG strand (Fig. S3A). The predominant products at 0.1 and 1 pmol of DinB1 had undergone one and two steps of dNMP addition, respectively; complete fill-in synthesis was observed at 10 pmol of DinB1 (Fig. S3A). The filled-in product of pDNAppG extension by DinB1 was hydrolyzed by alkali to pDNAppGp (Fig. S3B). Thus, cap-primed DNA synthesis pertains to exemplary DNA polymerases from two different families.
dNTP and Template Requirements for DNA Synthesis at a 3′ Cap Primer.
No extension of the pDNAppG primer-template by Klenow Pol1 or DinB1 was seen in the absence of exogenous dNTPs (Fig. 2C and Fig. S3C). By performing the reactions in the presence of single dNTPs, we found that dGTP was the sole effective substrate for nucleotide addition by Klenow Pol1 (Fig. 2C), indicating that the first steps of synthesis were templated by the cytosines in the single-strand tail (Fig. S2). The predominant product seen when Klenow Pol1 acted on the pDNAppG primer in the presence of only dGTP was extended by three nucleotide steps (Fig. 2C). When DinB1 reacted with the capped primer-template in the presence of dGTP, the predominant product was extended by two deoxyguanosine monophosphate (dGMP) addition steps, with a minority fraction extended by three steps. We suggest that the n + 2 product pDNApp(rG)GG can slip back on the template CCC sequence to allow incorporation of a third dGMP nucleotide when the “correct” dNTP is unavailable for addition at the next template position. DinB1 was less faithful than Klenow Pol1 with respect to its capacity to add nonguanine dNMPs to the pDNAppG primer-template (Fig. S3C). There was a hierarchy of single-step misincorporation efficiencies across from the template cytidines, with dATP (80%) > dTTP (45%) > dCTP (14%), compared with 96% extension with dGTP.
The role of the template sequence in cap primer utilization by Klenow Pol1 was addressed in the experiment shown in Fig. 2D, in which the identities of the three template strand nucleobases immediately flanking the primer terminus were varied as cytosine or adenine and the primer-templates were reacted with polymerase plus all four dNTPs. The key initial finding was that changing CCC to AAA abolished dNMP addition to the pDNAppG strand, signifying that the cap guanine, per se, must pair with a template cytosine to prime synthesis (as depicted in Fig. S2). However, to which of the three cytosine bases does it pair, and might the pyrophosphate linkage of the cap allow the guanosine primer the option to pair with more than one cytosine within the CCC triplet? By surveying all C/A combinations within the trinucleotide, we found that a template cytosine immediately opposite the cap guanosine was necessary and sufficient for effective priming of fill-in synthesis (i.e., CAC, CCA, and CAA template trinucleotides did prime), whereas any template containing adenine opposite the cap guanosine was inactive (i.e., ACC, AAC, and ACA did not prime) (Fig. 2D).
DinB1 also required pairing of the cap guanosine with the template strand (i.e., the AAA template did not support DNA synthesis above and beyond a low level of unfaithful single-nucleotide addition) (Fig. S3D). Extension was also limited to a single-nucleotide step when the template sequence was AAC or ACA (Fig. S3D). By contrast, efficient fill-in synthesis was observed in all cases when the template had a cytosine base opposite the cap guanosine (CAC, CCA, and CAA). The main difference between DinB1 and Klenow Pol1 was that DinB1 was able to perform fill-in synthesis when the template sequence was ACC (Fig. S3D), whereas Klenow Pol1 was not (Fig. 2D). The more open structure typical of Y family polymerases in complex with DNA primer-template and dNTPs may account for the lower fidelity and more liberal template preferences of DinB1 vs. Klenow polymerase.
Aprataxin Decaps DNA3′pp5′G Ends.
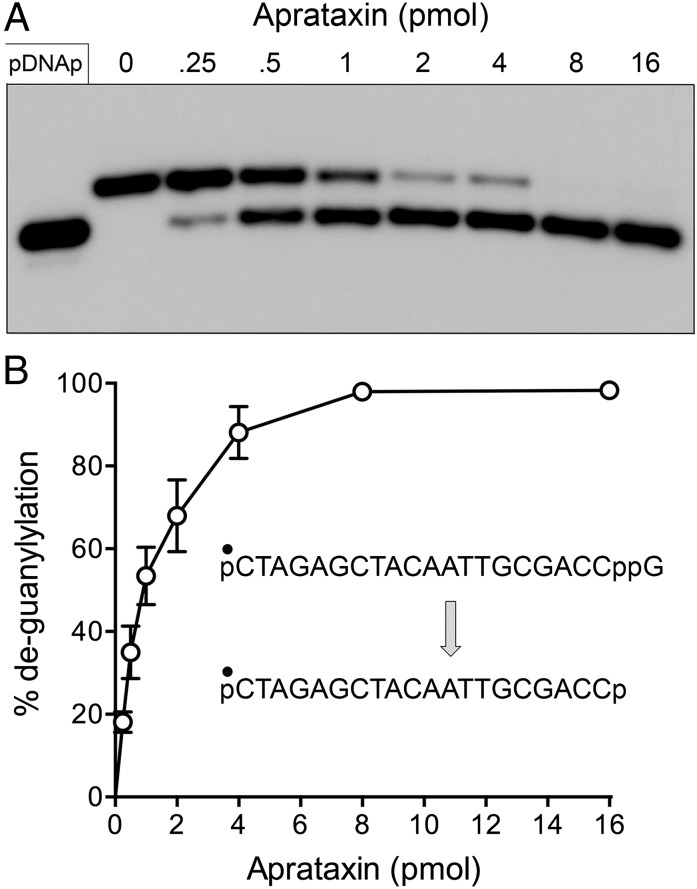
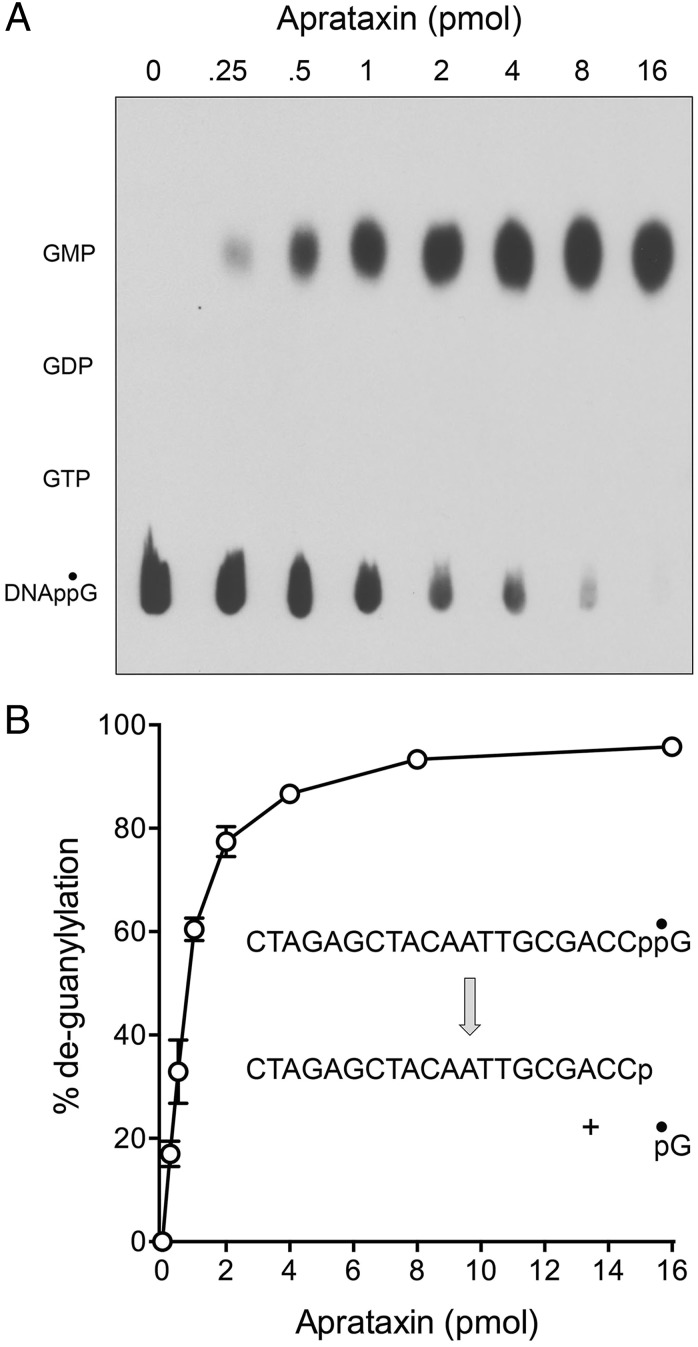
A pervasive theme in biology is that the covalent modification of macromolecules is typically a dynamic process in which the enzymes that install the modification (kinases, methyltransferases, acetyltransferases, protein conjugating systems, adenylyltransferases, etc.) are countered by enzymes that remove the covalent mark (phosphatases, demethylases, deacetylases, peptidases, deadenylases, etc.). Thus, we envisioned that DNA 3′ capping by RtcB enzymes might exist in dynamic equilibrium with a DNA 3′ decapping activity, much like mRNA 5′ modification reflects a dynamic between mRNA capping and decapping enzymes (12). Moreover, we hypothesized that the DNA repair enzyme aprataxin, a member of the histidine triad family of nucleotidyltransferases, is a plausible candidate to serve as a DNA 3′ decapping enzyme. Aprataxin has been the focus of much attention because inactivating mutations in the enzyme are the cause of the human neurological disorder ataxia oculomotor apraxia-1 (13, 14). Aprataxin deadenylates abortive A5′pp5′DNA intermediates that can accumulate when classic DNA ligases attempt to seal DNA 5′-PO4 ends at sites of damage or RNA 5′-PO4 ends embedded in DNA (15, 16). Aprataxin catalyzes the conversion of A5′pp5′DNA to AMP and pDNA via a covalent enzyme-(histidinyl)–AMP intermediate. Because the available crystal structures of Schizosaccharomyces pombe and human aprataxin with AMP bound in the active site are noteworthy for the absence of adenine-specific enzymic contacts to the nucleobase (16–18), we thought it plausible that aprataxin might accept a guanylylated substrate with a phosphoanhydride linkage between GMP and a polynucleotide. To test this idea, we produced and purified recombinant S. pombe aprataxin and reacted it with the 5′ 32P-labeled pDNAppG strand. We observed that aprataxin effected conversion of pDNAppG to a product that comigrated with pDNAp during PAGE (Fig. 3A). The extent of decapping was proportional to input aprataxin and was quantitative at saturating enzyme (Fig. 3 A and B). In parallel experiments, we reacted fission yeast aprataxin with [32P]GMP cap-labeled DNAppG and analyzed the products by polyethyleneimine (PEI) cellulose TLC. Whereas the DNAppG substrate remained at the origin after TLC, the reaction with aprataxin resulted in the release of a single 32P-labeled product that comigrated with GMP (Fig. 4A). Here again, the extent of deguanylylation of DNAppG was proportional to input aprataxin and quantitative at saturating enzyme (Fig. 4 A and B). The aprataxin titration curves in the two decapping assay formats, tracking the fate of the DNA and the cap, respectively, were virtually identical, thereby establishing the decapping reaction as: DNAppG → DNAp + GMP.
Fig. 3.
Aprataxin has DNA 3′ decapping activity. (A) Reaction mixtures (10 μL) containing 50 mM Tris⋅HCl (pH 8.0); 40 mM NaCl; 5 mM EDTA; 1 pmol of 5′ 32P-labeled pDNAppG; and 0, 0.25, 0.5, 1, 2, 4, 8, or 16 pmol of S. pombe aprataxin were incubated at 30 °C for 10 min. The reactions were quenched with formamide and EDTA, and the products were analyzed by urea-PAGE and visualized by autoradiography. The 32P-labeled pDNAp strand was analyzed in parallel in the leftmost lane. (B) Extents of conversion of pDNAppG to pDNAp are plotted as a function of input aprataxin. Each datum is the average of three independent enzyme titration experiments ± SEM.
Fig. 4.
Decapping by aprataxin liberates GMP. (A) Reaction mixtures (10 μL) containing 50 mM Tris⋅HCl (pH 8.0); 40 mM NaCl; 5 mM EDTA; 1 pmol 5′ 32P-labeled pDNAppG; and 0, 0.25, 0.5, 1, 2, 4, 8, or 16 pmol of S. pombe aprataxin as indicated were incubated at 30 °C for 10 min. The reaction products were analyzed by PEI-cellulose TLC (using 0.45 M ammonium sulfate as the mobile phase) in parallel with cold GTP, GDP, and GMP standards. An autoradiogram of the TLC plate is shown. (B) Extents of 32P-GMP release from DNAppG are plotted as a function of input aprataxin. Each datum is the average of three independent enzyme titration experiments ± SEM.
Conclusions, Implications, and Speculations.
The implications of DNA 3′ capping for DNA repair are potentially profound, given the wide phylogenetic distribution of RtcB enzymes (19–22) and the many biological settings in which DNA damage generates 3′-PO4 ends, either directly or as repair intermediates. We conclude from the results presented here that DNA capping prevents resection and healing of a 3′-PO4 end by exonucleases and phosphoesterase, while permitting templated DNA synthesis from the cap guanosine 3′-OH primer by polymerases. The price of cap-primed DNA repair synthesis is the embedding of a ribonucleotide and an unconventional pyrophosphate linkage in the repaired DNA strand. Conceptually, the DNA cap is a two-edged sword: advantageous in some scenarios but disadvantageous in others.
An upside to capping would be to protect damaged DNA 3′ ends from erosion by exonucleases. For example, (i) preserving double-strand breaks against 3′ exonucleases, although allowing action of 5′ exonucleases, could drive the formation of the 3′ single-strand tails that undergo synapsis during homologous recombination, and (ii) the capping of broken ssDNAs could help maintain the integrity of telomeres. Another upside of capping emerges from our finding that the cap guanylate is used as an efficient primer for DNA synthesis by at least two families of DNA polymerases. Thus, capping rescues a dirty 3′ end, without end-healing by a phosphatase, and allows progression of repair synthesis, provided, at least in the case of Klenow Pol1 and optimally in the case of DinB1, that the cap guanylate is positioned opposite a cytosine in the template strand. It is conceivable that other flavors of DNA polymerases, analogous to specialized bypass enzymes, can extend the 3′ cap when opposite other template nucleotides, and thereby broaden its priming capacity.
A downside to capping and cap-primed DNA synthesis is that it deposits a pyrophosphate linkage in the repaired DNA segment. The biological consequences of the pyrophosphate linkage are unknown, because there has heretofore been no inkling of a natural enzymatic pathway to generate such a structure. Shabarova and coworkers (23, 24) had previously exploited chemical methods to introduce an internal pyrophosphate linkage in the DNA backbone via chemical ligation of synthetic DNAp and pDNA strands annealed to a bridging template strand. This synthesis enabled a useful modification interference approach to gauge the impact of an internal pyrophosphate linkage on nucleic acid recognition by proteins and catalysis by nucleic acid-modifying enzymes (24, 25). Our work here establishes that internal pyrophosphate DNA linkages are not merely oddities of chemical manipulation but can be synthesized on DNAp ends by natural enzymes distributed widely (RtcB) or universally (DNA polymerase) among taxa. Molecular modeling studies suggest that the pyrophosphate backbone substitution has minimal effect on DNA helical structure, base pairing, or base stacking in aqueous solution (26). Moreover, a pyrophosphate internucleotide linkage in the template DNA strand had no apparent effect on DNA synthesis by Klenow Pol1 (26), suggesting that an embedded single pyrophosphate “lesion” might be fairly well tolerated with respect to replication.
The demonstration of a DNA 3′ decapping activity inherent in the DNA repair enzyme aprataxin affords a means to temper and regulate the effects of the cap on 3′ end metabolism. Indeed, our findings raise the prospect that some of the profound neurological consequences of aprataxin deficiency might arise because it is not available to deguanylylate 3′ capped ends deposited at sites of oxidative DNA damage that generate 3′-PO4 breaks.
The work presented here lays open a fresh field for study, which will require the development of methods to detect and map sites of 3′ capping and pyrophosphate linkages in cellular DNA (or RNA) undergoing a variety of genotoxic stresses. The issue of cellular stress is especially pertinent to the still-uncharted physiological function of E. coli RtcB, the focus of our biochemical analysis. The E. coli rtcB gene is in an operon that is transcriptionally silent under standard laboratory growth conditions (27). The expression of E. coli RtcB is under the control of RtcR, a coactivator for the stress-responsive σ54 transcription initiation factor (27), but the stress signals that induce RtcB expression are not known. It is attractive to speculate that (i) the RNA repair activity of RtcB might ameliorate the effects of bacterial endonuclease ribotoxins that are turned on in stress situations and (ii) the DNA capping activity of RtcB might aid a bacterial DNA damage response.
Methods
Enzymes.
RtcB was produced in E. coli and purified from a soluble bacterial extract as described (2). M. smegmatis DinB1 (Msmeg3172; GenBank accession no. YP_887486) was produced in E. coli as a His10Smt3–DinB1 fusion and isolated from a soluble bacterial extract by Ni-affinity chromatography. The tag was removed by treatment with Smt3 protease Ulp1, and the tag-free DinB1 was recovered in the flow-through during a second round of Ni-affinity chromatography. The DinB1 protein was further purified by gel filtration. T4 polynucleotide 3′ phosphatase, Pnkp-(149-301), was produced in E. coli and purified as described (10). The S. pombe hnt3 ORF encoding aprataxin (SPCC18.09c; GenBank accession no. CAA21423) was amplified by PCR from a cDNA library and inserted into pET16b. His10-tagged aprataxin was produced in E. coli and purified from a soluble bacterial extract by Ni-agarose chromatography. Protein concentrations were determined by the BioRad dye binding method with BSA as the standard. E. coli exonucleases I and III and DNA Pol1 Klenow fragment (3′ exo–) were purchased from New England Biolabs.
Substrates.
The 5′ 32P-labeled pDNAp and pDNAOH strands were prepared by enzymatic phosphorylation of 20-mer HOCTAGAGCTACAATTGCGACC oligonucleotides, with either 3′-PO4 or 3′-OH termini, by using [γ32P]ATP and the phosphatase-dead mutant T4 Pnkp-D167N. The radiolabeled pDNAp and pDNAOH strands were gel-purified. The 5′ 32P-labeled pDNAppG strand was prepared by incubating pDNAp with a 20-fold molar excess of RtcB in the presence of 2 mM MnCl2 and 1 mM GTP for 20 min at 37 °C, followed by gel purification. Primer-templates for DNA polymerase assays were prepared by annealing the radiolabeled pDNAOH, pDNAp, and pDNAppG primers to a fourfold molar excess of the unlabeled template DNA strands as specified in Fig. 2 and Fig. S3.
Supplementary Material
Acknowledgments
S.S. is an American Cancer Society Research Professor. This research was supported by National Institutes of Health Grant GM46330.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409203111/-/DCSupplemental.
References
- 1.Nair PA, Smith P, Shuman S. Structure of bacterial LigD 3′-phosphoesterase unveils a DNA repair superfamily. Proc Natl Acad Sci USA. 2010;107(29):12822–12827. doi: 10.1073/pnas.1005830107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanaka N, Chakravarty AK, Maughan B, Shuman S. Novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-Phosphate/5′-hydroxyl ligation reactions. J Biol Chem. 2011;286(50):43134–43143. doi: 10.1074/jbc.M111.302133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakravarty AK, Subbotin R, Chait BT, Shuman S. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc Natl Acad Sci USA. 2012;109(16):6072–6077. doi: 10.1073/pnas.1201207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das U, Chakravarty AK, Remus BS, Shuman S. Rewriting the rules for end joining via enzymatic splicing of DNA 3′-PO4 and 5′-OH ends. Proc Natl Acad Sci USA. 2013;110(51):20437–20442. doi: 10.1073/pnas.1314289110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA. 2011;2(2):277–298. doi: 10.1002/wrna.52. [DOI] [PubMed] [Google Scholar]
- 6.Lehman IR, Nussbaum AL. The deoxyribonucleases of Escherichia coli. V. On the specificity of exonuclease I (phosphodiesterase) J Biol Chem. 1964;239:2628–2636. [PubMed] [Google Scholar]
- 7.Thomas KR, Olivera BM. Processivity of DNA exonucleases. J Biol Chem. 1978;253(2):424–429. [PubMed] [Google Scholar]
- 8.Richardson CC, Kornberg A. A deoxyribonucleic acid phosphatase-exonuclease from Escherichia coli. I. Purification of the enzyme and characterization of the phosphatase activity. J Biol Chem. 1964;239:242–250. [PubMed] [Google Scholar]
- 9.Richardson CC, Lehman IR, Kornberg A. A deoxyribonucleic acid phosphatase-exonuclease from Escherichia coli. II. Characterization of the exonuclease activity. J Biol Chem. 1964;239:251–258. [PubMed] [Google Scholar]
- 10.Wang LK, Lima CD, Shuman S. Structure and mechanism of T4 polynucleotide kinase: An RNA repair enzyme. EMBO J. 2002;21(14):3873–3880. doi: 10.1093/emboj/cdf397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu H, Smith P, Wang LK, Shuman S. Structure-function analysis of the 3′ phosphatase component of T4 polynucleotide kinase/phosphatase. Virology. 2007;366(1):126–136. doi: 10.1016/j.virol.2007.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu M, Lima CD. Processing the message: Structural insights into capping and decapping mRNA. Curr Opin Struct Biol. 2005;15(1):99–106. doi: 10.1016/j.sbi.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 13.Date H, et al. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat Genet. 2001;29(2):184–188. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- 14.Moreira MC, et al. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet. 2001;29(2):189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 15.Ahel I, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443(7112):713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 16.Tumbale P, Williams JS, Schellenberg MJ, Kunkel TA, Williams RS. Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity. Nature. 2014;506(7486):111–115. doi: 10.1038/nature12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tumbale P, et al. Structure of an aprataxin-DNA complex with insights into AOA1 neurodegenerative disease. Nat Struct Mol Biol. 2011;18(11):1189–1195. doi: 10.1038/nsmb.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong Y, et al. Crystal structures of aprataxin ortholog Hnt3 reveal the mechanism for reversal of 5′-adenylated DNA. Nat Struct Mol Biol. 2011;18(11):1297–1299. doi: 10.1038/nsmb.2145. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka N, Shuman S. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J Biol Chem. 2011;286(10):7727–7731. doi: 10.1074/jbc.C111.219022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka N, Meineke B, Shuman S. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J Biol Chem. 2011;286(35):30253–30257. doi: 10.1074/jbc.C111.274597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Englert M, Sheppard K, Aslanian A, Yates JR, 3rd, Söll D. Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc Natl Acad Sci USA. 2011;108(4):1290–1295. doi: 10.1073/pnas.1018307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popow J, et al. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011;331(6018):760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- 23.Purmal AA, Drutsa VL, Shabarova ZA. [A new type of DNA modification. Specific introduction of 3′-5′-pyrophosphate internucleotide bonds] Bioorg Khim. 1984;10(3):394–400. Russian. [PubMed] [Google Scholar]
- 24.Purmal AA, Shabarova ZA, Gumport RI. A new affinity reagent for the site-specific, covalent attachment of DNA to active-site nucleophiles: Application to the EcoRI and RsrI restriction and modification enzymes. Nucleic Acids Res. 1992;20(14):3713–3719. doi: 10.1093/nar/20.14.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purmal AA, Wallace SS, Kow YW. The phosphodiester bond 3′ to a deoxyuridine residue is crucial for substrate binding for uracil DNA N-glycosylase. Biochemistry. 1996;35(51):16630–16637. doi: 10.1021/bi961774o. [DOI] [PubMed] [Google Scholar]
- 26.Rogacheva MV, Bochenkova AV, Kuznetsova SA, Saparbaev MK, Nemukhin AV. Impact of pyrophosphate and O-ethyl-substituted pyrophosphate groups on DNA structure. J Phys Chem B. 2007;111(2):432–438. doi: 10.1021/jp065947n. [DOI] [PubMed] [Google Scholar]
- 27.Genschik P, Drabikowski K, Filipowicz W. Characterization of the Escherichia coli RNA 3′-terminal phosphate cyclase and its σ54-regulated operon. J Biol Chem. 1998;273(39):25516–25526. doi: 10.1074/jbc.273.39.25516. [DOI] [PubMed] [Google Scholar]