Significance
The emergence of vascular tissues played a central role in the plant conquest of land. Both xylem and phloem are essential for the development of flowering plants, yet little is known about the molecular genetic control of phloem specification and differentiation. Here we show that delicate quantitative interplay between two opposing signaling pathways determines cellular commitment to protophloem sieve element fate in root meristems of the model plant Arabidopsis thaliana. Our data suggest that a recently described phloem-specific protein is a positive, quantitative master regulator of phloem fate.
Keywords: stem cell, division plane switching
Abstract
The phloem performs essential systemic functions in tracheophytes, yet little is known about its molecular genetic specification. Here we show that application of the peptide ligand CLAVATA3/EMBRYO SURROUNDING REGION 45 (CLE45) specifically inhibits specification of protophloem in Arabidopsis roots by locking the sieve element precursor cell in its preceding developmental state. CLE45 treatment, as well as viable transgenic expression of a weak CLE45G6T variant, interferes not only with commitment to sieve element fate but also with the formative sieve element precursor cell division that creates protophloem and metaphloem cell files. However, the absence of this division appears to be a secondary effect of discontinuous sieve element files and subsequent systemically reduced auxin signaling in the root meristem. In the absence of the formative sieve element precursor cell division, metaphloem identity is seemingly adopted by the normally procambial cell file instead, pointing to possibly independent positional cues for metaphloem formation. The protophloem formation and differentiation defects in brevis radix (brx) and octopus (ops) mutants are similar to those observed in transgenic seedlings with increased CLE45 activity and can be rescued by loss of function of a putative CLE45 receptor, BARELY ANY MERISTEM 3 (BAM3). Conversely, a dominant gain-of-function ops allele or mild OPS dosage increase suppresses brx defects and confers CLE45 resistance. Thus, our data suggest that delicate quantitative interplay between the opposing activities of BAM3-mediated CLE45 signals and OPS-dependent signals determines cellular commitment to protophloem sieve element fate, with OPS acting as a positive, quantitative master regulator of phloem fate.
The vascular tissues of higher plants are an important evolutionary invention that enabled their land conquest (1). At the heart of the plant vasculature, xylem tissue transports water and inorganic nutrients absorbed by the root system to aboveground organs, whereas phloem tissue distributes photosynthetic assimilates as well as systemic signals throughout the plant to coordinate its growth (1, 2). In Arabidopsis thaliana (Arabidopsis) roots, the vascular tissues are formed from stem cells located at the root tip, in the root meristem (1, 3). They create a bilaterally arranged vasculature, with central neighboring protoxylem and metaxylem cells that span the diameter of the stele, and two phloem poles that are located opposite of each other, flanking the xylem. Each phloem pole is composed of two distinct and intimately connected cell types: the sieve elements, which are the actual conductive cells of the phloem, and the companion cells, which provide essential metabolic functions for the enucleated sieve elements. Each phloem pole’s sieve elements are ultimately derived from a single stem cell, which produces a daughter cell by anticlinal division (Fig. 1A). This stem cell daughter (termed the sieve element procambium precursor cell here) then switches division plane to give rise to two cells by periclinal division. Whereas the inner cell forms a cell file of procambial character by subsequent anticlinal divisions, the outer cell (termed the sieve element precursor cell here) eventually divides periclinally once more. The cell file subsequently formed by the inner cell eventually differentiates into metaphloem further up in the root to functionally replace the protophloem (1), which differentiates closer to the tip from the cell file emerging from the outer cell. Finally, as these cell files differentiate into sieve elements, the immediately neighboring cell files (which originate from different stem cells) synchronously differentiate into companion cells, although with a delay of two to four cells with respect to the sieve elements. In the Arabidopsis root meristem, the protophloem strands can be identified in confocal microscopy through their enhanced propidium iodide cell wall staining (Fig. 1B) (4).
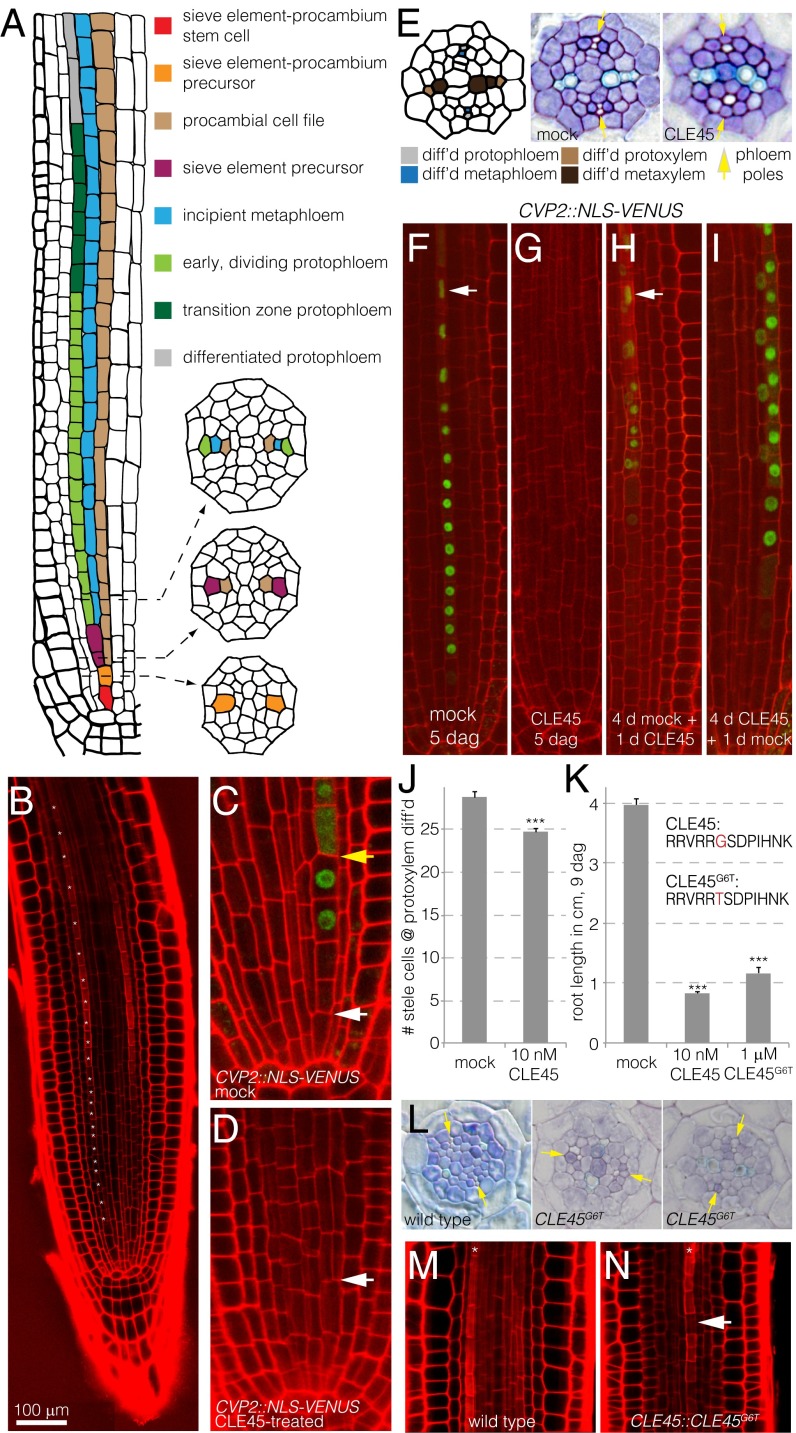
Fig. 1.
Elevated CLE45 activity suppresses protophloem sieve element differentiation and the formative cell division of the sieve element precursor cell. (A) Schematic presentation of protophloem sieve element formation in the Arabidopsis root meristem. (B) Confocal microscopy image of a propidium iodide-stained root meristem. The developing protophloem strands (one of them marked by asterisks) stand out because of their enhanced propidium iodide cell wall staining, allowing their unequivocal identification. (C and D) Expression pattern of the CVP2::NLS-VENUS fluorescent reporter in 5-d-old wild-type roots on normal media (C) or when shifted onto media containing 10 nM CLE45 for 24 h after 4 d (D). (E) Toluidine blue-stained histological cross sections of mock- and CLE45-treated roots, taken at the level of advanced metaxylem differentiation. Differentiated protophloem and metaphloem sieve elements can be recognized by their position and by the absent staining. Note differentiated metaphloem, but not protophloem, in the CLE45-treated sample. (F–I) Effects of transient CLE45 application on protophloem differentiation, monitored in CVP2::NLS-VENUS plants. White arrowheads (F and H): advanced nuclear degradation in the most apical transition zone cells. (I, composite image). (J) Reduced stele cell number in CLE45-treated plants resulting from missing cell files, counted at the position of differentiated xylem. (K) Root growth inhibition by application of CLE45 or the CLE45G6T variant. (L) Cross-sections illustrating representative phenotypes of plants expressing a CLE45::CLE45G6T transgene with delayed protophloem differentiation (Center) or delayed protophloem differentiation and missing cell file (Right). (M and N) Occurrence of gap cells (arrowhead) in the protophloem transition zone of CLE45::CLE45G6T (N), but not wild type plants (M). ***P < .001.
Mutant analyses in Arabidopsis have identified various genes involved in the formation of vascular tissues (1, 3, 5–9). In addition to upstream regulators required for overall vascular morphogenesis, these screens have also identified factors that are specifically involved in xylem or phloem development. However, whereas various signaling pathway components and transcriptional regulators required for xylem development have been found, comparatively little is known about phloem development (1). The few identified specific regulators of phloem formation include the transcription factor ALTERED PHLOEM DEVELOPMENT (APL) (5) and OCTOPUS (OPS), a polar localized plasma membrane-associated protein of unknown biochemical function (10). APL is required for completion of protophloem and metaphloem differentiation, and its loss of function results in the replacement of functional phloem with cells that display xylem characteristics (1, 5). OPS loss of function has been reported to delay protophloem differentiation because in a stochastic manner, individual ops protophloem cells fail to undergo the nuclear degradation and cell wall thickening that characterizes sieve element formation (10). These undifferentiated cells (termed gap cells here) interrupt the protophloem sieve element strand integrity and, thus, likely source-to-sink transfer of growth-limiting metabolites and developmental signals, which eventually compromises root growth (10). A similar phenotype is observed in loss-of-function mutants of BREVIS RADIX (BRX) (11), another polar localized plasma membrane-associated protein that is also thought to act in the nucleus (11, 12). The brx short root phenotype is rescued by second site mutation in BARELY ANY MERISTEM 3 (BAM3), which encodes a receptor-like kinase that is required for suppression of protophloem differentiation on treatment with the peptide ligand, CLAVATA3/EMBRYO SURROUNDING REGION 45 (CLE45) (13).
Results and Discussion
To better characterize CLE45 effects, we sought to establish a specific molecular marker for sieve element fate. The COTYLEDON VASCULAR PATTERN 2 (CVP2) gene, which is required for vascular morphogenesis (14), turned out to specifically mark the sieve element precursor cell and the subsequent protophloem cell file, as revealed by a nuclear localized fluorescent VENUS reporter protein expressed under control of the CVP2 promoter (CVP2::NLS-VENUS) (Fig. 1C). In seedlings shifted onto media containing CLE45, CVP2 expression disappeared concomitant with cell wall build-up (Fig. 1 D and G), confirming that CLE45 suppresses protophloem differentiation (Fig. 1 E–G) (13). Moreover, the periclinal division of the sieve element precursor cell did not occur, leading to a loss of the incipient metaphloem cell file (Fig. 1 D, E, and J). Transient treatments revealed that CLE45 only acted on cells that had not yet started the differentiation process (Fig. 1H) and that its effects were reversible; that is, cells formed during CLE45 treatment started to express CVP2 and differentiate into protophloem (as indicated by recovery of APL expression) once seedlings were shifted to CLE45-free media (Fig. 1I and Fig. S1A). Thus, the data suggest that CLE45 treatment locks the sieve element precursor cell in its preceding developmental state, and thereby prevents protophloem differentiation.
Because we could not recover transgenic lines with increased CLE45 dosage (13), we constructed a weak CLE45 variant to verify these effects in planta. The amino acid at position 6 of CLE peptides is crucial for their activity (15), and indeed, replacement of the corresponding glycine by threonine yielded a CLE45 variant (CLE45G6T) that had identical effects as the wild-type version but required application of higher peptide concentrations (Fig. 1K). Plants that expressed a corresponding CLE45::CLE45G6T transgene could be recovered, and in all of these independent lines, root growth was impaired (Fig. S1B), the periclinal division of the sieve element precursor cell was frequently abolished, and protophloem differentiation was frequently delayed (Fig. 1L and Fig. S1C). Moreover, CLE45::CLE45G6T transgenics displayed stochastic occurrence of nondifferentiating protophloem cells, which were morphologically similar to the gap cells observed in brx and ops mutants (Fig. 1 M and N). Thus, a mild increase in CLE45-like activity interfered both with the formative division that gives rise to the metaphloem and protophloem cell files and with protophloem differentiation.
These results supported the notion that the gap cell phenotype of brx mutants reflects stochastic above-threshold activity of the CLE45-BAM3 pathway (13). Further corroborating the phenotypic similarity between increased CLE45 dosage and BRX loss of function, closer inspection of brx mutants revealed a previously unrecognized phenotype; that is, the absence or severe delay of the sieve element precursor cell’s periclinal division in the majority of phloem poles (Fig. 2 A and C). The same phenotype, although at possibly lower penetrance, was also observed in ops mutants (Fig. 2 B and C). In summary, CLE45::CLE45G6T plants, brx mutants, and ops mutants all displayed a similar phenotype spectrum that included frequently absent sieve element precursor division, the occurrence of gap cells, and an associated reduction in the number of early dividing protophloem cells, in root meristem size and root growth (Fig. 2 D and E). Notably, even in the absence of the incipient metaphloem cell file in the three genotypes, cells with metaphloem morphology appeared in the correct position, possibly because the normally procambial cell file adopted metaphloem identity.
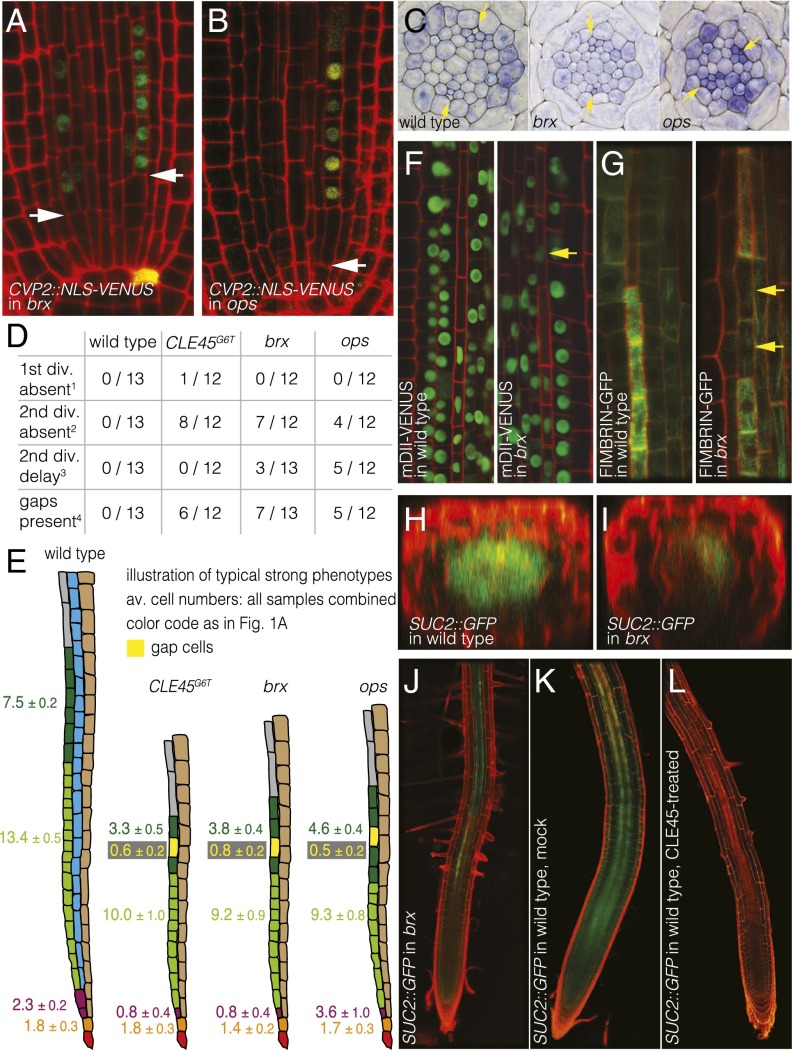
Fig. 2.
Perturbed phloem formation in CLE45::CLE45G6T transgenic plants, brx mutants, and ops mutants. (A and B) Absence of the periclinal sieve element precursor cell division, but not the periclinal sieve element procambium precursor cell division (arrowhead), in brx (A) and ops (B). (C) Cross-sections show differentiated protophloem (arrowheads) in both phloem poles of wild-type at the level of differentiated protoxylem, but no differentiation in at least one pole in brx and ops. (D) Summary statistics of protophloem development defects in one typical experiment. (E) Illustration of the maximum penetrance phloem pole phenotypes, with average cell numbers across samples indicated. (F) Persistence of the nucleus in brx gap cells (arrowhead), as indicated by the nuclear mDII-VENUS marker. (G) Absence of the typical strong actin cytoskeleton signal in brx gap cells (arrowheads), as indicated by the FIMBRIN-GFP marker (composite image on the left). (H and I) Optical sections showing local and diffused GFP signal expressed under control of the companion cell-specific SUC2 promoter, from both poles in wild-type (H) and from typically at most one pole in brx (I). (J–L) Patchy, interrupted SUC2::GFP expression in brx (J) compared with wild-type (K), and suppression of the signal in seedlings grown on CLE45-containing media (L).
Gap cells are located in the protophloem transition zone (Figs. 1A and 2E), and characterization of wild-type and the ops mutant has shown that this is where key events of protophloem sieve element differentiation take place, such as cell wall thickening and nuclear degradation (10, 16). To further characterize brx gap cells, we introduced a nuclear marker into the mutant, the stabilized fluorescent fusion protein mDII-VENUS, expressed under the constitutive 35S promoter (17). Investigation of these lines revealed that brx gap cells indeed displayed a persisting nucleus concomitant with the lack of cell wall thickening, similar to ops gap cells (Fig. 2F). Moreover, a fluorescent cytoskeleton marker, that is, FIMBRIN-GFP expressed under the constitutive 35S promoter (18), revealed a characteristic increase of actin filament abundance during protophloem differentiation, which did not occur in gap cells (Fig. 2G). Finally, the failure to form a continuous file of proper protophloem sieve elements possibly impinges on the differentiation of the neighboring companion cells because expression of the companion cell-specific SUCROSE TRANSPORTER 2 (SUC2) gene, monitored by a fluorescent reporter (SUC2::GFP) (19), was frequently patchy or absent in brx phloem poles (Fig. 2 H–K). Consistent with this observation, CLE45 treatment also suppressed SUC2::GFP expression (Fig. 2 K and L).
The close relation between the action of BRX and OPS in protophloem formation was also underlined by our finding that one of the loci emerging from a brx second site suppressor screen (13) mapped to the OPS locus. The corresponding ops allele carries a glutamate to lysine mutation in position 319 (E319K), which segregated as a dosage-dependent semidominant suppressor locus (Fig. 3A and Fig. S1D). In the F1 obtained from crosses between the ops loss-of-function and the opsE319K mutant, the ops root growth defects were fully restored, suggesting that opsE319K is a genuine, fully active gain-of-function allele (Fig. 3B). Moreover, we observed dosage-dependent OPS action in plants expressing extra copies of wild-type OPS fused to GFP under control of the native OPS promoter (OPS::OPS-GFP; this transgene was functional, as indicated by its ability to rescue the ops mutant phenotype) (Fig. 3C). When introduced into brx mutants, this construct rescued the root phenotype to varying extents, apparently correlating with OPS-GFP signal intensity in individual lines and frequently conferring near-wild-type levels of root growth (Fig. 3D). Concomitantly, the missing sieve element precursor division was restored (Fig. 3E) and gap cells were absent. When introduced into wild-type background, OPS::OPS-GFP transgene copies conferred resistance to CLE45 treatment (Fig. 3F), and the same was true for the opsE319K single mutant (Fig. 3 G and H). Thus, an increase in OPS activity was sufficient to overcome the inhibitory effects of CLE45 treatment. This CLE45-resistance through enhanced OPS activity largely depended on the presence of functional BRX (Fig. 3F and Fig. S1D), suggesting that BRX conditions OPS action by keeping CLE45-BAM3 activity below a certain threshold level. Finally, corroborating this dose-dependent opposition between CLE45 and OPS action, ops root growth and protophloem differentiation defects were perfectly rescued in bam3 ops double mutants (Fig. 3 I and J), similar to bam3 brx double mutants (13).
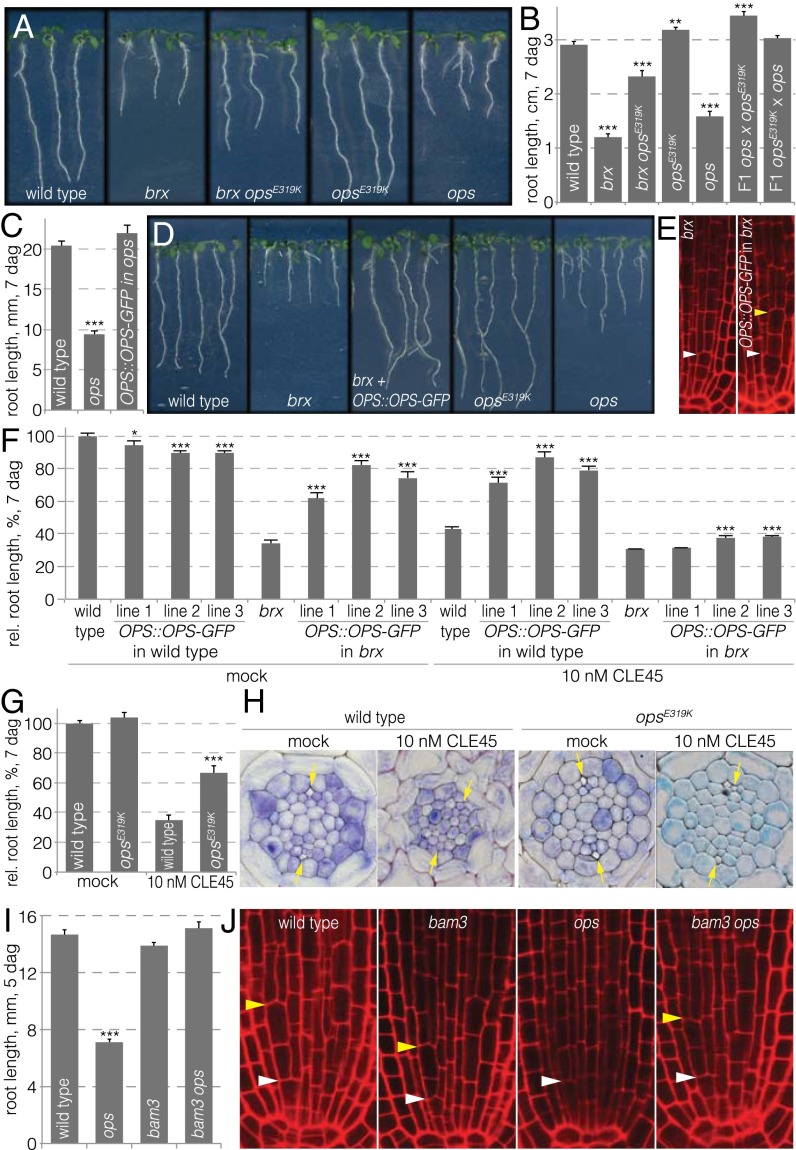
Fig. 3.
OPS dosage rescues brx defects and confers CLE45 resistance. (A) Representative 7-d-old seedlings for indicated genotypes. (B) Complementation of ops loss-of-function mutants by the opsE319K gain-of-function allele. (C) Complementation of the ops mutant by an OPS::OPS-GFP transgene. (D) Representative 7-d-old seedlings for indicated genotypes. (E) Restoration of the periclinal sieve element precursor cell division in brx by addition of an OPS::OPS-GFP transgene. (F) Rescue of brx root growth by increased OPS dosage through an OPS::OPS-GFP transgene and CLE45 resistance conferred by increased OPS dosage in wild-type. (G and H) CLE45 resistance conferred by the opsE319K gain-of-function allele, as indicated by root growth (G) as well as sieve element precursor division and protophloem differentiation (H). (I) Suppression of ops root growth defects on second-site loss-of-function mutation in bam3, the putative CLE45 receptor. (J) Restoration of the periclinal sieve element precursor division in ops bam3 double mutants. White arrowheads, position of sieve element-procambium precursor periclinal divisions; yellow arrowheads, position of sieve element precursor periclinal divisions. *P < 0.05; **P < 0.01; ***P < 0.001.
The highly specific meristematic reporter gene expression patterns of the five genes analyzed in this study matched a role in protophloem differentiation (Fig. 4A). OPS expression was also observed outside of the protophloem lineage, in the metaphloem, indicating that OPS may operate more generally during vascular development. The observation of the reporters’ expression across a number of samples that represented various stages in the differentiation process (e.g., samples in which two or three sieve element precursors could be observed because they had not yet divided compared with samples in which this was only the case for one) allowed us to establish a spatiotemporal hierarchy of expression (Fig. 4B). OPS was thus expressed earliest and was consistently present in the sieve element procambium precursor cell, whereas BRX, BAM3, CLE45, and CVP2 could only be detected in the sieve element precursor cell. However, although CLE45 expression was typically observed immediately after its formation, BRX and BAM3 were expressed with some delay, and CVP2 even later. These expression patterns are consistent with the observed defects in protophloem differentiation but are also consistent with the absent formative division of the sieve element precursor cell. However, it appears unlikely that the acquisition of sieve element precursor fate includes an obligatory periclinal cell division, because in roots grown on CLE45-containing media for 4 d and then shifted onto CLE45-free media, protophloem differentiation occurred within 20 h (as determined by CVP2 expression, APL expression, and cell wall build-up). In contrast, the missing cell division had not yet been recovered at this point in most samples (Fig. 1I). Conversely, in roots grown on CLE45-free media for 4 d and then shifted onto CLE45-containing media, protophloem differentiation was repressed within 20 h, whereas the periclinal sieve element precursor cell division was only fully suppressed in all samples after 30 h.
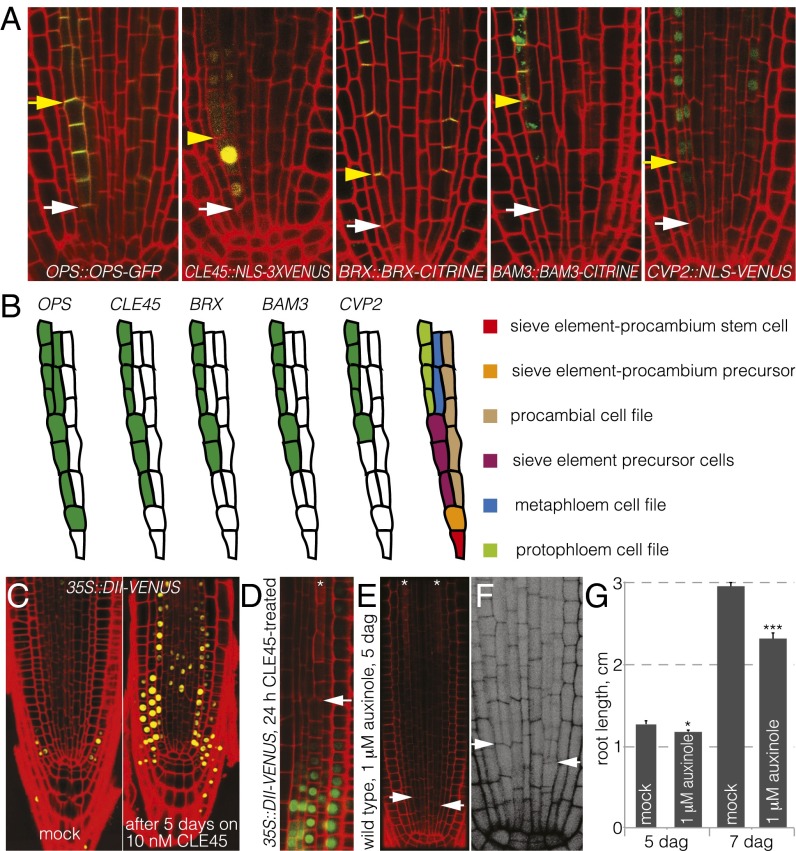
Fig. 4.
Expression patterns and auxin effects on the periclinal sieve element precursor division. (A) Expression patterns of indicated reporter transgenes in wild-type background. Note that OPS-GFP localizes to the shoot-ward plasma membrane of cells, whereas BRX-CITRINE localizes to the root-ward one. BAM3-CITRINE can be detected inside the cell (green signal), as well as at the plasma membrane (yellow signal resulting from overlay with propidium iodide staining). (B) Schematic summary of expression patterns determined from multiple observation. The rarer situation with three not-yet-divided sieve element precursor cells was chosen for illustration to allow us to plot this spatiotemporal hierarchy. (C) Auxin activity in the root meristem, as indicated by the inverse reporter DII-VENUS in mock conditions and on CLE45 treatment. (D) High auxin activity coincided with the last observed periclinal sieve element precursor division (arrowhead), whereas it was low in cells formed during CLE45 treatment. (E) Absence of the periclinal sieve element precursor division in wild-type roots treated with the auxin signaling inhibitor, auxinole. (F) Amplified detail of panel E in grayscale. (G) Effect of auxinole treatment on root growth. White arrowheads, position of sieve element-procambium precursor periclinal divisions; yellow arrowheads, position of sieve element precursor periclinal divisions; asterisks: protophloem cell files. *P < 0.05; ***P < 0.001.
The frequent lack of this division in the brx, ops, and CLE45::CLE45G6T genotypes might therefore be an indirect effect of the interruption of protophloem sieve element files by undifferentiated cells (Fig. S1E), which could interfere with the transport of metabolites and systemic signals, such as the plant hormone auxin, to the meristem (1, 2, 10, 13). Because it has been suggested that threshold levels of auxin are necessary for periclinal cell divisions in the root meristem (20), and because higher auxin activity was detected in developing protophloem compared with surrounding tissues (21) (Fig. S1F), we investigated whether a reduced meristematic auxin response, as indicated by the auxin sensor DII-VENUS (17), could be responsible for the absence of the sieve element precursor cell division. Consistent with these observations, CLE45 application resulted in reduced auxin response in the root meristem on prolonged, but not on short (i.e., a few hours), treatment (Fig. 4C). Observation of seedlings treated with CLE45 for 24 h also indicated coincidence of undetectable levels of DII-VENUS (i.e., high auxin activity) with the last periclinal sieve element precursor division before CLE45 application, and low auxin activity in cells formed thereafter (Fig. 4D). Moreover, treatment of seedlings with auxinole, a specific inhibitor of the nuclear auxin receptors, and thus auxin response (22, 23), abolished this division when applied at mild concentration (Fig. 4 E and F), suggesting it is particularly sensitive to changes in auxin activity. Given that BRX has been implicated in potentiating auxin response (11, 24), this would also explain why the penetrance of the observed phenotypes was highest in brx mutants. Remarkably, however, root growth rate and meristem size were only mildly impaired in auxinole-treated roots, despite the absence of the incipient metaphloem cell file (Fig. 4G), and protophloem differentiation was not affected. This underlines the proposed essential function of the protophloem for root meristem growth in the early seedling and systemic performance of the root system later on (13, 25). Systemically reduced auxin activity could also account for the observation that on CLE45 treatment, and in brx or ops loss-of-function mutants, overall cell number was sometimes slightly reduced in the radial dimension beyond the two missing incipient metaphloem cell files (e.g., Fig. 1J).
In summary, our data suggest opposing activities of BAM3-mediated CLE45 signals on one side and OPS-dependent signals on the other. Their quantitative interplay is sensitive to both CLE45 and OPS dosage and determines the proper timing of protophloem specification and the progression of its differentiation. Our analyses suggest these pathways act specifically on sieve element precursors, although based on expression pattern OPS might even have an earlier role. Notably, both brx and ops loss-of-function mutants are, in principal, able to form sieve elements, although at a reduced frequency. Considering that CLE45 specifically blocks precursor cells from adopting sieve element fate, the main role of BRX and OPS is therefore apparently to promote the transition to this state and to maintain it. The existence of independent positional cues for sieve element differentiation is also suggested by our observation that even in the absence of the incipient metaphloem cell file in brx and ops mutants, cells with metaphloem morphology differentiated in the correct position, possibly because the normally procambial cell file apparently adopted metaphloem identity. Because CLE45 can fully suppress any protophloem differentiation, the gap cells in the protophloem transition zone of brx and ops mutants might reflect incompetence to respond to such cues because of stochastic above-threshold activity of the CLE45-BAM3 module. Finally, we describe a genetic framework that is specific for protophloem sieve element differentiation, which, however, is possibly also necessary for parallel differentiation of companion cells, and thus the formation of functional protophloem poles. However, because loss-of-function brx bam3 or ops bam3 double mutants appear to be wild-type, our data also indicate that neither of these genes is strictly required for protophloem formation. Thus, they might constitute an additional regulatory layer, which could, for instance, serve to convey environmental signals in conditions in which a shutdown of primary root growth might be advantageous. The associated systemic effects, for example, ref. 25, would support such a scenario.
Materials and Methods
Plant tissue culture, CLE peptide treatments, plant transformation, histology, light microscopy, and confocal microscopy, as well as molecular biology experiments such as genomic DNA isolation, genotyping, or sequencing, were performed according to standard procedures, as described previously (11, 13). The OPS suppressor locus was identified by combined bulk segregant analysis and whole-genome sequencing, followed by confirmation through Sanger sequencing, as described (13).
Plant Materials.
The Arabidopsis wild-type line used in this study was Columbia (Col-0), which was also the genetic background for the mutants and transgenic lines. The following null mutant alleles were used throughout: brx-2 for BRX, bam3-2 for BAM3, and ops-2 for OPS. These mutants, as well as the transgenic reporter lines 35S::DII-VENUS, 35S::mDII-VENUS, and 35S::FIMBRIN-GFP, have been described previously (10, 13, 17, 18).
Transgenic Lines.
Reporter transgenes for plant transformation were created in suitable binary vectors and produced through standard molecular biology procedures and/or the Gateway Cloning Technology. The promoters or coding sequences for the BAM3, BRX, and CLE45 reporter constructs have been described previously (13). For cloning of the CVP2::NLS-VENUS construct, a 1.5-kb genomic promoter fragment upstream of the initiation codon was amplified using the 5′ to 3′ oligonucleotides GGT TTG TGG CAA TTT GTA TCC and GCT TTT AAA TTC CAT GAA GAT GGG C. For cloning of the SUC2::GFP construct, a 2.3-kb genomic promoter fragment upstream of the initiation codon was amplified using the 5′ to 3′ oligonucleotides AGT CAT TAT CAA CTA GGG GTG CAT and ATT TGA CAA ACC AAG AAA GTA AGA AAA. For cloning of the BAM3::BAM3-CITRINE construct, the BAM3 coding sequence was amplified using the 5′ to 3′ oligonucleotides ATG GCA GAC AAG ATC TTC AC and GAA AGT ATT AGG CTG TTT AG. For cloning of the OPS::OPS-GFP construct, a 1.9-kb OPS promoter fragment was amplified together with the (intron-free) OPS coding sequence using the 5′ to 3′ oligonucleotides GCG GTG TAA TCA TTA TTT CGT and TAT ACA GCC TCA TTA CAC TCC. To generate the CLE45::CLE45G6T construct, a replacement of the glycine in position 6 by threonine was introduced in the CLE45 peptide coding sequence by amplification with the 5′ to 3′ oligonucleotides ATG TTG GGT TCC AGT ACA AGA and AGG ATC TGA TGT TCG TCT, followed by secondary amplification with the 5′ to 3′ oligonucleotides ATG TTG GGT TCC AGT ACA AGA and TTA AGA AAA TGG CTG AGC TT to introduce the mutation. All binary constructs were introduced into Arabidopsis backgrounds by Agrobacterium-mediated transformation following standard procedures. At least three independent transgenic lines were used for each construct to perform experiments and verify reproducibility.
Genotyping.
Genotyping of mutant alleles was performed with the following 5′ to 3′ oligonucleotides: GTC AGT GTT TGC TTC CTC TCT ATG and TAT TTC CTT GTC TAG GTA AGA ATC C and TGA TCC ATG TAG ATT TCC CGG ACA TGA A in one reaction to genotype brx-2 (wild-type band 240 bp; T-DNA insertion band 200 bp); CTG CAA CTT CTT CTC CGT TTG with CTG CAA CTT CTT CTC CGT TTG to genotype the BAM3 wild-type allele (1.1 kb) and GAT TCC TTC GAA ACT CGG ATC with ATT TTG CCG ATT TCG GAA C (300 bp) to genotype the bam3-2 T-DNA insertion allele; CAC ACC GTT GGT TTG GTT AAC with TCT TCC TCT AAA AAG CCT CCG to genotype the OPS wild-type allele (1.1 kb) and TCT TCC TCT AAA AAG CCT CCG with ATT TTG CCG ATT TCG GAA C (600 bp) to genotype the ops-2 T-DNA insertion allele; and CTT CAG AAA TGG AGG CAG AAT and CAT ATC CGT AAT CAG CAA GCT to amplify a 145-bp OPS fragment that is cut into 124 and 21 bp on HinDIII digest if the opsE319K mutation is present.
Supplementary Material
Acknowledgments
We thank Prof. N. Geldner’s group for providing vectors and F. Misceo for BRX::BRX-CITRINE seeds. This work was supported by Swiss National Science Foundation Grant 310030B_147088 (to C.S.H.) and a European Molecular Biology Organization Long-Term Postdoctoral Fellowship (to A.R.-V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407337111/-/DCSupplemental.
References
- 1.Lucas WJ, et al. The plant vascular system: Evolution, development and functions. J Integr Plant Biol. 2013;55(4):294–388. doi: 10.1111/jipb.12041. [DOI] [PubMed] [Google Scholar]
- 2.Ruiz-Medrano R, Xoconostle-Cázares B, Lucas WJ. The phloem as a conduit for inter-organ communication. Curr Opin Plant Biol. 2001;4(3):202–209. doi: 10.1016/s1369-5266(00)00162-x. [DOI] [PubMed] [Google Scholar]
- 3.Helariutta Y. Cell signalling during vascular morphogenesis. Biochem Soc Trans. 2007;35(Pt 1):152–155. doi: 10.1042/BST0350152. [DOI] [PubMed] [Google Scholar]
- 4.Truernit E, et al. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell. 2008;20(6):1494–1503. doi: 10.1105/tpc.107.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonke M, Thitamadee S, Mähönen AP, Hauser MT, Helariutta Y. APL regulates vascular tissue identity in Arabidopsis. Nature. 2003;426(6963):181–186. doi: 10.1038/nature02100. [DOI] [PubMed] [Google Scholar]
- 6.De Rybel B, et al. A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev Cell. 2013;24(4):426–437. doi: 10.1016/j.devcel.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 7.Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17(5):1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mähönen AP, et al. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311(5757):94–98. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- 9.Mähönen AP, et al. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 2000;14(23):2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Truernit E, Bauby H, Belcram K, Barthélémy J, Palauqui JC. OCTOPUS, a polarly localised membrane-associated protein, regulates phloem differentiation entry in Arabidopsis thaliana. Development. 2012;139(7):1306–1315. doi: 10.1242/dev.072629. [DOI] [PubMed] [Google Scholar]
- 11.Scacchi E, et al. Spatio-temporal sequence of cross-regulatory events in root meristem growth. Proc Natl Acad Sci USA. 2010;107(52):22734–22739. doi: 10.1073/pnas.1014716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scacchi E, et al. Dynamic, auxin-responsive plasma membrane-to-nucleus movement of Arabidopsis BRX. Development. 2009;136(12):2059–2067. doi: 10.1242/dev.035444. [DOI] [PubMed] [Google Scholar]
- 13.Depuydt S, et al. Suppression of Arabidopsis protophloem differentiation and root meristem growth by CLE45 requires the receptor-like kinase BAM3. Proc Natl Acad Sci USA. 2013;110(17):7074–7079. doi: 10.1073/pnas.1222314110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carland FM, Nelson T. Cotyledon vascular pattern2-mediated inositol (1,4,5) triphosphate signal transduction is essential for closed venation patterns of Arabidopsis foliar organs. Plant Cell. 2004;16(5):1263–1275. doi: 10.1105/tpc.021030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song XF, Guo P, Ren SC, Xu TT, Liu CM. Antagonistic peptide technology for functional dissection of CLV3/ESR genes in Arabidopsis. Plant Physiol. 2013;161(3):1076–1085. doi: 10.1104/pp.112.211029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H, Zheng XF. Ultrastructural studies on the sieve elements in root protophleom of Arabidopsis thaliana. Acta Bot Sin. 2003;45(3):322–330. [Google Scholar]
- 17.Brunoud G, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482(7383):103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- 18.Wang YS, Motes CM, Mohamalawari DR, Blancaflor EB. Green fluorescent protein fusions to Arabidopsis fimbrin 1 for spatio-temporal imaging of F-actin dynamics in roots. Cell Motil Cytoskeleton. 2004;59(2):79–93. doi: 10.1002/cm.20024. [DOI] [PubMed] [Google Scholar]
- 19.Ivashikina N, et al. Isolation of AtSUC2 promoter-GFP-marked companion cells for patch-clamp studies and expression profiling. Plant J. 2003;36(6):931–945. doi: 10.1046/j.1365-313x.2003.01931.x. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida S, et al. Genetic control of plant development by overriding a geometric division rule. Dev Cell. 2014;29(1):75–87. doi: 10.1016/j.devcel.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Santuari L, et al. Positional information by differential endocytosis splits auxin response to drive Arabidopsis root meristem growth. Curr Biol. 2011;21(22):1918–1923. doi: 10.1016/j.cub.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9(1):109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi K, et al. Rational design of an auxin antagonist of the SCF(TIR1) auxin receptor complex. ACS Chem Biol. 2012;7(3):590–598. doi: 10.1021/cb200404c. [DOI] [PubMed] [Google Scholar]
- 24.Depuydt S, Hardtke CS. Hormone signalling crosstalk in plant growth regulation. Curr Biol. 2011;21(9):R365–R373. doi: 10.1016/j.cub.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 25.Gujas B, Alonso-Blanco C, Hardtke CS. Natural Arabidopsis brx loss-of-function alleles confer root adaptation to acidic soil. Curr Biol. 2012;22(20):1962–1968. doi: 10.1016/j.cub.2012.08.026. [DOI] [PubMed] [Google Scholar]