Significance
The tropomyosin-like protein, DivIVA, determines the site of growth and cell morphology in mycobacteria. Surprisingly, although DivIVA is located at the tip of the growing cell pole, cell wall addition is excluded from this site. Both late cell wall synthetic enzymes and new cell wall deposition occur at a subpolar space, distinct from the DivIVA-marked cell tip. Instead of directly recruiting terminal cell wall synthetic systems, DivIVA interacts with enzymes involved in the early steps of the cell wall precursor synthesis. These results suggest a unique organization of the polar elongasome, where cell wall precursors are concentrated at the cell tip by DivIVA and then incorporated into the nascent cell wall in an annular pattern at the subpolar zone.
Keywords: tuberculosis, polarity
Abstract
Mycobacteria are surrounded by a complex multilayered envelope and elongate at the poles. The principles that organize the coordinated addition of chemically diverse cell wall layers during polar extension remain unclear. We show that enzymes mediating the terminal cytosolic steps of peptidoglycan, arabinogalactan, and mycolic acid synthesis colocalize at sites of cell growth or division. The tropomyosin-like protein, DivIVA, is targeted to the negative curvature of the pole, is enriched at the growing end, and determines cell shape from this site. In contrast, cell wall synthetic complexes are concentrated at a distinct subpolar location. When viewed at subdiffraction resolution, new peptidoglycan is deposited at this subpolar site, and inert cell wall covers the DivIVA-marked tip. The differentiation between polar tip and cell wall synthetic complexes is also apparent at the biochemical level. Enzymes that generate mycolate precursors interact with DivIVA, but the final condensation of mycolic acids occurs in a distinct protein complex at the site of nascent cell wall addition. We propose an ultrastructural model of mycobacterial polar growth where new cell wall is added in an annular zone below the cell tip. This model may be broadly applicable to other bacterial and fungal organisms that grow via polar extension.
The growth and morphology of virtually all bacterial cells is determined by the ordered synthesis and cross-linking of the cell wall. However, the mechanism of growth can vary dramatically, even between organisms that superficially share the same shape. For example, many rod-shaped bacteria, such as Escherichia coli and Bacillus subtilis, extend via the intercalation of new peptidoglycan (PG) along the lateral cell surface (1, 2). This mode of growth requires the cylindrical shape of the cell body to be continuously maintained by the bacterial actin homolog, MreB, which promotes cell wall addition to regions of positive curvature on the cytosolic face of the cell membrane (3). Consequently, PG at the negatively curved pole is inert (1). In contrast, the focal addition of new cell wall at the pole is recognized to be necessary for the extension of specialized organelles, such as the stalk of Caulobacter crescentus, and the growth of an increasing number of rod-shaped, budding, and filamentous bacteria (4–6). The mechanisms that determine the morphology of these polar extensions remain unclear because the precise site and geometry of cell wall addition at the pole have not been defined.
Bacteria of the order Actinomycetales, such as mycobacteria, are recognized to grow by polar extension (7). Elongation occurs preferentially at the end distal to the division plane and proceeds at a rate that is proportional to pole age (8, 9). The multilayered structure of the mycobacterial cell envelope makes the elongation of this cell a particularly complex challenge (10). The mycobacterial cell wall is composed of three covalently linked layers. The innermost PG polymer that surrounds virtually all bacteria is linked to an intermediate layer of arabinogalactan (AG) (11). The AG polymer is subsequently esterified at its terminal nonreducing ends to an outer coat of mycolic acids (MA) (12), which form a nonfluid and relatively impermeable outer membrane-like structure, or “mycomembrane” (13–16). The chemical structures of PG, AG, and MA and the synthetic pathways that produce them are well understood. Each layer is composed of monomers synthesized in the cytosol as either glyco- or isoprenyl phosphate-conjugates, which are exported across the plasma membrane before polymerization and cross-linking (17). Although it is intuitive that the activities of cell wall synthetic systems must be coordinated at the site of polar elongation, it remains unclear whether this coordination is accomplished through the specific localization, organization, or regulation of these enzymatic activities.
Actinobacteria do not express an obvious MreB ortholog, indicating that alternative mechanisms are likely to direct focal cell wall synthesis in these organisms. A variety of data implicate the tropomyosin-like protein, DivIVA as a coordinator of polar growth complexes in these organisms. DivIVA marks the hyphal tip of the filamentous bacterium Streptomyces coelicolor, and alterations in the level of expression or phosphorylation of this protein can cause fragmentation of the DivIVA focus, branching, and altered cell curvature (6, 18). Similarly, mycobacterial and corynebacterial DivIVA have been localized at the pole and septum and are essential for cell growth (19–21). Despite marking the poles and/or septa of many Gram-positive bacteria, DivIVA plays distinct functional roles in different species. For example, B. subtilis DivIVA is not required for growth and is instead a component of the Min system involved in the correct positioning of the cell septum (22, 23). The functional differentiation of these orthologs appears to result from specific adaptations in the DivIVA protein itself, because only proteins from closely related species can functionally complement each other (21). Species-specific protein–protein interactions may be responsible for functional divergence, because different DivIVA orthologs have been found to interact with a variety of proteins involved in cell division, chromosomal partitioning, and growth (24, 25).
We have investigated the mechanisms underlying polar growth using the mycobacterial cell as a model. We found that asymmetric polar growth correlates with the uneven distribution of cell wall synthetic complexes between opposite poles and different levels of coordination at these sites. The site of growth is determined by the abundance of the DivIVA protein. However, although DivIVA marks the tip of the growing poles and interacts with enzymes required for cell wall precursor synthesis, the elongation machinery is located at a distinct subpolar site that corresponds to the site of nascent cell wall deposition. Thus, DivIVA nucleates the polar organelle at least in part by recruiting early cell wall synthetic enzymes whose products diffuse to a subpolar site where they are incorporated into the growing cell wall.
Results
Coordinated Localization of Cell Wall Synthetic Machinery During Mycobacterial Growth.
Both Mycobacterium tuberculosis and its saprophytic relative, Mycobacterium smegmatis, are surrounded by a complex cell wall skeleton composed of covalently linked layers of PG, AG, and MA (10, 17). To investigate the localization and coordination of the proteins responsible for the synthesis of each distinct cell wall layer, we fused a multifunctional protein tag to the enzymes involved in the terminal cytosolic steps of cell wall synthesis. We tagged MurG, GlfT2, and Pks13 of M. smegmatis, which are involved in PG, AG, and MA synthesis, respectively. The glycosyltransferase MurG catalyzes the transfer of N-acetylglucosamine to N-acetylmuamic acid-(pentapeptide)-pyrophosphoryl undecaprenol (lipid I), yielding lipid II, the complete PG precursor that is exported to the extracytoplasmic space (26). The galactofuranosyltransferase GlfT2 catalyzes the polymerization of the galactan domain of AG, using a polyprenol-bound oligosaccharide as the acceptor (27). The polyketide synthase Pks13 catalyzes the condensation between two activated fatty acids, meromycoloyl-AMP (C40–C60) and 2-carboxyacyl-CoA (C22–C26) to form mature MA after reduction by CmrA (28, 29).
All three fusions were generated at the endogenous gene locus to preserve native expression levels and ensure the functionality of the fusion protein. Along with green fluorescent protein (GFP), the tag also contained antibody epitopes and an inducible degradation (ID) tag, together allowing the fusion protein to be visualized, purified, and conditionally depleted via targeting to the ClpXP protease (30). The observed molecular weights of the tagged proteins were consistent with the predicted sequences of intact fusions (Fig. S1A). The C-terminal tag had no effect on growth, as measured by optical density of the culture or the elongation rate of single cells (Fig. S1 B and C), or on cellular morphology (Fig. 1A). Inducible depletion of MurG, GlfT2, and Pks13 via the addition of anhydrotetracycline (ATc) inhibited growth and induced morphological changes, consistent with the inhibition of cell wall synthesis (Fig. S1D). This characterization confirmed that these fusion proteins represented the only functional version of these gene products in the cell.
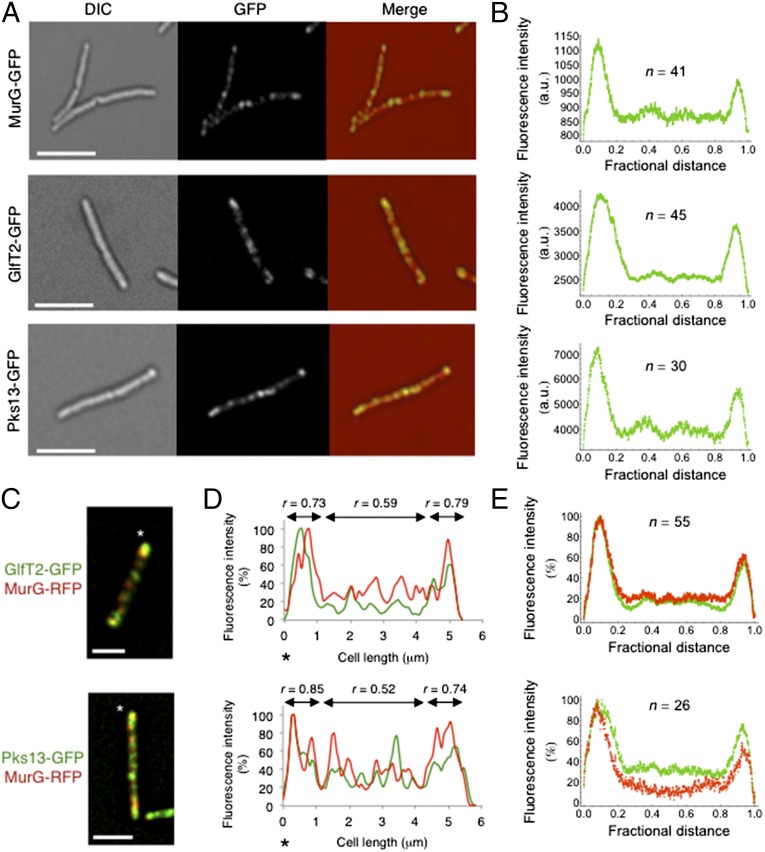
Fig. 1.
Coordination of cell wall synthetic enzymes during growth. Images are sum projections of deconvolved z-sections. (Scale bars, 5 μm.) (A) Differential interference contrast (DIC), GFP, and merged images of M. smegmatis cells expressing MurG–GFP, GlfT2–GFP, or Pks13–GFP at the endogenous gene locus. (B) Respective profiles of the moving average of fluorescence intensity along the length of the cells, show uneven distribution of the proteins between opposite poles (see also Fig. S1E). (C and D) Representative merged images of M. smegmatis cells coexpressing GlfT2–GFP (endogenous gene fusion) and MurG–RFP (pXM02) (C, Upper) or Pks13–GFP (endogenous gene fusion) and MurG–RFP (pXM02) (C, Lower) and the corresponding fluorescence intensity profiles (D). Asterisks show orientation of the cell. r value represents Pearson’s correlation coefficient. (E) Moving averages of fluorescence intensities show overlapping of the proteins mostly at the poles.
To investigate how the synthesis of chemically distinct cell wall layers is coordinated, we used wide-field deconvolution microscopy to determine the localization of MurG–GFP, GlfT2–GFP, and Pks13–GFP in actively growing cells. All three proteins were concentrated at the poles and septa with weak foci distributed along the lateral cell body (Fig. 1A). Upon quantifying the distribution of fluorescence, we observed an asymmetric localization of proteins at the poles (Fig. 1B). The fluorescence associated with a single pole was more intense and occupied a greater area.
To determine whether these cell wall synthetic systems were concentrated at the growing end of the cell, we used time-lapse imaging to track these proteins during growth (Movies S1–S3). All three proteins were more abundant at the old pole, the previously described preferential site of growth (8, 31). MurG, GlfT2, and Pks13 were also concentrated at the septum during cell division and remained associated with the new pole, albeit at lower abundance than the old pole. Unlike the relatively stable localization of these proteins at the poles, the fluorescent foci observed along the lateral cell body were highly mobile.
Mycobacteria extend through polar addition of new cell wall (7, 8, 32), indicating that the laterally localized synthetic enzymes are unlikely to be active. The ability to spatially differentiate active and inactive complexes allowed us to determine whether the colocalization of PG, AG, and MA systems was constitutive or induced at sites of growth. We found that Pks13–GFP or GlfT2–GFP did not coincide with MurG–RFP along the side of the cell body, but colocalized almost completely at the brighter, growing pole (Fig. 1 C–E). The correlation between colocalization and growth was also evident when the poles of individual cells were compared. The Pearson coefficient of correlation between the cell wall synthetic enzymes was significantly greater at the poles (Fig. 1D). These findings indicated that both the local concentration of cell wall synthetic enzymes and their degree of colocalization correlate with local elongation and that the spatial coordination of functionally distinct complexes is induced at sites of growth.
DivIVA Is Concentrated at the Site of Growth and Determines Cell Shape from This Location.
The DivIVA homologs of Mycobacteria and Streptomyces mark the cell poles and have been implicated in polar organization and extension (19, 20). These previous studies indicated that DivIVA might be involved in the formation of the inducible complexes that we found to be associated with sites of extension. To investigate whether DivIVAMsm localization correlated with growth, we created strains expressing tagged alleles of this protein either by integrating a second copy of the gene into the chromosome (divIVA+/divIVA–dendra, diviIVA+/divIVA–mrfp) or by inserting the multifunctional fluorescent tag sequence at the 3′ end of the endogenous copy of the gene (divIVA–egfp). The essentiality of DivIVAMsm and functionality of the C-terminal fusion proteins was verified by depleting the protein in the divIVA–egfp strain, which inhibited growth and caused cell rounding (Fig. S2A). Complementation of this strain with C-terminal, but not N-terminal, RFP fusion proteins reversed the growth defect observed upon depletion of the endogenous allele (Fig. S3A).
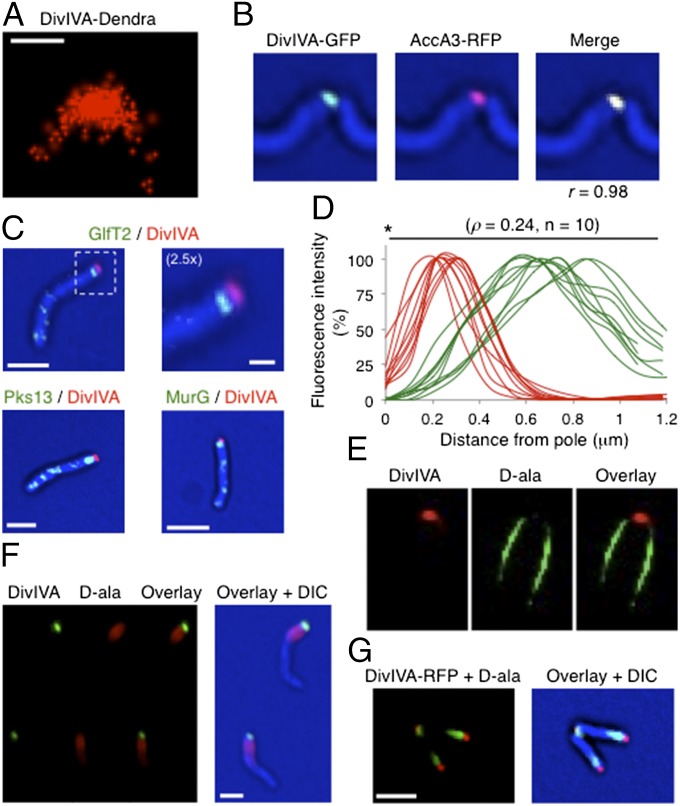
The association between DivIVA and sites of cell growth was then investigated. In contrast to previous overexpression studies that found mycobacterial DivIVA at both poles (20), we found that when expressed at native (divIVA–egfp) or similarly low (divIVA+/divIVA–dendra) levels, DivIVA was preferentially concentrated at a single pole. In merodiploid strains that express both tagged and wild-type (WT) DivIVA, the fluorescence was concentrated at the old pole (Fig. 2A and Movie S4), and the N- and C-terminal fusion proteins were found at an identical cellular location (Fig. S3B). This asymmetric localization was observed regardless of expression level, although expression from a very strong promoter resulted in a more even distribution (Fig. S4A). To determine whether this DivIVAMsm-marked pole was the site of elongation, we nonspecifically labeled the cell wall with an amine-reactive fluorophore. After a chase period, we found that the unlabeled end of the cell was preferentially marked by DivIVA–Dendra (Fig. 2 B and C), verifying the concentration of this protein at the old pole that represents the major site of elongation in WT mycobacteria (8).
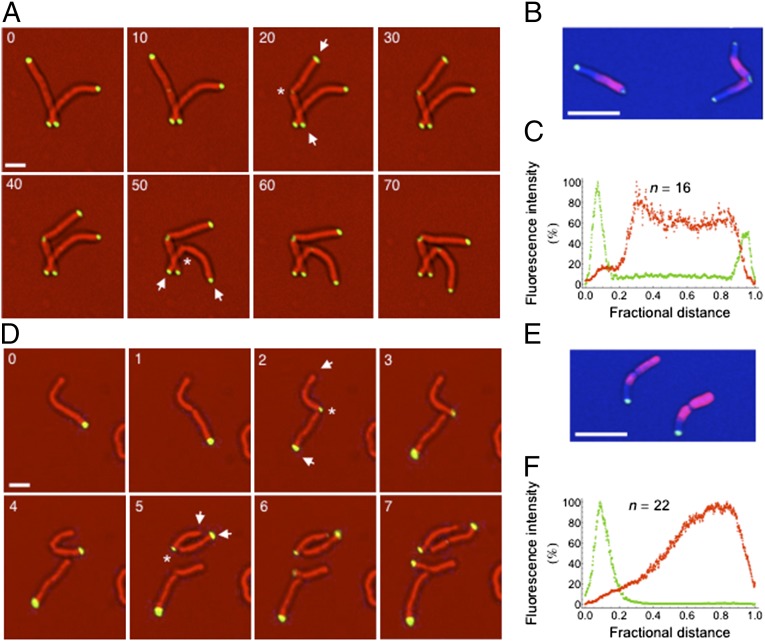
Fig. 2.
DivIVAMsm concentration correlates with the site of growth. (A) Time-lapse microscopy of the M. smegmatis strain expressing both DivIVA–Dendra (pXM05) and endogenous native DivIVA. Representative images were recorded at the indicated time (minutes). Asterisks show new poles, and arrows show old poles. (Scale bar, 2 μm.) (B and C) M. smegmatis cells expressing DivIVA–Dendra (pXM05) and endogenous DivIVA were nonspecifically labeled on their surface with the amine-reactive dye (AF-546), and unlabeled extensions were allowed to grow for 60 min. Representative image (B) and fluorescence quantification over the long axis of 16 cells (C) are shown (see also Fig. S2C). (Scale bar, 5 μm.) (D) Time-lapse microscopy of the divIVA–egfp M. smegmatis strain that only expresses the tagged copy of DivIVA at the gene locus. Representative images were recorded at the indicated time (hours). Asterisks show new poles, and arrows show old poles. (Scale bar, 2 μm.) (E and F) divIVA–egfp M. smegmatis cells were nonspecifically labeled on their surface with the amine-reactive dye (AF-546), and unlabeled extensions were allowed to grow for 60 min. Representative image (E), and fluorescence quantification over the long axis of 22 cells (F) are shown (see also Fig. S2C). (Scale bar, 5 μm.)
Unexpectedly, the sites of growth marked by fluorescent DivIVAMsm fusions differed depending on whether the strain expressed native untagged DivIVAMsm. When DivIVA–GFP was the sole allele expressed, new DivIVAMsm–foci only formed at the new pole. This pole became the preferential site of growth, as assessed by time-lapse microscopy and pulse/chase labeling (Fig. 2 D–F and Movie S5). Thus, in the absence of native DivIVAMsm, the allele tagged at the C-terminal end was targeted to the incorrect pole and appeared to drive growth at this ectopic site.
Although DivIVA concentration determined growth site preference, its abundance was not the sole determinant of elongation rate. To quantify the elongation rate at each pole, we used a d-alanine metabolic label that is incorporated into nascent PG (33). Pulse labeling of bacteria with this reagent produced the expected asymmetric labeling pattern (Fig. S4B). Although overexpression of DivIVA–RFP increased the amount of this protein at the growing pole, it did not influence the extension rate per unit of time (Fig. S4C).
Because the divIVA–egfp allele altered the site of elongation, we further assessed the growth and morphology of the divIVA–egfp strain. The mutant showed a very modest 1% increase in doubling time, as measured by OD of broth culture, but a more dramatic 40% decrease in single cell elongation rate (Fig. 3 A and B). These disparate observations were reconciled by the larger average diameter of the mutant cells (0.95 vs. 0.81 μm; Fig. 3C), which resulted in a larger proportional volume increase at a given extension rate. In addition to its greater width, the divIVA–egfp strain adopted a curved shape (Fig. 3C). The morphological abnormalities in the divIVA–egfp strain were reversed by complementation with a second copy of the gene (Fig. 3D). Thus, the C-terminal extension of DivIVAMsm did not significantly influence growth, but altered cell shape and the site of extension.
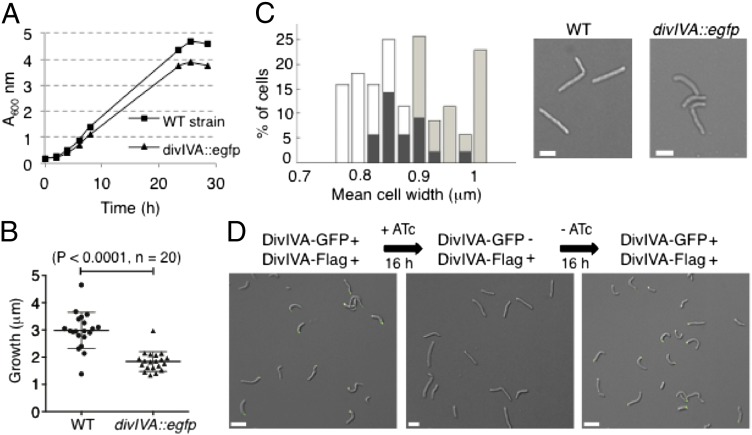
Fig. 3.
DivIVAMsm determines cell shape from sites of growth. (A) Growth of the WT strain (rectangles) and the divIVA–egfp strain that expresses only tagged DivIVA (triangles) in 7H9–Tween 80 medium at 37 °C. Data represent three experiments. (B) Elongation (micrometers) of the indicated strains between two consecutive cell-division events defined as membrane invagination observed on DIC images (P < 0.0001; n = 20). (C, Left) Mean cell width as percentage of cells of the WT (white bars, n = 36) and the divIVA–egfp (light gray bars, n = 44) strains (overlap, black bars). (Right) DIC images of the indicated strains are shown. (Scale bars, 2 μm.) (D) Cell shape of the divIVA–egfp strain expressing a second copy of DivIVA–Flag (pXM06) grown overnight without ATc (Left), after overnight incubation with ATc to deplete DivIVA–GFP protein (Center), and after overnight removal of ATc to allow reexpression (Right). (Scale bars, 5 μm.)
Negative Membrane Curvature Is Necessary for DivIVAMsm Localization and the Generation of a Growth Pole.
The DivIVA protein of B. subtilis is targeted to the nascent septum and poles at least partially through the recognition of the strong negative membrane curvature of these sites by the N-terminal domain of the protein (34–36). Because DivIVAMsm was found at different cellular locations than DivIVABsu, we investigated whether negative curvature is also a targeting cue for this protein in mycobacteria. Overnight depletion of the DivIVAMsm protein arrested growth and induced the formation of spherical cells that remained intact, contained DNA (Fig. S2A), and had no obvious local sites of high curvature (i.e., higher than average for the sphere). Newly expressed DivIVAMsm–GFP was detectable 4 h after ATc removal, and the protein formed mobile, transient foci (Fig. 4A and Movie S6). However, neither a stable focus of DivIVAMsm nor a new growth pole was formed 12 h later. Conversely, the generation of ectopic sites of curvature with the membrane-perturbing antibiotic, daptomycin (37), induced the formation of lateral branches in M. smegmatis that were marked with DivIVAMsm at their tip (Fig. 4B). As shown for the B. subtilis homolog, a DivIVAMsm mutant that lacked the putative N-terminal curvature-sensing domain (divIVAMsm∆1–15) was not capable of functionally complementing the depletion of the endogenous protein (Fig. 4C). In the presence of the endogenous allele, Δ1–15DivIVA is located at the pole, suggesting that the N-terminal truncated protein is able to oligomerize with the native protein, but is unable to form a functional polar focus by itself. Together, these observations indicate that recognition of negative curvature by the N terminus of DivIVAMsm is both necessary and sufficient for targeting mycobacterial DivIVA, although additional cues are likely to be involved in polarity determination.
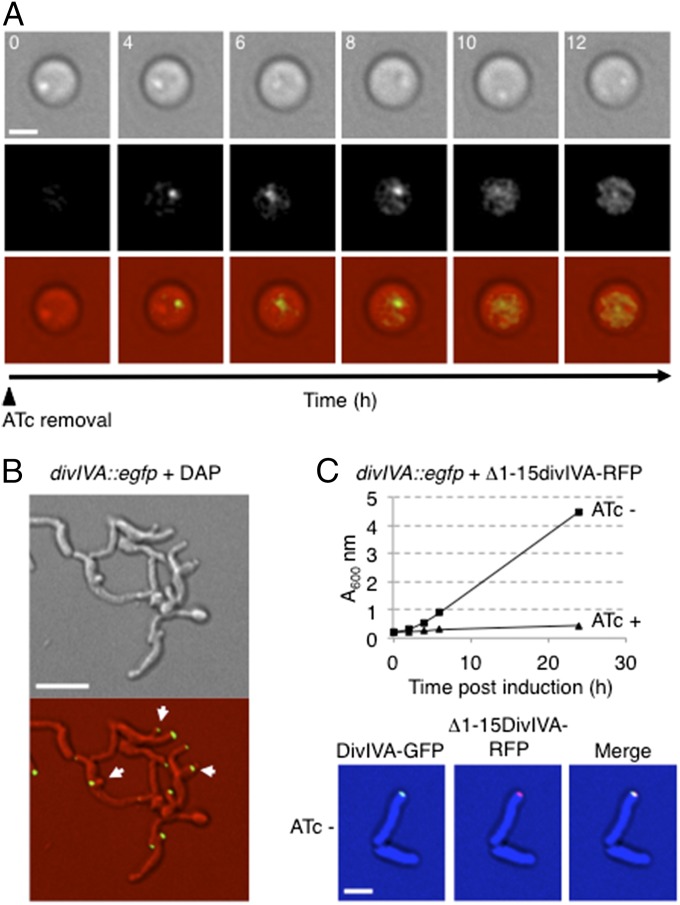
Fig. 4.
High membrane curvature is necessary and sufficient to generate a growing pole. (A) divIVA–egfp M. smegmatis strain was converted to spherical shape by 16 h of ATc incubation. Time-lapse microscopy was recorded after removal of ATc (Top, DIC; Middle, GFP; Bottom, overlays). (Scale bar, 2 μm.) (B) divIVA–egfp after treatment with daptomycin (DAP; 3 h at 250 μg/mL). Arrows show DivIVA–GFP marked branches. (Scale bar, 5 μm.) (C, Upper) Growth of the divIVA–egfp strain expressing Δ1–15DivIVA–RFP (pXM10) with or without ATc in 7H9–Tween 80 medium at 37 °C. The truncated protein is not capable to functionally complement depletion of the endogenous protein. (Lower) Imaging of the strain before inducible depletion shows expression of the truncated protein. (Scale bar, 2 μm.)
DivIVA Interacts with Enzymes Involved in the Early Stages of Mycolate Synthesis.
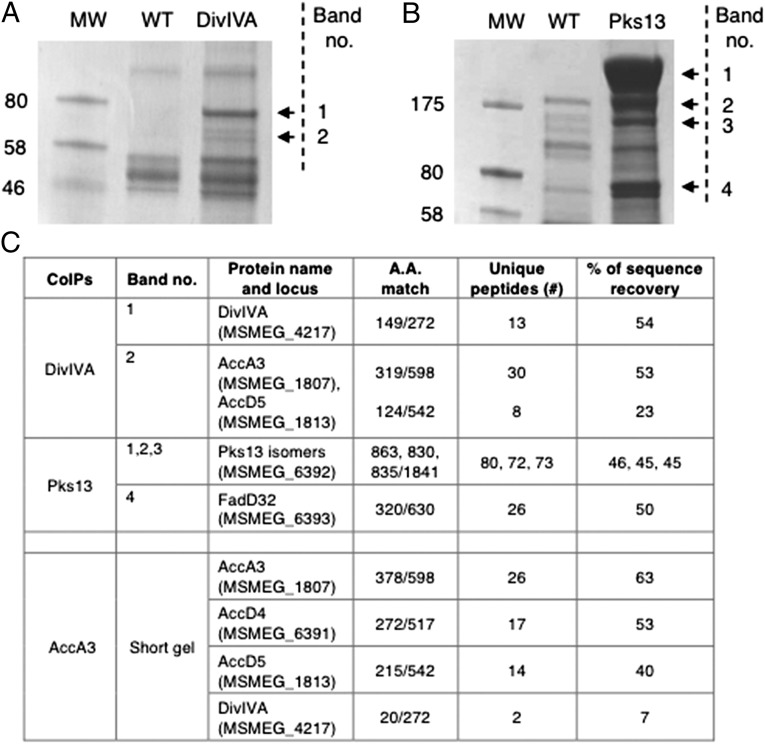
To understand whether DivIVA directs the formation of the growth pole through specific protein–protein interactions, we purified the endogenously tagged alleles of DivIVA, MurG, GlfT2, and Pks13 from M. smegmatis lysates and searched for associated proteins. We identified proteins that specifically copurified with DivIVAMsm and Pks13 (Fig. 5 A and B). Tandem mass spectrometry (MS/MS) analysis of the corresponding gel slices identified the DivIVAMsm-interacting proteins as AccA3 and AccD5, two members of the acyl-CoA carboxylase (ACC) complex (Fig. 5C). The Pks13 interactor was the Acyl–AMP ligase FadD32 (Fig. 5C). To exclude the possibility that the DivIVA–ACC complex interaction was mediated by the GFP tag, we also verified the presence of this interaction in WT M. smegmatis strains expressing different DivIVA alleles (FLAG or Dendra tag) expressed as a second copy in the chromosome (Fig. S5). In addition, analysis of the proteins found in the inverse pulldown of AccA3 identified DivIVAMsm, AccD4, and AccD5 (Fig. 5C), verifying that these four proteins stably associate in cell lysates.
Fig. 5.
DivIVA interacts with enzymes involved in mycolate precursor synthesis. (A and B) DivIVA–GFP (A) or Pks13–GFP (B) expressing the FLAG epitope was purified from bacterial lysates with anti-FLAG affinity beads. Interacting proteins were eluted with SDS, resolved by SDS/PAGE, and visualized by Coomassie Blue staining. MW, molecular weight standards; WT, WT M. smegmatis control strain MC2155. Numbered bands were excised and subjected to MS analysis for protein identification. (C) Interacting partners identified by Mascot searches in the M. smegmatis MC2155 database. As confidence in protein identification, the number of amino acids matched, the number of unique peptides found, and the percentage of sequence recovery are shown. For DivIVA and Pks13 coimmunoprecipitation (Co-IP) samples, numbers correspond to the specific bands identified on resolutive SDS/PAGE (top two rows). For the reverse Co-IP (bottom row), AccA3–RFP expressing the Myc epitope was purified from bacterial lysate with anti-Myc agarose beads. Interacting proteins were eluted with SDS and loaded on SDS/PAGE for a short run. A unique band was excised and subjected to MS for protein identification.
The ACC complex and FadD32 are involved in the activation of the two penultimate MA precursors (38–40). AccA3, AccD5, and AccD4 form the ACC complex that catalyzes the synthesis of the 2-carboxyacyl-CoA (C22–C26) (38), whereas the fatty acyl–AMP ligase FadD32 catalyzes the synthesis of the meromycoloyl–AMP (C40–C60) (39, 40). The two activated fatty acid chains are the substrates of the condensing enzyme, Pks13, which produces mature MA (28). Our biochemical data suggest that MA synthesis involves two different protein complexes, the DivIVA-associated ACC system and a distinct Pks13–FadD32 pair.
Cell Wall Synthesis Is Physically Separated from DivIVA.
The lack of a stable physical association between DivIVAMsm and the terminal cell wall synthetic enzymes suggested that new cell wall deposition might not occur precisely at the DivIVAMsm-marked tip of the cell. Superresolution photoactivated localization microscopy (PALM) verified that DivIVAMsm–Dendra was concentrated to the very tip of the cell pole when expressed in a merodiploid strain (Fig. 6A). Colocalization studies with cell wall synthetic enzymes confirmed our biochemical data. The ACC complex, typified by AccA3, was completely coincident with DivIVAMsm (Fig. 6B), but the more terminal enzymes, MurG, GlfT2, and Pks13, were found at a distinct site, renamed hereafter as the subpolar space (Fig. 6C). The lack of colocalization between terminal cell wall synthetic enzymes and DivIVAMsm (Fig. 6D) reinforces the conclusion that these proteins form two distinct structures at the growing pole.
Fig. 6.
DivIVAMsm marks the cell tip and new cell wall deposition occurs at a subpolar site. (A) PALM reconstituted image of the WT M. smegmatis strain expressing DivIVA–Dendra2 (pXM05) shows that DivIVA is located at the extreme cell tip. (Scale bar, 0.2 μm.) (B) Colocalization of DivIVA–GFP and AccA3–RFP (pXM09) in M. smegmatis using conventional microscopy. r, Pearson’s correlation coefficient between green and red fluorescence intensities. (C) Colocalization of MurG–GFP, GlfT2–GFP, and Pks13–GFP (endogenous gene fusions) with DivIVA–RFP (pXM01) in M. smegmatis using conventional microscopy. (Scale bars, 2 μm.) Upper Right shows an enlarged view of the dashed rectangle. (Scale bar, 0.5 μm.) (D) Intensity profiles of GlfT2–GFP and DivIVA–RFP fluorescence along the first 1.2 μm of 10 individual bacteria. Asterisk marks the cell tip. (E) Nascent cell wall of bacteria expressing DivIVA–RFP (pXM04) was labeled with alkyne d-ala for 20 min and stained with azido-PEG-Fluor 488, and a single optical section was obtained by structured illumination microscopy. One pole is shown. (F) Nascent cell wall of the divIVA–egfp strain was labeled with alkyne d-ala for 20 min and stained with azido-PEG-Fluor 545 (red), and cells were imaged by conventional microscopy. (Scale bar, 2 μm.) (G) Nascent cell wall of M. tuberculosis H37Rv expressing DivIVAMtb–RFP (pXM11) was labeled with alkyne d-ala for 20 min and stained with azido-PEG-Fluor 488 (green), and cells were imaged by conventional microscopy. (Scale bar, 2 μm.)
To verify that new cell wall deposition occurred at the same location as the terminal cell wall synthetic enzymes, we visualized nascent PG via the d-alanine chemical reporter. When viewed by using both wide-field deconvolution imaging and superresolution structured illumination microscopy, new cell wall synthesis was excluded from the DivIVAMsm-marked tip and occurred in the subpolar space where the terminal synthetic enzymes were found (Fig. 6 E and F). Different alleles of DivIVAMsm were tested to verify that the distinct location between the protein and the cell wall synthetic enzymes was not an artifact of DivIVA expression levels, DivIVA ectopic localization (divIVA–egfp strain), the sequence of the tag, or the position of the tag (Fig. S6 A and B). A similar labeling pattern was observed in M. tuberculosis (Fig. 6G), indicating that subpolar elongation is a general feature of mycobacterial growth. To verify this observation by an independent method, we nonspecifically labeled the cell wall with amine-reactive fluorophore and compared the relative stability of the label at the tip and subpolar sites. We found that a short chase was sufficient to remove the label from the subpolar area, whereas the tip remained inert (Fig. S7). Together, these results suggest a model in which DivIVAMsm initiates growth by recruiting early cell wall synthetic enzymes, whose products are then deposited at the subpolar space by the terminal cell wall synthetic systems.
Discussion
A growing number of rod-shaped and filamentous bacteria are recognized to grow or extend specialized appendages via the addition of new cell wall material at the pole. Despite the widespread use of this mode of elongation, both the precise site of cell wall addition and the mechanisms that maintain cellular morphology in these organisms have remained unclear. We have applied biochemistry, high-resolution microscopy, and bioorthogonal metabolic labeling to investigate the dynamics of cell wall synthesis in mycobacteria and develop an initial ultrastructural model of the growing pole (Fig. 7). The major tenets of this model are described below.
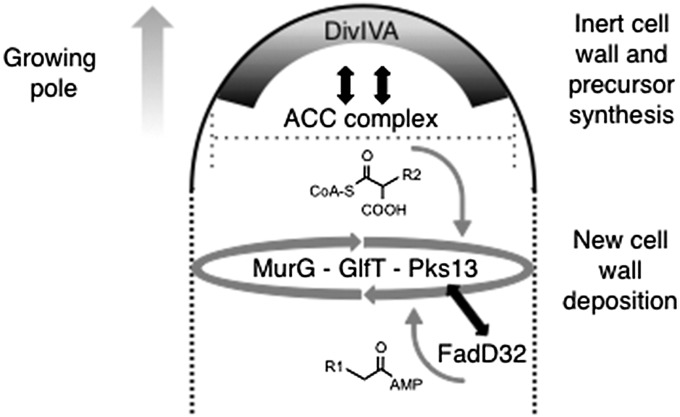
Fig. 7.
Model of the functional organization of the growing pole. DivIVA marks the tip of growing poles, and the terminal cell wall synthetic enzymes, MurG, GlfT2, and Pks13, are located at a subpolar space. Bioorthogonal labeling showed that new cell wall deposition occurs at this subpolar site (dashed lines). Double-headed arrows indicate protein–protein interactions identified in Co-IP experiments (Fig. 5C). Structures of the ACC complex and FadD32 products are shown. These products are condensed by Pks13 to form mature mycolates at the subpolar site. The dotted line crossing the cell tip depicts the role of DivIVA in determining cell diameter.
Terminal Cell Wall Synthetic Enzymes Colocalize at Sites of Growth.
Both the concentrations and colocalization of MurG, GlfT2, and Pks13 correlate with local cell wall synthesis. The asymmetry of the mycobacterial cell was evident in the distribution of these proteins, with the highest local concentration found at the old pole. This pole has been described as the preferential site of growth in mycobacteria (8). Although growth under certain conditions may produce a more symmetric growth pattern (9, 41), our time-lapse imaging, pulse–chase, and metabolic labeling studies all confirm the asymmetric growth pattern of M. smegmatis under nutrient-replete broth culture and in the microfluidic devices used in this study.
Terminal cell wall synthetic enzymes were also found as mobile foci along the lateral cell body. The apparent lack of cell wall synthesis at these sites and the low degree of colocalization between laterally localized synthetic systems suggested that these enzymes are not active and may simply be in transit to the final site of activity. Conversely, it is possible that these laterally localized systems might become active when remodeling of the cell wall is required. Indeed, stress can simultaneously inhibit mycobacterial growth and induce cell wall remodeling (42, 43). Laterally acting synthetic complexes would allow for more rapid adaptation than polar extension, which can only alter cell wall chemistry through growth. In the absence of an MreB homolog, the mycobacterial cell would presumably need another filament-like protein to maintain its rod shape. FtsZ has been shown to form a spiral-like structure during the intracellular growth of M. tuberculosis in macrophages (44), suggesting the possibility that this protein could substitute for MreB during stress-induced lateral remodeling.
DivIVA Specifically Marks Sites of Growth.
The DivIVA–GFP protein preferentially localizes to the growing pole in M. smegmatis, and the recognition of negative membrane curvature appears to play an important role in targeting the protein to this site. In B. subtilis, the first 22 amino acids of DivIVA form a curvature-sensing domain, and the exposed Phe-17 residue is proposed to interact directly with the membrane (34–36). Consistent with the importance of this domain, the critical Phe residue is conserved in DivIVAMsm, and truncation of this region produced a nonfunctional allele of DivIVA.
Although geometric constraints are likely to contribute to the polar association of DivIVA in mycobacteria, it is difficult to envision how this mechanism alone would preferentially target the protein to a single pole, and additional mechanisms may be involved. Several observations suggest that phosphosignaling could play a role. The Ser/Thr protein kinases, PknA and PknB, phosphorylate DivIVAMtb on Thr-73 (45), and mutation of this residue modestly affects polar localization (46). Similarly, in the filamentous bacterium Streptomyces coelicolor, constitutive expression of an active form of the Ser/Thr protein kinase AfsK induces hyperphosphorylation of DivIVA that leads to branching through the fragmentation of the apical DivIVASco focus (18). In addition, we cannot rule out the contribution of an uncharacterized landmark protein, such as Scy, which acts as a molecular assembler that influences polarized growth by interacting with DivIVA in S. coelicolor (47). The observation that DivIVAMsm–GFP localizes to the new pole and drives growth from this unusual site suggests that the addition of a bulky tag to the C terminus of the protein interferes with one of these targeting mechanisms.
Cell Wall Is Added at a Subpolar Site That Is Spatially Separated from DivIVA and Mycolate Precursor Synthesis.
We and others (48) have hypothesized that DivIVA determines growth site preference through physical interactions with cell elongation machinery. Copurification of DivIVAMsm and the ACC complex supported this general model and indicated that synthesis of the 2-carboxyacyl-CoA (C22–C26) mycolate precursor (38) occurs at the pole tip. A distinct Pks13 and FadD32 complex then condenses this acyl chain with the meromycoloyl–AMP (49, 50) to produce mature MA at the site of cell wall synthesis. It is possible that we have only identified a fraction of the DivIVAMsm-interacting proteins and that PG and AG precursors are also generated at this site. Despite its ability to determine the site of growth, overexpression of DivIVAMsm did not increase the rate of elongation. Thus, extension is not limited by the local concentration of this protein, and other factors, such as the abundance of precursors or synthetic enzymes, control elongation rate in mycobacteria.
To our knowledge, the precise site and geometry of cell wall addition during polar growth has not been reported in any organism. Our biochemical and imaging studies indicate that cell wall synthesis is excluded from the DivIVA-marked tip of the mycobacterial pole and occurs at a distinct subpolar location. These observations suggest that new cell wall could be added circumferentially at the border between the cylindrical cell body and the highly curved pole (Fig. 7) in a manner that is analogous to the lateral addition of PG that has been described extensively in B. subtilis and E. coli. The area occupied by cell wall synthetic enzymes was greater at the growing end of the cell (Fig. 1B), suggesting that the size of the zone in which synthesis occurs might be dynamically regulated based on elongation rate. These paradigms may be generally applicable to other organisms that produce polar extensions.
Although cell wall is not added at the same site in which DivIVA resides, this protein is still able to determine the site of growth, cell diameter, and cell curvature. A number of models can explain this protein’s ability to determine the location and geometry of cell wall deposition without directly interacting with the terminal synthetic complexes. DivIVA oligomers could influence cell width and curvature by providing the physical scaffolding that defines the dimensions of the pole or by nucleating the assembly of additional intermediate filaments, as it has been suggested in Streptomyces (51). In addition, our data suggest that the maintenance of precursor gradients could restrict cell wall addition to the subpolar space. The presence of presumably inactive MurG at the nongrowing pole devoid of DivIVA–GFP, in the divIVA–egfp strain (Fig. S6B), is consistent with a model in which DivIVA acts to concentrate cell wall precursors at the growing cell tip, but not to target terminal cell wall synthetic enzymes to this site.
Materials and Methods
Bacterial Strains and Culture Conditions.
M. smegmatis MC2155 was grown at 37 °C in Middlebrook 7H9 broth with 0.05% Tween 80, 0.2% glycerol, 0.5% BSA, 0.2% dextrose, and 0.085% NaCl plus hygromycin (50 μg/mL) and kanamycin (25 μg/mL) as needed. Lysine/pantothenate double auxotroph of M. tuberculosis (H37Rv) was grown in the presence of lysine and pantothenate as described (52). Transformants were plated on Middlebrook 7H10 agar with 0.5% glycerol, 0.5% BSA, 0.2% dextrose, and 0.085% NaCl. NEB-5α E. coli cells were grown at 37 °C in LB (Difco) with ampicillin (100 μg/mL), hygromycin (100 μg/mL), and kanamycin (50 μg/mL) as required. After growth overnight at 37 °C, strains were diluted to A600 ∼ 0.2 into fresh medium with or without ATc (50 ng/mL). Growth curves represent the mean of triplicate samples. Cell elongation (micrometers) was determined by measuring cell length increase during one doubling.
Inducible Protein Degradation, Plasmids, and Primers.
Degradation (ID) tags were introduced in the chromosome of a M. smegmatis recBCD- mutant strain at the 3′ end of murG, glfT2, pks13, and divIVA, and protein depletion was initiated at OD600 ∼ 0.2 by addition of ATc (50 ng/mL) as described (30, 32, 53). For the complementation assay of DivIVA depletion and colocalization experiments, divIVA was cloned into integrating and replicating vectors, respectively (gifts from Dirk Schnappinger, Weill Cornell Medical College, New York). Tables S1 and S2 list plasmids and primers.
Western Blot Analysis.
Mycobacteria were lysed by bead beating in 50 mM Hepes (pH 7.4), 50 mM NaCl, and 0.5% Triton X-100 with protease inhibitor (Roche). Lysates were quantified by BCA assay (Pierce), separated by SDS/PAGE (Bio-Rad), and immunoblotted with HRP-conjugated anti-6xHis antibodies (Abcam). Blots were visualized by using ECL reagents (GE Healthcare).
Coimmunoprecipitation Experiments.
Cell lysates were prepared as described above by using 0.5% dodecyl maltoside instead of Triton X-100. Coimmunoprecipitations (Co-IPs) were performed with Anti-FLAG M2 Affinity Gel (Sigma-Aldrich), separated by SDS/PAGE, and visualized with Coomassie Brilliant Blue. The AccA3 Co-IP sample was run 1 cm into the stacking gel [4% (wt/vol) polyacrylamide] before excision for MS/MS.
MS/MS Analysis of Tryptic Peptides.
Excised bands were in-gel trypsin-digested and then submitted to liquid chromatography (LC)/MS/MS by using an Agilent 6520 Accurate-Mass Quadrupole Time-of-Flight instrument. Peptides were separated on a POROSHELL 300SB-C18 (2.1 × 75 mm, 5 μm) at a 0.5 mL/min flow rate, by using a linear gradient of increasing acetonitrile in water. Protein identification was processed by using Spectrum Mill software (Agilent). For the AccA3 Co-IP sample, peptide analysis was performed by using the Thermo Scientific Q-exactive Mass spectrometer with Waters Nanoacquity Ultraperformance LC. Protein identification was processed by using Mascot data analysis software (Version 2.4.0; Matrix Science).
Cell Wall Labeling.
Cells were washed with PBS containing 0.2% Tween 20 (PBST) and resuspended in 1/10 of the culture volume. Alexa Fluor 546 carboxylic acid succinimidyl ester was added to a final concentration of 0.05 mg/mL (Invitrogen). The cells were immediately washed in PBST, resuspended in 7H9, and either imaged or incubated for a chase period before imaging. Click chemistry reactions were performed as described by using azido-PEG-Fluor 488 or azido-PEG-Fluor 545 (Click Chemistry Tools) following a 20-min incubation with 5 mM alkyne d-alaine (Acros) (33).
Conventional Wide-Field Microscopy.
Time-lapse imaging was performed with a DeltaVision Personal DV microscope equipped with an environmental chamber warmed to 37 °C (Applied Precision). Cells were imaged every 10 min for 15–17 h in a B04A Microfluidic bacteria plate (CellASIC) continuously perfused with 7H9 medium. In Fig. 4A, bacteria were grown on an agarose pad. Softworx software (Applied Precision) was used to deconvolve z-stacks of six images acquired every 200 nm in the z axis and project 2D images by using the sum method. Tagged-DivIVA foci intensity and cell wall extent were quantified from still images by using CellProfiler (54) and Excel. Data analysis to generate moving average fluorescence intensity profiles was performed by using Mathematica (Version 9.0; Wolfram Research). The number of cells per segment per experiment are 41 (MurG–GFP), 45 (GlfT2–GFP), 30 (Pks13–GFP), 55 (GlfT2–GFP/MurG–RFP), 26 (Pks13–GFP/MurG–RFP), 16 (WT + DivIVA–Dendra), and 22 (DivIVA–GFP). For each strain, individual cells showing no septum were picked manually and oriented according to the brightest pole by using the Softworx software. Data were extracted as fluorescence intensity per pixel along the cell lengths (or segments). Because of the variability in cell lengths, the segment lengths in each experiment were normalized to yield a fractional distance (i.e., between 0 and 1). The {fractional distance,fluorescence}-pairs for all segments were combined into one dataset and sorted on fractional distance. A moving average was calculated from the resulting dataset by using a window size equal to the number of segments. The fluorescence intensity for the green and red channels was normalized in colocalization experiments (Figs. 1 D and E and 2 C and F).
High-Resolution Microscopy.
Structured illumination microscopy images were acquired by using the Deltavision OMX Blaze microscope. Unprocessed image stacks were composed of 15 images per z-section. The z-sections were completed at 125-nm spacing. Raw data from the OMX Blaze was processed as for OMX V3 data. For PALM, Dendra-labeled cells mounted on an agarose pad were imaged by using the Nikon N-Storm system, and images were processed by using Nikon software.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants AI10954208 (to C.M.S.) and AI051622 (to C.R.B.); American Cancer Society Grant 119087-PF-10-258-01-MPC (to M.S.S.); and the Howard Hughes Medical Institute (C.M.S. and C.R.B.). R.O. is a Howard Hughes Medical Institute Fellow of the Damon Runyon Cancer Research Foundation, DRG-2114-12.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402158111/-/DCSupplemental.
References
- 1.de Pedro MA, Quintela JC, Höltje JV, Schwarz H. Murein segregation in Escherichia coli. J Bacteriol. 1997;179(9):2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: Two distinct ways to make a rod-shaped cell. Cell. 2003;113(6):767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 3.Salje J, van den Ent F, de Boer P, Löwe J. Direct membrane binding by bacterial actin MreB. Mol Cell. 2011;43(3):478–487. doi: 10.1016/j.molcel.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner JK, Galvani CD, Brun YV. Caulobacter crescentus requires RodA and MreB for stalk synthesis and prevention of ectopic pole formation. J Bacteriol. 2005;187(2):544–553. doi: 10.1128/JB.187.2.544-553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown PJ, et al. Polar growth in the Alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci USA. 2012;109(5):1697–1701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flärdh K. Growth polarity and cell division in Streptomyces. Curr Opin Microbiol. 2003;6(6):564–571. doi: 10.1016/j.mib.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Thanky NR, Young DB, Robertson BD. Unusual features of the cell cycle in mycobacteria: Polar-restricted growth and the snapping-model of cell division. Tuberculosis (Edinb) 2007;87(3):231–236. doi: 10.1016/j.tube.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Aldridge BB, et al. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science. 2012;335(6064):100–104. doi: 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santi I, Dhar N, Bousbaine D, Wakamoto Y, McKinney JD. Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria. Nat Commun. 2013;4:2470. doi: 10.1038/ncomms3470. [DOI] [PubMed] [Google Scholar]
- 10.Hett EC, Rubin EJ. Bacterial growth and cell division: A mycobacterial perspective. Microbiol Mol Biol Rev. 2008;72(1):126–156. doi: 10.1128/MMBR.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daffe M, Brennan PJ, McNeil M. Predominant structural features of the cell wall arabinogalactan of Mycobacterium tuberculosis as revealed through characterization of oligoglycosyl alditol fragments by gas chromatography/mass spectrometry and by 1H and 13C NMR analyses. J Biol Chem. 1990;265(12):6734–6743. [PubMed] [Google Scholar]
- 12.McNeil M, Daffe M, Brennan PJ. Location of the mycolyl ester substituents in the cell walls of mycobacteria. J Biol Chem. 1991;266(20):13217–13223. [PubMed] [Google Scholar]
- 13.Liu J, Barry CE, 3rd, Besra GS, Nikaido H. Mycolic acid structure determines the fluidity of the mycobacterial cell wall. J Biol Chem. 1996;271(47):29545–29551. doi: 10.1074/jbc.271.47.29545. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Rosenberg EY, Nikaido H. Fluidity of the lipid domain of cell wall from Mycobacterium chelonae. Proc Natl Acad Sci USA. 1995;92(24):11254–11258. doi: 10.1073/pnas.92.24.11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci USA. 2008;105(10):3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuber B, et al. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bacteriol. 2008;190(16):5672–5680. doi: 10.1128/JB.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daffé M, Draper P. The envelope layers of mycobacteria with reference to their pathogenicity. Adv Microb Physiol. 1998;39:131–203. doi: 10.1016/s0065-2911(08)60016-8. [DOI] [PubMed] [Google Scholar]
- 18.Hempel AM, et al. The Ser/Thr protein kinase AfsK regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proc Natl Acad Sci USA. 2012;109(35):E2371–E2379. doi: 10.1073/pnas.1207409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen L, et al. Antigen 84, an effector of pleiomorphism in Mycobacterium smegmatis. J Bacteriol. 2007;189(21):7896–7910. doi: 10.1128/JB.00726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang CM, Nyayapathy S, Lee JY, Suh JW, Husson RN. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology. 2008;154(Pt 3):725–735. doi: 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
- 21.Letek M, et al. DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum. J Bacteriol. 2008;190(9):3283–3292. doi: 10.1128/JB.01934-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards DH, Errington J. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol. 1997;24(5):905–915. doi: 10.1046/j.1365-2958.1997.3811764.x. [DOI] [PubMed] [Google Scholar]
- 23.Cha JH, Stewart GC. The divIVA minicell locus of Bacillus subtilis. J Bacteriol. 1997;179(5):1671–1683. doi: 10.1128/jb.179.5.1671-1683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ginda K, et al. ParA of Mycobacterium smegmatis co-ordinates chromosome segregation with the cell cycle and interacts with the polar growth determinant DivIVA. Mol Microbiol. 2013;87(5):998–1012. doi: 10.1111/mmi.12146. [DOI] [PubMed] [Google Scholar]
- 25.Donovan C, Sieger B, Krämer R, Bramkamp M. A synthetic Escherichia coli system identifies a conserved origin tethering factor in Actinobacteria. Mol Microbiol. 2012;84(1):105–116. doi: 10.1111/j.1365-2958.2012.08011.x. [DOI] [PubMed] [Google Scholar]
- 26.Salmond GP, Lutkenhaus JF, Donachie WD. Identification of new genes in a cell envelope-cell division gene cluster of Escherichia coli: Cell envelope gene murG. J Bacteriol. 1980;144(1):438–440. doi: 10.1128/jb.144.1.438-440.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kremer L, et al. Galactan biosynthesis in Mycobacterium tuberculosis. Identification of a bifunctional UDP-galactofuranosyltransferase. J Biol Chem. 2001;276(28):26430–26440. doi: 10.1074/jbc.M102022200. [DOI] [PubMed] [Google Scholar]
- 28.Portevin D, et al. A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc Natl Acad Sci USA. 2004;101(1):314–319. doi: 10.1073/pnas.0305439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lea-Smith DJ, et al. The reductase that catalyzes mycolic motif synthesis is required for efficient attachment of mycolic acids to arabinogalactan. J Biol Chem. 2007;282(15):11000–11008. doi: 10.1074/jbc.M608686200. [DOI] [PubMed] [Google Scholar]
- 30.Wei JR, et al. Depletion of antibiotic targets has widely varying effects on growth. Proc Natl Acad Sci USA. 2011;108(10):4176–4181. doi: 10.1073/pnas.1018301108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joyce G, et al. Cell division site placement and asymmetric growth in mycobacteria. PLoS ONE. 2012;7(9):e44582. doi: 10.1371/journal.pone.0044582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gee CL, et al. A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci Signal. 2012;5(208):ra7. doi: 10.1126/scisignal.2002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegrist MS, et al. (D)-amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem Biol. 2013;8(3):500–505. doi: 10.1021/cb3004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenarcic R, et al. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28(15):2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci USA. 2009;106(32):13541–13545. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliva MA, et al. Features critical for membrane binding revealed by DivIVA crystal structure. EMBO J. 2010;29(12):1988–2001. doi: 10.1038/emboj.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pogliano J, Pogliano N, Silverman JA. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J Bacteriol. 2012;194(17):4494–4504. doi: 10.1128/JB.00011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gande R, et al. The two carboxylases of Corynebacterium glutamicum essential for fatty acid and mycolic acid synthesis. J Bacteriol. 2007;189(14):5257–5264. doi: 10.1128/JB.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Portevin D, et al. The acyl-AMP ligase FadD32 and AccD4-containing acyl-CoA carboxylase are required for the synthesis of mycolic acids and essential for mycobacterial growth: Identification of the carboxylation product and determination of the acyl-CoA carboxylase components. J Biol Chem. 2005;280(10):8862–8874. doi: 10.1074/jbc.M408578200. [DOI] [PubMed] [Google Scholar]
- 40.Trivedi OA, et al. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature. 2004;428(6981):441–445. doi: 10.1038/nature02384. [DOI] [PubMed] [Google Scholar]
- 41.Wakamoto Y, et al. Dynamic persistence of antibiotic-stressed mycobacteria. Science. 2013;339(6115):91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- 42.Walters SB, et al. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol Microbiol. 2006;60(2):312–330. doi: 10.1111/j.1365-2958.2006.05102.x. [DOI] [PubMed] [Google Scholar]
- 43.Gonzalo Asensio J, et al. The virulence-associated two-component PhoP-PhoR system controls the biosynthesis of polyketide-derived lipids in Mycobacterium tuberculosis. J Biol Chem. 2006;281(3):1313–1316. doi: 10.1074/jbc.C500388200. [DOI] [PubMed] [Google Scholar]
- 44.Chauhan A, et al. Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J Bacteriol. 2006;188(5):1856–1865. doi: 10.1128/JB.188.5.1856-1865.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang CM, et al. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: Substrate identification and regulation of cell shape. Genes Dev. 2005;19(14):1692–1704. doi: 10.1101/gad.1311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jani C, et al. Regulation of polar peptidoglycan biosynthesis by Wag31 phosphorylation in mycobacteria. BMC Microbiol. 2010;10:327. doi: 10.1186/1471-2180-10-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holmes NA, et al. Coiled-coil protein Scy is a key component of a multiprotein assembly controlling polarized growth in Streptomyces. Proc Natl Acad Sci USA. 2013;110(5):E397–E406. doi: 10.1073/pnas.1210657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hempel AM, Wang SB, Letek M, Gil JA, Flärdh K. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. J Bacteriol. 2008;190(22):7579–7583. doi: 10.1128/JB.00839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Léger M, et al. The dual function of the Mycobacterium tuberculosis FadD32 required for mycolic acid biosynthesis. Chem Biol. 2009;16(5):510–519. doi: 10.1016/j.chembiol.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 50.Gavalda S, et al. The Pks13/FadD32 crosstalk for the biosynthesis of mycolic acids in Mycobacterium tuberculosis. J Biol Chem. 2009;284(29):19255–19264. doi: 10.1074/jbc.M109.006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fuchino K, et al. Dynamic gradients of an intermediate filament-like cytoskeleton are recruited by a polarity landmark during apical growth. Proc Natl Acad Sci USA. 2013;110(21):E1889–E1897. doi: 10.1073/pnas.1305358110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sampson SL, et al. Protection elicited by a double leucine and pantothenate auxotroph of Mycobacterium tuberculosis in guinea pigs. Infect Immun. 2004;72(5):3031–3037. doi: 10.1128/IAI.72.5.3031-3037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nat Methods. 2007;4(2):147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 54.Lamprecht MR, Sabatini DM, Carpenter AE. CellProfiler: Free, versatile software for automated biological image analysis. Biotechniques. 2007;42(1):71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.