Significance
For more than three decades since the discovery of HIV-1, AIDS remains a major public health problem affecting greater than 35.3 million people worldwide. Current antiretroviral therapy has failed to eradicate HIV-1, partly due to the persistence of viral reservoirs. RNA-guided HIV-1 genome cleavage by the Cas9 technology has shown promising efficacy in disrupting the HIV-1 genome in latently infected cells, suppressing viral gene expression and replication, and immunizing uninfected cells against HIV-1 infection. These properties may provide a viable path toward a permanent cure for AIDS, and provide a means to vaccinate against other pathogenic viruses. Given the ease and rapidity of Cas9/guide RNA development, personalized therapies for individual patients with HIV-1 variants can be developed instantly.
Keywords: CRISPR/Cas9, genome editing, latency, retrovirus, reservoir
Abstract
AIDS remains incurable due to the permanent integration of HIV-1 into the host genome, imparting risk of viral reactivation even after antiretroviral therapy. New strategies are needed to ablate the viral genome from latently infected cells, because current methods are too inefficient and prone to adverse off-target effects. To eliminate the integrated HIV-1 genome, we used the Cas9/guide RNA (gRNA) system, in single and multiplex configurations. We identified highly specific targets within the HIV-1 LTR U3 region that were efficiently edited by Cas9/gRNA, inactivating viral gene expression and replication in latently infected microglial, promonocytic, and T cells. Cas9/gRNAs caused neither genotoxicity nor off-target editing to the host cells, and completely excised a 9,709-bp fragment of integrated proviral DNA that spanned from its 5′ to 3′ LTRs. Furthermore, the presence of multiplex gRNAs within Cas9-expressing cells prevented HIV-1 infection. Our results suggest that Cas9/gRNA can be engineered to provide a specific, efficacious prophylactic and therapeutic approach against AIDS.
Infection with HIV-1 is a major public health problem affecting more than 35 million people worldwide (1). Current therapy for controlling HIV-1 infection and impeding AIDS development (highly active antiretroviral therapy; HAART) includes a mixture of compounds that suppress various steps of the viral life cycle (2). HAART profoundly reduces viral replication in cells that support HIV-1 infection and reduces plasma viremia to a minimal level but neither suppresses low-level viral genome expression and replication in tissues nor targets the latently infected cells that serve as a reservoir for HIV-1, including brain macrophages, microglia, and astrocytes, gut-associated lymphoid cells, and others (3, 4). HIV-1 persists in ∼106 cells per patient during HAART, and is linked to comorbidities including heart and renal diseases, osteopenia, and neurological disorders (5). Because current therapies are unable to suppress viral gene transcription from integrated proviral DNA or eliminate the transcriptionally silent proviral genomes, low-level viral protein production by latently infected cells may contribute to multiple illnesses in the aging HIV-1–infected patient population. Supporting this notion, pathogenic viral proteins including transactivator of transcription (Tat) are present in the cerebrospinal fluid of HIV-1–positive patients receiving HAART (6). To prevent viral protein expression and viral reactivation in latently infected host cells, new strategies are thus needed to permanently disable the HIV-1 genome by eradicating large segments of integrated proviral DNA.
Advances in the engineered nucleases including zinc finger nuclease (ZFN), transcription activator-like effector nuclease (TALEN), and clustered regularly interspaced short palindromic repeats (CRISPR) associated 9 (Cas9) that can disrupt target genes have raised prospects of selectively deleting HIV-1 proviral DNA integrated into the host genome (7–10). These approaches have been used to disrupt HIV-1 entry coreceptors C-C chemokine receptor 5 (CCR5) or C-C-C chemokine receptor 4 (CXCR4) and proviral DNA-encoding viral proteins (8, 9). CCR5 gene-targeting ZFNs are in phase II clinical trials for HIV-1/AIDS treatment (11). Also, various gene editing technologies have recently been shown to remove the proviral HIV-1 DNA from the host cell genome by targeting its highly conserved 5′ and 3′ long terminal repeats (LTRs) (12, 13). However, introduction of nucleases into cells via these nuclease-based genomic editing approaches remains inefficient and partially selective to remove the entire HIV-1 genome. Thus, the key barrier to their clinical translation is insufficient gene specificity to prevent potential off-target effects (toxicities). To achieve highly specific HIV-1 genome editing, we combined approaches to identify HIV-1 targets while circumventing host off-target effects. The resulting highly specific Cas9-based method proved capable of eradicating integrated HIV-1 DNA with high efficiency from latently infected human “reservoir” cell types, and prevented their infection by HIV-1.
Results
We assessed the ability of HIV-1–directed guide RNAs (gRNAs) to abrogate LTR transcriptional activity and eradicate proviral DNA from the genomes of latently infected myeloid cells that serve as HIV-1 reservoirs in the brain, a particularly intractable target population. Our strategy was focused on targeting the HIV-1 LTR promoter U3 region. By bioinformatic screening and efficiency/off-target prediction (14, 15), we identified four gRNA targets (protospacers; LTRs A−D) that avoid conserved transcription factor binding sites, minimizing the likelihood of altering host gene expression (Table S1 and Fig. S1). We inserted DNA oligonucleotides (Table S2) complementary to gRNAs A−D into a humanized Cas9 expression vector (A/B in pX260; C/D in pX330) (16) and tested their individual and combined abilities to alter the integrated HIV-1 genome activity. We first used the microglial cell line CHME5, which harbors integrated copies of a single round HIV-1 vector that includes the 5′ and 3′ LTRs, and a gene encoding an enhanced green fluorescent protein (EGFP) reporter replacing Gag (pNL4-3-ΔGag-d2EGFP) (17). Treating CHME5 cells with trichostatin A (TSA), a histone deacetylase inhibitor, reactivates transcription from the majority of the integrated proviruses and leads to expression of EGFP and the remaining HIV-1 proteome (17). Expressing of gRNAs plus Cas9 markedly decreased the fraction of TSA-induced EGFP-positive CHME5 cells (Fig. 1A and Fig. S2). We detected insertion/deletion gene mutations (indels) for LTRs A−D (Fig. 1B and Fig. S2B) using a Cel I nuclease-based heteroduplex-specific SURVEYOR assay. Similarly, expressing gRNAs targeting LTRs C and D in HeLa-derived TZM-bI cells, which contain stably incorporated HIV-1 LTR copies driving a firefly luciferase reporter gene (18), suppressed viral promoter activity (Fig. S3A), and elicited indels within the LTR U3 region (Fig. S3 B−D) demonstrated by SURVEYOR and Sanger sequencing. Moreover, the combined expression of LTR-C/D-targeting gRNAs in these cells caused excision of the predicted 302-bp viral DNA sequence, and emergence of the residual 194-bp fragment (Fig. S3 E and F).
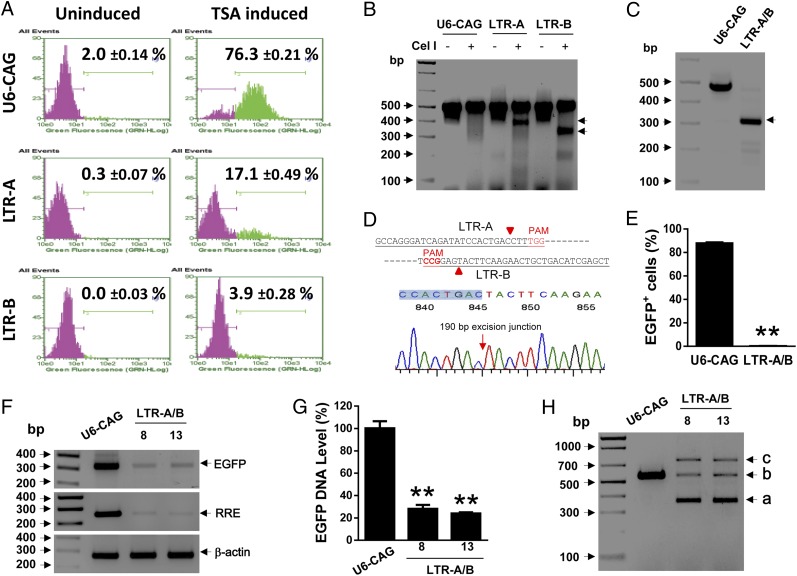
Fig. 1.
Cas9/LTR-gRNA suppresses HIV-1 reporter virus production in CHME5 microglial cells latently infected with HIV-1. (A) Representative gating diagram of EGFP flow cytometry shows a dramatic reduction in TSA-induced reactivation of latent pNL4-3-ΔGag-d2EGFP reporter virus by stably expressed Cas9 plus LTR-A or -B, vs. empty U6-driven gRNA expression vector (U6-CAG). (B) SURVEYOR Cel-I nuclease assay of PCR product (−453 to +43 within LTR) from selected LTR-A- or -B-expressing stable clones shows dramatic indel mutation patterns (arrows). (C and D) PCR fragment analysis shows a precise deletion of 190-bp region between LTR-A and -B cutting sites (red arrowhead and arrow), leaving 306-bp fragment (black arrow) validated by TA-cloning and sequencing results. (E−G) Subcloning of LTR-A/B stable clones reveals complete loss of reporter reactivation determined by EGFP flow cytometry (E) and elimination of pNL4-3-ΔGag-d2EGFP proviral genome detected by standard (F) and real-time (G) PCR amplification of genomic DNA for EGFP and HIV-1 Rev response element (RRE); β-actin is a DNA purification and loading control. (H) PCR genotyping of LTR-A/B subclones (#8, #13) using primers to amplify DNA fragment covering HIV-1 LTR U3/R/U5 regions (−411 to +129) shows indels (a, deletion; c, insertion) and “intact” or combined LTR (b).
Multiplex expression of LTR-A/B gRNAs in mixed clonal CHME5 cells caused deletion of a 190-bp fragment between A and B target sites and led to indels to various extents (Fig. 1 C and D). Among >20 puromycin-selected stable subclones, we found cell populations with complete blockade of TSA-induced HIV-1 proviral reactivation determined by flow cytometry for EGFP (Fig. 1E). PCR-based analysis for EGFP and HIV-1 Rev response element (RRE) in the proviral genome validated the eradication of HIV-1 genome (Fig. 1 F and G). Furthermore, sequencing of the PCR products revealed that the entire 5′−3′ LTR-spanning viral genome was deleted, yielding a 351-bp fragment via a 190-bp excision between cleavage sites A and B (Fig. 1H and Fig. S4), and a 682-bp fragment with a 175-bp insertion and a 27-bp deletion at the LTR-A and -B sites, respectively (Fig. S4C). The residual HIV-1 genome (Fig. 1 F−H) may reflect the presence of trace Cas9/gRNA-negative cells. These results indicate that LTR-targeting Cas9/gRNAs A/B eradicates the HIV-1 genome and blocks its reactivation in latently infected microglial cells.
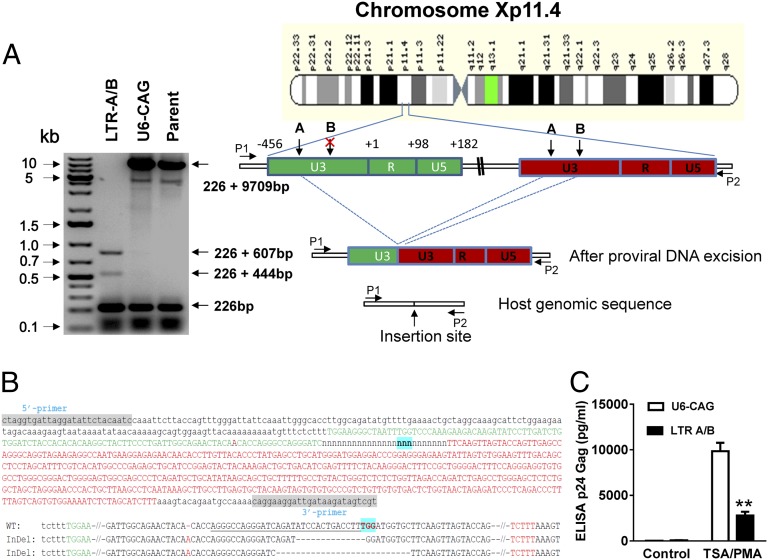
The promonocytic U-937 cell subclone U1, an HIV-1 latency model for infected perivascular macrophages and monocytes, is chronically HIV-1 infected and exhibits low-level constitutive viral gene expression and replication (19). GenomeWalker mapping detected two integrated proviral DNA copies at chromosomes Xp11-4 (Fig. 2A) and 2p21 (Fig. S5A) in U1 cells. A 9,935-bp DNA fragment representing the entire 9,709-bp proviral HIV-1 DNA plus a flanking 226-bp X-chromosome-derived sequence (Fig. 2A), and a 10,176-bp fragment containing 9,709-bp HIV-1 genome plus its flanking 2-chromosome-derived 467 bp (Fig. S5 A and B) were identified by the long-range PCR analysis of the parental control or empty-vector (U6-CAG) U1 cells. The 226-bp and 467-bp fragments represent the predicted segment from the other copy of chromosome X and 2, respectively, which lacked the integrated proviral DNA. In U1 cells expressing LTR-A/B gRNAs and Cas9, we found two additional DNA fragments of 833 and 670 bp in chromosome X and one additional 1,102-bp fragment in chromosome 2. Thus, gRNAs A/B enabled Cas9 to excise the HIV-1 5′−3′ LTR-spanning viral genome segment in both chromosomes. The 833-bp fragment includes the expected 226-bp from the host genome and a 607-bp viral LTR sequence with a 27-bp deletion around the LTR-A site (Fig. 2 A and B). The 670-bp fragment encompassed a 226-bp host sequence and residual 444-bp viral LTR sequence after 190-bp fragment excision (Fig. 1D), caused by gRNAs-A/B-guided cleavage at both LTRs (Fig. 2A). The additional fragments did not emerge via circular LTR integration, because it was absent in the parental U1 cells, and such circular LTR viral genome configuration occurs immediately after HIV-1 infection but is short lived and intolerant to repeated passaging (20). These cells exhibited substantially decreased HIV-1 viral load, shown by the functional p24 ELISA replication assay (Fig. 2C) and real-time PCR analysis (Fig. S5 C and D). The detectable but low residual viral load and reactivation may result from cell population heterogeneity and/or incomplete genome editing. We also validated the ablation of HIV-1 genome by Cas9/LTR-A/B gRNAs in latently infected J-Lat T cells harboring integrated HIV-R7/E-/EGFP (21) using flow cytometry analysis, SURVEYOR assay, and PCR genotyping (Fig. S6), supporting the results of previous reports on HIV-1 proviral deletion in Jurkat T cells by Cas9/gRNA (12) and ZFN (13). Taken together, our results suggest that the multiplex LTR-gRNAs/Cas9 system efficiently suppresses HIV-1 replication and reactivation in latently HIV-1–infected “reservoir” (microglial, monocytic, and T) cells typical of human latent HIV-1 infection, and in TZM-bI cells highly sensitive for detecting HIV-1 transcription and reactivation. Single or multiplex gRNAs targeting 5′ and 3′ LTRs effectively eradicated the entire HIV-1 genome.
Fig. 2.
Cas9/LTR-gRNA efficiently eradicates latent HIV-1 virus from U1 monocytic cells. (A) (Right) Diagram showing excision of HIV-1 entire genome in chromosome Xp11.4. HIV-1 integration sites were identified using a Genome-Walker link PCR kit. (Left) Analysis of PCR amplicon lengths using a primer pair (P1/P2) targeting chromosome X integration site-flanking sequence reveals elimination of the entire HIV-1 genome (9,709 bp), leaving two fragments (833 and 670 bp). (B) (Upper) TA cloning and sequencing of the LTR fragment (833 bp) showing the host genomic sequence (small letters, 226 bp) and the partial sequences (634 − 27 = 607 bp) of 5′ LTR (green) and 3′ LTR (red) with a 27-bp deletion around the LTR A targeting site (underlined). (Lower) Two indel alleles identified from 15 sequenced clonal amplicons. The 670-bp fragment consists of a host sequence (226 bp) and the remaining LTR sequence (634-190 = 444 bp) after 190-bp excision by simultaneous cutting at LTR-A and -B target sites. The underlined and green-highlighted sequences indicate the gRNA LTR-A target site and PAM. (C) Functional analysis of LTR-A/B-induced eradication of HIV-1 genome, showing substantial blockade of p24 virion release induced by TSA/phorbol myristate acetate (PMA) treatment. U1 cells were transfected with pX260-LTRs A, B, or A/B. After 2-wk puromycin selection, cells were treated with TSA (250 nM)/PMA for 2 d before p24 Gag ELISA was performed.
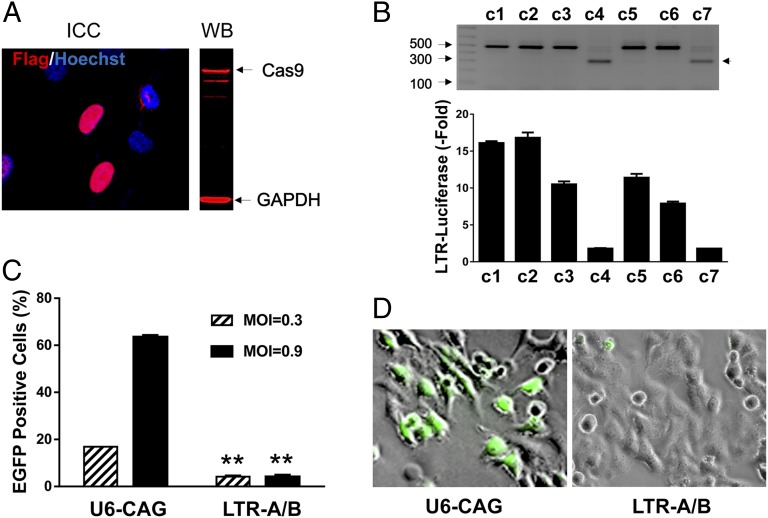
We next tested whether combined Cas9/LTR gRNAs can immunize cells against HIV-1 infection using stable Cas9/gRNAs-A and -B-expressing TZM-bI-based clones (Fig. 3A). Two of seven puromycin-selected subclones exhibited efficient excision of the 190-bp LTR-A/B site-spanning DNA fragment (Fig. 3B). However, the remaining five subclones exhibited no excision (Fig. 3B) and no indel mutations as verified by Sanger sequencing. PCR genotyping using primers targeting Cas9 and U6-LTR showed that none of these ineffective subclones retained the integrated copies of Cas9/LTR-A/B gRNA expression cassettes. (Fig. S7 A and B). As a result, no expression of full-length Cas9 was detected (Fig. S7 C and D). The long-term expression of Cas9/LTR-A/B gRNAs did not adversely affect cell growth or viability, suggesting a low occurrence of off-target interference with the host genome or Cas9-induced toxicity in this model. We assessed de novo HIV-1 replication by infecting cells with the VSV-G-pseudotyped pNL4-3-ΔE-EGFP reporter virus (22), with EGFP positivity by flow cytometry indicating HIV-1 replication. Unlike the control U6-CAG cells, the cells stably expressing Cas9/LTR-A/B gRNAs failed to support HIV-1 replication at 2 d postinfection, indicating that they were immunized effectively against new HIV-1 infection (Fig. 3 C and D). A similar immunity against HIV-1 was observed in Cas/LTR-A/B gRNA expressing cells infected with native T-tropic X4 strain pNL4-3-ΔE-EGFP reporter virus (Fig. S8A) or native M-tropic R5 strains such as SF162 and JRFL (Fig. S8 B−D).
Fig. 3.
Stable expression of Cas9 plus LTR-A/B vaccinates TZM-bI cells against new HIV-1 virus infection. (A) Immunocytochemistry (ICC) and Western blot (WB) analyses with anti-Flag antibody confirm the expression of Flag-Cas9 in TZM-bI stable clones puromycin (1 µg/mL) selected for 2 wk. (B) PCR genotyping of Cas9/LTR-A/B stable clones (c1−c7) reveals a close correlation of LTR excision with repression of LTR luciferase reporter activation. Fold changes represent TSA/PMA-induced levels over corresponding noninduction levels. (C) Stable Cas9/LTR-A/B-expressing cells (c4) were infected with pseudotyped-pNL4-3-Nef-EGFP lentivirus at indicated multiplicity of infection (MOI), and infection efficiency was measured by EGFP flow cytometry, 2 d postinfection. (D) Representative phase-contrast/fluorescence micrographs show that LTR-A/B stable but not control (U6-CAG) cells are resistant to new infection by pNL4-3-ΔE-EGFP HIV-1 reporter virus (green).
The appeal of Cas9/gRNA as an interventional approach rests on its highly specific on-target indel-producing cleavage (15, 16), but multiplex gRNAs could potentially cause host genome mutagenesis and chromosomal disorders, cytotoxicity, genotoxicity, or oncogenesis. Fairly low viral-human genome homology reduces this risk, but the human genome contains numerous endogenous retroviral genomes that are potentially susceptible to HIV-1–directed gRNAs. Therefore, we assessed off-target effects of selected HIV-1 LTR gRNAs on the human genome. Because the 12- to 14-bp seed sequence nearest the protospacer-adjacent motif (PAM) region (NGG) is critical for cleavage specificity (14, 23), we searched >14-bp seed+NGG, and found no off-target candidate sites by LTR gRNAs A−D (Table S1). It is not surprising that progressively shorter gRNA segments yielded increasing off-target cleavage sites 100% matched to corresponding on-target sequences (i.e., NGG+13 bp yielded 6, 0, 2, and 9 off-target sites, respectively, whereas NGG+12 bp yielded 16, 5, 16, and 29) (Table S1). From human genomic DNA, we obtained a 500- to 800-bp sequence covering one of the predicted off-target sites using high-fidelity PCR, and analyzed the potential mutations by SURVEYOR and Sanger sequencing. We found no mutations (see representative off-target sites #1, 5, and 6 in TZM-bI and U1 cells; Fig. 4A).
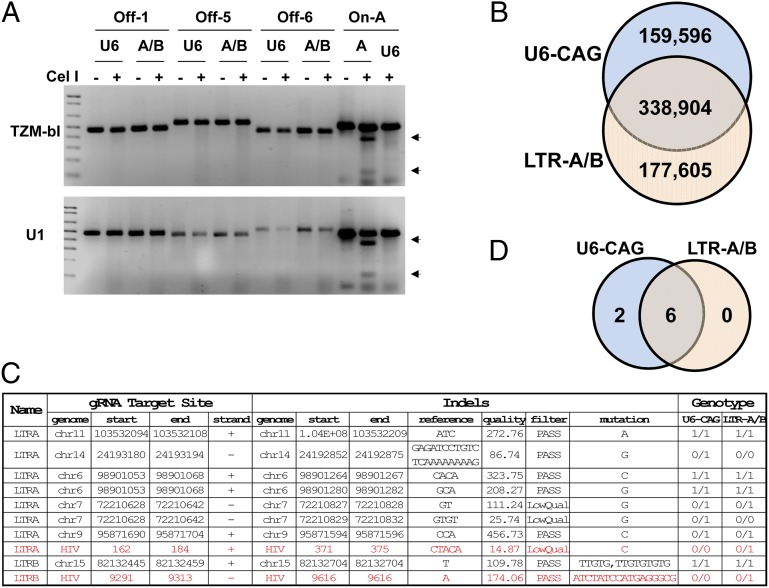
Fig. 4.
Off-target effects of Cas9/LTR-A/B on human genome. (A) SURVEYOR assay shows no indel mutations in predicted/potential off-target regions in human TZM-bI and U1 cells. LTR-A on-target region (A) was used as a positive control and empty U6-CAG vector (U6) as a negative control. (B−D) Whole-genome sequencing of LTR-A/B stable TZM-bI subclone showing the numbers of called indels in the U6-CAG control and LTR-A/B samples (B), detailed information on 10 called indels near gRNA target sites in both samples (C), and distribution of off-target called indels (D).
To assess risk of off-target effects comprehensively, we performed whole-genome sequencing (WGS) using the stable Cas9/gRNA A/B-expressing and control U6-CAG TZM-bI cells (Fig. 4 B−D). We identified 676,105 indels, using a genome analysis toolkit (GATK, v.2.8.1) with human (hg19) and HIV-1 genomes as reference sequences. Among the indels, 24% occurred in the U6-CAG control, 26% in LTR-A/B subclone, and 50% in both (Fig. 4B). Such substantial intersample indel-calling discrepancy suggests the probable off-target effects but most likely results from its limited confidence, limited WGS coverage (15−30×), and cellular heterogeneity. GATK reported only confidently identified indels: some found in the U6-CAG control but not in the LTR-A/B subclone, and others in the LTR-A/B but not in the U6-CAG. We expected abundant missing indel calls for both samples due to the limited WGS coverage. Such limited indel-calling confidence also implies the possibility of false negatives: missed indels occurring in LTR-A/B but not U6-CAG controls. Cellular heterogeneity may reflect variability of Cas9/gRNA editing efficiency and effects of passaging. Therefore, we tested whether each indel was LTR-A/B gRNA-induced, by analyzing ±300 bp flanking each indel against LTRs-A/-B-targeted sites of the HIV-1 genome and predicted/potential gRNA off-target sites of the host genome (Table S3). For sequences 100% matched to one containing the seed (12 bp) plus NRG, we identified only 8 overlapped regions of 92 potential off-target sites against 676,105 indels: 6 indels occurring in both samples, and 2 only in the U6-CAG control (Fig. 4 C and D). We also identified two indels on HIV-1 LTR that occurred only in the LTR-A/B subclone but, as expected, not in the U6-CAG control (Fig. 4C). The results suggest that LTR-A/B gRNAs induce the indicated on-target indels but no off-target indels, consistent with prior findings using deep sequencing of PCR products covering predicted/potential off-target sites (14, 24–27).
Discussion
The Cas9/gRNA technology platform is facile, versatile and improving rapidly (23), and clinical application is anticipated, particularly in the fields of virus infection, genetic diseases, and cancer (9, 28, 29). Here, we found that LTR-directed gRNA/Cas9 eradicates the HIV-1 genome and effectively immunizes target cells against HIV-1 reactivation and infection with high specificity and efficiency. These properties may provide a viable path toward a permanent or “sterile” HIV-1 cure, and perhaps provide a means to eradicate and vaccinate against other pathogenic viruses. In the current study, we have mainly focused our efforts on myeloid lineage cells (microglia/macrophage), which are the primary cell types that harbor HIV-1 in the brain. However, this proof of concept is certainly applicable to any other cell type, including T-lymphoid cells (Fig. S6) (12, 13), astrocytes, and neural stem cells.
Our combined approaches minimized off-target effects while achieving high efficiency and complete ablation of the genomically integrated HIV-1 provirus. In addition to an extremely low homology between the foreign viral genome and host cellular genome including endogenous retroviral DNA, the key design attributes in our study included: bioinformatic screening using the strictest 12-bp+NGG target selection criteria to exclude off-target human transcriptome or (even rarely) untranslated genomic sites; avoiding transcription factor binding sites within the HIV-1 LTR promoter (potentially conserved in the host genome); selection of LTR-A- and -B-directed, 30-bp protospacer and also precrRNA system reflecting the original bacterial immune mechanism to enhance specificity/efficiency vs. 20-bp protospacer-based, chimeric crRNA-tracRNA system (16, 30); and WGS, Sanger sequencing, and SURVEYOR assay, to identify and exclude potential off-target effects. Indeed, the use of newly developed Cas9 double-nicking (23) and RNA-guided FokI nuclease (31, 32) may further assist identification of new targets within the various conserved regions of HIV-1 with reduced off-target effects.
More recently, a clinical trial using the ZFN gene editing strategy was launched to disrupt the gene encoding the HIV-1 coreceptor, CCR5 (8, 9, 11). Functional knockout of CCR5 in autologous CD4 T cells of a small cohort of patients revealed that in one out of four enrolled subjects, the viral load remained undetectable at the time of treatment (33). Similarly, TALEN and Cas9 have been tested experimentally for efficient disruption of CCR5 and CXCR4 (9, 28, 34–37); therefore, taking them into consideration for clinical trials is anticipated. Whether or not the strategies targeting HIV-1 entry can reach the “sterile” cure of AIDS remains to be seen. Our results show that the HIV-1 Cas9/gRNA system has the ability to target more than one copy of the LTR, which are positioned on different chromosomes, suggesting that this genome-editing system can alter the DNA sequence of HIV-1 in latently infected patient’s cells harboring multiple proviral DNAs. To further ensure high editing efficacy and consistency of our technology, one may consider the most stable region of HIV-1 genome as a target to eradicate HIV-1 in patient samples, which may not harbor only one strain of HIV-1. Alternatively, one may develop personalized treatment modalities based on the data from deep sequencing of the patient-derived viral genome before engineering therapeutic Cas9/gRNA molecules.
Our results also demonstrate, for the first time to our knowledge, that Cas9/gRNA genome editing can be used to immunize cells against HIV-1 infection. The preventative vaccination is independent of HIV-1 strain’s diversity because the system targets genomic sequences regardless of how the viruses enter the infected cells. Interestingly, the preexistence of the Cas9/gRNA system in cells leads to a rapid elimination of the new HIV-1 before it integrates into the host genome, just like the way by which the bacteria defense system evolved to combat phage infection (38). Similarly, a gene-editing-based vaccine strategy may be effective in eradicating postintegrated HIV-1 genome and newly packaged proviruses in cells. Therefore, investigation of such HIV-1 vaccination in various latent reservoir cells and animal models with stable expression of Cas9/LTR-gRNAs presents an important next step to assess the ability of Cas9 to eradicate viral reservoirs in vivo. Moreover, in light of recent data illustrating efficient in vitro genome editing using a mixture of Cas9/gRNA and DNA (39–42), one may explore various systems for delivery of Cas9/LTR-gRNA via various routes for immunizing high-risk subjects. Once advanced, one may use gene therapies (viral vector and nanoparticle) and transplantation of autologous Cas9/gRNA-modified bone marrow stem/progenitor cells (43, 44) or inducible pluripotent stem cells for eradicating HIV-1 infection.
Here, we demonstrated the high specificity of Cas9/gRNAs in editing HIV-1 target genome. Results from subclone data revealed the strict dependence of genome editing on the presence of both Cas9 and gRNA. Moreover, only one nucleotide mismatch in the designed gRNA target will disable the editing potency. In addition, all four of our designed LTR gRNAs worked well with different cell lines, indicating that the editing is more efficient in the HIV-1 genome than the host cellular genome, wherein not all designed gRNAs are functional, which may be due to different epigenetic regulation, variable genome accessibility, or other reasons. Given the ease and rapidity of Cas9/gRNA development, even if HIV-1 mutations confer resistance to one Cas9/gRNA-based therapy, as described above, HIV-1 variants can be genotyped to enable another personalized therapy for individual patients (10).
Materials and Methods
Plasmid Preparation.
Vectors containing human Cas9 and gRNA expression cassette, pX260, and pX330 (Addgene) were used to create various constructs, LTR-A, -B, -C, and -D (for details, see SI Materials and Methods).
Cell Culture and Stable Cell Lines.
TZM-bI reporter and U1 cell lines were obtained from the National Institutes of Health (NIH) AIDS Reagent Program, and CHME5 microglial cells were described previously (17). The detailed procedure for cell growth or preparation of stable cell lines is described in SI Materials and Methods.
Immunocytochemistry and Western Blot.
Standard methods for immunocytochemical observation of the cells and evaluation of protein expression by Western blot were used as described in detail in SI Materials and Methods.
Firefly Luciferase Assay.
Cells were lysed 24 h posttreatment using Passive Lysis Buffer (Promega) and assayed with a Luciferase Reporter Gene Assay kit (Promega) according to the manufacturer’s protocol. Luciferase activity was normalized to the number of cells determined by a parallel MTT assay (Vybrant; Invitrogen).
p24 ELISA.
After infection or reactivation, the levels of HIV-1 viral load in supernatant were quantified by p24 Gag ELISA (Advanced BioScience Laboratories, Inc.) following the manufacturer’s protocol. To assess cell viability upon treatments, MTT assay was performed in parallel according to the manufacturer’s manual (Vybrant; Invitrogen).
EGFP Flow Cytometry.
Cells were trypsinized, washed with PBS, and fixed in 2% (wt/vol) paraformaldehyde for 10 min at room temperature, then washed twice with PBS and analyzed using a Guava EasyCyte Mini flow cytometer (Guava Technologies).
HIV-1 Reporter Virus Preparation and Infections.
HEK293T cells were transfected using Lipofectamine 2000 reagent (Invitrogen) with pNL4-3-ΔE-EGFP (NIH AIDS Research and Reference Reagent Program). After 48 h, the supernatant was collected, 0.45-μm filtered and titered in HeLa cells using EGFP as an infection marker. For viral infection, stable Cas9/gRNA TZM-bI cells were incubated 2 h with diluted viral stock, and then washed twice with PBS. At 2 and 4 d postinfection, cells were collected, fixed, and analyzed by flow cytometry for EGFP expression, or genomic DNA purification was performed for PCR and WGS.
Genomic DNA Amplification, PCR, TA Cloning, Sanger Sequencing, and GenomeWalker Link PCR.
Standard methods for DNA manipulation for cloning and sequencing were used (see SI Materials and Methods). For identification of the integration sites of HIV-1, we used a Lenti-X integration site analysis kit as detailed in SI Materials and Methods.
Some PCR products were used for restriction fragment length polymorphism analysis. Equal amounts of the PCR products were digested with BsaJI. Digested DNA was separated on an ethidium bromide-contained agarose gel [2% (wt/vol)]. For sequencing, PCR products were cloned using a TA Cloning Kit Dual Promoter with pCRII vector (Invitrogen). The insert was confirmed by digestion with EcoRI, and positive clones were sent to Genewiz for Sanger sequencing.
SURVEYOR Assay.
The presence of mutations in PCR products was examined using a SURVEYOR Mutation Detection Kit (Transgenomic) according to the protocol from the manufacturer. Briefly, heterogeneous PCR product was denatured for 10 min in 95 °C and hybridized by gradual cooling using a thermocycler. Next, 300 ng of hybridized DNA (9 μL) was subjected to digestion with 0.25 μL of SURVEYOR Nuclease in the presence of 0.25 μL SURVEYOR Enhancer S and 15 mM MgCl2 for 4 h at 42 °C. Then, Stop Solution was added and samples were resolved in 2% (wt/vol) agarose gel together with equal amounts of undigested PCR product controls.
Selection of LTR Target Sites, WGS, Bioinformatics, and Statistical Analysis.
We used Jack Lin's CRISPR/Cas9 gRNA finder tool for initial identification of potential target sites within the LTR. Detailed WGS, bioinformatic, and statistical analyses are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Jessica Otte for technical support; Jennifer Gordon, Shohreh Amini, and Xuebin Qin for helpful comments; and Jeffrey B. Tatro and Cynthia Papaleo for editorial assistance. This work was supported by National Institutes of Health Grants R01MH093271 (to K.K.), R01NS087971 (to W.H. and K.K.), and P30MH092177 (to K.K.).
Footnotes
Conflict of interest statement: A patent application has been filed relating to this work.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405186111/-/DCSupplemental.
References
- 1.UNAIDS . Global Report: UNAIDS Report on the Global AIDS Epidemic 2012. Geneva: Joint United Nations Programme on HIV/AIDS; 2012. [Google Scholar]
- 2. Taylor BS, Wilkin TJ, Shalev N, & Hammer SM (2013) CROI 2013: Advances in antiretroviral therapy. Top Antiviral Med 21(2):75–89. [PMC free article] [PubMed]
- 3.Eisele E, Siliciano RF. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity. 2012;37(3):377–388. doi: 10.1016/j.immuni.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun TW, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387(6629):183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 5.Chun TW, Fauci AS. HIV reservoirs: Pathogenesis and obstacles to viral eradication and cure. AIDS. 2012;26(10):1261–1268. doi: 10.1097/QAD.0b013e328353f3f1. [DOI] [PubMed] [Google Scholar]
- 6.Johnson TP, et al. Induction of IL-17 and nonclassical T-cell activation by HIV-Tat protein. Proc Natl Acad Sci USA. 2013;110(33):13588–13593. doi: 10.1073/pnas.1308673110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Crumpacker C. Eradication of HIV and cure of AIDS, now and how? Front Immunol. 2013;4:337. doi: 10.3389/fimmu.2013.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stone D, Kiem HP, Jerome KR. Targeted gene disruption to cure HIV. Curr Opin HIV AIDS. 2013;8(3):217–223. doi: 10.1097/COH.0b013e32835f736c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manjunath N, Yi G, Dang Y, Shankar P. Newer gene editing technologies toward HIV gene therapy. Viruses. 2013;5(11):2748–2766. doi: 10.3390/v5112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10(10):957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofer U, et al. Pre-clinical modeling of CCR5 knockout in human hematopoietic stem cells by zinc finger nucleases using humanized mice. J Infect Dis. 2013;208(Suppl 2):S160–S164. doi: 10.1093/infdis/jit382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu X, et al. Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res. 2013;41(16):7771–7782. doi: 10.1093/nar/gkt571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wires ES, et al. Methamphetamine activates nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and induces human immunodeficiency virus (HIV) transcription in human microglial cells. J Neurovirol. 2012;18(5):400–410. doi: 10.1007/s13365-012-0103-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derdeyn CA, et al. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol. 2000;74(18):8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Folks TM, Justement J, Kinter A, Dinarello CA, Fauci AS. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science. 1987;238(4828):800–802. doi: 10.1126/science.3313729. [DOI] [PubMed] [Google Scholar]
- 20.Pace MJ, Graf EH, O’Doherty U. HIV 2-long terminal repeat circular DNA is stable in primary CD4+T Cells. Virology. 2013;441(1):18–21. doi: 10.1016/j.virol.2013.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003;22(8):1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, et al. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J Virol. 2004;78(4):1718–1729. doi: 10.1128/JVI.78.4.1718-1729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho SW, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24(1):132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gabriel R, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011;29(9):816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 27.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31(9):839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 29.Zhang F, Wen Y, Guo X. 2014. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum Mol Genet, 10.1093/hmg/ddu125.
- 30.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32(6):569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32(6):577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tebas P, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370(10):901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41(20):9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L, et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41(19):9049–9061. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mussolino C, et al. TALENs facilitate targeted genome editing in human cells with high specificity and low cytotoxicity. Nucleic Acids Res. 2014;42(10):6762–6773. doi: 10.1093/nar/gku305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Gaj T, Patterson JT, Sirk SJ, Barbas CF., 3rd Cell-penetrating peptide-mediated delivery of TALEN proteins via bioconjugation for genome engineering. PLoS ONE. 2014;9(1):e85755. doi: 10.1371/journal.pone.0085755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 39.Chen H, Choi J, Bailey S. Cut site selection by the two nuclease domains of the Cas9 RNA-guided endonuclease. J Biol Chem. 2014;289(19):13284–13294. doi: 10.1074/jbc.M113.539726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim JM, Kim D, Kim S, Kim JS. Genotyping with CRISPR-Cas-derived RNA-guided endonucleases. Nat Commun. 2014;5:3157. doi: 10.1038/ncomms4157. [DOI] [PubMed] [Google Scholar]
- 41.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karvelis T, Gasiunas G, Siksnys V. Programmable DNA cleavage in vitro by Cas9. Biochem Soc Trans. 2013;41(6):1401–1406. doi: 10.1042/BST20130164. [DOI] [PubMed] [Google Scholar]
- 43.Li L, et al. Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther. 2013;21(6):1259–1269. doi: 10.1038/mt.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Younan P, Kowalski J, Kiem HP. Genetically modified hematopoietic stem cell transplantation for HIV-1-infected patients: Can we achieve a cure? Mol Ther. 2014;22(2):257–264. doi: 10.1038/mt.2013.264. [DOI] [PMC free article] [PubMed] [Google Scholar]