Significance
Cancer cells release from their cell surface membrane-lined microvesicles (MVs), which contain proteins, mRNAs, and microRNAs that can be taken up by other cells. We report that breast cancer cells exposed to decreased oxygen availability (hypoxia) increase their production of MVs, which stimulate invasion and metastasis by recipient breast cancer cells. Increased MV shedding by hypoxic cells requires expression of hypoxia-inducible factors (HIFs), which activate transcription of the RAB22A gene, and expression of the small GTPase RAB22A, which is a protein that localizes to budding MVs. Our results delineate a molecular mechanism by which hypoxia increases invasion and metastasis by stimulating MV shedding and provide further evidence that addition of HIF inhibitors to current treatment regimens may improve clinical outcome.
Keywords: orthotopic transplantation, triple negative breast cancer, oxygen, tumor microenvironment, mammary fat pad implantation
Abstract
Extracellular vesicles such as exosomes and microvesicles (MVs) are shed by cancer cells, are detected in the plasma of cancer patients, and promote cancer progression, but the molecular mechanisms regulating their production are not well understood. Intratumoral hypoxia is common in advanced breast cancers and is associated with an increased risk of metastasis and patient mortality that is mediated in part by the activation of hypoxia-inducible factors (HIFs). In this paper, we report that exposure of human breast cancer cells to hypoxia augments MV shedding that is mediated by the HIF-dependent expression of the small GTPase RAB22A, which colocalizes with budding MVs at the cell surface. Incubation of naïve breast cancer cells with MVs shed by hypoxic breast cancer cells promotes focal adhesion formation, invasion, and metastasis. In breast cancer patients, RAB22A mRNA overexpression in the primary tumor is associated with decreased overall and metastasis-free survival and, in an orthotopic mouse model, RAB22A knockdown impairs breast cancer metastasis.
The release of extracellular vesicles represents a recently appreciated mechanism of intercellular communication by which human cancer cells promote disease progression by altering the phenotype of stromal cells and other cancer cells that take up these membrane-bound particles and/or their cargo of proteins, mRNAs, and microRNAs (1–3). At least two distinct types of extracellular vesicles have been identified, based on particle size and mechanism of biogenesis (1). Microvesicles (MVs) form by outward budding and fission of the plasma membrane mediated by contraction of the actin cytoskeleton, which generates extracellular vesicles with a broad range of sizes from ∼100 nm to ∼1 µm in diameter that are pelleted by centrifugation at 10,000 × g. In contrast, exosomes are preformed within multivesicular bodies, which fuse with the plasma membrane to discharge their contents into the extracellular space; exosomes vary in size between ∼50 nm and ∼200 nm and are pelleted by centrifugation at 100,000 × g after discarding the 10,000 × g pellet. Unfortunately, this nomenclature has not been uniformly adopted in the field, with some investigators using either term to refer generically to all extracellular vesicles (4) or using MVs to refer to particles collected by centrifugation at 15,000 × g (5), 20,000 × g (6), or even 100,000 × g (7, 8). Other methods of isolation, such as flow cytometry using fluorescent microbeads as size standards, add further uncertainty regarding the nature of the extracellular vesicles being studied (1, 6).
Depending on the cancer type and method of isolation, extracellular vesicles have been shown to promote primary tumor growth, angiogenesis, immune escape, multidrug resistance, invasion, and metastasis (1, 4, 9). In patients with melanoma, the abundance of exosomes in peripheral blood was correlated with mortality; in mouse models, injection of exosomes isolated from melanoma cells into the peripheral blood of mice induced prometastatic behavior of bone marrow cells, and the small GTPase RAB27A was required for exosome biogenesis in melanoma cells (10). In HeLa cells, RAB27A and RAB27B were each required for exosome biogenesis, based on different loss-of-function phenotypes (11). RAB27A loss of function in 4T1 breast cancer cells inhibited both primary tumor growth and lung metastasis (12). Elevated concentrations of MVs have been reported in the peripheral blood of patients with breast cancer (5).
Intratumoral hypoxia is a common microenvironmental stimulus that drives cancer progression and is associated with metastasis and patient mortality (13–18). Reduced oxygen availability triggers changes in gene expression mediated by hypoxia-inducible factors (HIFs) that impact on many critical aspects of cancer biology, including angiogenesis, metabolic reprogramming, invasion, and metastasis (19). HIFs are heterodimeric proteins consisting of an O2-regulated HIF-1α or HIF-2α subunit and a constitutively expressed HIF-1β subunit (20). In breast cancer, overexpression of HIF-1α or HIF-2α is associated with metastasis, treatment failure, and patient mortality (21–26). HIFs are required for lymph node and lung metastasis in both autochthonous (27) and orthotopic (28–30) mouse models of breast cancer and activate the transcription of genes encoding proteins that are required for multiple steps in the invasion and metastasis of breast cancer cells (28–38).
Hypoxia has been reported to induce increased release of proangiogenic extracellular vesicles (6, 39–41). Hypoxia-induced exosome release was shown to be HIF dependent (41, 42), although specific downstream target genes required for exosome release have not been identified. Effects of hypoxia on MV formation by cancer cells have not been reported. In the present study, we have specifically investigated the mechanisms and consequences of MV shedding by hypoxic breast cancer cells.
Results
Hypoxia Induces Increased MV Formation in a HIF-Dependent Manner.
The median pO2 in advanced breast cancers is 10 mmHg (∼1.4% O2), compared with 65 mmHg (∼9.3% O2) in normal breast tissue, and pO2 measurements of <10 mmHg are associated with metastasis and patient mortality (15). We analyzed the effect of hypoxia (1% O2 for 24 h) on the production of MVs by three human breast cancer cell lines: MCF-7 expresses the estrogen and progesterone receptors, but not HER2, and is classified as luminal subtype based on its gene expression pattern, whereas MDA-MB-231 and MDA-MB-435 cells (hereafter designated MDA-231 and MDA-435) do not express estrogen or progesterone receptors or HER2 (i.e., triple negative) and are classified as basal-like/claudin-low subtype by gene expression criteria (43, 44). To analyze the role of HIFs in responses to hypoxia, MDA-231 and MDA-435 cells were stably transfected with lentiviral vectors encoding short hairpin RNAs (shRNAs) targeting both HIF-1α and HIF-2α (double knockdown, DKD) or with either empty vector (EV) or vector encoding a scrambled control shRNA (SC). An MCF-7-SC subclone was also established for comparison with MDA-231-SC. We have previously demonstrated that MDA-231-DKD and MDA-435-DKD have impaired metastasis compared with MDA-231-SC and MDA-435-EV, respectively (29–31).
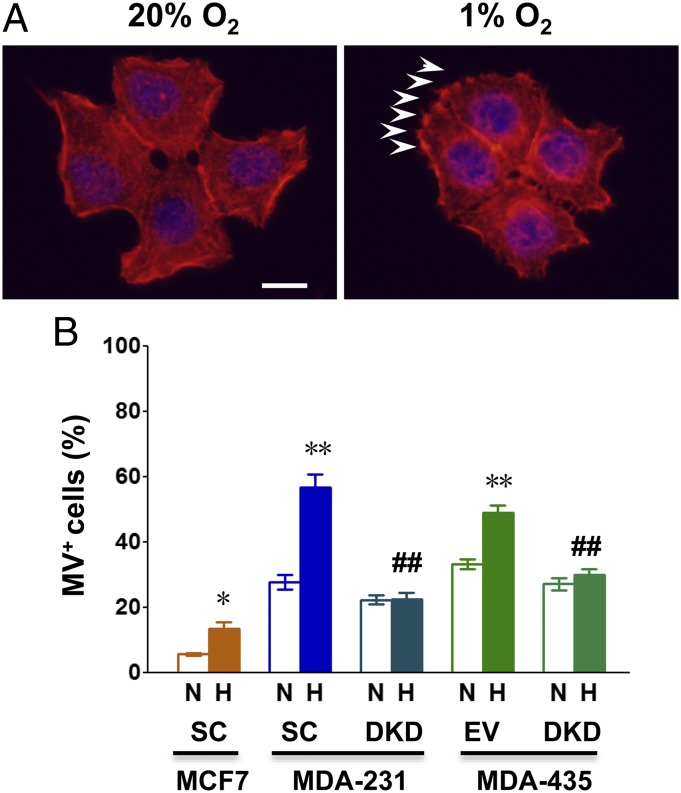
We used three different methods to quantify MV formation. First, we directly imaged budding MVs by fluorescence microscopy of cells incubated with Alexa Fluor 568-conjugated phalloidin, which stains F-actin, and scored the percentage of cells with MVs at the cell surface (Fig. 1A and Fig. S1). Exposure of MCF-7-SC, MDA-231-SC, and MDA-435-EV cells to hypoxia significantly increased the number of cells with MVs at the cell surface, whereas hypoxia had no effect on MV formation by MDA-231-DKD or MDA-435-DKD cells (Fig. 1B).
Fig. 1.
Hypoxia induces the generation of microvesicles (MVs) in breast cancer cell lines. Double knockdown (DKD) subclones of MDA-MB-231 and MDA-MB-435 cells were stably transfected with lentiviral vectors encoding short hairpin RNAs (shRNAs) directed against both HIF-1α and HIF-2α. MCF-7, MDA-MB-231, and MDA-MB-435 cells were also stably transfected with a lentiviral vector encoding a scrambled control shRNA (SC) or with an empty vector (EV). Cells were exposed to serum-free Dulbecco’s modified Eagle medium (DMEM) under either 20% O2 (nonhypoxic conditions, N) or 1% O2 (hypoxic conditions, H) for 24 h. (A) Representative immunofluorescent images of MCF-7 SC breast cancer cells stained with Alexa Fluor 568-conjugated phalloidin (to detect F-actin; red) and DAPI (to detect nuclei; blue) are shown. MVs are denoted by arrowheads. (Scale bar, 10 µm.) (B) The percentage of cells containing five or more MVs (MV+) is shown. Data in bar graphs represent mean ± SEM (n = 3). *P < 0.05, **P < 0.01 for H vs. N by Student's t test; ##P < 0.01 for DKD subclone (H) vs. corresponding SC or EV subclone (H) by ANOVA.
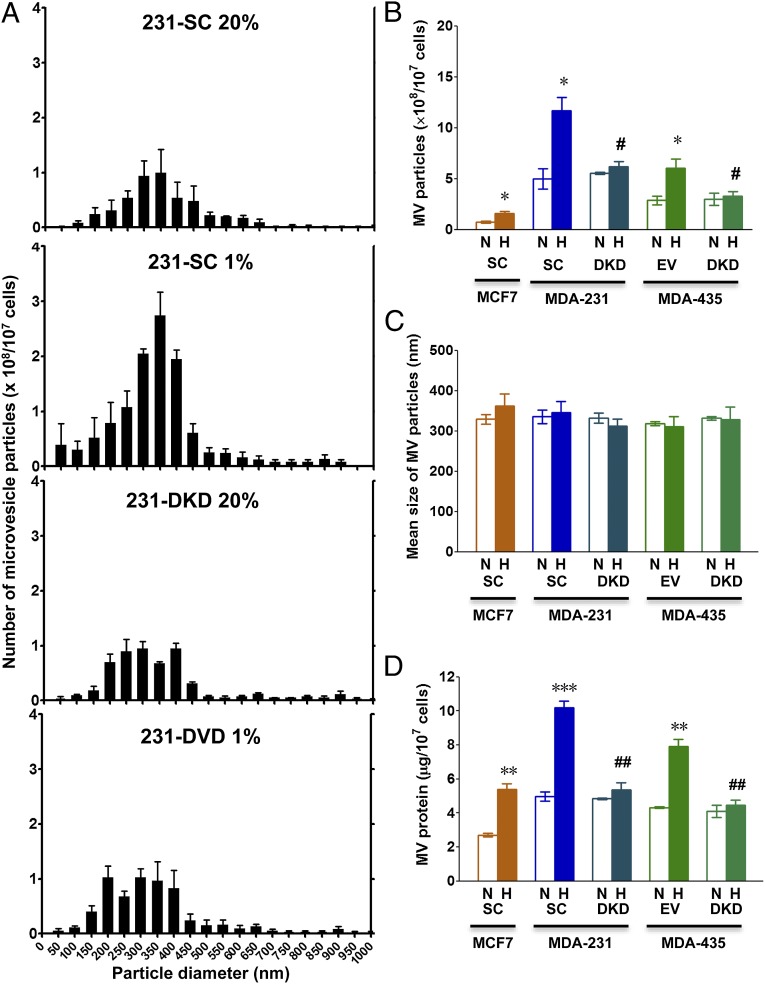
Next, we isolated MVs, which were shed into the culture medium, by serial centrifugation of conditioned media at 2,000 × g for 20 min to remove cell debris and then at 10,000 × g for 70 min to pellet MVs, which were resuspended in PBS. Nanoparticle tracking analysis was performed using the NanoSight system and revealed a distribution of particles of varying abundance with diameters ranging from 100 to 900 nm (Fig. S2). The mean particle size distribution (Fig. 2A), mean number of MV particles (Fig. 2B), and mean particle size (Fig. 2C) were determined based on three independent experiments. Hypoxia increased the number of MV particles in a HIF-dependent manner across the entire size distribution, such that there was no change in the mean particle diameter.
Fig. 2.
Analysis of MVs shed from breast cancer cells. Breast cancer subclones were exposed to serum-free DMEM under either 20% O2 (nonhypoxic conditions, N) or 1% O2 (hypoxic conditions, H) for 24 h. Shed MVs were collected from conditioned media by centrifugation. (A–C) MVs were subjected to nanoparticle tracking analysis (NanoSight). Histograms of particle size distribution for MVs from three independent experiments are shown for MDA-231 SC and DKD subclones (A). The total number of MVs (× 108 per 107 cells) (B) and mean MV particle diameter (C) are shown for each condition. (D) MVs were isolated as described above and total MV protein (micrograms per 107 cells) for each condition is shown. Data in all bar graphs represent mean ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 for H vs. N by Student's t test; #P < 0.05, ##P < 0.01 for DKD (H) vs. SC or EV (H) by ANOVA.
Finally, we quantified the total protein content of MVs pelleted by centrifugation at 10,000 × g per 107 cells. This assay revealed that the mass of shed MVs was greater in conditioned media from hypoxic compared with nonhypoxic MCF-7-SC, MDA-231-SC, and MDA-435-EV cells, whereas there was no effect of hypoxia on the mass of MVs shed from MDA-231-DKD or MDA-435-DKD cells (Fig. 2D). In addition, the mass of MVs was greater in conditioned media from the metastatic breast cancer cell lines (MDA-231 and MDA-435) than from the nonmetastatic line (MCF-7), results that were all consistent with the fluorescent microscopy and NanoSight data. Taken together, the results in Figs. 1 and 2 indicate that hypoxia induces increased MV formation in breast cancer cells and that this response is HIF dependent.
RAB22A Overexpression in Breast Cancer Is Associated with Patient Mortality.
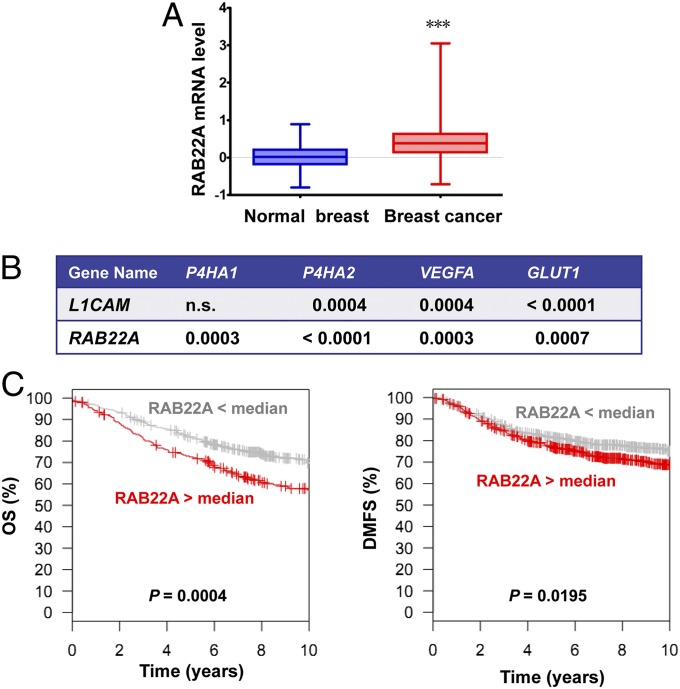
RAB proteins are membrane-bound GTPases that are critical for vesicle formation, trafficking, and membrane fusion. We searched two large collections of mRNA expression data from human breast cancers (45, 46) for a RAB family member that satisfied three criteria: overexpression in breast cancer compared with normal breast, coexpression with HIF target genes known to be expressed in breast cancer, and association of overexpression in breast cancer with decreased patient survival. RAB22A gene expression was significantly increased in breast cancers (n = 530) compared with normal breast tissue (n = 63; P < 0.001; Fig 3A). RAB22A expression showed highly significant correlations with the expression of four representative HIF target genes (29, 35, 36) analyzed, P4HA1, P4HA2, VEGFA, and GLUT1 (P ≤ 0.0007; Fig. 3B). For comparison, expression of L1CAM, which is another known HIF target gene expressed in breast cancer (28), was significantly correlated with only three of the four reference genes. RAB22A mRNA levels in primary breast cancers that were greater than the mean were associated with significantly decreased overall survival (P = 0.0004; n = 737 patients) and distant metastasis-free survival (P = 0.0195; n = 1,379 patients) in 10-y Kaplan–Meier analyses (Fig. 3C).
Fig. 3.
RAB22A expression in breast cancer is correlated with HIF target gene expression and decreased survival of breast cancer patients. (A) Relative levels of RAB22A mRNA from microarray analysis (normalized log2 ratios) of primary tumor samples (n = 530) relative to adjacent normal tissue (n = 63) from breast cancer patients (TCGA database) are shown as median, minimum, and maximum; ***P < 0.001 vs. normal adjacent breast tissue by ANOVA. (B) Expression of RAB22A mRNA and mRNAs encoded by four known HIF target genes (P4HA1, P4HA2, VEGFA, and GLUT1) in 596 tissue samples from breast cancer patients (530 primary tumors, 63 adjacent normal tissue, and 3 metastases; TCGA database) was compared by Pearson’s correlation test; P values for statistically significant correlations are shown; n.s., not significant. Expression of another known HIF target gene (L1CAM) is shown as a reference for comparison with RAB22A. (C) Kaplan–Meier curves were constructed to analyze the association of RAB22A mRNA levels in the primary tumor with overall survival (OS, Left, n = 737) and distant metastasis-free survival (DMFS, Right, n = 1,379) of patients with breast cancer using the GOBO database. Statistical analysis was performed using log-rank tests.
Hypoxia Induces HIF-Dependent RAB22A Expression.
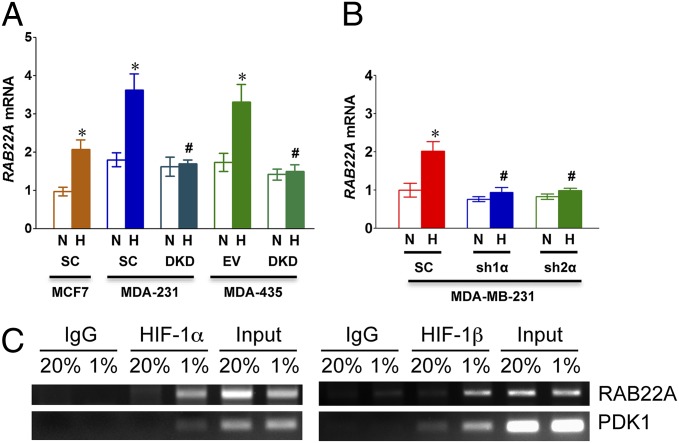
Reverse transcription and quantitative real-time PCR (RT-qPCR) assays revealed that expression of RAB22A mRNA was significantly induced by hypoxia in MCF-7-SC, MDA-231-SC, and MDA-435-EV cells, but not in MDA-231-DKD or MDA-435-DKD cells (Fig. 4A), indicating that the induction was HIF dependent. To further analyze the role of HIFs in regulating RAB22A expression, MDA-231 cells were transduced with a lentiviral vector encoding shRNA targeting HIF-1α (sh1α) or HIF-2α (sh2α). Knockdown of either HIF-1α or HIF-2α was sufficient to block hypoxia-induced RAB22A mRNA expression (Fig. 4B). To determine whether RAB22A was a direct HIF target gene, we analyzed the RAB22A locus for the presence of consensus HIF binding site sequences (5′-RCGTG-3′) located in DNase I-hypersensitive chromatin domains. We identified five candidate sites that met these criteria (Fig. S3) and chromatin immunoprecipitation assays revealed hypoxia-inducible binding of HIF-1α and HIF-1β to one site, which was located in the 5′-untranslated region of exon 1 (Fig. 4C).
Fig. 4.
Hypoxia-induced RAB22A expression is HIF-1α- and HIF-2α dependent. (A) MDA-MB-231 and MDA-MB-435 cells were stably transfected with lentiviral vectors encoding shRNAs directed against both HIF-1α and HIF-2α (DKD); a lentiviral encoding a scrambled control shRNA (SC); or an empty vector (EV). The subclones were exposed to serum-free DMEM under 20% (N) or 1% (H) O2 for 24 h and total RNA was isolated. Reverse transcription and quantitative real-time PCR (RT-qPCR) analysis of RAB22A mRNA was performed and normalized to MCF-7-SC (N). Mean ± SEM (n = 3) is shown; *P < 0.05 for H vs. N by Student's t test; #P < 0.05 for DKD (H) vs. corresponding SC (H) or EV (H) by ANOVA. (B) MDA-MB-231 cells were stably transfected with a lentiviral vector encoding shRNA directed against HIF-1α (sh1α) or HIF-2α (sh2α) or a scrambled control shRNA (SC). Cells were cultured in serum-free DMEM under 20% O2 (N) or 1% O2 (H) for 24 h. RAB22A mRNA levels were analyzed by RT-qPCR. Mean ± SEM (n = 3) are shown; *P < 0.05 for H vs. N by Student's t test; #P < 0.05 for sh1α or sh2α (H) vs. SC (H) by ANOVA. (C) MDA-MB-231 cells were exposed to 20% or 1% O2 for 16 h, chromatin was immunoprecipitated with IgG or antibody against HIF-1α or HIF-1β, and PCR was performed with primers flanking a candidate HIF binding site in the RAB22A gene or a known HIF binding site in the PDK1 gene. As a positive control for PCR, DNA was isolated directly from an aliquot of the lysate before immunoprecipitation (input).
Hypoxia-Induced MV Formation Requires RAB22A Expression.
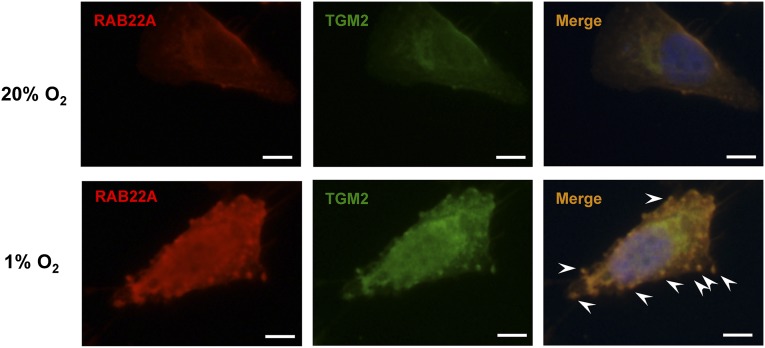
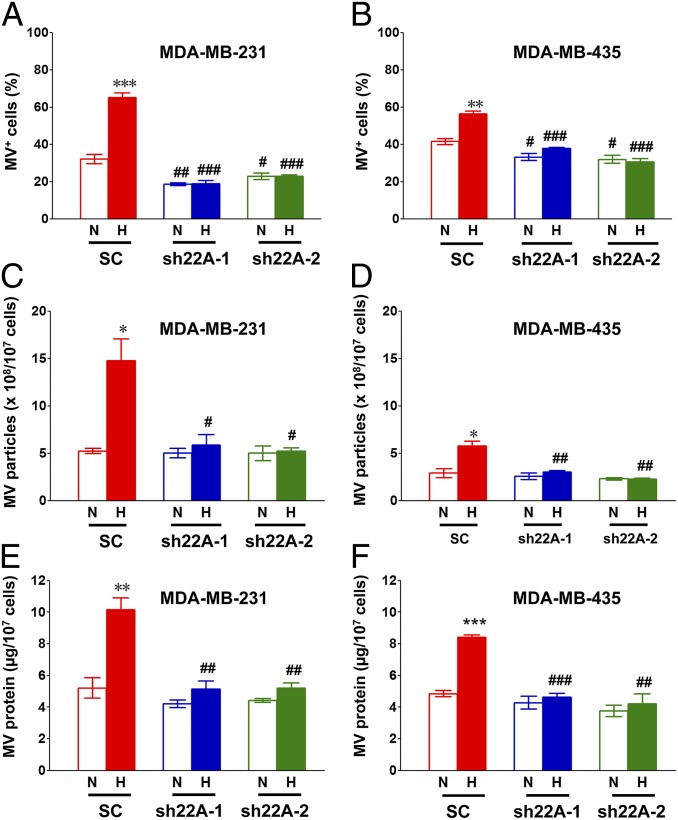
Transglutaminase 2 (TGM2) is a cross-linking enzyme that was enriched in the membranes of MVs shed by MDA-MB-231 cells, but inhibitors of TGM2 enzymatic activity had no effect on MV formation (7), indicating that TGM2 is a cargo protein rather than a regulator of MV biogenesis. Dual immunofluorescence with antibodies against RAB22A and TGM2 demonstrated that both RAB22A and TGM2 colocalize with budding MVs in hypoxic MDA-231 cells (Fig. 5). To determine whether RAB22A expression was required for MV formation, MDA-231 and MDA-435 cells were stably transfected with lentiviral vectors encoding two different shRNAs targeting RAB22A (sh22A-1 and sh22A-2). The efficient knockdown of RAB22A mRNA and protein was demonstrated by RT-qPCR and immunocytochemistry, respectively (Fig. S4). The MDA-231 and MDA-435 subclones were exposed to 20% or 1% O2 and MV formation was analyzed by immunofluorescence (Fig. 6 A and B), nanoparticle tracking analysis (Fig. 6 C and D), and total protein content of MVs (Fig. 6 E and F). By all three assays, RAB22A knockdown completely eliminated the increase in MV production under hypoxic conditions, whereas under nonhypoxic conditions, only the percentage of MV-positive cells was significantly decreased by RAB22A loss of function. These data indicate that HIF-dependent induction of RAB22A expression is required for hypoxia-induced MV production.
Fig. 5.
RAB22A expression is induced by hypoxia and colocalizes with MVs in breast cancer cells. MDA-MB-231-SC cells were cultured for 24 h in serum-free DMEM under 20% or 1% O2 and analyzed by immunofluorescence using polyclonal rabbit anti-human RAB22A antibody with Cy3-conjugated goat anti-rabbit IgG (red) and a monoclonal mouse anti-human TGM2 antibody with FITC-conjugated goat anti-mouse IgG2a (green) to label budding MVs on the surface of the cells. To visualize the nucleus, cells were also stained with DAPI. Merged images demonstrate colocalization of RAB22A and TGM2 (yellow) with MVs (arrowheads) in the hypoxic cell. (Scale bar in all panels, 10 µm.)
Fig. 6.
RAB22A knockdown impairs the generation of MVs in response to hypoxia. (A–F) MDA-MB-231 (A, C, and E) and MDA-MB-435 (B, D, and F) subclones were stably transfected with lentiviral vectors encoding shRNAs directed against RAB22A (sh22A-1 or sh22A-2) or a scrambled control shRNA (SC). The subclones were cultured in serum-free DMEM under either 20% (N) or 1% (H) O2 for 24 h and then stained with Alexa Fluor 568-conjugated phalloidin and DAPI. The percentage of cells with budding MVs (MV+) was determined (A and B). Shed MVs were isolated from conditioned medium by differential centrifugation, the number of MV particles per 107 cells was determined by nanoparticle tracking analysis (C and D), and total MV protein per 107 cells was determined (E and F). In all bar graphs, mean ± SEM (n = 3) are shown; *P < 0.05, **P < 0.01, ***P < 0.001 for H vs. N by Student's t test; #P < 0.05, ##P < 0.01, ###P < 0.001 for sh22A (H) vs. SC (H), or sh22A (N) vs. SC (N), by ANOVA.
RAB22A Deficiency Impairs Breast Cancer Cell Invasion.
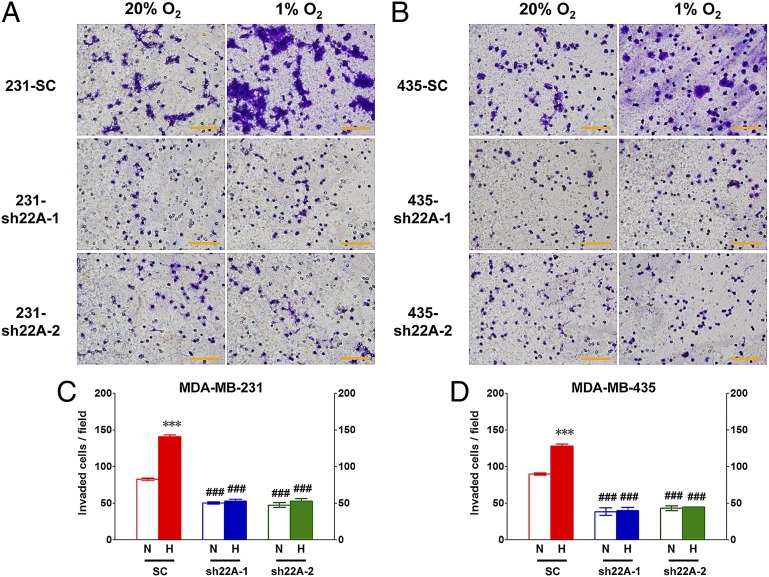
We next analyzed the ability of SC, sh22A-1, and sh22A-2 subclones of MDA-231 (Fig. 7 A and C) or MDA-435 (Fig. 7 B and D) to invade through Matrigel, a tumor-derived extracellular matrix (ECM) preparation, under nonhypoxic or hypoxic conditions. Compared with SC cells, RAB22A knockdown subclones had decreased invasive properties and the stimulatory effect of hypoxia on invasion was completely eliminated by RAB22A knockdown.
Fig. 7.
RAB22A knockdown impairs breast cancer cell invasion. (A and B) A total of 2 × 105 cells of the MDA-MB-231 (A) and MDA-MB-435 (B) SC, sh22A-1, and sh22A-2 subclones were seeded on top of Matrigel-coated chamber inserts and incubated in serum-free DMEM under either 20% (N) or 1% (H) O2 for 24 h. The number of cells that invaded through the Matrigel to the underside of the filter was determined by staining with crystal violet and counting under bright field microscopy. (C and D) The number of invaded cells per field was determined from 10 fields per filter. Mean ± SEM (n = 3) are shown; ***P < 0.001 for H vs. N by Student's t test; ###P < 0.001 for sh22A (H) vs. SC (H), or sh22A (N) vs. SC (N), by ANOVA. (Scale bar, 500 µm.)
Hypoxia-Induced MVs Stimulate Focal Adhesion Formation, ECM Invasion, and Lung Colonization in a RAB22A-Dependent Manner.
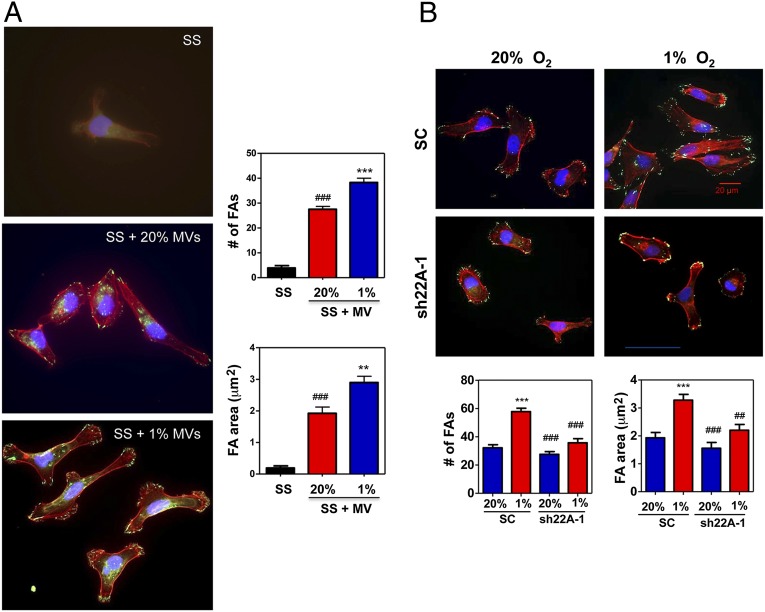
We have previously demonstrated that exposure of MDA-231 cells to hypoxia induces focal adhesion formation, which stimulates cell motility that is required for invasion and metastasis, as a result of HIF-dependent RHOA and ROCK1 gene expression, leading to activation of focal adhesion kinase (37). To determine whether these properties can be transferred via MVs, parental MDA-231 cells were incubated at 20% or 1% O2 for 24 h, MVs were collected from the tissue culture media and incubated with serum-starved MDA-231 cells at 20% O2 for 24 h, and the cells were stained with Alexa 568-conjugated phalloidin to detect F-actin stress fibers and anti-vinculin antibodies to detect focal adhesions. Whereas very few focal adhesions were observed in serum-starved control cells, MDA-231 cells exposed to MVs from conditioned medium of nonhypoxic cells had a significantly increased number of focal adhesions and total focal adhesion area, which were further increased when cells were incubated with MVs from hypoxic cells (Fig. 8A).
Fig. 8.
MVs from hypoxic cells stimulate increased focal adhesion formation. (A, Left) Parental MDA-MB-231 cells were exposed to 20% or 1% O2 for 24 h and shed MVs were collected from the conditioned media. Naïve MDA-MB-231 cells were incubated in serum-free DMEM, either alone [serum starved (SS)] or supplemented with MVs from nonhypoxic cells (SS + 20% MVs) or MVs from hypoxic cells (SS + 1% MVs) for 24 h, and the cells were stained with Alexa Fluor 568-conjugated phalloidin to detect F-actin stress fibers (red), vinculin antibody to detect focal adhesions (FAs; green), and DAPI to detect nuclei (blue). (Right) Image analysis was performed to determine the number of FAs (Upper graph) and total FA area (Lower graph). Mean ± SEM (n = 30 cells) are shown; ###P < 0.001 vs. SS; **P < 0.01, ***P < 0.001 vs. SS + MV (20%), two-way ANOVA with Bonferroni posttest to compare all conditions. (B) MDA-231 SC and sh22A-1 subclones were exposed to 20% or 1% O2 for 24 h. Shed MVs were collected from the conditioned media, incubated with naïve MDA-MB-231 cells at 20% O2 for 24 h, and the cells were stained as described above (Upper) and scored for number of FAs (Lower Left graph) and total FA area (Lower Right graph). Mean ± SEM (n = 30 cells) are shown; ***P < 0.001 vs. SC (20%); ##P < 0.01, ###P < 0.001 vs. SC (1%), two-way ANOVA with Bonferroni posttest to compare all conditions.
To determine the role of RAB22A in MV-induced focal adhesion formation, SC and sh22A-1 subclones were incubated at 20% or 1% O2 for 24 h, MVs were collected from the tissue culture media and incubated with naïve MDA-231 cells at 20% O2 for 24 h. MVs shed from hypoxic SC cells induced significantly greater focal adhesion formation compared with MVs shed from nonhypoxic SC cells, whereas there was no significant difference between MVs from hypoxic and nonhypoxic sh22A-1 cells (Fig. 8B). Thus, MVs from hypoxic breast cancer cells can induce complex cytoskeletal alterations in a RAB22A-dependent manner.
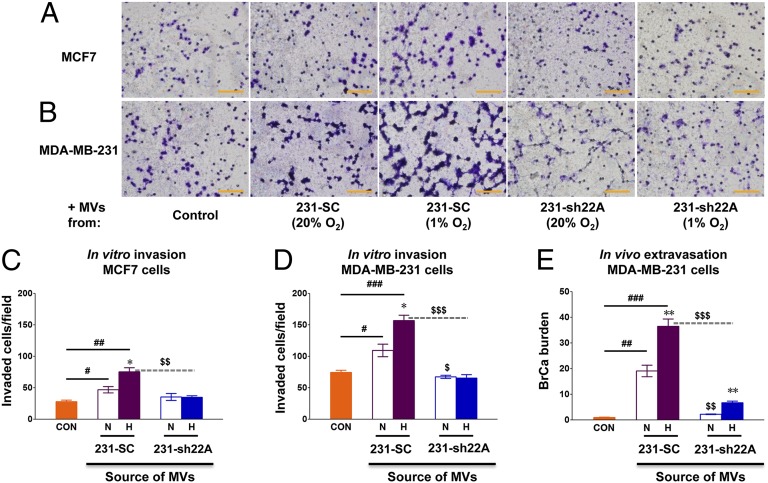
To determine whether the RAB22A-dependent effect on invasion (Fig. 7) was due to the RAB22A-dependent effect on MV formation (Fig. 6), we incubated SC and sh22A-1 cells under nonhypoxic or hypoxic conditions for 24 h, isolated MVs from the tissue culture media, incubated the MVs with naïve MCF-7 (Fig. 9 A and C) or MDA-231 (Fig. 9 B and D) cells, and measured their ability to invade through Matrigel. Compared with control cells not incubated with MVs, breast cancer cells incubated with MVs from SC cells were increased in their ability to invade through Matrigel, and MVs from hypoxic cells had a greater effect than those from nonhypoxic cells (Fig. 9 C and D). MVs shed from the sh22A-1 subclone had decreased ability to stimulate breast cancer cell invasion and the increased stimulatory effect of MVs from hypoxic SC cells was completely eliminated by RAB22A knockdown. These data indicate that MVs stimulate breast cancer cell invasion and that the reduced number of MVs shed from RAB22A knockdown cells account for the reduced stimulation of invasion.
Fig. 9.
MVs shed from MDA-MB-231-SC cells promote invasion in vitro and extravasation in vivo. (A–D) MDA-MB-231 subclones expressing sh22A-1 (231-sh22A) or scrambled control shRNA (231-SC) were cultured in serum-free DMEM under either 20% (nonhypoxic, N) or 1% (hypoxic, H) O2 for 24 h. MVs were isolated from cell culture supernatants by centrifugation and resuspended in PBS. A total of 2 × 105 MCF-7 (A and C) or MDA-MB-231 (B and D) cells were seeded on top of Matrigel-coated chamber inserts and incubated in serum-free DMEM with PBS (control) or with MVs shed from equal numbers of N or H 231-SC or 231-sh22A cells. Invasion of the cells through Matrigel to the underside of the filter was assessed 24 h later by staining with crystal violet and counting under bright field microscopy (Scale bar in A and B, 500 µm.) The mean number of invaded MCF-7 (C) or MDA-MB-231 (D) cells per field was determined from 10 fields per filter. (E) A total of 1 × 106 MDA-MB-231 cells were incubated for 24 h with PBS (control, CON) or with MVs, which were shed from equal numbers of N or H 231-SC or 231-sh22A cells, and then injected into the tail vein of 6-wk-old female SCID mice (n = 3 mice per group). Three weeks later, lungs were harvested, genomic DNA was isolated, and qPCR was performed with human-specific HK2 primers to determine the number of extravasated breast cancer cells relative to CON. Data are presented as mean ± SEM; *P < 0.05, **P < 0.01 for H vs. N by Student's t test; #P < 0.05, ##P < 0.01, ###P < 0.001 for 231-SC vs. CON by ANOVA; and $P < 0.05, $$P < 0.01, $$$P < 0.001 for 231-sh22A (H) vs. 231-SC (H), or 231-sh22A (N) vs. 231-SC (N), by ANOVA.
To determine whether MVs affect the ability of breast cancer cells to colonize the lung, we incubated MDA-231-SC and MDA-231-sh22A-1 cells under nonhypoxic or hypoxic conditions for 24 h, isolated MVs from the conditioned media, and incubated the MVs with naïve MDA-231 cells, which were then injected into the tail vein of severe combined immunodeficiency (SCID) mice. Three weeks later, the lungs were harvested and the total breast cancer cell burden was determined by qPCR using human-specific primers. Compared with control cells not treated with MVs, lung colonization was markedly increased by exposure of cells to MVs from nonhypoxic and, to a greater extent, hypoxic MDA-231-SC cells (Fig. 9E). Compared with MVs isolated from SC cells, MVs from sh22A cells were significantly deficient in their ability to stimulate lung colonization by breast cancer cells.
RAB22A Deficiency Impairs Spontaneous Metastasis of Breast Cancer Cells to the Lungs.
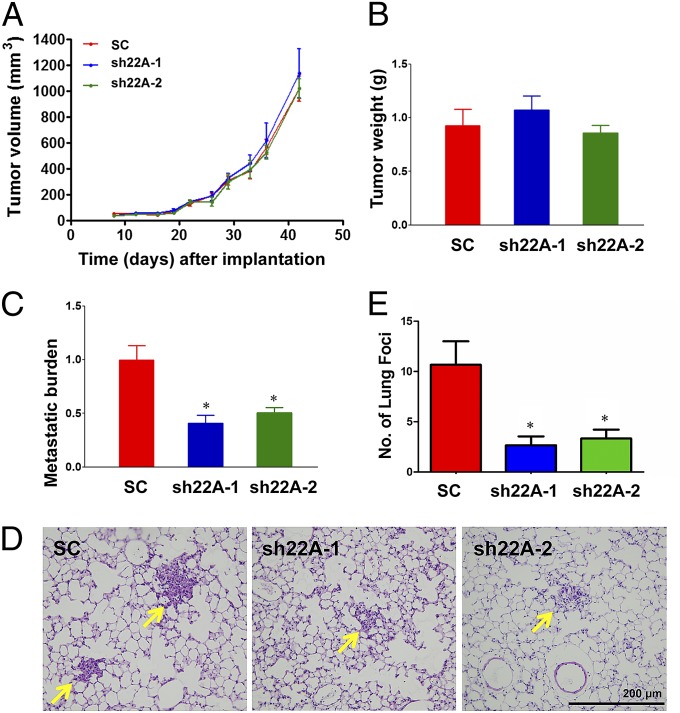
We next implanted SC, sh22A-1, or sh22A-2 subclones of MDA-231 into the mammary fat pad of female SCID mice. RAB22A knockdown had no effect on primary tumor growth (Fig. 10 A and B) but significantly decreased the lung metastatic burden, as determined by qPCR using human-specific primers (Fig. 10C), and decreased the number of metastatic foci in the lungs, as determined by analysis of hematoxylin- and eosin-stained lung sections (Fig. 10 D and E). These results are consistent with, and provide a mechanistic basis for, the observed correlation between increased RAB22A expression in breast cancer and decreased patient survival (Fig. 3C).
Fig. 10.
RAB22A knockdown impairs the spontaneous metastasis of breast cancer cells to the lungs. (A–E) MDA-MB-231 subclones (2 × 106 cells) were implanted into the mammary fat pad of 6-wk-old female SCID mice. Primary tumor volume (A) was determined from day 8 to day 43. On day 43, the primary tumor was harvested and weighed (B). To analyze lung metastasis, genomic DNA was purified from one lung for qPCR using human-specific HK2 primers and the results were normalized to the SC group (C); the other lung was fixed under inflation, paraffin embedded, and sections were stained with hematoxylin and eosin (D) to count the number of metastatic foci (arrows) per section (E). For all bar graphs, mean ± SEM (n = 3) are shown; *P < 0.05 vs. SC by Student's t test.
Discussion
There is a growing appreciation that extracellular vesicles provide an important molecular mechanism by which cancer cells can export their phenotype to neighboring and even distant cells. As a result, a minority population of cancer cells, such as those present in regions of intratumoral hypoxia, may alter the behavior of the bulk population, and cancer cells can reprogram stromal cells, through the intercellular transfer of proteins, mRNAs, and miRNAs by exosomes and MVs. Exosomes (10) and MVs (5) are increased in the peripheral blood of patients with cancer, suggesting that both types of extracellular vesicles can exert both local and systemic effects.
There is considerable evidence that the number of extracellular vesicles shed by cancer cells increases with disease progression (4), but the molecular mechanisms regulating the biogenesis of exosomes and MVs have not been delineated. In this study, we demonstrate that hypoxia, a critical microenvironmental stimulus in advanced breast cancers, stimulates HIF-dependent RAB22A gene expression and increases MV generation. RAB22A protein colocalizes with budding MVs in hypoxic breast cancer cells, suggesting that it plays a direct role in their generation. It is interesting that RAB22A knockdown completely eliminated increased MV generation under hypoxic conditions, but had only a modest effect on MV formation under nonhypoxic conditions, suggesting that another RAB protein may play a predominant role in MV biogenesis under nonhypoxic conditions. Nanoparticle tracking analysis indicated that hypoxia induced an increase in MVs across a broad range of particle sizes. However, further molecular studies are required to determine whether there are qualitative differences regarding the specific cargo carried by MVs generated in hypoxic vs. nonhypoxic cells. One interesting possibility is that RAB22A might specifically package cargo proteins whose expression is induced by hypoxia in a HIF-dependent manner. The observation that MVs from both nonhypoxic and hypoxic RAB22A knockdown cells were markedly impaired in their ability to stimulate lung colonization (Fig. 9E) suggests that RAB22A may do more than simply stimulate MV formation in hypoxic cells.
Although extracellular vesicles have been shown to affect many different aspects of cancer biology, loss of RAB22A expression in MDA-231 cells had no effect on primary tumor growth after mammary fat pad injection, whereas defects in focal adhesion formation and ECM invasion in vitro, lung colonization after i.v. injection, and spontaneous metastasis to the lungs after mammary fat pad implantation were observed. Further studies are required to determine whether RAB22A promotes breast cancer metastasis to other sites, such as bone, in which involvement of HIFs has also been implicated (47, 48). The association of RAB22A overexpression in the primary tumor with decreased overall and distant metastasis-free survival of women with breast cancer indicates that these findings are clinically relevant.
Women with triple negative breast cancer and those with metastatic disease require treatment with cytotoxic chemotherapy that has limited efficacy and thus there is an unmet clinical need for novel therapeutics (49, 50). Small molecule inhibitors of RAB proteins have not been identified (51). A large and rapidly growing body of experimental data indicates that in hypoxic breast cancer cells, HIFs function as master regulators by mediating the expression of a large battery of genes that promote multiple steps in the metastatic process (52). Treatment of SCID mice bearing orthotopic transplants of human breast cancer cells with drugs that inhibit HIF activity results in a dramatic reduction of primary tumor growth, local metastasis to axillary lymph nodes, and distant metastasis to the lungs (28, 29, 53, 54). These preclinical data provide a scientific foundation for clinical trials that test the safety and efficacy of adding HIF inhibitors to current chemotherapy regimens.
Methods
Maintenance of Cell Lines.
MCF-7, MDA-MB-231, and MDA-MB-435 human breast cancer cells were maintained in high-glucose (4.5 mg/mL) DMEM supplemented with 10% (vol/vol) FBS and penicillin–streptomycin (Invitrogen). All cells were maintained at 37 °C in a 5% CO2, 95% air incubator (20% O2). Hypoxic cells (1% O2) were maintained at 37 °C in a modular incubator chamber (Billups-Rothenberg) flushed with a gas mixture containing 1% O2, 5% CO2, and 94% N2.
Construction and Transduction of Lentiviral Vectors Encoding shRNA.
Expression vectors encoding shRNA targeting HIF-1α and HIF-2α were described previously (28). Vectors encoding shRAB22A were purchased from Sigma-Aldrich (sh22A-1 clone: NM_020673.2–303s1c1; sh22A-2 clone: NM_020673.2–8213s21c1). Lentiviruses were packaged in 293T cells by cotransfection with plasmid pCMV-dR8.91 and plasmid encoding vesicular stomatitis virus G protein using PolyJet (SignaGen). Viral supernatant was harvested 48 h posttransfection, filtered (0.45-µm pore size), and transduced into MDA-MB-231 or MDA-MB-435 cells in the presence of 8 µg/mL of polybrene (Sigma-Aldrich). After 24 h, cells were maintained in medium containing 1 μg/mL puromycin (Sigma-Aldrich) for selection.
Isolation of MVs.
MVs were isolated from the conditioned medium of breast cancer cells. Cells were seeded in tissue culture flasks at 50% confluency. Media was changed to serum-free DMEM when cells reached 70% confluence. Twenty-four hours later, the media was carefully removed from the flasks and centrifuged consecutively at 500 × g for 10 min, 2,000 × g for 20 min, and 10,000 × g for 70 min. The resulting pellet was washed and resuspended in PBS. Protein content was determined by the Bradford assay using a commercial kit (Bio-Rad).
Nanoparticle Tracking Analysis.
Samples of MVs were diluted in PBS and analyzed in real time using the NanoSight NS500 instrument equipped with a green laser (532 nm) and Nanoparticle Tracking Analysis software version 2.3, Build 0033 (NanoSight). Postacquisition settings were based on the manufacturer’s recommendations and kept constant between samples. Each video was analyzed to obtain particle size distribution profiles and concentration measurements.
Analysis of MVs by Fluorescence Microscopy.
Breast cancer cells plated on Lab-Tek II chamber slides (Thermo Fisher Scientific) were fixed with 3.7% paraformaldehyde, permeabilized with 0.2% Triton X-100, and blocked with 10% BSA. The cells were then incubated with primary antibodies (polyclonal rabbit anti-human RAB22A and monoclonal mouse anti-human transglutaminase 2 antibodies; Novus Biologicals) followed by incubation with secondary antibodies (Cy3-conjugated goat anti-rabbit IgG and FITC-conjugated goat anti-mouse; Novus Biologicals). Alexa Fluor 568-conjugated phalloidin (Life Technologies) was used to label F-actin and 4,6-diamidino-2-phenylindole (DAPI; Invitrogen) was used to stain nuclei. The samples were visualized using an Olympus fluorescence microscope and software. When staining for F-actin to visualize surface-associated MVs, cells with five or more MVs at the surface were scored as positive, and the percentage of MV-positive cells was determined. Two hundred fifty cells were scored for each experiment, and data from three independent experiments were plotted as mean ± SEM.
Microarray Data.
Level 3 data from the Breast Invasive Carcinoma dataset of The Cancer Genome Atlas (TCGA) (45) were obtained from http://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp. Pearson’s correlation coefficient was used to determine P values for coexpression. Prognostic significance of RAB22A mRNA expression in breast cancer was analyzed in the GOBO database (http://co.bmc.lu.se/gobo) (46). Survival plots were created by the survival analysis function in GOBO (Kaplan–Meier methods and log-rank tests).
RT-qPCR.
Total RNA was extracted from cells using TRIzol (Invitrogen) and treated with DNase I (Ambion). One microgram of total RNA was used for first-strand DNA synthesis using the iScript cDNA Synthesis kit (BioRad). qPCR was performed on the iCycler Real-Time PCR Detection system (BioRad) using SYBR Green qPCR master mix (Fermentas). For each primer pair, annealing temperature was optimized by gradient PCR. The hypoxia-induced expression (E) of each target mRNA, normalized to 18S rRNA in each sample, was calculated based on the threshold cycle (Ct) as E = 2−∆(∆Ct), where ∆Ct = Ct(target) – Ct(18S) and ∆(∆Ct) = ∆Ct(20%) − ∆Ct(1%). Primer sequences are as follows: RAB22A mRNA, CACCTTACCGTCTCTGTGATTG and GTCTGTGGCCATGTGTCTTTA and 18S rRNA, GAGGATGAGGTGGAACGTGT and AGAAGTGACGCAGCCCTCTA.
Chromatin Immunoprecipitation Assays.
MDA-MB-231 cells were treated with 1% formaldehyde for 10 min at 37 °C, quenched with 125 mM glycine, lysed with SDS lysis buffer, and sheared by sonication. Lysates were precleared with salmon sperm DNA/protein A-agarose slurry (Millipore) and incubated with IgG (Novus Biologicals) or antibody against HIF-1α (Santa Cruz) or HIF-1β (Novus Biologicals). The precipitates were resuspended in 80 µL of water, and 2-µL aliquots were used for PCR with primers for amplification of sequences in the PDK1 (ACCAGGACAGCCAATACAAG and CCTCGGTCACTCATCTTCAC) or RAB22A (CCTCTCAGCGAAGGTGGCTG and GCCGTAGGTCCGGCCTGTC) gene. The PCR products were analyzed by 2% agarose gel electrophoresis.
Focal Adhesion Assays.
Cells were incubated at 20% or 1% O2 for 24 h in serum-free DMEM. MVs were collected from the tissue culture media as described above and added to an equivalent number of naïve MDA-231 cells in serum-free DMEM, which were incubated for 24 h at 20% O2. Focal adhesions were analyzed as previously described (37). Cells were fixed with 4% paraformaldehyde (Sigma), permeabilized for 5 min with 0.1% Triton X-100 (Fisher), blocked with PBS supplemented with 10% FBS for 20 min, and stained with anti-vinculin antibody (Sigma), Alexa Fluor 568-conjugated phalloidin (Invitrogen), and 300 nM DAPI (Invitrogen). Fluorescent imaging was performed by confocal microscopy (A1; Nikon) through a 60× plan or water immersion lens (NA = 1.2). Morphometric analysis of focal adhesion number and area was performed using NIS-Elements software (Nikon).
Invasion Assays.
A total of 2 × 105 cells were seeded onto filters of a 24-well Transwell chamber (Fisher Scientific) that were coated with Matrigel (BD Biosciences). Invasion of the cells through Matrigel to the underside of the filter was assessed 24 h later by staining with crystal violet and counting under bright field microscopy. The mean number of invaded cells per field was determined from 10 fields per filter. To analyze the effect of MVs on invasion, 2 × 105 cells were seeded on top of Matrigel-coated chamber inserts and incubated in serum-free DMEM with MVs shed from equal numbers of 231-SC and 231-sh22A cells that were cultured under nonhypoxic or hypoxic conditions. The vehicle for MV suspension (PBS) served as the control.
Animal Studies.
Animal protocols were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Johns Hopkins University Animal Care and Use Committee. Mycoplasma-free MDA-231 subclones were harvested from tissue culture by trypsinization, resuspended at 107 cells/mL in a 50:50 mix of PBS:Matrigel, and 2 × 106 cells were injected into the mammary fat pad of 6-wk-old female SCID mice (National Cancer Institute). Primary tumors were measured in three dimensions (a, b, and c), and volume was calculated as abc × 0.52. Tumors were excised and weighed. Lungs were perfused with PBS. One lung was inflated for formalin fixation, paraffin embedding, and hematoxylin-and-eosin histology; the other lung was harvested for genomic DNA isolation as previously described (28).
For lung colonization assays, 1 × 106 MDA-MB-231 cells were incubated for 24 h with PBS or MVs isolated from equal numbers of 231-SC or 231-sh22A cells that were cultured under nonhypoxic or hypoxic conditions. PBS- or MV-exposed cells were injected into the tail vein of 6-wk-old female SCID mice. After 3 wk, lungs were harvested and genomic DNA was isolated for qPCR as previously described (28).
Analysis of Lung Genomic DNA.
Lungs were digested with proteinase K at 55 °C and genomic DNA was extracted with phenol–chloroform, precipitated with isopropanol, and washed with ethanol. A 200-ng aliquot of genomic DNA was used for qPCR with human-specific HK2 gene primers (CCAGTTCATTCACATCATCAG and CTTACACGAGGTCACATAGC) and 18S rRNA gene primers (CGGCGACGACCCATTCGAAC and GAATCGAACCCTGATTCCCCGTC).
Statistical Analysis.
Data are presented as mean ± SEM unless otherwise noted. All experiments were performed at least in triplicate. Statistical significance (P < 0.05) was assessed by Student's t test or ANOVA, followed by Bonferroni posttest for multiple comparisons.
Supplementary Material
Acknowledgments
We thank Karen Padgett of Novus Biologicals for providing IgG, Cy3-conjugated goat anti-rabbit IgG, FITC-conjugated goat anti-mouse, and RAB22A and transglutaminase 2 antibodies. This work was supported in part by the American Cancer Society and by Public Health Service Grants U54-CA143868 from the National Cancer Institute (NCI) and R01-EB016721 from the National Institute of Biomedical Imaging and Bioengineering. T.W. was supported by the National Natural Science Foundation of China (Grant 81172253) and the Pu Jiang Scholar Program (Grant 10PJ1407100). D.M.G. was supported by the Susan G. Komen Foundation. N.T. was supported by the Japan Science and Technology Agency. L.X. was supported by the China Scholarship Council. W.L. was supported by NCI (K99-CA168746). G.L.S. is an American Cancer Society Research Professor and the C. Michael Armstrong Professor at The Johns Hopkins University School of Medicine.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 11230.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410041111/-/DCSupplemental.
References
- 1.D’Souza-Schorey C, Clancy JW. Tumor-derived microvesicles: Shedding light on novel microenvironment modulators and prospective cancer biomarkers. Genes Dev. 2012;26(12):1287–1299. doi: 10.1101/gad.192351.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl) 2013;91(4):431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts CT, Jr, Kurre P. Vesicle trafficking and RNA transfer add complexity and connectivity to cell-cell communication. Cancer Res. 2013;73(11):3200–3205. doi: 10.1158/0008-5472.CAN-13-0265. [DOI] [PubMed] [Google Scholar]
- 4.Muralidharan-Chari V, Clancy JW, Sedgwick A, D’Souza-Schorey C. Microvesicles: Mediators of extracellular communication during cancer progression. J Cell Sci. 2010;123(Pt 10):1603–1611. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galindo-Hernandez O, et al. Elevated concentration of microvesicles isolated from peripheral blood in breast cancer patients. Arch Med Res. 2013;44(3):208–214. doi: 10.1016/j.arcmed.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Wysoczynski M, Ratajczak MZ. Lung cancer secreted microvesicles: Underappreciated modulators of microenvironment in expanding tumors. Int J Cancer. 2009;125(7):1595–1603. doi: 10.1002/ijc.24479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antonyak MA, et al. Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc Natl Acad Sci USA. 2011;108(12):4852–4857. doi: 10.1073/pnas.1017667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li B, Antonyak MA, Zhang J, Cerione RA. RhoA triggers a specific signaling pathway that generates transforming microvesicles in cancer cells. Oncogene. 2012;31(45):4740–4749. doi: 10.1038/onc.2011.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang HG, Grizzle WE. Exosomes: A novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am J Pathol. 2014;184(1):28–41. doi: 10.1016/j.ajpath.2013.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ostrowski M, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12(1):19–30. doi: 10.1038/ncb2000. 1–13. [DOI] [PubMed] [Google Scholar]
- 12.Bobrie A, et al. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012;72(19):4920–4930. doi: 10.1158/0008-5472.CAN-12-0925. [DOI] [PubMed] [Google Scholar]
- 13.Harris AL. Hypoxia—a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2(1):38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 14.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 15.Vaupel P, Mayer A, Höckel M. Tumor hypoxia and malignant progression. Methods Enzymol. 2004;381:335–354. doi: 10.1016/S0076-6879(04)81023-1. [DOI] [PubMed] [Google Scholar]
- 16.Brahimi-Horn MC, Chiche J, Pouysségur J. Hypoxia and cancer. J Mol Med (Berl) 2007;85(12):1301–1307. doi: 10.1007/s00109-007-0281-3. [DOI] [PubMed] [Google Scholar]
- 17.Gillies RJ, Gatenby RA. Hypoxia and adaptive landscapes in the evolution of carcinogenesis. Cancer Metastasis Rev. 2007;26(2):311–317. doi: 10.1007/s10555-007-9065-z. [DOI] [PubMed] [Google Scholar]
- 18.Melillo G, editor. Hypoxia and Cancer: Biological Implications and Therapeutic Opportunities. New York: Springer; 2014. [Google Scholar]
- 19.Semenza GL. Hypoxia-inducible factors: Mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci. 2012;33(4):207–214. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prabhakar NR, Semenza GL. Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Physiol Rev. 2012;92(3):967–1003. doi: 10.1152/physrev.00030.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schindl M, et al. Austrian Breast and Colorectal Cancer Study Group Overexpression of hypoxia-inducible factor 1α is associated with an unfavorable prognosis in lymph node-positive breast cancer. Clin Cancer Res. 2002;8(6):1831–1837. [PubMed] [Google Scholar]
- 22.Bos R, et al. Levels of hypoxia-inducible factor-1α independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer. 2003;97(6):1573–1581. doi: 10.1002/cncr.11246. [DOI] [PubMed] [Google Scholar]
- 23.Giatromanolaki A, et al. c-erbB-2 related aggressiveness in breast cancer is hypoxia inducible factor-1α dependent. Clin Cancer Res. 2004;10(23):7972–7977. doi: 10.1158/1078-0432.CCR-04-1068. [DOI] [PubMed] [Google Scholar]
- 24.Dales JP, et al. Overexpression of hypoxia-inducible factor HIF-1α predicts early relapse in breast cancer: Retrospective study in a series of 745 patients. Int J Cancer. 2005;116(5):734–739. doi: 10.1002/ijc.20984. [DOI] [PubMed] [Google Scholar]
- 25.Generali D, et al. Hypoxia-inducible factor-1α expression predicts a poor response to primary chemoendocrine therapy and disease-free survival in primary human breast cancer. Clin Cancer Res. 2006;12(15):4562–4568. doi: 10.1158/1078-0432.CCR-05-2690. [DOI] [PubMed] [Google Scholar]
- 26.Helczynska K, et al. Hypoxia-inducible factor-2α correlates to distant recurrence and poor outcome in invasive breast cancer. Cancer Res. 2008;68(22):9212–9220. doi: 10.1158/0008-5472.CAN-08-1135. [DOI] [PubMed] [Google Scholar]
- 27.Liao D, Corle C, Seagroves TN, Johnson RS. Hypoxia-inducible factor-1α is a key regulator of metastasis in a transgenic model of cancer initiation and progression. Cancer Res. 2007;67(2):563–572. doi: 10.1158/0008-5472.CAN-06-2701. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, et al. HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs. Oncogene. 2012;31(14):1757–1770. doi: 10.1038/onc.2011.365. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Schito L, et al. Hypoxia-inducible factor 1-dependent expression of platelet-derived growth factor B promotes lymphatic metastasis of hypoxic breast cancer cells. Proc Natl Acad Sci USA. 2012;109(40):E2707–E2716. doi: 10.1073/pnas.1214019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong CC, et al. Hypoxia-inducible factor 1 is a master regulator of breast cancer metastatic niche formation. Proc Natl Acad Sci USA. 2011;108(39):16369–16374. doi: 10.1073/pnas.1113483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erler JT, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440(7088):1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 32.Luo W, Chang R, Zhong J, Pandey A, Semenza GL. Histone demethylase JMJD2C is a coactivator for hypoxia-inducible factor 1 that is required for breast cancer progression. Proc Natl Acad Sci USA. 2012;109(49):E3367–E3376. doi: 10.1073/pnas.1217394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Regan Anderson TM, et al. Breast tumor kinase (Brk/PTK6) is a mediator of hypoxia-associated breast cancer progression. Cancer Res. 2013;73(18):5810–5820. doi: 10.1158/0008-5472.CAN-13-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaturvedi P, et al. Hypoxia-inducible factor-dependent breast cancer-mesenchymal stem cell bidirectional signaling promotes metastasis. J Clin Invest. 2013;123(1):189–205. doi: 10.1172/JCI64993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilkes DM, et al. Collagen prolyl hydroxylases are essential for breast cancer metastasis. Cancer Res. 2013;73(11):3285–3296. doi: 10.1158/0008-5472.CAN-12-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilkes DM, et al. Procollagen lysyl hydroxylase 2 is essential for hypoxia-induced breast cancer metastasis. Mol Cancer Res. 2013;11(5):456–466. doi: 10.1158/1541-7786.MCR-12-0629. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Gilkes DM, et al. Hypoxia-inducible factors mediate coordinated RhoA-ROCK1 expression and signaling in breast cancer cells. Proc Natl Acad Sci USA. 2014;111(3):E384–E393. doi: 10.1073/pnas.1321510111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Lee JS, et al. Negative regulation of hypoxic responses via induced Reptin methylation. Mol Cell. 2010;39(1):71–85. doi: 10.1016/j.molcel.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park JE, et al. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol Cell Proteomics. 2010;9(6):1085–1099. doi: 10.1074/mcp.M900381-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svensson KJ, et al. Hypoxia triggers a proangiogenic pathway involving cancer cell microvesicles and PAR-2-mediated heparin-binding EGF signaling in endothelial cells. Proc Natl Acad Sci USA. 2011;108(32):13147–13152. doi: 10.1073/pnas.1104261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kucharzewska P, et al. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc Natl Acad Sci USA. 2013;110(18):7312–7317. doi: 10.1073/pnas.1220998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421. doi: 10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russnes HG, Navin N, Hicks J, Borresen-Dale AL. Insight into the heterogeneity of breast cancer through next-generation sequencing. J Clin Invest. 2011;121(10):3810–3818. doi: 10.1172/JCI57088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timmerman LA, et al. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell. 2013;24(4):450–465. doi: 10.1016/j.ccr.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ringnér M, Fredlund E, Häkkinen J, Borg Å, Staaf J. GOBO: Gene expression-based outcome for breast cancer online. PLoS ONE. 2011;6(3):e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunn LK, et al. Hypoxia and TGF-β drive breast cancer bone metastases through parallel signaling pathways in tumor cells and the bone microenvironment. PLoS ONE. 2009;4(9):e6896. doi: 10.1371/journal.pone.0006896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bendinelli P, et al. Hypoxia inducible factor-1 is activated by transcriptional co-activator with PDZ-binding motif (TAZ) versus WWdomain-containing oxidoreductase (WWOX) in hypoxic microenvironment of bone metastasis from breast cancer. Eur J Cancer. 2013;49(11):2608–2618. doi: 10.1016/j.ejca.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 49.Polyak K. Heterogeneity in breast cancer. J Clin Invest. 2011;121(10):3786–3788. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marino N, et al. Breast cancer metastasis: Issues for the personalization of its prevention and treatment. Am J Pathol. 2013;183(4):1084–1095. doi: 10.1016/j.ajpath.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spiegel J, et al. Direct targeting of Rab-GTPase-effector interactions. Angew Chem Int Ed Engl. 2014;53(9):2498–2503. doi: 10.1002/anie.201308568. [DOI] [PubMed] [Google Scholar]
- 52.Gilkes DM, Semenza GL. Role of hypoxia-inducible factors in breast cancer metastasis. Future Oncol. 2013;9(11):1623–1636. doi: 10.2217/fon.13.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong CC, et al. Inhibitors of hypoxia-inducible factor 1 block breast cancer metastatic niche formation and lung metastasis. J Mol Med (Berl) 2012;90(7):803–815. doi: 10.1007/s00109-011-0855-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiang L, et al. Ganetespib blocks HIF-1 activity and inhibits tumor growth, vascularization, stem cell maintenance, invasion, and metastasis in orthotopic mouse models of triple-negative breast cancer. J Mol Med (Berl) 2014;92(2):151–164. doi: 10.1007/s00109-013-1102-5. [DOI] [PMC free article] [PubMed] [Google Scholar]