Significance
Delivery of nucleotide sugar substrates into the Golgi apparatus and endoplasmic reticulum for processes such as cell wall biosynthesis and protein glycosylation is critical for plant growth and development. Plant genomes encode large families of uncharacterized nucleotide sugar transporters that are specifically presumed to deliver the diverse array of nucleotide sugars found in plants. This study has developed a novel approach that enabled functional characterization of six bifunctional UDP-rhamnose (Rha)/UDP-galactose (Gal) transporters from Arabidopsis. An analysis of loss-of-function and overexpression lines for two of these transporters identified biochemical alterations supporting their roles in the biosynthesis of Rha- and Gal-containing polysaccharides. Thus, cell wall polysaccharide biosynthesis in the Golgi apparatus of plants is likely also regulated by substrate transport mechanisms.
Keywords: membrane transport, proteoliposomes, glycan biosynthesis, galactan
Abstract
Plant cells are surrounded by a cell wall that plays a key role in plant growth, structural integrity, and defense. The cell wall is a complex and diverse structure that is mainly composed of polysaccharides. The majority of noncellulosic cell wall polysaccharides are produced in the Golgi apparatus from nucleotide sugars that are predominantly synthesized in the cytosol. The transport of these nucleotide sugars from the cytosol into the Golgi lumen is a critical process for cell wall biosynthesis and is mediated by a family of nucleotide sugar transporters (NSTs). Numerous studies have sought to characterize substrate-specific transport by NSTs; however, the availability of certain substrates and a lack of robust methods have proven problematic. Consequently, we have developed a novel approach that combines reconstitution of NSTs into liposomes and the subsequent assessment of nucleotide sugar uptake by mass spectrometry. To address the limitation of substrate availability, we also developed a two-step reaction for the enzymatic synthesis of UDP–l-rhamnose (Rha) by expressing the two active domains of the Arabidopsis UDP–l-Rha synthase. The liposome approach and the newly synthesized substrates were used to analyze a clade of Arabidopsis NSTs, resulting in the identification and characterization of six bifunctional UDP–l-Rha/UDP–d-galactose (Gal) transporters (URGTs). Further analysis of loss-of-function and overexpression plants for two of these URGTs supported their roles in the transport of UDP–l-Rha and UDP–d-Gal for matrix polysaccharide biosynthesis.
Plant cell walls are largely composed of polysaccharides, and with the exception of cellulose and callose, these polymers are synthesized in the Golgi apparatus by families of glycosyltransferases (1, 2). Many glycan structures of glycoproteins and glycolipids are likewise assembled in the Golgi apparatus (3). The nucleotide sugar substrates necessary for the assembly of these polysaccharides and glycans are predominantly synthesized in the cytosol. This subcellular partitioning of substrates and enzymes necessitates transmembrane transport into the lumen of the Golgi apparatus and endoplasmic reticulum (ER). Consequently, nucleotide sugar transporters (NSTs) have evolved to enable the transfer of nucleotide sugars into these organelles. Given the essential roles of NSTs in the glycosylation of proteins, lipids, and proteoglycans, as well as in the biosynthesis of plant cell wall polysaccharides, it is not surprising that these transporters have been identified in all eukaryotes (4). NSTs consist of 300- to 400-aa residues and possess six to 10 transmembrane domains (5, 6). Plant NSTs belong to the NST/triose phosphate translocator (TPT) superfamily, and phylogenetic analyses have identified more than 50 members that are distributed in six clades in the reference plant, Arabidopsis thaliana (7) (Fig. 1).
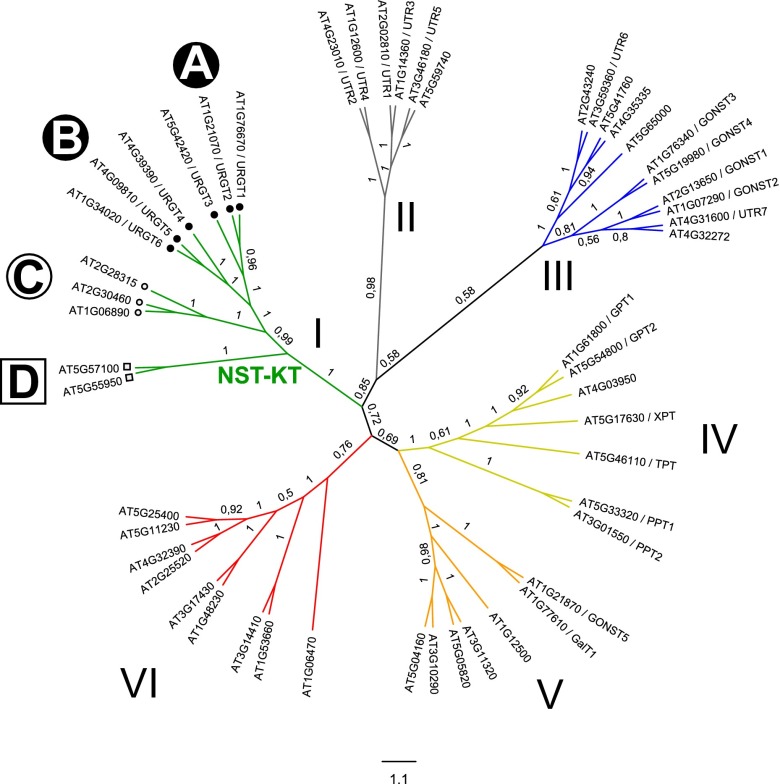
Fig. 1.
Phylogenetic tree of the Arabidopsis NST-TPT superfamily. Full-length amino acid sequences were aligned using the Clustal Omega program, and the phylogenetic tree was generated using Molecular Evolutionary Genetics Analysis (MEGA) 5. Clades were assigned arbitrarily.
In the past decade, few plant NSTs have been functionally characterized at the molecular level, and those characterized only account for the transport of GDP-mannose (Man), UDP-galactose (Gal), UDP-glucose (Glc), and CMP-sialic acid (Sia), although the latter sugar is unlikely to be part of any plant glycan (8–18). Considering the diverse composition of polysaccharide structures in plants, this collection of nucleotide sugars only represents a fraction of required substrates. Thus, there must be additional NSTs mediating the transport of other key nucleotide sugars, such as UDP-rhamnose (Rha), UDP-glucuronic acid (GlcA), GDP-fucose (Fuc), UDP-xylose (Xyl), and UDP-arabinose (Ara), all of which are essential for plant development and function. Plants harboring mutations in the biosynthesis of nucleotide sugar substrates display developmental and biochemical defects (e.g., GDP-Fuc biosynthesis) (19). Alterations to plant NSTs have only resulted in observable phenotypes for a GDP-Man transporter (Arabidopsis thaliana Golgi nucleotide sugar transporter 1) (14) and a UDP-Glc transporter (Oryza sativa nucleotide sugar transporter 1) (8). In contrast, studies in other eukaryotic systems have shown that mutations in a number of NSTs result in various pathological phenotypes (4, 20) [e.g., Schneckenbecken dysplasia in humans and mice resulting from mutations in a UDP–N-acetyl-α-d-galactosamine (GalNAc)/UDP-GlcA transporter (21)]. Attempts to predict the function and substrate specificity of NSTs by protein sequence similarity have been unsuccessful (22); therefore, studies in other systems have provided minimal insight into predicting activity of the transporters in plants. Thus, the characterization of plant NSTs usually requires biochemical approaches, which are often limited by the inadequate availability of nucleotide sugar substrates.
To overcome the above limitations, we sought to develop a biochemical approach that is not limited by the availability of radiolabeled substrates or the complexities of genetic complementation experiments. Using this approach, we show for the first time, to our knowledge, that six of 11 NSTs derived from the Arabidopsis NST-lysine/threonine (KT) clade of the NST/TPT superfamily are capable of transporting UDP-Rha as well as UDP-Gal with different preferences. We designate these six proteins as UDP-Rha/UDP-Gal transporters (URGTs) 1–6. Localization analyses showed that all six NSTs are targeted to the Golgi, and mutants had altered levels of Rha and Gal in cell wall polysaccharides.
Results
NST-KT Clade of the NST/TPT Family of Arabidopsis.
The Arabidopsis NST-KT clade comprised two members that had previously been described as monospecific UDP-Gal transporters (10, 17). Thus, we initially sought to test our approach on these functionally characterized NSTs. The previously defined NST-KT subfamily is characterized by a highly conserved KT motif (7) and forms clade I of the NST/TPT family, encompassing 11 putative NSTs. They share 25–93% identity in their amino acid sequences and include the two characterized NSTs, namely, At1g76670 (URGT1/UDP-galactose transporter 2) (10) and At4g39390 (URGT4/NST-KT1) (17). URGT1 shows the highest sequence similarity to At1g21070 (URGT2) and At5g42420 (URGT3). These three proteins form subclade A, which is clearly distinguishable from another subclade (B) also consisting of three proteins, namely, URGT4 (17), At4g09810 (URGT5), and At1g34020 (URGT6). Members of the subclades C and D share 25–36% identity in their amino acid sequences compared with all other members of the NST-KT family (Fig. 1 and Table S1).
URGTs Are Ubiquitously Expressed and Localize to the Golgi.
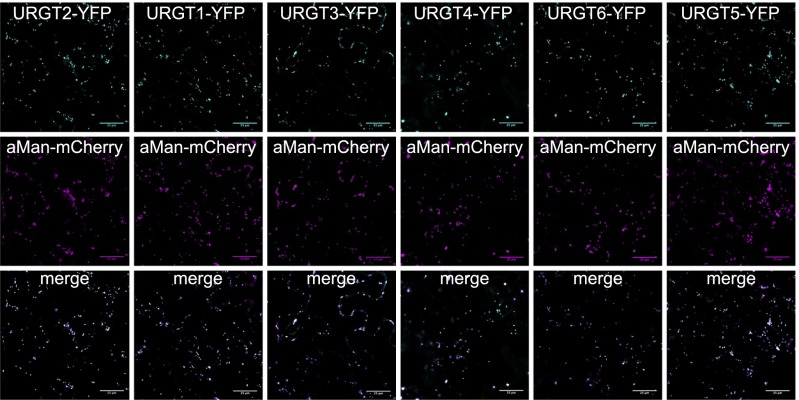
Analysis of publicly available microarray expression data from the developmental dataset (23) for the NST-KT clade revealed ubiquitous expression for all six NSTs throughout plant development, with URGT1 showing the highest level in virtually all organs and URGT2 showing the highest levels in developing seeds (Fig. S1). Notably, the expression patterns appeared to be largely overlapping, suggesting a high degree of functional redundancy. To examine the subcellular localizations of members of the NST-KT clade, we fused their coding sequences to YFP and transiently expressed them in tobacco leaves (Fig. 2). All six candidates localized to moving, dot-like structures typical of the Golgi apparatus, which also colocalized with the cis-Golgi marker α-Man I (24). This observation is consistent with their putative roles as NSTs of the Golgi apparatus.
Fig. 2.
Subcellular localizations of subclade A and B NST proteins. NST coding sequences were fused at their C terminus to the YFP and transiently expressed in tobacco leaves. All six candidates localized to moving dot-like structures typical for the Golgi apparatus and colocalized with the cis-Golgi marker α-Man consistent with a role in nucleotide sugar transport across the Golgi membrane. (Scale bars: 25 μm.)
Functional Determination of the NST-KT Clade.
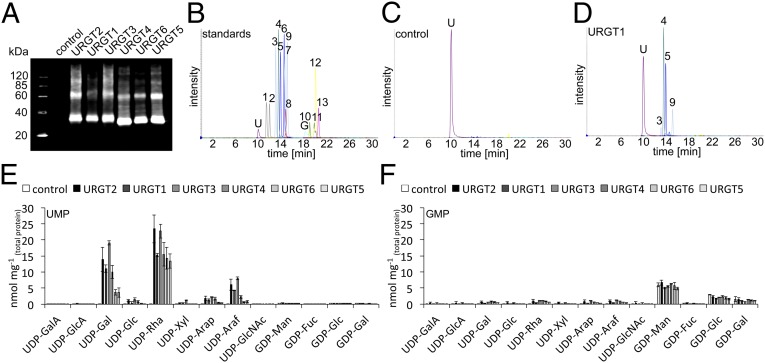
To investigate the substrate specificity of the URGTs fully, we first needed to obtain all potential nucleotide sugar substrates in plants. Neither UDP-Rha nor GDP-Gal was commercially available; therefore, we synthesized both substrates enzymatically (Materials and Methods). Enzymatic synthesis of UDP-Rha had not been previously described, and we developed a two-step reaction using UDP-Glc as a substrate and expression of the individual domains of UDP-Rha synthase. The synthesized and commercially available nucleotide sugars comprised a set of 13 potential substrates. To confirm that URGT1 transports UDP-Gal, and to test whether this NST is monospecific, URGT1 was heterologously expressed in yeast and microsomal proteins were obtained. Immunoblot analysis confirmed the presence of the URGT1 protein in yeast microsomal extracts (Fig. 3A). Subsequently, proteoliposomes preloaded with UMP were incubated with a mixture of the 13 nucleotide sugars. Nonincorporated nucleotide sugars were removed by gel filtration, and the content of the liposomes was analyzed by LC-tandem MS (MS/MS) (Table S2). Our data confirmed URGT1 as a UDP-Gal transporter; however, URGT1 was not monospecific for UDP-Gal because it was also capable of transporting UDP-Rha with an even higher efficiency and transported comparatively low amounts of UDP-arabinopyranose (Arap)/arabinofuranose (Araf). Subsequently, we analyzed the closest homologs of URGT1, namely, URGT2, URGT3, URGT4, URGT5, and URGT6. All these NSTs showed high transport activity of UDP-Rha and UDP-Gal when UMP was provided as a counterexchange substrate (Fig. 3 D and E). In contrast, when liposomes were preloaded with GMP, only GDP-sugar transport was observed, resulting from endogenous yeast GDP-Man transport activity, because the incorporation levels were similar to those observed in the control (yeast transformed with the empty vector, pYES-DEST52) (Fig. 3 C and F). Analysis of the remaining five NST-KT subfamily C and D members (Fig. 1) did not reveal any UDP-Rha or UDP-Gal transport activity.
Fig. 3.
Detection and quantification of URGT transport activities. (A) Immunoblot of URGT1–6 expression in yeast microsomal fractions used for reconstitution into liposomes using an anti–V5- tag antibody. Expressed URGTs are present at the ∼35-kDa band. The bands at ∼60 kDa likely represent URGT dimers. (B) Separation and MS/MS detection of a 13-nucleotide sugar standard mix: 1, UDP-GalA; 2, UDP-GlcA; 3, UDP-Arap; 4, UDP-Rha; 5, UDP-Gal; 6, UDP-Glc; 7, UDP-xylose; 8, UDP–N-acetyl-α-d-glucosamine (GlcNAc); 9, UDP-Araf; 10, GDP-Man; 11, GDP-Gal; 12, GDP-Glc; 13, GDP-Fuc. (C) Chromatogram of an empty vector control assay with UMP-loaded liposomes. (D) Representative chromatogram of URGT1 proteoliposomes revealing selective transport activities after feeding 13 nucleotide sugars (50 μM) for 20 min at 25 °C. Quantification of nucleotide sugar uptake into proteoliposomes containing each of the URGTs preloaded with UMP (E) or GMP (F) is shown. Quantitative data are mean and SD of two independent experiments and are normalized to the total protein content reconstituted into liposomes.
Enzyme Kinetics of the Six Arabidopsis URGTs.
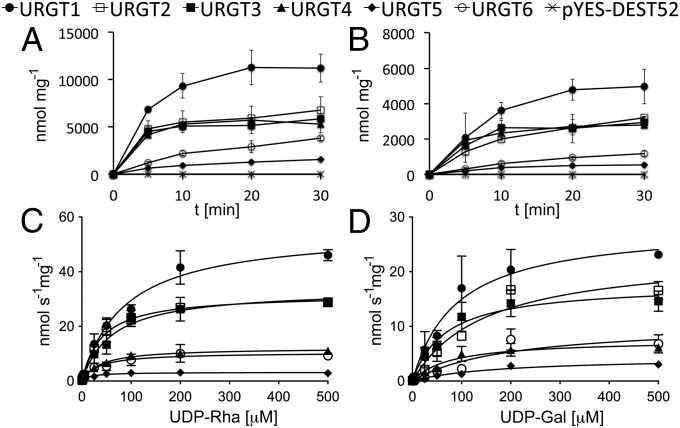
Determination of the kinetic parameters indicated a higher affinity for UDP-Rha than for UDP-Gal for all six URGTs. The apparent Km values for UDP-Rha were in the range of 17–87 μM, with turnover rates between 0.13 and 2.35 s−1, and those for UDP-Gal were in the range of 68–196 μM, with turnover rates of 0.18–1.21 s−1 (Fig. 4 and Table 1). To determine whether the Km values were within the range of the physiological nucleotide sugar concentrations, we determined the nucleotide sugar pools in different Arabidopsis organs and at different developmental stages. The UDP-Rha and UDP-Gal contents are in the range of 40–120 pmol⋅mg−1 of dry weight and 130–400 pmol⋅mg−1 of dry weight, respectively (Table S3). In plants, the central vacuole can make up 90% of the total cell volume, depending on cell type and developmental stage (25). These measurements indicate that the cellular levels of UDP-Rha and UDP-Gal are in the micromolar range. Thus, we estimate that the Km values for all six NSTs are within physiological range. To investigate the dual functionality of the URGT transporters further, we undertook a competition analysis using both substrates (Fig. S2). These results clearly indicate that the addition of excess UDP-Gal reduced the amount of UDP-Rha transported in vitro for all members of the URGT family and demonstrate that both substrates compete for the same transport mechanism.
Fig. 4.
Time- and concentration-dependent nucleotide sugar uptake into proteoliposomes. Time courses of UDP-Rha (A) and UDP-Gal (B) uptake. Proteoliposomes, preloaded with UMP, were incubated with the respective nucleotide sugar at a concentration of 50 μM for the indicated time points at 25 °C. Kinetics of UDP-Rha (C) and UDP-Gal (D) transport. Proteoliposomes, preloaded with UMP, were incubated with their respective nucleotide sugar at varying concentrations (0.5–400 μM) for 3 min at 25 °C. Values were normalized to the actual NST protein content present in proteoliposome preparations and represent the mean and SD of two independent experiments. pYES-DEST52, yeast transformed with the empty vector.
Table 1.
Enzyme kinetics for UDP-Rha and UDP-Gal transport into proteoliposomes
| URGT1 | URGT2 | URGT3 | URGT4 | URGT5 | URGT6 | ||
| UDP-Rha* | Km, μM | 87 (12) | 61 (11) | 40 (7) | 37 (8) | 17 (5) | 33 (8) |
| Vmax, pmol⋅min−1 | 742 (36) | 1,411 (79) | 1,023 (48) | 631 (38) | 188 (12) | 264 (16) | |
| Vmax, nmol⋅s−1⋅mg−1 | 56 (2) | 34 (2) | 32 (2) | 12 (1) | 4 (0) | 10 (0) | |
| Kcat, s−1 | 2.35 | 1.44 | 1.36 | 0.51 | 0.13 | 0.44 | |
| UDP-Gal | Km, μM | 90 (22) | 68 (16) | 149 (37) | 77 (19) | 177 (53) | 196 (96) |
| Vmax, pmol⋅min−1 | 381 (33) | 742 (57) | 751 (78) | 392 (32) | 253 (33) | 267 (59) | |
| Vmax, nmol⋅s−1⋅mg−1 | 28 (2) | 19 (2) | 24 (2) | 8 (0) | 4 (0) | 10 (1) | |
| Kcat, s−1 | 1.21 | 0.76 | 1.00 | 0.32 | 0.18 | 0.44 |
Role of URGT1 and URGT2 in Planta.
To assess the in vivo functions of URGTs, we analyzed plants with altered levels of URGT1 (the NST with the highest expression relative to the others) and URGT2 (an NST that has a more restricted pattern of expression during seed development). To examine loss-of-function mutants, transfer DNA (T-DNA) insertion lines were obtained for URGT1 and URGT2, and to explore URGT function further, we also overexpressed URGT1 or URGT2 under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter.
URGT1 is expressed throughout plant development and shows the highest expression levels compared with the other five URGTs (Fig. S1). Analysis of leaf cell wall composition of urgt1 mutants showed a slight reduction in Gal content, whereas other monosaccharides, including Rha, were unchanged (Fig. S3A). When URGT1 was overexpressed under the control of the CaMV 35S promoter (35Spro:URGT1-YFP), the Gal content of leaf cell walls increased up to 50% (Fig. S3B). The reduction or increase in Gal did not affect the morphology of the plants compared with the WT. Most of the accumulating Gal in cell walls of URGT1-overexpressing plants was found in a fraction that could be digested with β-1,4-galactanase (Fig. 5E), indicating that excess cell wall Gal was mostly deposited in the pectic polymer rhamnogalacturonan-I (RG-I). The main hemicellulose in Arabidopsis primary walls is xyloglucan, which also contains galactosyl residues. To test if xyloglucan was altered in urgt1 mutants or the 35Spro:URGT1-YFP plants, we performed an oligo mass profiling analysis (26, 27). No differences in xyloglucan structure or the abundance of galactosylated xyloglucan oligosaccharides compared with control plants were observed (Fig. S3D).
Fig. 5.
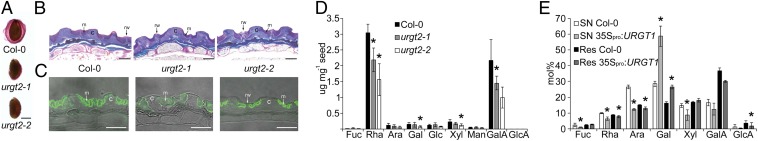
URGT1 and URGT2 are involved in pectic galactan and seed mucilage formation, respectively. (A) Ruthenium red staining of dry WT and urgt2 seeds. (Scale bar: 200 μm.) (B) Sections of dry seeds stained with toluidine blue. WT seeds accumulate more mucilage in the apoplast in comparison to the URGT2 mutant lines. c, columella; m, mucilage; rw, radial wall. (Scale bars: 25 μm.) (C) Immunolabeling of dehydrated mucilage from dry seed sections using the antibody CCRC-M36, specific to unbranched RG-I. The labeling with CCRC-M36 antibody of WT seed mucilage is stronger in comparison to urgt2 lines. (D) Monosaccharide composition of hydrolyzed soluble mucilage derived from WT, urgt2-1, and urgt2-2 dry seeds. Data represent the mean and SD of n = 4 biological replicates. (E) Monosaccharide composition of hydrolysate of endo-β1,4-galactanase digested cell wall material derived from 35Spro:URGT1 plants. Res, residue; SN, supernatant. Data are the mean and SD of n = 4 replicates. *Significantly different (t test at P < 0.05).
URGT2 is highly expressed during seed development at 6–8 d after pollination (Fig. S1), specifically in the seed coat (28). This is a period when large amounts of pectic polysaccharides, especially RG-I, are synthesized and released to the apoplast of seed coat epidermal cells. Upon imbibition of the dry seeds, these polysaccharides form the mucilage. We observed that the mucilage of urgt2 mutants showed an altered pattern when stained with the pectin-staining dye ruthenium red (Fig. 5A). In addition, sections stained with toluidine blue or labeled with the CCRC-M36 antibody (Carbosource Services, Complex Carbohydrate Research Center), which recognizes unbranched parts of the RG-I backbone (29), showed that urgt2 mutants have a lower content of mucilage (Fig. 5 B and C). Monosaccharide analysis of hydrolyzed seed mucilage confirmed that there was a decrease in the total amount of mucilage in both urgt2 mutant alleles but no change in mucilage composition (Fig. 5D). Polysaccharide analysis using carbohydrate gel electrophoresis (30) of RG-I from mucilage also showed no change in the fine structure of the polymer (Fig. S4), suggesting that urgt2 mutants have either fewer or shorter RG-I chains. No change was observed in the cell wall composition of leaf material derived from urgt2 mutants. Only minor changes in cell wall sugars were detected in leaf material from plants overexpressing URGT2 (35Spro:URGT2-YFP) (Fig. S3C).
Discussion
NSTs play an important role in delivering nucleotide sugar substrates for a variety of glycosylation reactions that occur in the Golgi and ER lumen. However, despite being essential for polysaccharide biosynthesis in the Golgi apparatus, NSTs have historically been poorly characterized. Thus far, only plant NSTs that transport the nucleotide sugars GDP-Man, UDP-Gal, and UDP-Glc to the Golgi have been reported (10–13, 15–17). Given the number and variety of nucleotide sugars required for the synthesis of plant cell wall polysaccharides, there is an expectation that a range of plant NSTs with various substrate specificities will be required.
Characterizing Plant NSTs.
In the past, a variety of approaches have been used to assess the function and substrate specificities of plant NSTs. One approach was to use mutants from yeast or other organisms deficient in a specific NST and to use heterologous complementation (9–11, 13, 31). These approaches resulted in the characterization of plant NSTs with substrate specificities for UDP-Gal, GDP-Man, and CMP-Sia. However, the method is limited by the availability of relevant mutant backgrounds, especially for plant-specific nucleotide sugars. Furthermore, an NST may have sufficient activity with a given nucleotide sugar to complement a mutation even though the nucleotide sugar is not the substrate in planta. For example, the Arabidopsis NST that was found to transport CMP-Sia is unlikely to have that function in the plant because there is strong evidence that sialic acid is not present in plants (9). Multiple studies have also presented data on nucleotide sugar transport measured in microsomes or Golgi-enriched vesicles isolated from plants or yeast upon overexpressing specific NSTs (8, 9, 12, 15, 16, 31). These approaches were used to characterize plant UDP-Glc, UDP-Gal, GDP-Man, and CMP-Sia transport. Because the transport of nucleotide sugars requires the presence of a counterexchange substrate (e.g., UMP, GMP), isolated vesicles are likely to be depleted of these compounds. Yeast microsomes will incorporate Glc, N-acetyl-α-d-glucosamine, and Man into yeast glycans (32), thereby generating UMP or GMP. Consequently, this could result in the apparent transport of nucleotide sugars but may not reflect the actual substrate specificity of an expressed NST. There is the possibility that this may have led to incorrect assignments for the activity of some plant NSTs. The reconstitution of transporters into liposomes has proven to be a more reliable approach to demonstrate transport activity (14, 17, 33). The possibility of preloading liposomes with counterexchange substrates at a desired concentration and reduction of background activity from endogenous transporters and glycosyltransferases provides an essential advantage over preexisting methods.
NST-KT Clades A and B Are UDP-Rha/UDP-Gal Transporters.
The NST-KT clade of Arabidopsis comprises two previously characterized NSTs. Both URGT1 and URGT4 have previously been shown to transport UDP-Gal by complementation of a UDP-Gal–deficient CHO cell line (10) or by reconstitution of a heterologous expressed protein in liposomes (17), respectively. Although only a limited set of nucleotide sugar substrates was tested, in both cases, the authors argued that URGT1 and URGT4 are monospecific for UDP-Gal (10, 17). However, to date, demonstrated transporter activities have largely relied on the uptake of radiolabeled nucleotide sugar substrates that are commercially available. Important substrates for biosynthesis of plant cell walls, such as UDP-GalA, UDP-Arap, UDP-Araf, and UDP-Rha, have not been assessed. Consequently, to characterize the URGT family of Arabidopsis fully, we enzymatically synthesized nucleotide sugars, such as UDP-Rha and GDP-Gal, which are not currently commercially available in either the unlabeled or radiolabeled form. To overcome the limitations of using radiolabeled substrates, we adapted and optimized a recently developed LC-MS method, which facilitates the separation and quantification of all major nucleotide sugars (34). In conjunction with this MS-based assay technique, we reconstituted each member of the NST-KT clade into preloaded liposomes to determine its substrate specificity using a mix of 13 nucleotide sugars. This novel approach enabled us to identify a family of six bifunctional UDP-Rha/UDP-Gal transporters from Arabidopsis. An assessment of cellular nucleotide sugar levels indicated that their determined Km values were within physiological range.
URGT Functions in Planta.
The subcellular localization studies of the URGT family indicated that they are located in the Golgi apparatus, which underlines their function as Golgi resident NSTs. All six URGTs are expressed ubiquitously throughout plant development, with URGT1 showing the highest expression levels and URGT2 preferentially expressed during seed development. To characterize the function of URGT1 and URGT2 in planta, we identified loss-of-function mutants and generated plants overexpressing URGT1 and URGT2.
The urgt1 mutants and plants overexpressing URGT1 confirmed its role as a UDP-Gal transporter in vivo because both mutants showed a reduction in cell wall Gal and the overexpressors accumulated Gal in the leaf cell wall. Recently, it was demonstrated that the Gal content of the cell wall could be easily manipulated (35); this is probably due to the extension of galactan side chains on the RG-I backbone. In contrast, no changes in the levels of Rha were observed in either the urgt1 mutants or overexpression plants. Although it is likely that functional redundancy among the URGT family may account for the lack of a Rha phenotype in the urgt1 mutants, the unchanged levels of Rha in the cell walls of URGT1 overexpression lines are still not explained. Overexpression of UDP-Rha synthase (RHM1) can increase the amount of Rha in leaf cell wall material of Arabidopsis (36), indicating that UDP-Rha synthesis, rather than UDP-Rha transport, is limiting. This could explain the inability of URGT1 overexpression lines to increase cell wall Rha content in leaves. Our results demonstrate that URGT1 mainly affects pectic galactan, whereas the Gal content in xyloglucan is unchanged. The specificity may be due to channeling of UDP-Gal to specific galactosyltransferases or to different galactosyltransferases having different Km values for UDP-Gal. In a classic study of mammalian UDP-Gal transport, some proteoglycans were affected by a 98% reduction in transporter activity, whereas other proteoglycans were unaffected, and differences in Km were suggested as the most likely explanation (37).
Analysis of the urgt2 mutants revealed a reduction in the amount of seed mucilage, reflecting the role of URGT2 in providing UDP-Rha for RG-I biosynthesis. Although the data indicate that URGT2 transports UDP-Rha in the seed coat, it is unclear whether it also transports UDP-Gal, because the Gal level is already very low in seed mucilage (Fig. 5D). The specific expression of URGT2 in the seed coat likely explains its distinct function in this tissue. However, no difference was observed in the Rha content of leaf cell wall preparations from either the URGT2 overexpression or mutant lines. Only a minor increase in cell wall Gal was measured in URGT2 overexpression lines, but this also coincided with slight changes to other sugars. Why the URGT2 overexpression lines do not have increased leaf Rha is not clear; however, as outlined above, overexpression of UDP-RHM1 alone results in up to 40% higher Rha content in leaf cell walls (36), indicating that UDP-Rha, rather than transport, is limiting in leaves. Furthermore, synthesis of RG-I also requires a sufficient supply of UDP-GalA, which would be provided by different NSTs and may be limiting. Overexpression of URGT2 led to a smaller increase in leaf cell wall Gal than overexpression of URGT1. This could be due to different levels of overexpressed protein in the plants or to a channeling effect.
Our data indicate that despite overlapping functions in vitro, plant NSTs seem to have more restricted functions in vivo. This might suggest that nucleotide sugars are channeled by NSTs to specific glycosyltransferases, although different Km values for the glycosyltransferases that use the same substrate may also play a role. Channeling of UDP-Gal to different glycosyltransferases has been suggested previously based on the apparent specificity of cytosolic UDP-Glc epimerase isoforms (38). Similar to our results, the GDP-Man transporter GONST1 has been shown specifically to supply GDP-Man to the Golgi lumen for glycosphingolipid mannosylation but not for processes such as glucomannan biosynthesis (14). Given that many more glycosyltransferases exist in plants than NSTs, it is evident that although a specific NST may preferentially be involved in the synthesis of specific polymers, each NST is generally providing substrate for multiple glycosyltransferases.
Conclusion
We have developed a novel approach to analyze NST activities and characterized a family of six bifunctional UDP-Rha/UDP-Gal transporters from Arabidopsis. The assay enables the determination of substrate specificity for putative NSTs whose functions are currently unknown or incompletely characterized. The method described here will allow rapid progress in determining the functional role of NSTs in plants and other organisms. This study represents the first characterized NST, to our knowledge, with specificity for UDP-Rha.
Materials and Methods
Enzymatic Synthesis of Nucleotide Sugars.
GDP–α-l-Gal (GDP-Gal) was enzymatically synthesized according to previously outlined methods (39) and HPLC-purified using a linear ammonium formate gradient (40). UDP–β-l-Rha was enzymatically synthesized by a two-step reaction using UDP–α-d-Glc (UDP-Glc) as a substrate. For this purpose, the dehydrogenase (RHM1-D) and the epimerase reductase (RHM1-ER) domains of Arabidopsis UDP-Rha synthase (RHM1, At1g78570) were fused at their C termini with a 6× His-tag and expressed in Escherichia coli Rosetta 2 (DE3) strain (EMD Millipore) and purified from the cell lysates by Talon resins (Clontech). For the first step of the reaction, UDP–d-quinov-4-ulose was synthesized using 1 mM UDP-Glc, 4 μg of RHM1-D, 0.5 mM NAD+, and 10 mM Tris⋅HCl (pH 8.5) in a total volume of 0.4 mL. After overnight incubation at 25 °C, the reaction was stopped by heat inactivation for 10 min at 95 °C. The volume of the reaction mixture was then adjusted to 2 mL, containing 1 mM NADPH, 8 μg of RHM1-ER, and 10 mM Tris⋅HCl (pH 8.5). After overnight incubation at 25 °C, the reaction was stopped as described previously. The obtained UDP-Rha product was subsequently purified using a Carbo column (Envi-Carb SPE column; Sigma–Aldrich) as described below, and the concentration was determined based on the absorbance at 254 nm. To verify the identity of the nucleotide sugar, an aliquot was hydrolyzed by incubation with 2 N of TFA at 90 °C for 30 min in a final volume of 200 μL. Monosaccharides in this reaction mixture were separated and quantitated by GC-MS of alditol acetates using authentic standards to determine retention times and mass spectra (41).
Heterologous Expression, Reconstitution, and in Vitro Assay of Transport Activities.
The uracil-auxotrophic yeast strain INVScI was transformed according to the manufacturer’s instructions (Life Technologies). Microsomal membranes were isolated from 500-mL cultures (SI Materials and Methods). Reconstitution of yeast microsomal proteins into liposomes was done by detergent solubilization and rapid removal essentially according to previous techniques (42), using acetone-washed soybean l-α-phosphatidylcholine (Avanti Polar Lipids), a lipid/protein ratio of 13, and octyl-β-glucoside as the detergent (SI Materials and Methods). The obtained liposomes were diluted in reconstitution buffer and subsequently used for transporter activity assays with the appropriate nucleotide sugar substrates at 25 °C for the indicated time points. The reaction was terminated by gel filtration on Sephadex G50 (GE Healthcare) columns. Trapped nucleotide sugars were extracted as previously described (43). The freeze-dried extracts were dissolved in 1 mL of 10 mM ammonium bicarbonate before using the Carbo column purification protocol as previously described (44). Purified extracts were lyophilized and resuspended in 200 μL of LC-MS grade water.
Analysis of Nucleotide Sugars by MS.
Nucleotide sugar separations were undertaken using porous graphitic carbon as the stationary phase using an Agilent 1100 Series HPLC instrument coupled to a mass spectrometer (34) (SI Materials and Methods). Detection was undertaken using a 4000 QTRAP LC-MS/MS system (AB Sciex) equipped with a TurboIonSpray ion source (AB Sciex) (SI Materials and Methods).
Plant Material.
T-DNA insertion mutants urgt1-1 (SAIL_768_C08), urgt1-2 (SAIL_1296_A09), urgt2-1 (SALK_125196), urgt2-2 (SALK_071647), and the A. thaliana (L.) Heynh. (Col-0) were obtained from the Arabidopsis Biological Resource Center (SI Materials and Methods).
Cytochemical Staining and Immunolabeling Procedures.
Mucilage released from mature dry seeds was stained with ruthenium red. For immunolabeling, seeds were fixed with formaldehyde and embedded in LR White resin (London Resin Company). Immunolabeling was performed with an anti–RG-I antibody (CCRC-M36; Carbosource Services, Complex Carbohydrate Research Center) (SI Materials and Methods).
Mucilage Extraction, Cell Wall Preparation, Fractionation, and Monosaccharide Analysis.
Dry seeds were imbibed in extraction buffer and filtered, and polysaccharides were precipitated with ethanol and lyophilized. Extracted mucilage was incubated in 2 N of TFA for 4 h at 120 °C, dried, and resuspended in water. High-performance anion exchange chromatography with pulsed amperometric detection was performed as previously described (14) (SI Materials and Methods). Alcohol-insoluble residue from leaves of 6-wk-old plants was prepared as described earlier (45) (SI Materials and Methods). Cell wall preparations from URGT1-overexpressing plants were analyzed by digestion with an endo-β-1,4-galactanase from Aspergillus niger (Megazyme International) as described previously (35).
Supplementary Material
Acknowledgments
This work was supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, through Contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy. Funding for W.-D.R. was provided by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy through Grant DE-FG02-08ER20203. This work was also supported by CONICYT fellowships (I.M., H.T., and A.M.) and by Fondo de Areas Prioritarias-Centro de Regulación del Genoma-15090007, PFB-16, ICM-Millenium Nucleus P10-062-F, DI-365-13/R, and Fondecyt 3140415 grants (to A.O.). J.C.M. was supported by Biotechnology and Biological Sciences Research Council Grant BB/G016240/1 (to P.D.). Both A.S. and M.P. were supported by Fred E. Dickinson Chair funds. B.E. was supported in part by the Danish Research Council for Strategic Research. The substrates obtained from Carbosource Services were supported, in part, by National Science Foundation-Research Coordination Network Grant 0090281.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KJ667158, KJ667159, KJ667160, KJ667161, and KJ667162).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406073111/-/DCSupplemental.
References
- 1.Liepman AH, Wightman R, Geshi N, Turner SR, Scheller HV. Arabidopsis—A powerful model system for plant cell wall research. Plant J. 2010;61(6):1107–1121. doi: 10.1111/j.1365-313X.2010.04161.x. [DOI] [PubMed] [Google Scholar]
- 2.Scheible WR, Pauly M. Glycosyltransferases and cell wall biosynthesis: Novel players and insights. Curr Opin Plant Biol. 2004;7(3):285–295. doi: 10.1016/j.pbi.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Schoberer J, Strasser R. Sub-compartmental organization of Golgi-resident N-glycan processing enzymes in plants. Mol Plant. 2011;4(2):220–228. doi: 10.1093/mp/ssq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, Hirschberg CB. Developmental diseases caused by impaired nucleotide sugar transporters. Glycoconj J. 2013;30(1):5–10. doi: 10.1007/s10719-012-9375-4. [DOI] [PubMed] [Google Scholar]
- 5.Handford M, Rodriguez-Furlán C, Orellana A. Nucleotide-sugar transporters: Structure, function and roles in vivo. Braz J Med Biol Res. 2006;39(9):1149–1158. doi: 10.1590/s0100-879x2006000900002. [DOI] [PubMed] [Google Scholar]
- 6.Ishida N, Kawakita M. Molecular physiology and pathology of the nucleotide sugar transporter family (SLC35) Pflugers Arch. 2004;447(5):768–775. doi: 10.1007/s00424-003-1093-0. [DOI] [PubMed] [Google Scholar]
- 7.Knappe S, Flügge UI, Fischer K. Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol. 2003;131(3):1178–1190. doi: 10.1104/pp.016519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B, et al. Golgi nucleotide sugar transporter modulates cell wall biosynthesis and plant growth in rice. Proc Natl Acad Sci USA. 2011;108(12):5110–5115. doi: 10.1073/pnas.1016144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakker H, et al. A CMP-sialic acid transporter cloned from Arabidopsis thaliana. Carbohydr Res. 2008;343(12):2148–2152. doi: 10.1016/j.carres.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Bakker H, et al. Molecular cloning of two Arabidopsis UDP-galactose transporters by complementation of a deficient Chinese hamster ovary cell line. Glycobiology. 2005;15(2):193–201. doi: 10.1093/glycob/cwh159. [DOI] [PubMed] [Google Scholar]
- 11.Baldwin TC, Handford MG, Yuseff MI, Orellana A, Dupree P. Identification and characterization of GONST1, a golgi-localized GDP-mannose transporter in Arabidopsis. Plant Cell. 2001;13(10):2283–2295. doi: 10.1105/tpc.010247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Handford M, et al. Arabidopsis thaliana AtUTr7 encodes a golgi-localized UDP-glucose/UDP-galactose transporter that affects lateral root emergence. Mol Plant. 2012;5(6):1263–1280. doi: 10.1093/mp/sss074. [DOI] [PubMed] [Google Scholar]
- 13.Handford MG, Sicilia F, Brandizzi F, Chung JH, Dupree P. Arabidopsis thaliana expresses multiple Golgi-localised nucleotide-sugar transporters related to GONST1. Mol Genet Genomics. 2004;272(4):397–410. doi: 10.1007/s00438-004-1071-z. [DOI] [PubMed] [Google Scholar]
- 14.Mortimer JC, et al. Abnormal glycosphingolipid mannosylation triggers salicylic acid-mediated responses in Arabidopsis. Plant Cell. 2013;25(5):1881–1894. doi: 10.1105/tpc.113.111500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norambuena L, et al. Transport of UDP-galactose in plants. Identification and functional characterization of AtUTr1, an Arabidopsis thaliana UDP-galactos/UDP-glucose transporter. J Biol Chem. 2002;277(36):32923–32929. doi: 10.1074/jbc.M204081200. [DOI] [PubMed] [Google Scholar]
- 16.Norambuena L, et al. AtUTr2 is an Arabidopsis thaliana nucleotide sugar transporter located in the Golgi apparatus capable of transporting UDP-galactose. Planta. 2005;222(3):521–529. doi: 10.1007/s00425-005-1557-x. [DOI] [PubMed] [Google Scholar]
- 17.Rollwitz I, Santaella M, Hille D, Flügge UI, Fischer K. Characterization of AtNST-KT1, a novel UDP-galactose transporter from Arabidopsis thaliana. FEBS Lett. 2006;580(17):4246–4251. doi: 10.1016/j.febslet.2006.06.082. [DOI] [PubMed] [Google Scholar]
- 18.Seino J, et al. Characterization of rice nucleotide sugar transporters capable of transporting UDP-galactose and UDP-glucose. J Biochem. 2010;148(1):35–46. doi: 10.1093/jb/mvq031. [DOI] [PubMed] [Google Scholar]
- 19.O’Neill MA, Eberhard S, Albersheim P, Darvill AG. Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science. 2001;294(5543):846–849. doi: 10.1126/science.1062319. [DOI] [PubMed] [Google Scholar]
- 20.Song Z. Roles of the nucleotide sugar transporters (SLC35 family) in health and disease. Mol Aspects Med. 2013;34(2-3):590–600. doi: 10.1016/j.mam.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Hiraoka S, et al. Nucleotide-sugar transporter SLC35D1 is critical to chondroitin sulfate synthesis in cartilage and skeletal development in mouse and human. Nat Med. 2007;13(11):1363–1367. doi: 10.1038/nm1655. [DOI] [PubMed] [Google Scholar]
- 22.Berninsone PM, Hirschberg CB. Nucleotide sugar transporters of the Golgi apparatus. Curr Opin Struct Biol. 2000;10(5):542–547. doi: 10.1016/s0959-440x(00)00128-7. [DOI] [PubMed] [Google Scholar]
- 23.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37(5):501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 24.Strasser R, et al. Molecular cloning and characterization of Arabidopsis thaliana Golgi alpha-mannosidase II, a key enzyme in the formation of complex N-glycans in plants. Plant J. 2006;45(5):789–803. doi: 10.1111/j.1365-313X.2005.02648.x. [DOI] [PubMed] [Google Scholar]
- 25.Reisen D, Marty F, Leborgne-Castel N. New insights into the tonoplast architecture of plant vacuoles and vacuolar dynamics during osmotic stress. BMC Plant Biol. 2005;5:13. doi: 10.1186/1471-2229-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lerouxel O, et al. Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiol. 2002;130(4):1754–1763. doi: 10.1104/pp.011965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pauly M, et al. A xyloglucan-specific endo-beta-1,4-glucanase from Aspergillus aculeatus: Expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology. 1999;9(1):93–100. doi: 10.1093/glycob/9.1.93. [DOI] [PubMed] [Google Scholar]
- 28.Le BH, et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA. 2010;107(18):8063–8070. doi: 10.1073/pnas.1003530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pattathil S, et al. A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 2010;153(2):514–525. doi: 10.1104/pp.109.151985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goubet F, Dupree P, Johansen KS. Carbohydrate gel electrophoresis. Methods Mol Biol. 2011;715:81–92. doi: 10.1007/978-1-61779-008-9_5. [DOI] [PubMed] [Google Scholar]
- 31.Takashima S, et al. Analysis of CMP-sialic acid transporter-like proteins in plants. Phytochemistry. 2009;70(17-18):1973–1981. doi: 10.1016/j.phytochem.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 32.Lehle L, Strahl S, Tanner W. Protein glycosylation, conserved from yeast to man: A model organism helps elucidate congenital human diseases. Angew Chem Int Ed Engl. 2006;45(41):6802–6818. doi: 10.1002/anie.200601645. [DOI] [PubMed] [Google Scholar]
- 33.Hanke G, Bowsher C, Jones MN, Tetlow I, Emes M. Proteoliposomes and plant transport proteins. J Exp Bot. 1999;50(341):1715–1726. [Google Scholar]
- 34.Pabst M, et al. Nucleotide and nucleotide sugar analysis by liquid chromatography-electrospray ionization-mass spectrometry on surface-conditioned porous graphitic carbon. Anal Chem. 2010;82(23):9782–9788. doi: 10.1021/ac101975k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liwanag AJ, et al. Pectin biosynthesis: GALS1 in Arabidopsis thaliana is a β-1,4-galactan β-1,4-galactosyltransferase. Plant Cell. 2012;24(12):5024–5036. doi: 10.1105/tpc.112.106625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, et al. Overexpression of a cytosol-localized rhamnose biosynthesis protein encoded by Arabidopsis RHM1 gene increases rhamnose content in cell wall. Plant Physiol Biochem. 2009;47(2):86–93. doi: 10.1016/j.plaphy.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 37.Toma L, Pinhal MA, Dietrich CP, Nader HB, Hirschberg CB. Transport of UDP-galactose into the Golgi lumen regulates the biosynthesis of proteoglycans. J Biol Chem. 1996;271(7):3897–3901. doi: 10.1074/jbc.271.7.3897. [DOI] [PubMed] [Google Scholar]
- 38.Seifert GJ. Nucleotide sugar interconversions and cell wall biosynthesis: How to bring the inside to the outside. Curr Opin Plant Biol. 2004;7(3):277–284. doi: 10.1016/j.pbi.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Major LL, Wolucka BA, Naismith JH. Structure and function of GDP-mannose-3′,5′-epimerase: An enzyme which performs three chemical reactions at the same active site. J Am Chem Soc. 2005;127(51):18309–18320. doi: 10.1021/ja056490i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rautengarten C, et al. The interconversion of UDP-arabinopyranose and UDP-arabinofuranose is indispensable for plant development in Arabidopsis. Plant Cell. 2011;23(4):1373–1390. doi: 10.1105/tpc.111.083931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reiter WD, Chapple CC, Somerville CR. Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science. 1993;261(5124):1032–1035. doi: 10.1126/science.261.5124.1032. [DOI] [PubMed] [Google Scholar]
- 42.Lanfermeijer FC, Venema K, Palmgren MG. Purification of a histidine-tagged plant plasma membrane H+-ATPase expressed in yeast. Protein Expr Purif. 1998;12(1):29–37. doi: 10.1006/prep.1997.0788. [DOI] [PubMed] [Google Scholar]
- 43.Arrivault S, et al. Use of reverse-phase liquid chromatography, linked to tandem mass spectrometry, to profile the Calvin cycle and other metabolic intermediates in Arabidopsis rosettes at different carbon dioxide concentrations. Plant J. 2009;59(5):826–839. doi: 10.1111/j.1365-313X.2009.03902.x. [DOI] [PubMed] [Google Scholar]
- 44.Ito J, et al. Analysis of plant nucleotide sugars by hydrophilic interaction liquid chromatography and tandem mass spectrometry. Anal Biochem. 2014;448:14–22. doi: 10.1016/j.ab.2013.11.026. [DOI] [PubMed] [Google Scholar]
- 45.Harholt J, et al. ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol. 2006;140(1):49–58. doi: 10.1104/pp.105.072744. [DOI] [PMC free article] [PubMed] [Google Scholar]