Significance
Plants sense long-wavelength and low-intensity UV-B light as a signal for the induction of photomorphogenesis. Recently, a number of regulatory factors involved in this developmental process have been uncovered through the use of conventional genetic and molecular approaches. A systematic understanding of the whole photomorphogenic UV-B signaling network, however, has not been achieved to date. Thus, in our study, we use a combination of mathematical modeling and biological experiments to capture the plausible mechanisms coordinating the different pathways in UV-B light signal transduction, which are difficult to define accurately in traditional wet laboratory studies.
Keywords: light signaling, ordinary differential equation
Abstract
Long-wavelength and low-fluence UV-B light is an informational signal known to induce photomorphogenic development in plants. Using the model plant Arabidopsis thaliana, a variety of factors involved in UV-B–specific signaling have been experimentally characterized over the past decade, including the UV-B light receptor UV resistance locus 8; the positive regulators constitutive photomorphogenesis 1 and elongated hypocotyl 5; and the negative regulators cullin4, repressor of UV-B photomorphogenesis 1 (RUP1), and RUP2. Individual genetic and molecular studies have revealed that these proteins function in either positive or negative regulatory capacities for the sufficient and balanced transduction of photomorphogenic UV-B signal. Less is known, however, regarding how these signaling events are systematically linked. In our study, we use a systems biology approach to investigate the dynamic behaviors and correlations of multiple signaling components involved in Arabidopsis UV-B–induced photomorphogenesis. We define a mathematical representation of photomorphogenic UV-B signaling at a temporal scale. Supplemented with experimental validation, our computational modeling demonstrates the functional interaction that occurs among different protein complexes in early and prolonged response to photomorphogenic UV-B.
Among natural environmental factors, light is an informational cue of paramount significance for plant life. Seedling photomorphogenesis is one of the earliest responses to light in higher plants. Physically, this developmental response is featured by short hypocotyls, open and expanded cotyledons, and the maturation of green chloroplasts for photosynthesis. At the mechanistic level, this process is regulated by intricate molecular signaling networks from light input to gene expression output. In addition to far-red, red, and blue/UV-A light, UV-B light (280–315 nm) of long wavelength and low intensity has also been found to direct photomorphogenic development in various plant species. Seedlings that fail to develop normal UV-B–induced photomorphogenesis suffer from the defects in hypocotyl growth, flavonoid accumulation, and stress tolerance (1–4).
Arabidopsis thaliana UV resistance locus 8 (UVR8) is a recently identified chromoprotein that detects UV-B light using its tryptophan chromophores. Upon perception of UV-B light, UVR8 is activated by undergoing a dimer-to-monomer conformational change (5–7). The monomerization of UVR8 allows it to associate with the tetrameric constitutively photomorphogenic 1-suppressor of PHYA (COP1-SPA) core complex (8). This association, in turn, drives the release of COP1-SPA from the cullin 4-damaged DNA-binding protein 1 (CUL4-DDB1) E3 apparatus that represses light signaling under various light conditions (8, 9). The UV-B–dependent UVR8–COP1-SPA complex then acts as a molecular switch to turn on downstream signaling, including positively modulating the stability and activity of a central photomorphogenesis-promoting transcription factor, elongated hypocotyl 5 (HY5) (8). As a feedback response, HY5 activates the COP1 transcription, whereas another transcription factor, far-red elongated hypocotyl 3 (FHY3), functions genetically upstream of both COP1 and HY5 to directly activate COP1 (10). Taken together, these results have largely shaped our current understanding of the positive signaling framework of UV-B–induced photomorphogenesis.
In addition to these positive regulators of UV-B–induced photomorphogenesis, several negative regulators of this UV-B–specific pathway have also been uncovered in recent years. For example, two WD40-repeat proteins, repressor of UV-B photomorphogenesis 1 (RUP1) and RUP2, are accumulated in a UV-B–induced manner. Genetically downstream of UVR8 and COP1, RUP1 and RUP2 are capable of mediating the redimerization of UVR8 monomers through their physical interaction with UVR8, which forms a negative feedback on UVR8-dependent UV-B signaling (11, 12). A B-box protein, salt tolerance (STO), originally identified as being involved in plant salt tolerance, has been found to impinge HY5 in the transcriptional activation of the UV-B–inducible genes (13). CUL4, a scaffold protein of multimeric cullin-ring E3 ligases (CRL), has been recently characterized to play a negative role in this pathway, based on the elevated anthocyanin accumulation, the enhanced UV-B–responsive gene expression, and the increased HY5 protein abundance in UV-B–treated cul4 mutant seedlings (8). Therefore, the various functions of these negative regulators enable a multilevel guarantee of a balanced photomorphogenic UV-B signaling.
Although these positive and negative regulatory mechanisms have been accumulated by a variety of genetic and molecular studies, no information to date has systematically presented how multiple signaling events take place and correlate with one another over the course of photomorphogenic UV-B signal transduction. Here, we develop an ordinary differential equation (ODE)-based mathematical framework with which to investigate the photomorphogenic UV-B signaling at the level of the whole signaling network. More importantly, we decipher the dynamic correlations among multiple signaling factors in the shape of protein complexes in response to photomorphogenic UV-B light.
Results and Discussion
Building a Mathematical Model of a Photomorphogenic UV-B Signaling Network.
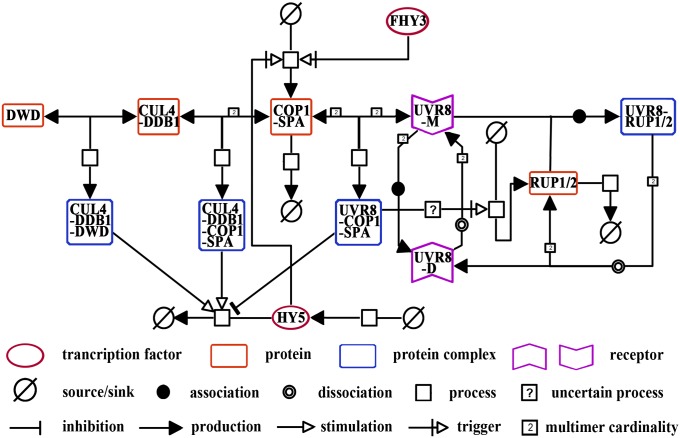
To investigate the positive and negative regulation in UV-B–induced photomorphogenesis, we first constructed a signaling network based on published experimental data (Fig. S1). As HY5 is a hub transcription factor to promote UV-B–responsive transcriptomic programming (14, 15), we introduced the following primary signaling events that directly or indirectly regulate the protein abundance of HY5 into this network: in response to photomorphogenic UV-B light, (i) the dimer-to-monomer switch of UVR8 (5–7), (ii) the UVR8–COP1 direct interaction (7, 16), (iii) the UVR8–COP1-SPA association and the CUL4-DDB1–COP1-SPA dissociation (8), (iv) HY5 stabilization by the UVR8–COP1-SPA complex (8), and (v) the UVR8–RUP1/RUP2 direct interaction (12, 17); in response to the elimination of photomorphogenic UV-B light, (i) UVR8 dimerization mediated by RUPs (11, 18), (ii) the UVR8–COP1-SPA dissociation and the CUL4-DDB1–COP1-SPA association (8), and (iii) HY5 degradation by the CUL4-DDB1–COP1-SPA complex (9). In addition, we postulated the following regulatory action toward HY5, according to the indication derived from previous results. Under photomorphogenic UV-B, overexpressed HY5 protein was found to be unstable in cop1-4, and this process could be restored by the treatment of proteasome inhibitor MG132 (8). This result has indicated that there exists an E3 ligase other than COP1, the known E3 ligase for HY5 in darkness (19), to prevent the overaccumulation of HY5 under photomorphogenic UV-B. Meanwhile, CUL4 has been shown to take a negative part in UV-B–induced photomorphogenesis. Specifically, more HY5 proteins, rather than HY5 transcripts, were observed in cul4cs, the cosuppression mutant of CUL4, than in wild type. Recombinant HY5 protein was stabilized in the cell extracts of UV-B–treated cul4cs seedlings in vitro (8). Thus, this CUL4-based E3 ligase is likely to act as one, if not the only, machinery responsible for the degradation of HY5. In most cases, CUL4 recruits targets via an adaptor protein, DDB1, and substrate receptors such as DDB1 binding WD40 (DWD) proteins (20, 21). Therefore, we postulated that CUL4-DDB1 works in concert with an unidentified DWD protein to degrade HY5 under photomorphogenic UV-B. In addition to the above regulation that relied on protein–protein interactions, we also included the following photomorphogenic UV-B–dependent transcriptional regulation: (i) the activation of COP1 by FHY3 (10), (ii) the positive feedback of HY5 on COP1 (10), (iii) RUP1/2 activation downstream of the COP1–UVR8 module (12), and (iv) relatively steady expression of UVR8 and CUL4 (Fig. S2). We then constructed a system of ODEs to describe each of these signaling events. Consequently, a scheme of the full model reactions was set up in the Systems Biology Graphical Notation (SBGN) format (Fig. 1).
Fig. 1.
The reaction scheme of the photomorphogenic UV-B signaling network. Entity pool nodes in colors and shapes represent functional entities involved in UV-B–induced photomorphogenesis, including transcription factors, single proteins, protein complexes, photoreceptors, and their source and sink. Process nodes describe the way in which functional entities are transformed into different states, namely the association and dissociation of protein complexes and identified and uncertain processes. Connecting arcs indicate the effects of inhibition, production, stimulation, and trigger on processes exerted by functional entities. Multimer cardinality shows that several copies of the same function entities are involved in the process.
Dynamics of UVR8 Monomerization.
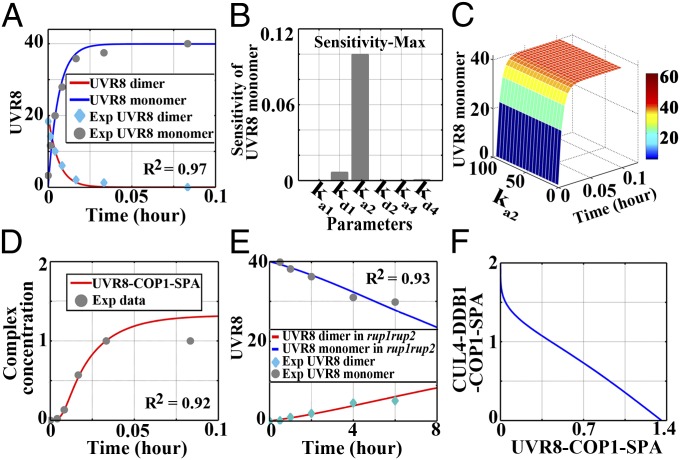
Upon UV-B light irradiation, UVR8 monomerization occurs at a rapid rate (7). This process reverses, however, when the source of irradiation is removed (11, 18). To determine the kinetics of the UV-B–induced UVR8 monomerization, we simulated the dimer-to-monomer switch of UVR8 plotted against published experimental data based on submodel 1 (Fig. 2A and Fig. S3A). The in vivo association between UVR8 and COP1 was detectable within 1 min of UV-B irradiation (7). It has likewise been suggested that both the UVR8 monomerization and the formation of the UVR8–COP1-SPA complex take place rapidly in an early response to UV-B. However, it is unclear whether the formation of the UVR8–COP1-SPA complex facilitates or hinders the process of UVR8 monomerization. Thus, we performed a sensitivity analysis to investigate the influence exerted by the association/dissociation of the UVR8–COP1-SPA complex (with the rate constant ka1/kd1), the CUL4-DDB1–COP1-SPA complex (with the rate constant ka2/kd2), and the CUL4-DDB1–DWD complex (with the rate constant ka4/kd4) on UVR8 monomerization. For each of these six parameters, we observed a low sensitivity of no more than 0.12, with respect to the abundance of UVR8 monomers (Fig. 2B). Furthermore, we scanned the parameter space ranging from 0 to 100 for ka2 and found that the production of UVR8 monomers is insensitive to ka2 variation under UV-B (Fig. 2C). This result thus mathematically substantiates the notion that UVR8 is strongly and quickly photoactivated even when plants are subjected to variable light environments that induce alternations in protein complex formation.
Fig. 2.
Dynamic changes in UVR8 conformation and COP1-containing complexes. (A) Simulation of UVR8 monomerization in response to photomorphogenic UV-B in wild-type seedlings. Exp, experiment. (B) Parameter sensitivity of the dynamic constants with respect to the concentration of UVR8 monomers. The parameters indicate the association/dissociation rate of the UVR8–COP1-SPA complex (with the rate constant ka1/kd1), the CUL4-DDB1–COP1-SPA complex (with the rate constant ka2/kd2), and the CUL4-DDB1–DWD complex (with the rate constant ka4/kd4). (C) The effect of the dissociation of the UVR8–COP1-SPA complex (with the rate constant ka2) on UVR8 monomerization. (D) Simulation of the formation of the UVR8–COP1-SPA complex in response to photomorphogenic UV-B in wild-type seedlings. (E) Simulation of UVR8 redimerization in rup1 rup2 seedlings during the transition from +UV-B to white light. (F) The correlation of the concentrations of the UVR8–COP1-SPA complex and the CUL4-DDB1–COP1-SPA complex.
The UVR8–COP1-SPA Association at the Initiation of Photomorphogenic UV-B Signaling.
To study the UV-B–responsive complex organization in our model, we simulated the association of the UVR8–COP1-SPA complex and the dissociation of the CUL4-DDB1–COP1-SPA complex. First, a systematic scan of the parameter space was performed to reproduce the following two sets of published experimental data: (i) the kinetics of the UVR8–COP1 association at the very beginning of UV-B treatment (7), as shown in submodel 1 (Fig. S3A and Fig. 2D), and (ii) the kinetics of UVR8 redimerization in rup1 rup2 recovered under white light from UV-B treatment (11), as shown in submodel 2 (Fig. S3B and Fig. 2E). Second, to constrain the model parameters, a qualitative scan was used to include the input experimental condition that the steady amount of COP1-SPA associating with CUL4-DDB1 under UV-B is ∼5% of that without UV-B (8). This simulation showed that there was a negative correlation between the accumulation of the CUL4-DDB1–COP1-SPA complex and that of the UVR8–COP1-SPA complex under photomorphogenic UV-B. Namely, the CUL4-DDB1–COP1-SPA concentration was reduced whereas the UVR8–COP1-SPA concentration was increased with the photomorphogenic UV-B treatment (Fig. 2F). It was difficult to determine these rapid changes accurately using in vivo coimmunoprecipitation assays in previous studies, due to the fact that whereas the experimental manipulations take hours, the endogenous molecular responses to light happen in seconds. Our computational modeling suggests, however, that at the very early stage of photomorphogenic UV-B signaling when COP1 has not been intensively induced yet, there is only a low level of free COP1-SPA to associate with monomerized UVR8, and thus CUL4-DDB1–COP1-SPA tends to dissociate to release COP1-SPA for UVR8 binding.
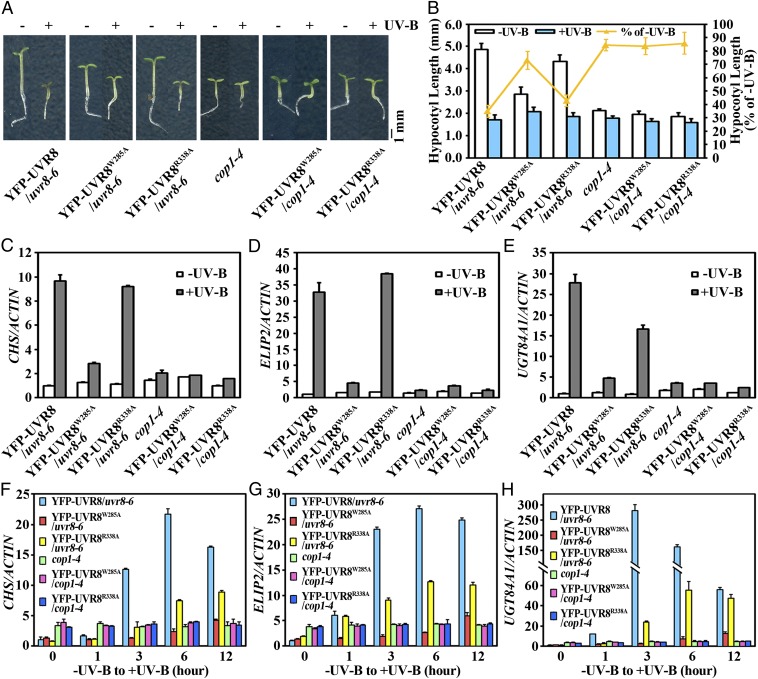
To further study how COP1 contributes to photomorphogenic UV-B signaling, we examined the effect on seedling photomorphogenesis when COP1 was loss-of-function mutated but UVR8 was constitutively active. One of our recent studies has engineered two monomeric variants of Arabidopsis UVR8, UVR8W285A and UVR8R338A. These two variants have been shown to both exist in a constant monomeric state and induce constitutive photomorphogenesis under UV-B (8, 17, 22). When introduced into the cop1-4 background that is barely sensitive to photomorphogenic UV-B, these two variants phenocopied cop1-4 by exhibiting obviously reduced sensitivity in UV-B–induced hypocotyl shortening (Fig. 3 A and B). Molecularly, compared with their –UV-B–grown counterparts, 4-d-old +UV-B–grown YFP-UVR8W285A/cop1-4 and YFP-UVR8R338A/cop1-4 resembled cop1-4 in displaying impaired transcript accumulation of three typical UV-B–inducible genes, chalcone synthase (CHS), early light-inducible protein 2 (ELIP2), and UDP-glycosyltransferase 84A1 (UGT84A1) (Fig. 3 C–E). Moreover, YFP-UVR8W285A/uvr8-6 and YFP-UVR8R338A/uvr8-6 showed UV-B–induced expression of these genes upon a short-term exposure to photomorphogenic UV-B, although at a retarded and reduced level compared to YFP-UVR8/uvr8-6. However, similarly to cop1-4, YFP-UVR8W285A/cop1-4 and YFP-UVR8R338A/cop1-4 suffered from completely abolished induction of these genes (Fig. 3 F–H). Therefore, COP1 is required for both prolonged and early responses to photomorphogenic UV-B. Together with the fact that UVR8–COP1-SPA associates in quick succession after UVR8 monomerization (Fig. 2 A and D), these observations indicate that COP1-centered COP1-SPA acts as an essential chaperone for UVR8 in UVR8-initiated photomorphogenic UV-B signaling.
Fig. 3.
COP1 is essential for active photomorphogenic UV-B signaling. (A) Phenotypes of 4-d-old seedlings of transgenic UVR8 variant lines grown under –UV-B and +UV-B. (B) Hypocotyl length of the seedlings shown in A. Data are shown as mean ± SD; n > 30. (C–E) qRT-PCR analysis of UV-B–responsive gene expression of CHS (C), ELIP2 (D), and UGT84A1 (E) in 4-d-old seedlings grown under –UV-B and +UV-B. Data are shown as mean ± SD; n = 3. (F–H) qRT-PCR analysis of UV-B–responsive gene expression of CHS (F), ELIP2 (G), and UGT84A1 (H) in 4-d-old seedlings grown under –UV-B and then transferred to +UV-B for the indicated time period. Data are shown as mean ± SD; n = 3.
The RUP1/RUP2 Molecular Brake in Photomorphogenic UV-B Signaling.
Given their role in assisting the redimerization of UVR8 following the removal of UV-B irradiation, RUP1 and RUP2 have been identified as negative regulators of UV-B–induced photomorphogenesis (11, 12). However, the exact functional form of RUP1 and RUP2 under UV-B remains unclear.
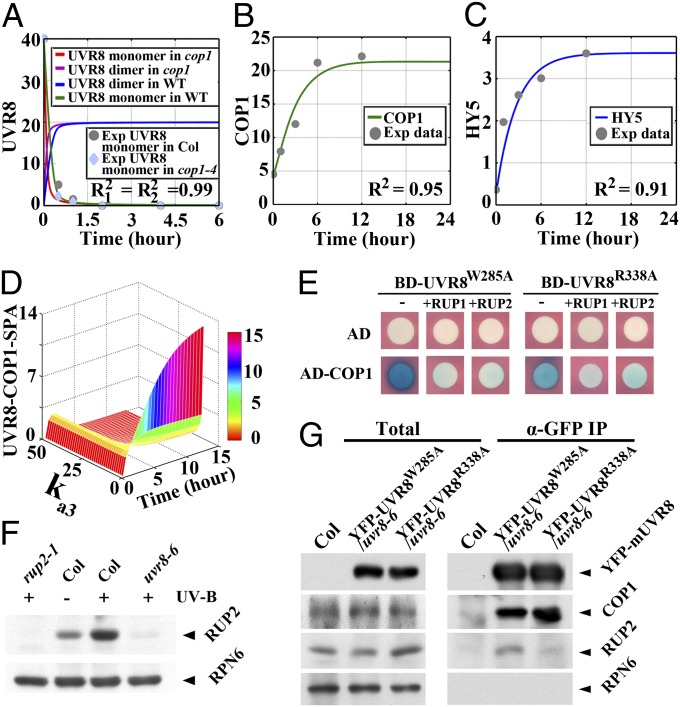
To parameterize the function of RUP1 and RUP2 under photomorphogenic UV-B, we started with a model simulation of RUP1 and RUP2’s role in the absence of UV-B. As RUP1 and RUP2 share similar protein sequence, expression pattern, and potentially regulatory function (12), these two proteins were represented together by a single variable in our model. First, the profile of UVR8 redimerization was computed over the course of white-light recovery from UV-B in wild type and cop1-4, as outlined in submodel 3 and submodel 4, respectively (Fig. 4A and Fig. S3 C and D). Second, the protein accumulation of COP1 and HY5 has been shown to be induced by photomorphogenic UV-B (8, 10). Consequently, we simulated the accumulation kinetics of these two proteins in response to photomorphogenic UV-B (Fig. 4 B and C), according to submodel 5 (Fig. S3E). Third, based on the outcomes of the above simulations, the association between RUP1/RUP2 and UVR8 (with the rate constant ka3) was found to affect the formation of the UVR8–COP1-SPA complex in a time- and dose-dependent manner (Fig. 4D). In the first few hours of UV-B irradiation, the UVR8–COP1-SPA complex continued to accumulate as a result of the UV-B signaling initiation, which then induced RUP1/RUP2. As the amount of RUP1/RUP2 increased, the abundance of the UVR8–COP1-SPA complex was reduced. Specifically, when there was no UVR8–RUP1/RUP2 interaction (ka3 = 0), the UVR8–COP1-SPA complex exhibited a constitutive association, which is in line with the previous observations (11). Therefore, our model outlines RUP1/RUP2’s role in controlling the progression of UV-B signaling at the photoreceptor level via differential regulation of the UVR8–COP1-SPA homeostasis in real time.
Fig. 4.
Negative regulation of photomorphogenic UV-B signaling by RUP1 and RUP2. (A) Simulation of UVR8 redimerization in wild-type and cop1-4 seedlings during the transition from +UV-B to white light. Exp, experiment. (B and C) Simulations of COP1 (B) and HY5 (C) protein accumulation in wild-type seedlings during the transition from –UV-B to +UV-B. (D) The effect of the UVR8–RUP1/RUP2 association (with the rate constant ka3) on the formation of the UVR8–COP1-SPA complex in wild-type seedlings during the transition from –UV-B to +UV-B. (E) The effects of RUP1 and RUP2 on the COP1–UVR8 interaction in yeast. (F) Immunoblot assay of RUP2 protein in 4-d-old seedlings grown under –UV-B or +UV-B. Anti-RPN6 was used as a loading control. (G) In vivo co-IP assays showing the association of UVR8 variants with COP1 and RUP2 in 4-d-old seedlings grown under +UV-B. Anti-RPN6 was used as an unimmunoprecipitated and loading control.
UVR8W285A and UVR8R338A have been shown to strongly interact with COP1 in yeast and Arabidopsis (17, 22). Similarly, these two proteins have also been shown to physically interact with RUP1 and RUP2 in yeast (17). Thus, it is possible that RUP1 and RUP2 might regulate the UVR8–COP1-SPA complex by directly modulating its formation. As expected, when RUP1 and RUP2 were introduced into our yeast two-hybrid system, we found that the interaction between COP1 and UVR8 variants was obviously attenuated (Fig. 4E). In Arabidopsis, we detected much more endogenous RUP2 proteins in 4-d-old wild-type seedlings grown under +UV-B than in either the wild-type counterparts grown under –UV-B or the uvr8-6 counterparts grown under +UV-B (Fig. 4F). It is suggested that RUP2 protein accumulation and probably in turn its function are dependent on photomorphogenic UV-B signaling. Further, in 4-d-old seedlings grown under +UV-B, YFP-UVR8W285A was found to coimmunoprecipitate less COP1 and more RUP2 than YFP-UVR8R338A (Fig. 4G). Collectively, these results demonstrate that the UVR8–RUP1/RUP2 affinity antagonizes the UVR8–COP1 affinity that determines the level of UV-B–induced photomorphogenesis. They experimentally verify the claim that the UVR8–RUP1/RUP2 association and the UVR8–COP1 association have a negative correlation at a balanced state of the photomorphogenic UV-B signaling. Therefore, the RUP1/RUP2-directed molecular brake of UV-B–induced photomorphogenesis is defined not only by its promotion of UVR8 redimerization at the removal of UV-B light, but also by its dynamic control of the UVR8–COP1-SPA abundance under photomorphogenic UV-B.
Regulation of HY5 by Photomorphogenic UV-B Signaling.
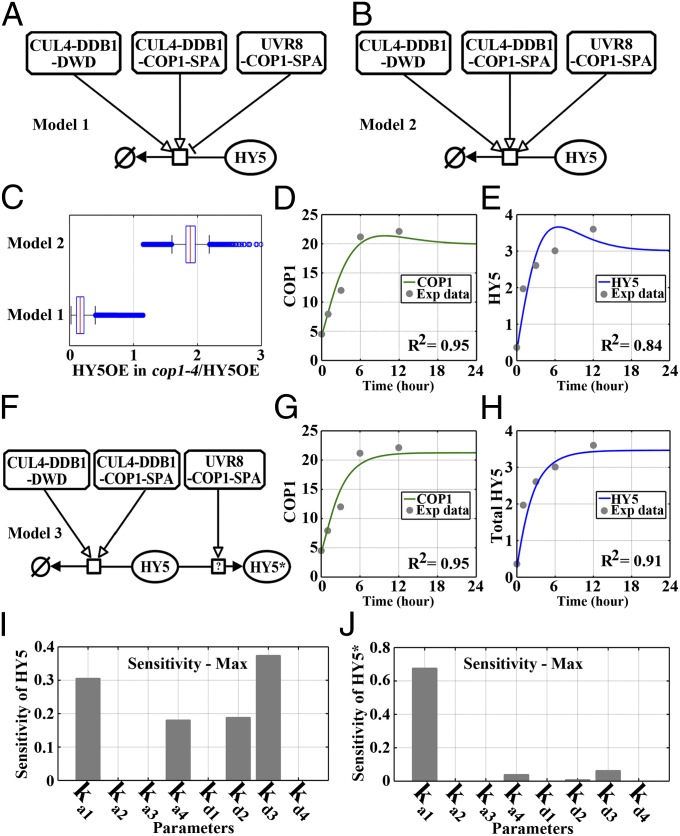
Whereas COP1 is a known repressor in the photomorphogenesis triggered by far-red, red, and blue/UV-A light, it functions as a promoter of UV-B–induced photomorphogenesis through its participation in the UVR8–COP1-SPA complex. It is noteworthy that COP1 antagonizes HY5 in darkness whereas both COP1 and HY5 positively regulate UV-B–induced photomorphogenesis. To computationally illustrate the regulatory mechanism of HY5 protein accumulation, we hypothesized two models: one in which HY5 degradation is promoted by CUL4-DDB1–DWD and CUL4-DDB1–COP1-SPA, but repressed by UVR8–COP1-SPA (Fig. 5A), and another in which HY5 degradation is promoted by CUL4-DDB1–DWD, CUL4-DDB1–COP1-SPA, and UVR8–COP1-SPA (Fig. 5B). Model 2, however, failed to simulate the impaired accumulation of overexpressed HY5 (HY5OE) that has been experimentally observed following the loss of COP1 function under photomorphogenic UV-B (8) (Fig. 5C).
Fig. 5.
The regulation of HY5 by photomorphogenic UV-B signaling. (A and B) Models 1 and 2 showing two possibilities for the regulation of HY5 by the UVR8–COP1-SPA complex. (C) Simulations of the accumulation of overexpressed HY5 (HY5OE) proteins in wild-type and cop1-4 seedlings based on the models shown in A and B. (D and E) Simulations of COP1 (D) and HY5 (E) protein accumulation in wild-type seedlings during the transition from –UV-B to +UV-B based on model 1. Exp, experiment. (F) Model 3 showing the possibility of a stabilized form of HY5, HY5*, promoted by the UVR8–COP1-SPA complex. (G and H) Simulations of COP1 (G) and HY5 (H) protein accumulation in wild-type seedlings during the transition from –UV-B to +UV-B based on model 3. (I and J) Parameter sensitivity of the dynamic constants with respect to the concentrations of HY5 (I) and HY5* (J). The parameters indicate the association/dissociation rate of the UVR8–COP1-SPA complex (with the rate constant ka1/kd1), the CUL4-DDB1–COP1-SPA complex (with the rate constant ka2/kd2), the UVR8–RUP1/RUP2 complex (with the rate constant ka3/kd3), and the CUL4-DDB1–DWD complex (with the rate constant ka4/kd4).
Next, the regulation of HY5 by these three complexes, as depicted in model 1, was integrated into our simulation of the time-course accumulation of COP1 and HY5 under photomorphogenic UV-B. Intriguingly, we found that the protein levels of both COP1 and HY5 obviously fell back, suggesting a robust negative feedback regulation right after the sufficient initiation of photomorphogenic UV-B signaling. Likewise, because HY5 is susceptible to the ubiquitin-proteasome–mediated degradation in cop1-4 under photomorphogenic UV-B (8) (Fig. 5C), it is possible that the UVR8–COP1-SPA complex is directly involved in the production of a stabilized form of HY5, HY5*, as shown in model 3 (Fig. 5F). The possibility of HY5* was found to cause a plateau in its protein accumulation with an increased fitting accuracy (Fig. 5 G and H), which suggests that a balance between the positive and negative signaling branches of photomorphogenic UV-B signaling is achieved at a fairly late stage. Based on this simulation, we next performed sensitivity analysis to examine the effects that the organization of protein complexes exhibited on the abundance of HY5 and HY5*. HY5 was found to be sensitive to a wide range of parameters, including the UVR8–COP1-SPA association (with the rate constant ka1), the CUL4-DDB1–DWD association (with the rate constant ka4), the CUL4-DDB1–COP1-SPA dissociation (with the rate constant kd2), and the UVR8–RUP1/RUP2 dissociation (with the rate constant kd3). In contrast, HY5* was found to be influenced principally by the formation of the pivotal positive molecular switch, the UVR8–COP1-SPA complex (with the rate constant ka1). Taken together, as a signaling center for downstream transcriptomic programming, HY5 is subjected to multilayer regulation governed by distinct protein complexes. HY5 stability is indispensable for seedling photomorphogenesis triggered by a variety of light conditions. On one hand, under far-red and visible light, the repressive role of COP1 is relieved by its association with phytochromes and cryptochromes and its nuclear exclusion. Dependent on these derepression mechanisms, HY5 is prevented from proteolysis to accumulate (23, 24). HY5 phosphorylation has also been found to promote its stability but disturb its transcription factor activity (25). On the other hand, in the UV-B–induced photomorphogenesis that is independent of phytochromes and cryptochromes (10, 26), both UVR8 and COP1 are positive regulators and colocalize in the nucleus (16). The UV-B–induced organization of the UVR8–COP1-SPA complex enables the stabilization of HY5 (8). The production of the possible stabilized form of HY5, HY5*, might be associated with posttranslational regulation of HY5, e.g., protein modification, the inhibition of proteolysis, and protein complex alteration.
In conclusion, we integrated computational and experimental approaches to conduct a systematic study of UV-B–induced photomorphogenesis in Arabidopsis. We have established a data-consistent mathematical model of photomorphogenic UV-B signaling that ranges from UVR8 monomerization to transcriptional feedback regulation. Thus, a quantitative understanding has been gained in our study for the time evolution of the regulatory behaviors of key signaling factors, including the molecular switch, the UVR8–COP1-SPA module, and the molecular brake, RUP1/RUP2. In future mechanistic explorations, it will be interesting to optimize and extend this model by including the effects of various light environments and the crosstalk of light with other environmental stimuli.
Materials and Methods
Computational Method.
The methods to build mathematical models are described in detail in SI Text.
Growth Conditions.
The Arabidopsis seeds were surface sterilized and sown on solid Murashige and Skoog medium supplemented with 1% sucrose for biochemical assays or with 0.3% sucrose for phenotypic analysis and cold treated at 4 °C for 4 d. Then for photomorphogenic UV-B treatment, seedlings were grown at 22 °C under continuous white light (3 μmol·m−2·s−1, measured by LI-250 Light Meter; LI-COR Biosciences) supplemented with Philips TL20W/01RS narrowband UV-B tubes (1.5 μmol·m−2·s−1, measured by TN-340 UV-B Light Meter) under a 350-nm cutoff (half-maximal transmission at 350 nm) filter, ZUL0350 (−UV-B; Asahi Spectra), or a 300-nm cutoff (half-maximal transmission at 300 nm) filter, ZUL0300 (+UV-B; Asahi Spectra).
Hypocotyl Measurement.
Hypocotyl length was measured as previously described (17). For each line grown under −UV-B or +UV-B for 4 d, hypocotyl length was analyzed in three biological replicates. In each replicate, at least 30 Arabidopsis seedlings were measured. The relative hypocotyl length was presented as the percentage of the hypocotyl length under +UV-B with respect to that under −UV-B (percentage of −UV-B).
Quantitative Real-Time PCR.
Total RNA was extracted from 4-d-old Arabidopsis seedlings grown under –UV-B or +UV-B, using the RNeasy plant mini kit (QIAGEN). Reverse transcription was performed using the SuperScript II first-strand cDNA synthesis system (Invitrogen) according to the manufacturer’s instructions. Real-time qPCR analysis was performed using SYBR Premix Ex Taq (Takara) with the Applied Biosystems 7500 Real-Time PCR System. Each experiment was repeated with three independent samples, and RT-PCR reactions were performed in three technical replicates for each sample. The primers are listed in Table S1.
Yeast Three-Hybrid Assay.
The LexA-based yeast two-hybrid system (Clontech) was modified by introducing a third plasmid expressing RUP1 or RUP2. To express RUP1 or RUP2 without any domain fusion in yeast, a KpnI/EcoRI fragment containing the full-length ORF of RUP1 or RUP2 was cloned into pGADT7 (Clontech). The respective combinations were cotransformed into the yeast strain EGY48 (Clontech) containing the reporter plasmid p8op::LacZ. Transformants were grown on proper dropout plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) for blue color development.
Coimmunoprecipitation Assays and Immunoblot Analysis.
Coimmunoprecipitation assays were performed as previously described (8). Total proteins were extracted from 4-d-old Arabidopsis seedlings in protein extraction buffer containing 50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% Tween 20, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1× complete protease inhibitor mixture (Roche). The extracts were incubated with anti-GFP antibodies (Invitrogen) coupled with Dynabeads Protein G (Invitrogen) for 3 h at 4 °C under the same conditions (−UV-B or +UV-B) as those where the seedlings were grown. Then the Dynabeads was washed and eluted before immunoblot analysis. Primary antibodies used in this study were anti-GFP (Invitrogen), anti-COP1 (27), and anti-RPN6 (27). The recombinant protein 6× His-tagged RUP2 was expressed and purified to generate the polyclonal antibodies against RUP2 in a rabbit.
Supplementary Material
Acknowledgments
We thank Abigail Coplin for her critical reading of the manuscript. This work is supported by the National Natural Science Foundation [Grant 31330048 to (X.W.D.)], the National Basic Research Program of China [Grant 2012CB910900 (to X.W.D.)], the Natural Science Foundation (MCB-0929100), the National Institutes of Health [Grant GM47850 to (X.W.D.)], the China Postdoctoral Science Foundation [Grants 2012M510266 and 2013T60032 (to X.O.) and 2013M530009 (to X.H.)], the State Key Laboratory of Protein and Plant Gene Research at Peking University, and the Peking–Tsinghua Center for Life Sciences. X.O. and X.H. are supported by postdoctoral fellowships at the Peking–Tsinghua Center for Life Sciences.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412050111/-/DCSupplemental.
References
- 1.Jenkins GI. Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol. 2009;60:407–431. doi: 10.1146/annurev.arplant.59.032607.092953. [DOI] [PubMed] [Google Scholar]
- 2.Hectors K, Prinsen E, De Coen W, Jansen MA, Guisez Y. Arabidopsis thaliana plants acclimated to low dose rates of ultraviolet B radiation show specific changes in morphology and gene expression in the absence of stress symptoms. New Phytol. 2007;175(2):255–270. doi: 10.1111/j.1469-8137.2007.02092.x. [DOI] [PubMed] [Google Scholar]
- 3.Ulm R, Nagy F. Signalling and gene regulation in response to ultraviolet light. Curr Opin Plant Biol. 2005;8(5):477–482. doi: 10.1016/j.pbi.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Frohnmeyer H, Staiger D. Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol. 2003;133(4):1420–1428. doi: 10.1104/pp.103.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu D, et al. Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484(7393):214–219. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- 6.Christie JM, et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012;335(6075):1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rizzini L, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332(6025):103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 8.Huang X, et al. Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc Natl Acad Sci USA. 2013;110(41):16669–16674. doi: 10.1073/pnas.1316622110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H, et al. Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell. 2010;22(1):108–123. doi: 10.1105/tpc.109.065490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X, et al. Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell. 2012;24(11):4590–4606. doi: 10.1105/tpc.112.103994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heijde M, Ulm R. Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc Natl Acad Sci USA. 2013;110(3):1113–1118. doi: 10.1073/pnas.1214237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruber H, et al. Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci USA. 2010;107(46):20132–20137. doi: 10.1073/pnas.0914532107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang L, et al. Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res. 2012;22(6):1046–1057. doi: 10.1038/cr.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulm R, et al. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA. 2004;101(5):1397–1402. doi: 10.1073/pnas.0308044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown BA, et al. A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA. 2005;102(50):18225–18230. doi: 10.1073/pnas.0507187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Favory JJ, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009;28(5):591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang X, Yang P, Ouyang X, Chen L, Deng XW. Photoactivated UVR8-COP1 module determines photomorphogenic UV-B signaling output in Arabidopsis. PLoS Genet. 2014;10(3):e1004218. doi: 10.1371/journal.pgen.1004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heilmann M, Jenkins GI. Rapid reversion from monomer to dimer regenerates the ultraviolet-B photoreceptor UV RESISTANCE LOCUS8 in intact Arabidopsis plants. Plant Physiol. 2013;161(1):547–555. doi: 10.1104/pp.112.206805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405(6785):462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 20.Lee J, Zhou P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell. 2007;26(6):775–780. doi: 10.1016/j.molcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Lee JH, et al. Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell. 2008;20(1):152–167. doi: 10.1105/tpc.107.055418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heijde M, et al. Constitutively active UVR8 photoreceptor variant in Arabidopsis. Proc Natl Acad Sci USA. 2013;110(50):20326–20331. doi: 10.1073/pnas.1314336110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17(10):584–593. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Yi C, Deng XW. COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 2005;15(11):618–625. doi: 10.1016/j.tcb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Hardtke CS, et al. HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 2000;19(18):4997–5006. doi: 10.1093/emboj/19.18.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oravecz A, et al. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell. 2006;18(8):1975–1990. doi: 10.1105/tpc.105.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, et al. Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell. 2006;18(8):1991–2004. doi: 10.1105/tpc.106.043224. [DOI] [PMC free article] [PubMed] [Google Scholar]