Significance
Newly synthesized proteins undergo a strict quality-control checkpoint, and misfolded secretory proteins are targeted across the endoplasmic reticulum membrane back to the cytosol for proteasome degradation. This process requires tagging errant proteins with ubiquitin by an E3 ubiquitin ligase. In a genetic screen we identified TMEM129 as a novel and unusual E3 ligase. TMEM129 is hijacked by the human cytomegalovirus to degrade MHC-I signaling molecules and avert immune recognition of the infected cell. We suggest TMEM129 is an important ligase in the turnover of misfolded secretory proteins within a novel endoplasmic reticulum-associated degradation complex.
Abstract
The US11 gene product of human cytomegalovirus promotes viral immune evasion by hijacking the endoplasmic reticulum (ER)-associated degradation (ERAD) pathway. US11 initiates dislocation of newly translocated MHC I from the ER to the cytosol for proteasome-mediated degradation. Despite the critical role for ubiquitin in this degradation pathway, the responsible E3 ligase is unknown. In a forward genetic screen for host ERAD components hijacked by US11 in near-haploid KBM7 cells, we identified TMEM129, an uncharacterized polytopic membrane protein. TMEM129 is essential and rate-limiting for US11-mediated MHC-I degradation and acts as a novel ER resident E3 ubiquitin ligase. TMEM129 contains an unusual cysteine-only RING with intrinsic E3 ligase activity and is recruited to US11 via Derlin-1. Together with its E2 conjugase Ube2J2, TMEM129 is responsible for the ubiquitination, dislocation, and subsequent degradation of US11-associated MHC-I. US11 engages two degradation pathways: a Derlin-1/TMEM129–dependent pathway required for MHC-I degradation and a SEL1L/HRD1-dependent pathway required for “free” US11 degradation. Our data show that TMEM129 is a novel ERAD E3 ligase and the central component of a novel mammalian ERAD complex.
Proteins inserted into the endoplasmic reticulum (ER) must fold and acquire their native state before further trafficking through the secretory pathway (1, 2). To avoid the toxicity associated with misfolded gene products, all proteins must pass the ER quality-control checkpoint. Misfolded proteins are rejected and dislocated across the ER-membrane for cytosolic proteasome degradation in a process known as ER-associated degradation (ERAD). ERAD degrades misfolded and unassembled proteins and regulates turnover of ER-resident proteins (3).
Ubiquitination of the protein substrate provides a critical step in protein dislocation (4). The RING family constitutes the largest family of E3 ligases, including those involved in ERAD (5). The RING domain creates a binding platform for the E2 conjugase and consists of two zinc atoms coordinated in a cross-brace motif via interspersed cysteine (C) and histidine (H) residues in a C3HC4, C3H2C3, or C4HC3 conformation (5). Most ERAD E3 ligases are integral membrane proteins; they form the core of the ERAD machinery, nucleating functionally distinct ERAD complexes. The mammalian system is more complex than yeast and has undergone an expansion of the ERAD E3 ligase family, with the Hrd1p homologs Hrd1 and Gp78, the Doa10p homolog MARCH6, RNF5, TRC8, and CHIP (3, 4).
Many pathogens appropriate the ubiquitin-proteasome system, particularly to degrade components of the host immune system (6, 7). The human cytomegalovirus (HCMV) US2 and US11 gene products have been instrumental in studies of the mammalian ERAD system. These viral proteins hijack separate components of the ERAD system to degrade MHC-I, thus preventing cytotoxic T lymphocyte recognition of infected cells (8, 9).
US2 appropriates the TRC8 E3 ubiquitin ligase (10), whereas dislocation induced by US11 is reported to be dependent on components of the canonical “HRD1 retrotranslocation machinery,” including the rhomboid pseudoprotease Derlin-1 (11, 12), SEL1L (13), AUP1, and UBXD8 (14). Despite recruiting these components HRD1 is not itself required for US11-mediated degradation, and the ERAD E3 ligase central to US11-mediated degradation remains undefined.
To further understand how US11 works, we used a forward genetic screen in US11-expressing KBM7 cells which are haploid for all chromosomes except chromosome 8, allowing gene disruption by targeting of a single gene copy (15, 16). We identified TMEM129, an uncharacterized membrane protein, as essential and rate-limiting for US11-mediated MHC-1 degradation. TMEM129 contains a novel and atypical RING domain with intrinsic protein E3 ubiquitin ligase activity. The loss of TMEM129 abolishes MHC-I ubiquitination in the presence of US11 and prevents MHC-I retrotranslocation and proteasomal degradation. TMEM129 is therefore a novel ERAD E3 ligase hijacked by HCMV US11 and critical for HCMV immune modulation.
Results
A Haploid Genetic Screen Identifies TMEM129 as an Essential Cellular Gene in the US11-Mediated Down-Regulation of MHC Class I Molecules.
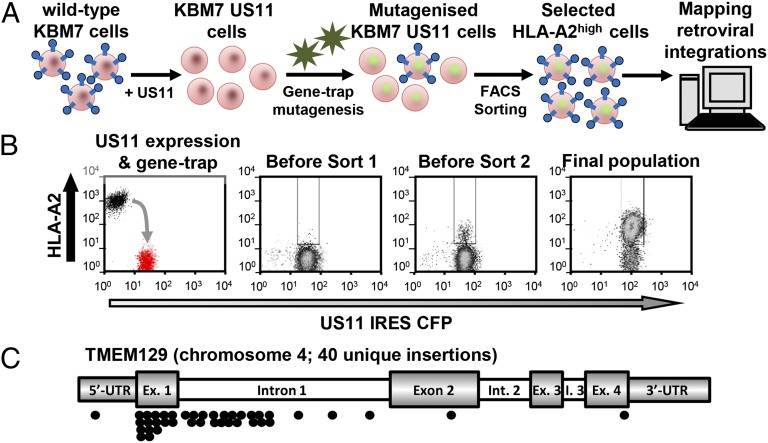
To identify novel host factors required for the US11-mediated degradation of MHC-I molecules, we screened near-haploid KBM7 cells. Our rationale was that disruption of a gene essential for US11 activity would rescue MHC-I from degradation and increase cell surface MHC-I (Fig. 1A). We screened on a KBM7 US11 internal ribosome entry site (IRES) CFP clone with low cell-surface endogenous HLA-A2 (Fig. 1B) and also selected for CFP as a surrogate marker of US11. After retroviral mutagenesis of US11 KBM7 cells, we enriched for rare HLA-A2high, CFP+ cells by iterative FACS (Fig. 1B). Following the second sort, a population of >90% HLA-A2high CFP+ cells was identified. Retroviral integration sites, mapped by splinkerette-PCR and 454 pyrosequencing, revealed 40 independent retroviral insertions in the uncharacterized TMEM129 gene on chromosome 4 (Fig. 1C). No other clusters of retroviral insertion sites were found; TMEM129 therefore represents the only bona fide hit from the screen.
Fig. 1.
A haploid genetic screen identifies TMEM129 as an essential component for US11-mediated MHC-I degradation. (A) Schematic overview of genetic screen. (B) Selection of mutant MHC-Ihigh cells by FACS. KBM7 US11 cells were mutagenised with gene-trap retrovirus and rare HLA-A2high CFPhigh cells selected by two sequential rounds of FACS, with sorting gates indicated. (C) Sequencing of retroviral insertion sites in the selected HLA-A2low CFPhigh population revealed 40 independent insertions in the TMEM129 gene on chromosome 4. The majority of gene-trap insertions cluster around the TMEM129 start site (5′ UTR, exon 1, intron 1).
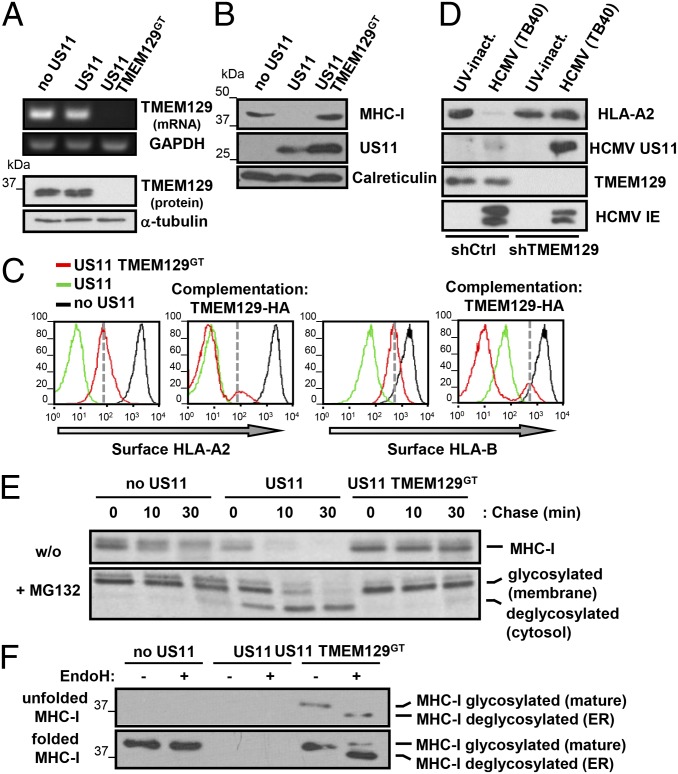
To verify the requirement for TMEM129 in US11-mediated degradation, we isolated a US11+, TMEM129 gene-trap clone (TMEM129GT) and confirmed the absence of TMEM129 mRNA (Fig. 2A, Upper), and protein (Fig. 2A, Lower) with a TMEM129-specific antibody that detects a 36-kDa band, absent from TMEM129GT cells (Fig. 2A, Lower). Immunoblot analysis of US11+ TMEM129GT cells showed a full restoration of MHC-I (Fig. 2B), although only a partial cell surface rescue of HLA-A2 (Fig. 2C, first panel, red line). Exogenous HA-tagged TMEM129 in US11 TMEM129GT cells restored US11 activity with a complete loss of cell surface HLA-A2 (Fig. 2C, second panel, red line), proving an absolute requirement for TMEM129 for US11 function. US11 targets the endogenous HLA-B allele of KBM7 (HLA-B60) less effectively than HLA-A2, causing only a partial cell surface loss of HLA-B (Fig. 2C, third panel, green line), which is also dependent on TMEM129 (Fig. 2C, third panel, red line). Exogenous TMEM129-HA induced further down-regulation of cell surface HLA-B (Fig. 2C, fourth panel, red line) beyond that seen in the original US11 cell (Fig. 2C, fourth panel, red line), suggesting TMEM129 is both essential and rate-limiting for US11-mediated MHC-I degradation. This finding was confirmed by showing that TMEM129 overexpression in KBM7 US11 cells further down-regulated HLA-B (Fig. S1A). This requirement for TMEM129 on US11 activity was not limited to KBM7 cells. siRNA TMEM129 depletion rescued GFP–HLA-A2 expression in HeLa-US11 GFP–HLA-A2 cells (Fig. S1B), suggesting a general requirement for TMEM129 in US11 activity. The specificity of TMEM129 for the US11 dislocation pathway was confirmed, as TMEM129 depletion did not rescue GFP–HLA-A2 in US2-expressing HeLa cells (Fig. S1C).
Fig. 2.
TMEM129 is essential and rate-limiting for US11-mediated MHC-I dislocation. (A–D) Characterization of a TMEM129 gene-trapped (US11+ TMEM129GT) KBM7 clone. (A and B) MHC-I is restored in US11 TMEM129GT cells. Control, US11, and US11 TMEM129GT KBM7 cells were examined by RT-PCR (Upper), immunoblot (TMEM129-specific mAb) (Lower) or (B) immunoblotted for MHC-I (HC10), US11, or calreticulin. (C) TMEM129 rescues the US11 phenotype in US11 TMEM129GT cells. Cytofluorometric analysis of HLA-A2 and HLA-B in TMEM129GT (red), US11 (green), and control (black) KBM7 cells (panels 1 and 3) and complemented with HA-tagged TMEM129 (panels 2 and 4). (D) TMEM129 is required for the US11-induced MHC-I down-regulation in HCMV-infected cells. Human foreskin fibroblast primary fibroblasts were stably transduced with control or TMEM129-specific shRNA, then infected with HCMV TB40Bac4 (multiplicity of infection, MOI 25) or UV-inactivated virus and after 48 h lysates immunoblotted for HLA-A2 (HCA2), US11, TMEM129, HCMV immediate early protein (IE1/2), or calreticulin. (E) US11 TMEM129GT cells do not dislocate or degrade MHC-I. Control, US11, and US11 TMEM129GT KBM7 cells were [35S]methionine pulse-labeled for 10 min, and MHC-I HC immunoprecipitated from lysates at the indicated times in the absence (Upper) or presence (Lower) of the MG132 proteasome inhibitor. (F) US11 TMEM129GT cells retain MHC-I in the ER in a partially folded state. KBM7, KBM7 US11, and KBM7 US11 TMEM129GT cells were immunoprecipitated with nonconformational (HC10, Upper) or conformational (W6/32, Lower), incubated ± EndoH, and immunoblotted for MHC-1 (HC10).
We wanted to ascertain whether TMEM129 is also required for the down-regulation of MHC-I in HCMV-infected cells. We therefore depleted TMEM129 from fibroblasts that were then infected with the HCMV TB40Bac4 strain, which lacks the IRS1-US9 genes and is reliant on US11 for MHC-I down-regulation. Depletion of TMEM129 prevented the loss of HLA-A2 in HCMV infection (Fig. 2D) and also increased US11 expression. TMEM129 is therefore responsible for the US11-induced down-regulation of MHC-I in HCMV infection.
TMEM129 depletion rescued endogenous MHC-1 (Fig. 2B), but failed to completely restore cell surface HLA-A2 expression (Fig. 2C), implying MHC-I must be retained within the cell. We therefore used pulse-chase analysis to determine the fate of MHC-I. In KBM7 US11 cells, nonconformational MHC-I is rapidly degraded with a half-life of ∼5 min (Fig. 2E, Upper, lanes 4–6) with little conformational MHC-I formed (Fig. S1D, lanes 4–6). Upon proteasome inhibition, these dislocated MHC-I molecules are visualized as faster migrating deglycosylated species (Fig. 2E, Lower, lanes 4–6) (8), resulting from cytosolic N-glycanase removal of the MHC I glycan (17). In KBM7 US11 TMEM129GT cells, MHC-I degradation is prevented (Fig. 2E, lanes 7–9) and no deglycosylated MHC-I is detected. Conformational MHC-I remains mainly EndoH-sensitive, indicating ER residency (Fig. 2F, Lower, lanes 5–6), and associates with a lower molecular-weight protein that comigrates with US11 (Fig. S1D, lanes 7–9). We conclude that TMEM129 is a novel, essential, rate-limiting protein for US11-mediated MHC-I dislocation and degradation. In the absence of TMEM129 the majority of MHC-I are retained in the ER bound to US11, explaining the partial rescue of cell surface MHC-I in TMEM129GT cells.
TMEM129 Is an ERAD RING E3 Ligase and Recruits Ube2J2 to Ubiquitinate MHC-I Before US11-Induced Degradation.
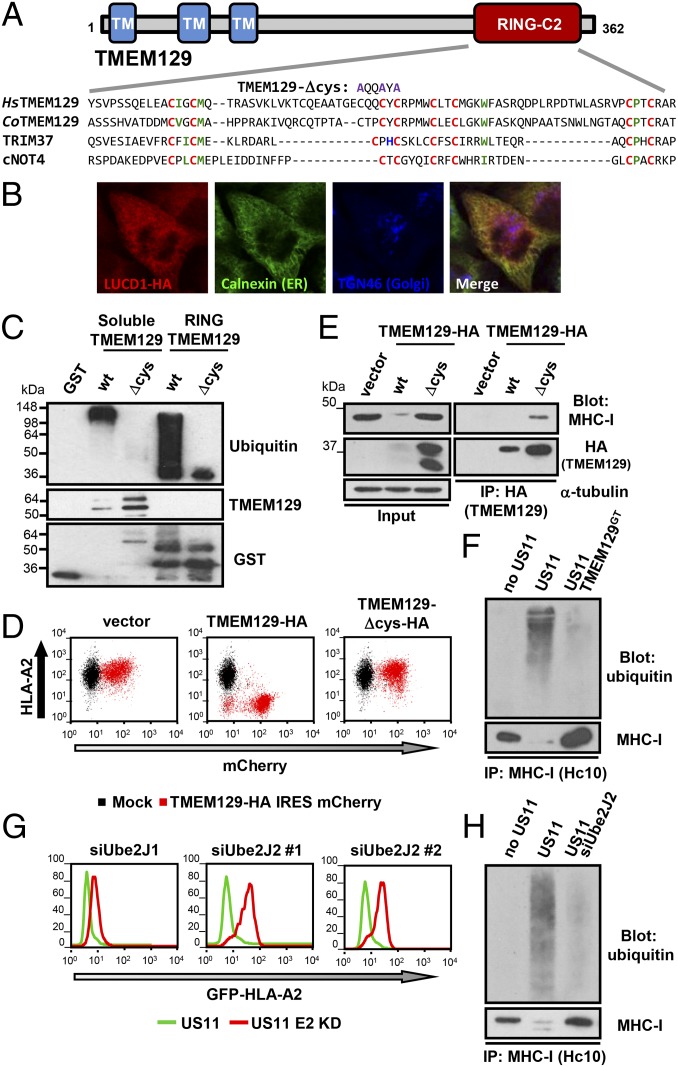
TMEM129 encodes an uncharacterized 362-aa protein. Bioinformatic analysis suggests an evolutionarily conserved, polytopic membrane protein with three transmembrane domains and a long C-terminal tail. Although lacking any yeast ortholog, TMEM129 can be traced back to the unicellular metazoan ancestor Capsaspora owczarzaki (Fig. 3A). Exogenous TMEM129-HA colocalized with the calnexin ER marker, consistent with ER residence (Fig. 3B). The predicted C terminus contains conserved cysteine residues whose spacing was reminiscent of the RING family of E3 ligases (Fig. 3A). In the classic RING (RING-CH), cysteine and histidine residues coordinate the binding of two zinc atoms, which together form the cross-brace structural motif of the RING and stabilizes the hydrophobic E2 binding site (5). The RING-CH arranges these zinc-binding residues in a C3HC4 conformation, whereas in the RING-H2 variant the sequence is C3H2C3. The putative RING of TMEM129 is unusual because it contains only cysteine, and no histidine residues for zinc coordination, and has additional extended loops between its zinc-coordinating cysteines (AA289-308 and AA323-345). This potential TMEM129 RING also contains a canonical E2 binding site, with an isoleucine residue between the first two cysteines, a tryptophan in position 4 of the second loop and a proline between the final two cysteine residues (Fig. 3A) (5).
Fig. 3.
TMEM129 is an ER resident E3 ubiquitin ligase that with Ube2J2 ubiquitinates MHC-I before US11-induced dislocation. (A) Schematic of TMEM129, with the indicated RING domain alignments. Zinc-coordinating cysteine (red), histidine (blue), and conserved residues of the E2 binding site (green) are indicated. (HsTMEM129, Homo sapiens; CoTMEM129, C. owczarzaki). (B) TMEM129-HA colocalizes with the ER marker calnexin. HeLa cells were transfected with TMEM129-HA, fixed, and immunostained for TMEM129-HA (HA, red), calnexin (ER, green), and TGN46 (Golgi, blue). (Magnification: 64×.) (C) The TMEM129 RING shows in vitro E3 ligase activity. GST-tagged wild-type or Δcys RING TMEM129 or soluble TMEM129 was purified and incubated in an in vitro ubiquitination reaction with UbcH5, E1, ubiquitin, and ATP. Immunoblots were analyzed for ubiquitin (P4D1), TMEM129, and GST. (D) Wild-type but not RING-mutant (Δcys) TMEM129-HA restores HLA-A2 down-regulation in US11+ TMEM129GT cells. Cytofluorometric analysis of HLA-A2 in US11+ TMEM129GT cells expressing wild-type or Δcys TMEM129-HA upstream of an IRES mCherry. (E) Catalytically inactive TMEM129 binds its MHC-I substrate. mCherry+ cells from expressing vector, wild-type or Δcys TMEM129-HA were isolated by FACS and directly immunoblotted (Left), or following HA-tag immunoprecipitations, eluted proteins probed for MHC-I and HA (Right). (F) TMEM129 is required for US11-induced MHC-I ubiquitination. MHC-I (HC10) was immunoprecipitated from 1% Triton X-100 lysates of MG132-treated KBM7, KBM7 US11, and KBM7 US11 TMEM129GT cells and immunoblotted for ubiquitin (VU-1) or MHC-I (HC10). (G and H) The E2 Ube2J2 is essential for US11-induced MHC-I ubiquitination and degradation. (G) Cytofluorometric analysis of GFP–HLA-A2 in HeLa GFP–HLA-A2 US11 cells depleted of Ube2J1 or Ube2J2. (H) MHC-I ubiquitination was assessed as in F on Ube2J2 or mock-depleted HeLa US11 cells.
The presence of this “cysteine-only” RING domain suggested TMEM129 is an E3 ligase. We therefore tested whether the TMEM129 C terminus contains a functional ubiquitin RING with autoubiquitylation E3 ligase activity. GST fusion constructs of the minimal RING (TMEM129 RING) or the whole TMEM129 C terminus (soluble TMEM129) were purified (Fig. S2). Incubation with the E2 ubiquitin conjugase (UbcH5) and E1 activating enzyme showed robust autoubiquitination of both the soluble and “RING-only” TMEM129 (Fig. 3C, lanes 2 and 4). Mutation of the zinc-coordinating cysteine residues abolished autoubiquitination in both constructs (Fig. 3C, Δcys lanes 3 and 5).
Because TMEM129 shows E3 ligase activity in vitro, we tested whether it is indeed the cellular E3 ligase appropriated by US11 for MHC-I degradation. We initially determined whether an active RING was required for its in vivo activity. Reconstitution of TMEM129GT cells with wild-type TMEM129-HA, but not the mutant RING (Δcys TMEM129-HA), restored US11-mediated MHC-I cell surface down-regulation (Fig. 3D) and degradation (Fig. 3E, Left). Because of MHC-I degradation, we could not identify MHC-I bound to wild-type TMEM129, but took advantage of the catalytically inactive mutant (Δcys TMEM129) and showed that in the presence of US11 it bound MHC-I (Fig. 3E). Lower expression of wild-type vs. Δcys TMEM129 (Fig. 3E, Left, lanes 2 and 3) is because of toxicity of wild-type reexpression in gene-trap cells.
To show that TMEM129 was required for US11-mediated MHC-I ubiquitination we initially immunoprecipitated MHC-I and found the robust MHC-I ubiquitination induced by US11 (Fig. 3F, lane 2) was abrogated in the TMEM129GT cells (Fig. 3F, lane 3). To confirm TMEM129 ubiquitination occurs in other cells and that ubiquitination is specific to the MHC-I heavy chain, we repeated the experiment in TMEM129-depleted HeLa US11 cells. Using more stringent, denaturing conditions, we showed that MHC-I ubiquitin species were still detected (Fig. S3A, lanes 1–3) and disappeared upon TMEM129 depletion, suggesting TMEM129 is the ligase recruited by US11 for MHC-I ubiquitination.
As TMEM129 has an atypical RING-C2 domain, and US11-induced MHC-I ubiquitination is not lysine-dependent (18, 19), we screened a custom siRNA library targeting the 39 annotated E2 conjugating enzymes for the E2 used by TMEM129 (10). Only the ER-membrane tail-anchored E2 ubiquitin conjugase Ube2J2 gave a significant rescue in fluorescence signal (Fig. S3B) (z-score 5.48). We confirmed that siRNA-mediated depletion of Ube2J2, but not Ube2J1, rescued endogenous MHC-1 in HeLa US11 cells (Fig. 3G) and abolished US11-induced MHC-I ubiquitination (Fig. 3H, Upper), with effective Ube2J1/2 depletions confirmed (Fig. S3C). Therefore, the RING-C2 E3 ligase TMEM129 recruits Ube2J2 for the US11-induced MHC-I ubiquitination that drives ER dislocation and proteasomal degradation.
US11 Associates with TMEM129 via the Rhomboid Pseudoprotease Derlin-1.
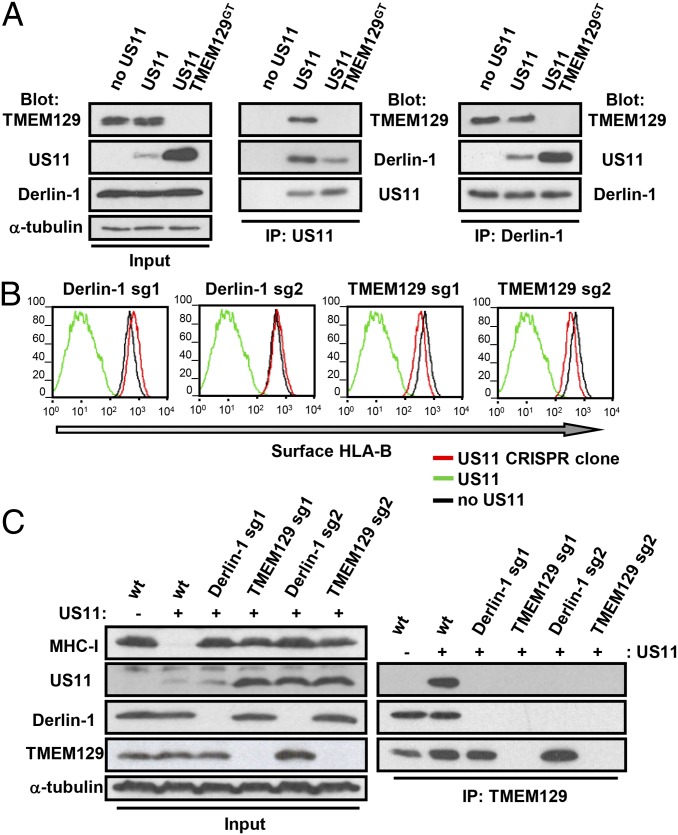
Derlin-1 is a component of the US11 retrotranslocation complex (11, 12) and immunoprecipitation of both US11 (Fig. 4A, Center) and Derlin-1 (Fig. 4A, Right) showed that TMEM129 is also a member of this complex. The TMEM129–US11 interaction is dependent on a polar glutamine residue in the US11 transmembrane (Q192) (Fig. S4) as reported for the US11–Derlin-1 interaction (12). TMEM129 binding to Derlin-1 is US11-independent (Fig. 4A, Right), suggesting TMEM129 forms part of an endogenous Derlin-1–containing ERAD complex. Conversely, the interaction between Derlin-1 and US11 is independent of TMEM129 (Fig. 4A, Right).
Fig. 4.
The rhomboid pseudoprotease Derlin-1 recruits US11 to TMEM129. (A) TMEM129 is recruited to a Derlin-1–US11 complex. KBM7, KBM7 US11, and KBM7 US11 TMEM129GT cells were lysed in 1% digitonin, immunoprecipitated with US11 (Center) or Derlin-1 (Right) antisera and probed for endogenous TMEM129, Derlin-1, or US11. (B) CRISPR-Cas9 knockout clones of Derlin-1 and TMEM129 restore cell surface HLA-B in US11-expressing cells. Cytofluorometric analysis of cell surface HLA-B on HeLa GFP-HLA-A2.US11 clones targeted with two independent short guide RNAs (sg) for Derlin-1 or TMEM129. (C) Derlin-1 bridges US11 to TMEM129. Endogenous TMEM129 was immunoprecipitated from CRISPR Cas9 generated TMEM129 knockout (TMEM129 sg1/2) or Derlin-1 knockout (Derlin-1 sg1/2) HeLa US11 clones and eluted proteins probed for US11, Derlin-1, and TMEM129 (Right). Immunoblotting of whole-cell lysates is shown (Left).
A functional role for Derlin-1 in US11-mediated degradation was previously shown using a dominant-negative Derlin-1–GFP construct (12). Because Derlin-1 siRNA depletion failed to inhibit US11 activity, we generated Derlin-1–deficient HeLa US11 cells using CRISPR (clustered regulatory interspaced short palindromic repeats) technology. Derlin-1 and TMEM129 knockout clones from independent sgRNAs showed full MHC-I restoration (Fig. 4 B and C, Left). Furthermore, whereas TMEM129 immunoprecipitated US11 in Derlin-1–sufficient cells, this interaction is lost in Derlin-1–deficient cells (Fig. 4C, Right). The TMEM129 E3 ligase is therefore recruited to US11 via Derlin-1.
US11 Avoids TMEM129-Mediated Degradation and Is Regulated by the HRD1/SEL1L Complex.
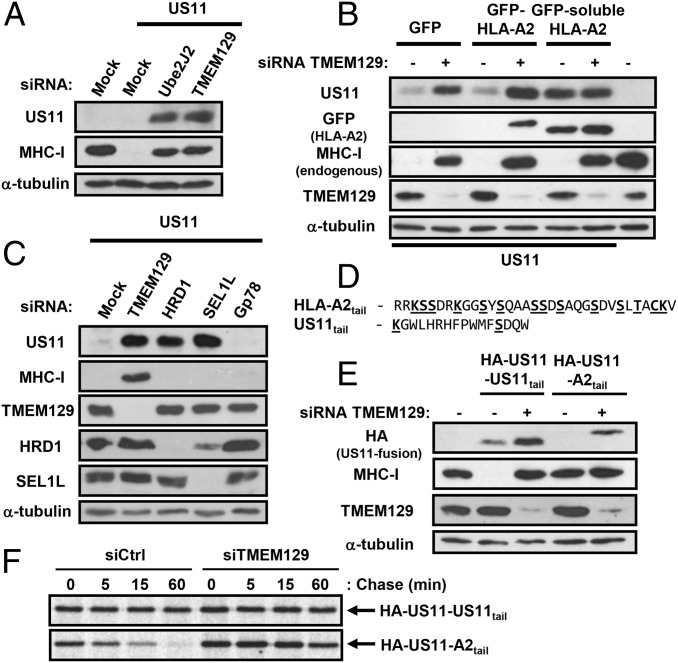
US11 levels are consistently elevated upon TMEM129 and Ube2J2 depletion in cell models as well as in HCMV infection (Figs. 2 B and D, 4 A and C, and 5A), suggesting US11 might also be degraded in a TMEM129 pathway. However, the 1-h half-life of US11 (Fig. S5A) is much longer than the very short half-life of MHC I (<5 min) (Fig. 2E), arguing against codegradation in a TMEM129-dependent pathway. We therefore asked whether the increase in steady-state US11 following TMEM129 depletion was indirect (i.e., secondary to the increased MHC-I levels that, following TMEM129 depletion, accumulate in the ER bound to US11) (Fig. S1D, lanes 7–9). To test this hypothesis, we examined whether soluble HLA-A2, which is not a substrate of US11 (Fig. 5B, lanes 5 and 6), affected US11 levels and activity. A GFP-tagged soluble HLA-A2 increased US11 levels (Fig. 5B, lane 5), which still retained activity against endogenous MHC-I (Fig. 5B, lane 5). Under these conditions, TMEM129 depletion did not further increase US11 levels (Fig. 5B, lanes 5 and 6). It therefore seems unlikely that a significant fraction of US11 is degraded by TMEM129. Instead, US11 stability seems dependent on stable binding to its MHC-I client. Consistent with this notion, the time course of US11 degradation is more reminiscent of the cellular degradation of endogenous misfolded proteins (20). We therefore asked whether US11 is targeted by known components of the cellular quality-control machinery. We found that depletion of HRD1, and its luminal substrate recruitment factor SEL1L, but not Gp78, stabilized US11 (Fig. 5C) without affecting MHC-I levels. We conclude that US11 is regulated by substrate availability and is itself degraded by the HRD1/SEL1L complex. Therefore, although US11 is targeted for slow HRD1-mediated degradation by the ER quality-control machinery, the MHC-I–US11 complex triggers rapid recognition and MHC-I dislocation via TMEM129.
Fig. 5.
US11 is stabilized by its MHC-I substrate but escapes degradation by TMEM129. (A) US11 is stabilized by TMEM129 or Ube2J2 depletion. HeLa US11 cells were siRNA-depleted of TMEM129 (96 h) or Ube2J2 (72 h) and immunoblots probed for US11 and MHC-I. (B) Soluble HLA-A2 rescues US11 expression. TMEM129 was siRNA depleted from HeLa US11 cells expressing GFP, GFP–HLA-A2, or GFP-soluble HLA-A2, and lysates probed for US11, GFP (HLA-A2), MHC-I (HC10), and TMEM129. (C) US11 is degraded via a HRD1/SEL1L pathway. HeLa US11 cells were siRNA depleted of TMEM129, HRD1, SEL1L, or Gp78 and lysates probed for US11, MHC-I, TMEM129, HRD1, and SEL1L. (D) Schematic of HLA-A2 and US11 cytosolic tail. Putative ubiquitination sites are shown in bold. (E and F) Transfer of the HLA-A2 cytosolic tail to US11 induces its rapid TMEM129-dependent degradation, leaving MHC-I unaffected. TMEM129 was siRNA-depleted from HeLa cells expressing HA-US11 or HA-US11-A2tail. Cells were (E) probed by immunoblot for HA, MHC-I (HC10), or TMEM129 or (F) [35S]methionine pulse-labeled and analyzed by autoradiography following HA-tag immunoprecipitations at the indicated time points.
US11 Tagged with a MHC-I Cytosolic Tail Is Rapidly Degraded by TMEM129 and Unable to Catalyze MHC-I Degradation.
How does US11 catalyze MHC-I degradation by TMEM129 while escaping rapid degradation itself? Although the luminal domain of US11 binds MHC-I, and its transmembrane domain recruits the dislocation machinery (21), the cytosolic tail of US11 is dispensable for its activity. In contrast, degradation by US11 requires the cytosolic tail of MHC-I, which is the most likely site of TMEM129-mediated ubiquitination (22). MHC-I’s tail contains multiple potential ubiquitin acceptor sites (Fig. 5D), whereas the cytoplasmic tail of US11 is short and devoid of ubiquitination sites, with a membrane proximal lysine residue buried just beyond the transmembrane domain and a single exposed serine residue (Fig. 5D). We therefore asked whether replacing the cytosolic tail of US11 with the tail of HLA-A2 (US11-A2tail) might target US11 for rapid, TMEM129-mediated degradation. Strikingly, the US11-A2tail is rapidly degraded in a TMEM129-dependent pathway (Fig. 5E), with its half-life decreasing to ∼8 min (US11-A2tail) (Fig. 5F), comparable to the half-life of US11-degraded MHC-I. These data imply that the tail of HLA-A2, but not US11, constitutes a substrate for rapid TMEM129-mediated degradation. Additional functional consequences of the HLA-A2 tail swap with US11 were also striking. Whereas endogenous MHC-I is rapidly degraded by wild-type US11 or US11Δtail mutant (23), in the presence of US11-A2tail, MHC-I remains stable and is no longer degraded (Fig. 5E), even though US11-A2tail shows MHC-I binding (Fig. S5B). Therefore, the US11-A2tail fusion becomes the preferred TMEM129 substrate, and endogenous MHC-I is ignored.
Discussion
US11 is one of HCMV’s most potent immune modulators and herein we have shown that TMEM129 is a novel membrane-bound RING E3 ligase that, together with its cognate E2 UbE2J2, is both essential and rate-limiting for US11-mediated degradation of MHC-1. By uncoupling US11 from the dislocation machinery we show that US11 retains its substrate in the ER, a common mechanism used by many viral proteins to prevent substrate expression. Linking up with the TMEM129 cellular E3 ligase provides a very efficient way to down-regulate MHC-I, which cannot be overcome by increased MHC-1 expression. US11’s cytosolic tail protects it from rapid degradation by TMEM129, and US11 is instead turned over by the HRD1/SEL1L ligase complex.
The TMEM129 RING domain is unusual in containing only cysteine residues for zinc coordination and extended loops. “Cysteine-only” RINGs, previously referred to as RING-C2 domains, are rare and difficult to predict because of their resemblance to other zinc-finger motifs. cNOT4 is the only human E3 ligase with a characterized RING-C2 (24), and other cysteine-only ligases might remain unidentified. A PROSITE search identified RING-C2 domains in the family of RFPL proteins and the RBR ligases HOIP (RING1) and RNF216 (RING2). Because the TMEM129 RING shows closer homology to RING-HC domains than cNOT4, we suggest that TMEM129 originated from a classic RING E3 ligase rather than being part of a larger RING-C2 ligase family. Despite the lack of TMEM129-related mammalian proteins, TMEM129 and its unusual RING-C2 domain are conserved throughout holozoa, including the unicellular C. owczarzaki, implying an important and conserved cellular function.
Although TMEM129 depletion stabilized US11 as well as MHC-I, our initial assumption that US11 and MHC-I were codegraded by the TMEM129 E3 ligase was inconsistent with the very different half-life of the proteins. The stabilization of US11 upon TMEM129 depletion was mimicked by exogenous expression of a soluble, US11-insensitive MHC-I, suggesting that US11 is stabilized by direct binding to its client MHC-I protein. In the absence of MHC-I, US11 is degraded in a TMEM129-independent pathway, presumably via a degron overlapping with the MHC-I binding site. Consistent with this result we found US11 degradation to be dependent on the HRD1 E3 ligase and its luminal substrate recruitment factor SEL1L.
We therefore suggest HCMV US11 engages two degradation pathways: a Derlin-1/TMEM129/Ube2J2-dependent pathway involved in the degradation of MHC-I and a SEL1L/HRD1-dependent pathway involved in the degradation of “free” US11. The TMEM129 pathway is prevented from ubiquitinating US11 by a short cytosolic tail with few ubiquitination sites. HRD1 does not ubiquitinate US11-associated MHC-I, even though disassembled MHC-I is a potent HRD1 client (20). Remarkably, a tail swap between US11 and MHC-I allowed US11 to be rapidly degraded in a TMEM129-dependent manner, leaving endogenous MHC-I untouched. Clearly a separation between both pathways is essential for US11 function, achieved by deflecting TMEM129’s ubiquitination onto the MHC-I tail with US11 acting as a pseudosubstrate. Both pathways are, however, intimately linked because TMEM129 depletion rescues MHC-I, which in turn stabilizes US11. Conversely HRD1/SEL1L-mediated degradation of free US11 might enhance TMEM129-mediated degradation of MHC-I by limiting the number of US11 molecule TMEM129 screens to find a US11 loaded with MHC-I. Such a flexible degradation system based on stabilization of the degradation machinery by a client protein, might allow US11 expression to rapidly adapt to changes in MHC-I expression, as might occur under the influence of IFN or other viral immune modulators.
TMEM129’s strong interaction with Derlin-1 and the Derlin-1 knockout confirmed the central role of this protein in US11 activity. Derlin-1 bridges US11 to TMEM129 but interacts with other ERAD E3 ligases, including HRD1, Gp78, and RMA-1, making it surprising that TMEM129 is the sole ligase able to catalyze US11-induced protein dislocation. Other ERAD E3 ligases cannot substitute for TMEM129, as depletions of neither Hrd1/Gp78 (Fig. 5C) nor TRC8 (Fig. S1C) prevented MHC-I degradation, implying that TMEM129 is the sole ligase responsible for US11-mediated MHC-I degradation. We therefore predict that TMEM129 plays an important role in the ubiquitination and degradation of Derlin-1–specific protein substrates. Derlin-1’s role within the US11 system is difficult to study because Derlin-1 depletion dissociates viral US11 from TMEM129, disrupting ubiquitination, recognition, and dislocation. If Derlin-1 is part of the retrotranslocon (25, 26), US11 binding might directly impact on channel opening and substrate dislocation.
The role of ubiquitin in US11-induced protein dislocation has been difficult to establish, not least because US11 is able to degrade a “lysine-less” MHC-I heavy chain (19). Ubiquitin can be conjugated to nonlysine residues, including serine, threonine, or cysteine residues and the protein N terminus (27, 28). TMEM129-dependent ubiquitination of MHC-I might proceed on a combination of lysine and nonlysine residues, consistent with the identification of Ube2J2 as TMEM129’s cognate E2. Ube2J2 is also recruited by the MHVγ68 mK3 viral E3 ligase for ubiquitination of MHC-I on nonlysine residues (28).
Paradoxically, it is US11’s potent ability to reduce MHC-I levels and alter antigen presentation that recently attracted attention in HIV vaccination studies. Simian Immunodeficiency Virus (SIV) vaccination, in the context of rhesus CMV (RhCMV), gives a unique long-term protection against subsequent SIV infection, that is exclusively dependent on the RhCMV US11 homolog (Rh189) (29). SIV normally evades immune recognition by mutation of its dominant antigens (30). US11 likely suppresses the dominant canonical T-cell response, allowing immune detection of protective SIV epitopes. Our characterization of TMEM129 as a novel E3 ligase and the rate-limiting component of HCMV US11-induced protein dislocation, therefore not only uncovers a novel mammalian ERAD pathway, but might aid in future targeting of viral diseases and vaccine design.
Materials and Methods
Details of experimental procedures, constructs, and reagents can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank all members of the P.J.L. laboratory. This work was supported by the Wellcome Trust and the National Institute for Health Research Cambridge Biomedical Research Centre.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409099111/-/DCSupplemental.
References
- 1.Guerriero CJ, Brodsky JL. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol Rev. 2012;92(2):537–576. doi: 10.1152/physrev.00027.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hegde RS, Ploegh HL. Quality and quantity control at the endoplasmic reticulum. Curr Opin Cell Biol. 2010;22(4):437–446. doi: 10.1016/j.ceb.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olzmann JA, Kopito RR, Christianson JC. The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb Perspect Biol. 2013;5(9) doi: 10.1101/cshperspect.a013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kostova Z, Tsai YC, Weissman AM. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin Cell Dev Biol. 2007;18(6):770–779. doi: 10.1016/j.semcdb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 6.Randow F, Lehner PJ. Viral avoidance and exploitation of the ubiquitin system. Nat Cell Biol. 2009;11(5):527–534. doi: 10.1038/ncb0509-527. [DOI] [PubMed] [Google Scholar]
- 7.Isaacson MK, Ploegh HL. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe. 2009;5(6):559–570. doi: 10.1016/j.chom.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiertz EJ, et al. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996;84(5):769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 9.Loureiro J, Ploegh HL. Antigen presentation and the ubiquitin-proteasome system in host-pathogen interactions. Adv Immunol. 2006;92:225–305. doi: 10.1016/S0065-2776(06)92006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stagg HR, et al. The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J Cell Biol. 2009;186(5):685–692. doi: 10.1083/jcb.200906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429(6994):841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 12.Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429(6994):834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- 13.Mueller B, Lilley BN, Ploegh HL. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J Cell Biol. 2006;175(2):261–270. doi: 10.1083/jcb.200605196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc Natl Acad Sci USA. 2008;105(34):12325–12330. doi: 10.1073/pnas.0805371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326(5957):1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 16.Timms RT, et al. Haploid genetic screens identify an essential role for PLP2 in the downregulation of novel plasma membrane targets by viral E3 ubiquitin ligases. PLoS Pathog. 2013;9(11):e1003772. doi: 10.1371/journal.ppat.1003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blom D, Hirsch C, Stern P, Tortorella D, Ploegh HL. A glycosylated type I membrane protein becomes cytosolic when peptide: N-glycanase is compromised. EMBO J. 2004;23(3):650–658. doi: 10.1038/sj.emboj.7600090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shamu CE, Flierman D, Ploegh HL, Rapoport TA, Chau V. Polyubiquitination is required for US11-dependent movement of MHC class I heavy chain from endoplasmic reticulum into cytosol. Mol Biol Cell. 2001;12(8):2546–2555. doi: 10.1091/mbc.12.8.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassink GC, Barel MT, Van Voorden SB, Kikkert M, Wiertz EJ. Ubiquitination of MHC class I heavy chains is essential for dislocation by human cytomegalovirus-encoded US2 but not US11. J Biol Chem. 2006;281(40):30063–30071. doi: 10.1074/jbc.M602248200. [DOI] [PubMed] [Google Scholar]
- 20.Burr ML, et al. HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc Natl Acad Sci USA. 2011;108(5):2034–2039. doi: 10.1073/pnas.1016229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lilley BN, Ploegh HL. Viral modulation of antigen presentation: Manipulation of cellular targets in the ER and beyond. Immunol Rev. 2005;207:126–144. doi: 10.1111/j.0105-2896.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 22.Barel MT, et al. Amino acid composition of alpha1/alpha2 domains and cytoplasmic tail of MHC class I molecules determine their susceptibility to human cytomegalovirus US11-mediated down-regulation. Eur J Immunol. 2003;33(6):1707–1716. doi: 10.1002/eji.200323912. [DOI] [PubMed] [Google Scholar]
- 23.Furman MH, Ploegh HL, Tortorella D. Membrane-specific, host-derived factors are required for US2- and US11-mediated degradation of major histocompatibility complex class I molecules. J Biol Chem. 2002;277(5):3258–3267. doi: 10.1074/jbc.M109765200. [DOI] [PubMed] [Google Scholar]
- 24.Albert TK, et al. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 2002;21(3):355–364. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenblatt EJ, Olzmann JA, Kopito RR. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant α-1 antitrypsin from the endoplasmic reticulum. Nat Struct Mol Biol. 2011;18(10):1147–1152. doi: 10.1038/nsmb.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehnert M, Sommer T, Jarosch E. Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat Cell Biol. 2014;16(1):77–86. doi: 10.1038/ncb2882. [DOI] [PubMed] [Google Scholar]
- 27.Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309(5731):127–130. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, et al. Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J Cell Biol. 2009;187(5):655–668. doi: 10.1083/jcb.200908036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen SG, et al. Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science. 2013;340(6135):1237874. doi: 10.1126/science.1237874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picker LJ, Hansen SG, Lifson JD. New paradigms for HIV/AIDS vaccine development. Annu Rev Med. 2012;63:95–111. doi: 10.1146/annurev-med-042010-085643. [DOI] [PMC free article] [PubMed] [Google Scholar]