Significance
When oxygen gets incompletely reduced, reactive oxygen species (ROS) are generated. These ROS molecules can harm the building blocks of the cell but are also important signaling molecules. Until now, the ROS language of the cell has not been understood and a clear view is needed on how the cell differentiates metabolic ROS noise from ROS that allows signaling, regulation, and protection. To address this question, we focused on Arabidopsis thaliana and identified the proteins that react with hydrogen peroxide on the thiol of the amino acid cysteine, which after reaction forms a sulfenic acid. The characterization of the plant sulfenome improves the understanding of important ROS signaling pathways.
Keywords: oxidative stress, redox regulation, cysteine oxidation
Abstract
Reactive oxygen species (ROS) have been shown to be potent signaling molecules. Today, oxidation of cysteine residues is a well-recognized posttranslational protein modification, but the signaling processes steered by such oxidations are poorly understood. To gain insight into the cysteine thiol-dependent ROS signaling in Arabidopsis thaliana, we identified the hydrogen peroxide (H2O2)-dependent sulfenome: that is, proteins with at least one cysteine thiol oxidized to a sulfenic acid. By means of a genetic construct consisting of a fusion between the C-terminal domain of the yeast (Saccharomyces cerevisiae) AP-1–like (YAP1) transcription factor and a tandem affinity purification tag, we detected ∼100 sulfenylated proteins in Arabidopsis cell suspensions exposed to H2O2 stress. The in vivo YAP1-based trapping of sulfenylated proteins was validated by a targeted in vitro analysis of DEHYDROASCORBATE REDUCTASE2 (DHAR2). In DHAR2, the active site nucleophilic cysteine is regulated through a sulfenic acid-dependent switch, leading to S-glutathionylation, a protein modification that protects the protein against oxidative damage.
Numerous posttranslational modifications (PTMs) have been discovered within proteomes, creating a complex landscape of protein diversity and function (1). One of the recognized reversible redox-based PTMs is the oxidation of a cysteine thiol group to a sulfenic acid (Cys-SOH) (2) that acts as regulatory switch in several oxidative stress signal transduction pathways (3). Sulfenic acids, unless they are stabilized into the protein environment, can react rapidly with other protein thiols or with low-molecular weight thiols to form intramolecular and intermolecular disulfides. These mechanisms protect the sulfenic acids against overoxidation to sulfinic (SO2H) or sulfonic (SO3H) acid and allow sulfur oxygen signaling (2).
In plants, the best-known redox regulation mechanisms are the light-dependent thiol-disulfide exchange switches in chloroplast proteins (4). Examples of other redox-regulated proteins are the transcription coactivator NONEXPRESSER OF PR GENES 1 (5), the vacuolar H+-ATPase (6), and several transcription factors (TFs), such as the AP2-type RAP2.4a (7), the G-group of basic leucine zipper TFs (8), and the TEOSINTEBRANCHED1/CYLOIDEA/PROLIFERATING CELL FACTOR class I TFs (9). The redox relay mechanisms that bridge the signal perception to the final oxidative stress response are largely unknown.
Some thiol peroxidases have an H2O2-dependent signaling function and can act as receptor and transducer (10). In yeast (Saccharomyces cerevisiae), the H2O2 sensor oxidant receptor peroxidase1 (ORP1/glutathione peroxidase 3) controls, together with the transcription factor YAP1 (yeast AP-1–like), a redox regulon via a sulfenic acid thiol-disulfide relay mechanism (11). Upon reaction with H2O2, the peroxidatic cysteine of ORP1 is oxidized to a sulfenic acid that reacts with the YAP1 C-terminal cysteine-rich domain (cCRD) and forms a disulfide (Fig. S1A). This specific mixed disulfide formation has prompted us to develop a YAP1-based probe for trapping plant sulfenylated proteins in vivo (12).
To categorize the sulfenome, which is the set of proteins with at least one sulfenic acid, and its dynamics at the proteome level in Arabidopsis thaliana cells upon oxidative stress, we implemented the YAP1-based sulfenic acid trapping method coupled to a tandem affinity purification (TAP) tag (13). We identified 97 sulfenylated proteins during the early and late oxidative stress responses, of which 67 had previously not been recognized to undergo oxidative PTMs. Validation of sulfenylation on DEHYDROASCORBATE REDUCTASE2 (DHAR2) demonstrates the importance of a glutathione (GSH)-dependent redox switch on its sulfenylated nucleophilic cysteine that reversibly regulates the DHAR activity.
Results and Discussion
H2O2 Triggers the Formation of YAP1C Heterocomplexes in a Time- and Dose-Dependent Manner.
To apply the YAP1-TAP approach, we synthesized a YAP1-cCRD construct with adapted codons for proficient expression in plants and mutated Cys620 and Cys629 to alanine and threonine (Fig. S1B and Table S1), retaining only the redox-active cysteine Cys598. Then, we fused this construct at its N terminus to a GS tag that combines two IgG-binding domains of protein G with a streptavidin-binding peptide (SBP), separated by a tobacco etch virus (TEV) protease cleavage site (13). The Cys598 of YAP1C-GS is essential for the formation of mixed disulfides with sulfenylated proteins (12). In addition, we constructed a similar control version, YAP1A-GS, in which all cysteines were mutated (Fig. S1B and Table S1). The cauliflower mosaic virus 35S promoter-driven constructs were transformed in Arabidopsis cell suspensions. Western blot analysis with a specific antibody complex to detect the G moiety of the tag [peroxidase-antiperoxidase (PAP) antibody complex] revealed that the yield of the two fusion proteins YAP1C-GS and YAP1A-GS is comparable and that they migrate as a single band at 35 kDa (Fig. S1C).
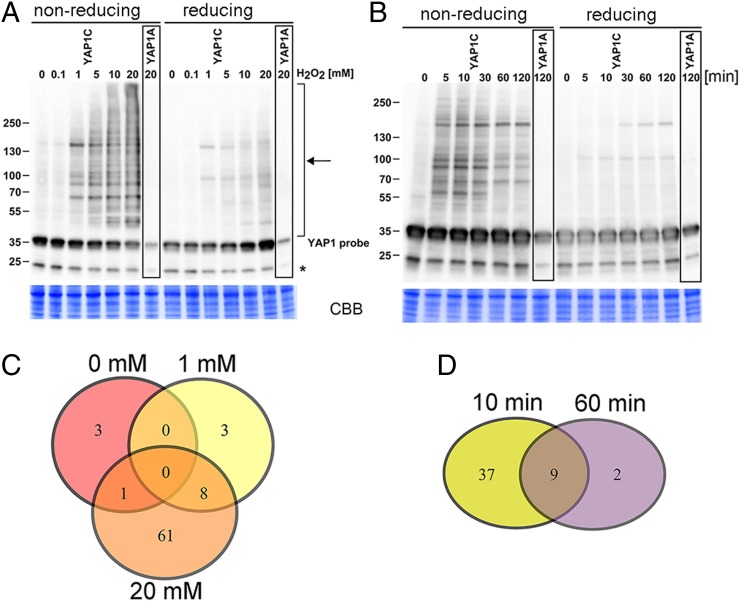
Previously, a 20-mM H2O2 treatment of Arabidopsis cells had been found to provoke oxidative stress signaling (14). We treated the transformed Arabidopsis cell suspension cultures with 0, 0.1, 1, 5, 10, and 20 mM H2O2 at the mid-log phase for 1 h. To block all free thiols, we extracted the soluble protein in the presence of iodoacetamide and N-ethylmaleimide (NEM) (Fig. S2) and analyzed the disulfide bond formation on a nonreducing Western blot with the PAP antibody complex. In untreated cells and in cells treated with 0.1 mM H2O2, YAP1C-GS migrated at 35 kDa, but H2O2 treatments ranging between 1 and 20 mM resulted in a proportional increase in the number of high-molecular weight YAP1C-GS complexes (Fig. 1A). The disulfide nature of the interactions has been proven by the disappearance of most of the high-molecular weight bands on a reducing Western blot (Fig. 1A). In cells producing YAP1A-GS, only the 35-kDa monomer is detected, even after a 20-mM H2O2 treatment, strengthening that Cys598 is essential for the YAP1-disulfide formation in plant cells under stress.
Fig. 1.
Dose- and time-dependent formation of YAP1C-involving complexes. (A) Cell cultures overproducing the YAP1C/YAP1A-GS probe treated with 0, 0.1, 1, 5, 10, and 20 mM H2O2 for 1 h. Complexes (marked with an arrow) are visualized with the PAP antibody complex. The H2O2 concentration and the signal intensity clearly correlate. Treatment of protein samples with 50 mM tris(2-carboxyethyl)phosphine led to reduction of the complexes. (B) Cell cultures treated with 1 mM H2O2. The time course was taken after 0, 5, 10, 30, 60, and 120 min. The initial signal intensity peak returns to a near basal level after 120 min of treatment. The asterisk denotes an unknown protein recognized by the antiserum. (C and D) Schematic comparison of datasets identified after treatment of cultures with 0, 1, and 20 mM H2O2 for 1 h (C) and 1 mM H2O2 for 10 min (early response) and 1 h (late response) (D).
Next, we checked the transient dynamic character of the intermolecular disulfide bond formation by YAP1C in a time-course experiment. Cell cultures were pulse-treated with 1 mM H2O2 and harvested 5, 10, 30, 60, and 120 min after treatment. The YAP1C complexes were most abundant after 10 min (Fig. 1B), but almost undetectable after 2 h. This decreased number of mixed disulfide bonds between YAP1C and target proteins could result from the protection by a resolving cysteine present in the target proteins, from the activation of the reducing systems [glutaredoxin (Grx)/GSH/glutathione reductase (GR), or thioredoxin (Trx)/Trx reductase system], or from the proteolytic degradation of proteins inactivated by sulfonic acid formation. Taken together, these results indicate that a YAP1-based sulfenic acid-trapping methodology is a solid tool to study time- and dose-dependent H2O2 stress responses in plant cells.
Unique Sulfenylated Proteins Are Selectively Trapped with YAP1C-GS.
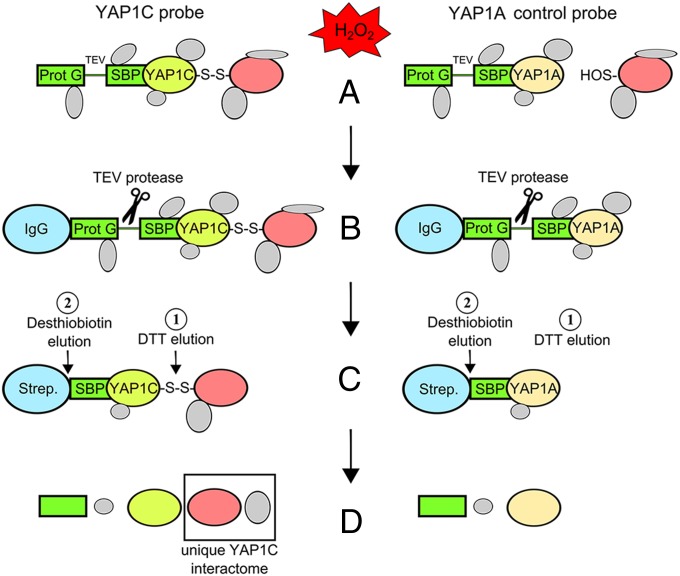
To evaluate the early signaling events in the presence of 1 mM H2O2, we decided to focus on cysteine oxidation 10 min after stress. Protein extracts of cells containing YAP1C-GS and YAP1A-GS were purified by TAP (Fig. 2). Briefly, first, YAP1-GS complexes were captured on IgG-Sepharose. Second, after a TEV protease cleavage step, the YAP1 fused to the SBP tag together with several mixed disulfide complexes and all possible interacting proteins were eluted. In a subsequent SBP purification step, the mixed disulfide YAP1 complexes were enriched. The sulfenylated proteins were released by selective elution of the disulfide-bonded proteins with 5 mM DTT followed by a 20-mM desthiobiotin elution. The majority of interactors (41 of 46) eluted with DTT (Dataset S1). To guarantee a complete interactor recovery, we decided to use desthiobiotin for all of the TAP purifications.
Fig. 2.
Experimental set-up for in vivo identification of the Arabidopsis sulfenome. (A) Cell cultures overproducing the YAP1C/YAP1A probes treated with H2O2 as described (see main text). (B and C) Proteins isolated and subjected to a two-step purification procedure based on IgG-protein G and SBP affinity. Numbers indicate the sequence of elution steps. (D) LC-MS/MS analysis of proteins after elution. Comparison of interactors between the negative control probes of YAP1C and YAP1A potentially undergoing cysteine sulfenic acid (-SOH) formation under oxidative stress.
Hereafter, we focused on the late-response sulfenome observed 1 h after an oxidative stress pulse with 0, 1, and 20 mM H2O2. In agreement with the increased mixed-disulfide complex formation that positively correlates with the H2O2 dose (Fig. 1A), we identified 4, 11, and 70 YAP1C-specific interactors (Fig. 1C, Table 1, and Dataset S1). Based on the background YAP1A datasets, 215 of the interactors can be regarded as nonspecific. Furthermore, almost all of the specific interactors contain at least one cysteine, except the histone superfamily protein, which might be a nondisulfide interactor of one of the YAP1C mixed-disulfide target proteins. The majority (8 of 11) of the specific interactors detected after treatment with 1 mM H2O2 are also present in the set of 70 proteins identified with 20 mM H2O2 (Fig. 1C). Nine of the 11 interactors derived from cells treated with 1 mM H2O2 for 1 h occur also at the 10-min early time point (Fig. 1D), emphasizing their sensitivity toward oxidation and suggesting an important function in oxidative stress sensing.
Table 1.
YAP1C-specific interactors within the first 10 min after the oxidative stress trigger
| AGI code | Description | H2O2 dynamics after 1 h |
Cys residues |
Cys PTM |
||
| 0 mM | 1 mM | 20 mM | ||||
| Signal perception and transduction | ||||||
| AT1G59580 | Map kinase 2 (MPK2) | 1 | 1 | 8 | ||
| AT4G01370 | Map kinase 4 (MPK4) | 1 | 8 | |||
| AT2G18170 | Map kinase 7 (MPK7) | 1 | 1 | 8 | ||
| AT3G50500 | SNF1-related protein kinase 2-2 (SNRK2-2) | 7 | ||||
| AT1G60940 | SNF1-related protein kinase 2-10 (SNRK2-10) | 1 | 6 | |||
| AT2G18790 | Phytochrome B (PHYB) | 25 | ||||
| AT2G43980 | Inositol 1,3,4-trisphosphate 5/6-kinase 4 (ITPK4) | 1 | 9 | |||
| AT1G71860 | Protein tyrosine phosphatase 1 (PTP1) | 7 | ||||
| AT1G51690 | Protein phosphatase 2A 55-kDa regulatory subunit (PP2A-b55α) | 2 | 2 | 15 | ||
| AT2G46900 | bHLH protein | 2 | 6 | |||
| Protein degradation | ||||||
| AT1G64520 | Regulatory particle non-ATPase 12A (RPN12A) | 4 | Trxs target | |||
| AT1G75950 | S phase kinase-associated protein 1 (SKP1) | 2 | 2 | 3 | ||
| AT2G47790 | CUL4-RING ubiquitin ligase complex subunit (GTS1) | 12 | ||||
| AT5G06600 | Ubiquitin-specific protease 12 (UBP12) | 11 | ||||
| AT3G11910 | Ubiquitin-specific protease 13 (UBP13) | 10 | ||||
| AT4G30890 | Ubiquitin-specific protease 24 (UBP24) | 3 | ||||
| AT4G17510 | Ubiquitin C-terminal hydrolase 3 (UCH3) | 1 | 4 | SOH | ||
| AT5G50870 | Ubiquitin-conjugating enzyme 27 (UBC27) | 1 | 4 | |||
| AT4G17830 | Peptidase M20/M25/M40 family protein | 1 | 8 | S-SG | ||
| AT5G14250 | COP9 signalosome subunit 3 (CSN3) | 1 | 8 | |||
| AT5G42190 | SKP-like protein 1B (SKP1B) | 1 | 2 | 3 | ||
| Redox related | ||||||
| AT1G65980 | Thioredoxin-dependent peroxidase 1 (TPX1) | 1 | 2 | SNO | ||
| AT1G75270 | Dehydroascorbate reductase 2 (DHAR2) | 1 | 2 | S-SG; Trxs target; S-S | ||
| AT4G04950 | Monothiol glutaredoxin 17 (GRXS17) | 2 | 2 | 6 | ||
| AT3G44190 | FAD/NAD(P)-binding oxidoreductase family protein | 2 | 3 | |||
| AT1G01800 | NAD(P)-binding Rossmann-fold superfamily protein | 4 | ||||
| RNA Binding translation | ||||||
| AT4G27000 | RNA-binding family protein (ATRBP45C) | 3 | ||||
| AT4G31120 | Protein arginine methyltransferase 5 (PRMT5) | 12 | ||||
| Primary metabolism | ||||||
| AT1G78900 | Vacuolar ATP synthase subunit A (VHA-A) | 2 | 6 | S-S | ||
| AT5G11670 | NADP-malic enzyme 2 (NADP-ME2) | 2 | 7 | S-SG | ||
| Amino acid metabolism | ||||||
| AT5G01410 | Pyridoxine biosynthesis 1.3 (PDX1.3) | 2 | 5 | |||
| AT5G17330 | Glutamate decarboxylase 1 (GAD1) | 1 | 2 | 7 | ||
| Protein transport | ||||||
| AT3G56190 | α-Soluble NSF attachment protein 2 (ASNAP) | 8 | ||||
| AT4G30550 | γ-Glutamyl peptidase 3 (GGP3) | 8 | ||||
| Miscellaneous and unknown functions | ||||||
| AT1G69800 | Cystathionine β-synthase (CBS) protein | 2 | 6 | |||
| AT2G42910 | Phosphoribosyltransferase family protein | 1 | 2 | 7 | ||
| AT3G07720 | Galactose oxidase/kelch repeat superfamily protein | 6 | ||||
| AT3G16520 | UDP-glucosyl transferase 88A1 (UGT88A1) | 1 | 9 | |||
| AT3G53180 | Nodulin/glutamine synthase-like protein (NODGS) | 9 | S-SG | |||
| AT3G63000 | NPL4-like protein 1 (NPL41) | 5 | ||||
| AT4G14710 | Acireductone dioxygenase 2 (ATARD2) | 5 | ||||
| AT4G29350 | Profilin 2 (PFN2) | 2 | Trxs target | |||
| AT5G13050 | 5-Formyltetrahydrofolate cycloligase (5-FCL) | 5 | ||||
| AT5G17270 | Protein prenylyltransferase superfamily protein | 2 | 20 | |||
| AT4G27450 | Unknown protein | 7 | Trxs target | |||
| AT3G29280 | Unknown protein | 6 | ||||
Numbers indicate the occurrence of proteins in two independent experiments after 1 h.
Functional Categorization of the Arabidopsis H2O2-Dependent Sulfenome.
In the Arabidopsis sulfenome we detected 67 proteins that, until now, had not been classified as sensitive to H2O2 and, additionally, 30 proteins that had previously been reported to have oxidative modifications, such as disulfides, S-glutathiolynation, S-nitrosylation, and sulfenic acids, and some to be Trx/Grx substrates (Table S2). A first step in the redox-dependent signaling pathway involves reversible sulfenic acid formation that later rapidly reacts with other thiols to form intra- or intermolecular disulfides, for example by S-glutathionylation. In a next step, specific redox enzymes, such as Trxs and Grxs, reduce these disulfides. The identification of S-glutathionylated proteins or Trx/Grx target proteins in the Arabidopsis sulfenome opens an interesting route for further investigation of the physiological consequence related to the pathways in which these enzymes operate.
Sixty-six of the proteins in the identified sulfenome could be functionally categorized: 13 are involved in signal perception and transduction, 19 in protein degradation, 7 in RNA-binding and translation, 6 in primary metabolism, 4 in hormone homeostasis, 5 in protein transport, 5 in amino acid metabolism, 7 are redox-related enzymes, and 31 have miscellaneous and unknown functions. For the complete list of interactors and the corresponding reference classification, see Table S2. When we focus on the 46 sulfenylated proteins within the first 10 min after the oxidative stress (Table 1), we find several signal perception and transduction proteins, of which almost one-fourth has proteasomal activities; moreover, some of several identified redox-related enzymes are detected at an early (10 min) as well as a late (1 h) oxidative stress response, whereas additional members of these functional classes are present within the late response (Table S2).
Signal Perception and Transduction.
Three different MAPKs have been found to be sulfenylated: MAPK2, MAPK4, and MAPK7. The MAPK signaling cascades, including MAPKs, are integral parts of plant biotic and abiotic stress signaling pathways and are activated by H2O2 (15, 16). In yeast and human model systems, MAPKs are redox-regulated through upstream regulators and through direct cysteine oxidation events (17). A cysteine oxidation event in the human p38 MAPK had been shown to act as a functional regulatory switch (18), whereas in plant MAPK modules, such a thiol modification has not yet been reported. The fast sulfenylation after an oxidative stress stimulus suggests that MAPK2, MAPK4, and MAPK7 could function as redox sensors downstream of a reactive oxygen species (ROS)-producing event. The MAPK activity is also controlled through dephosphorylation by redox-sensitive phosphatases (17). Sulfenylation of the catalytic nucleophilic cysteine leads to the inhibition of protein tyrosine phosphatases (PTPs) (19). The identified Arabidopsis AtPTP1 undergoes cysteine-dependent inhibition by H2O2 and negatively regulates the MAPKs (20), suggesting that the oxidation-dependent AtPTP1 inhibition might be a primary step in the oxidative stress response leading to MAPK signaling derepression (21). Furthermore, at least two members of the plant-specific SNF1-related protein kinase2 (SnRK2) family are rapidly sulfenylated. The plant stress response SnRK2 pathways are regulated by direct phosphorylation of various downstream targets, including the respiratory burst oxidase homolog RbohF and TFs, which are required for the expression of numerous stress response genes (22). Only recently, a redox-regulated rapeseed (Brassica napus) SnRK2 was shown to be involved in guard cell signaling (23).
Protein Degradation.
Approximately 20% of the YAP1C-GS interactors are involved in proteolysis, with a clear enrichment for proteins participating in proteasome-mediated degradation, among which five control ubiquitination, such as the UBIQUITIN-CONJUGATING ENZYME27 (UBC27), two subunits of the Skp/Cullin/F-box (SCF) E3-ubiquitin ligase complex (ASK1 and ASK2), and the 3 and 5A subunits of the constitute photomorphogenic9 (COP9) signalosome. In the 26S proteasome, we identified the REGULATORY PARTICLE NON-ATPASE 12A (RPN12A) as a potentially H2O2-modified protein. Interestingly, RPN12A has been established as a cytoplasmic thioredoxin h (Trxh) target protein (24) that acts as a potential cross-talk point in cytokinin signaling (25). The removal of oxidatively damaged proteins is an important event in the stress responses (26), but oxidative modifications of the proteasome and the ubiquitin-proteasome system itself trigger changes in activities in such a manner that it manages both the removal of oxidized proteins and the adaptation of the cellular metabolism to the stress situation (27). In maize (Zea mays), sugar starvation-triggered oxidative stress leads to oxidative modifications of the 20S proteasome, modulating its proteolytic activity (28). In addition, our dataset includes a number of de-ubiquitinating enzymes (DUBs), such as UBIQUITIN C-TERMINAL HYDROLASE 3 (UCH3) and the ubiquitin-specific proteases UBP12, UBP13, and UBP24. Redox regulation of multiple ovarian tumor DUBs has been reported to occur via reversible sulfenylation of a catalytic cysteine residue (29), but until now this inhibition mode has not been described for plant DUBs.
Redox Proteins.
At least four redox-related proteins have been detected: MONOTHIOL GLUTAREDOXIN17 (GRXS17), THIOREDOXIN-DEPENDENT PEROXIDASE1 (TPX1), GLUTAREDOXIN C2 (GRXC2), and a DHAR2. Other well-documented ROS-scavenging enzymes of plants, such as glutathione peroxidases, peroxiredoxins, and methionine sulfoxide reductases are not present in the list, possibly because of their highly specialized redox mechanisms that often involve resolving cysteines for the rapid conversion of sulfenic acids (2), hence impeding the formation of mixed disulfides with YAP1C. TPX1 had been identified as a target of cytosolic Trxh3 (30) and also in a subset of early-responsive redox-sensitive proteins (31). GRXC2 and GRXS17 function in early plant development during embryonic (32) and temperature-dependent postembryonic (33) growth, respectively, but their specific substrate proteins are unknown. DHAR2 was sulfenylated in both the early and late oxidative stress responses (Table S2). DHAR plays an important role in counterbalancing oxidative stress by catalyzing the regeneration of ascorbate, a major antioxidant in plants. In Arabidopsis, three isoforms are present: the mitochondrial DHAR1, the cytosolic DHAR2, and a chloroplastic DHAR3. In planta, perturbation of DHAR has been shown to limit ascorbate recycling, consequently influencing the rate of plant growth and leaf aging due to ROS-mediated damage (34).
DHAR2 Kinetics Are Affected by H2O2 Treatment.
DHAR2 catalyzes the reduction of oxidized ascorbate with a concomitant oxidation of GSH to GSSG; therefore, it is one of the core enzymes of the GSH/ascorbate cycle that maintains reduced ascorbate pools (35). We recombinantly produced and purified His-tagged DHAR2. Recombinant DHAR2 eluted as a monomer from the Ni2+-immobilized metal-affinity column and migrated as a single band at 25 kDa on a SDS-PAGE gel. Its molecular weight was confirmed by MS with a total mass of 26,748 Da after loss of the N-terminal methionine (Fig. S3).
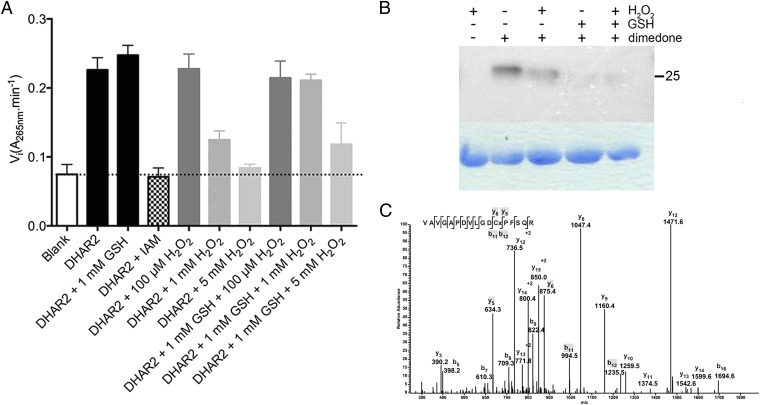
We analyzed the activity of recombinant DHAR2 by following the ascorbate formation in progress curves in function of time at 265 nm. The initial velocities were measured at varying dehydroascorbate (DHA) concentrations in a 5-mM excess of GSH. Plots of the initial velocities versus the DHA concentrations revealed a sigmoidal curve with a Hill factor of 2.65, indicative for positive cooperativity and hinting at the positive influence of the GSH binding at increasing DHA concentrations. For DHAR2, a kcat/K0.5 value of 9.3 × 105 M-1⋅s−1 was determined with a K0.5 of 23.8 ± 1.2 µM, whereas for DHAR1 and DHAR3, KM values of 260 µM and 500 µM had been reported (36). To understand the possible role of the cysteine thiols of DHAR2 in its catalytic cycle, we modified the free thiols with 1 mM iodoacetamide and oxidized DHAR2 with increasing concentrations of H2O2 (Fig. 3A). In both cases, the activity is affected, indicating that cysteines are essential for catalytic DHAR2 activity, as observed previously for DHAR1 (36).
Fig. 3.
Requirement of DHAR2 cysteines and GSH protection against overoxidation. (A) Mean initial velocities ± SD of three independent measurements of DHAR2 determined on progress curves under different conditions. The reaction was started under Vmax conditions. (B) Dimedone labeling of DHAR2-SOH in vitro. DHAR2 (20 µM) nontreated or treated with 1 mM GSH either not or incubated with 100 µM H2O2 in the presence or absence of dimedone (1 mM). DHAR2-SOH formation was analyzed by immunoblot with an anticysteine sulfenic acid antibody. (C) Identification of the dimedone modification Cys20 of DHAR2. The LC-MS/MS spectrum shows data obtained from a +2 parent ion with m/z 935.5. The cysteine residue corresponds to a dimedone-modified sulfenic acid, which produces a +138-Da mass increment.
DHAR2 Is Sulfenylated and S-glutathionylated on Its Nucleophilic Cysteine.
DHAR2 contains two cysteines (Cys6 and Cys20). To prove the sulfenylation of DHAR2 by H2O2, we used 5,5-dimethyl-1,3-cyclohexadione (dimedone), a chemical compound that forms a thioether bond with the electrophilic sulfur atom of sulfenylated proteins (37). Previously, dimedone and its derivatives have successfully been applied to study sulfenylation in many important physiological pathways in various organisms and in recombinant proteins (38). We analyzed the H2O2-induced DHAR2 sulfenylation in the presence and absence of GSH on Western blots with antibodies that specifically recognize dimedone-tagged sulfenic acids (Fig. 3B). After a 30-min treatment with 100 µM H2O2 in the presence of dimedone, DHAR2 was sulfenylated. Remarkably, sulfenylation was lower in H2O2-treated than in nontreated samples, possibly because of a rapid rescue of sulfenic acids by resolving cysteines or overoxidation to sulfinic or sulfonic acids (2). In the presence of 1 mM GSH, the sulfenylation signal did not depend on the H2O2 treatment, suggesting that the majority of the formed sulfenic acid is unavailable for dimedone because a mixed disulfide is formed with GSH. To confirm this observation, we analyzed H2O2-treated DHAR2 in the presence of dimedone or GSH by LC-MS/MS (Table S3). After treatment with 100 µM H2O2 in the presence of dimedone, we blocked all free thiols. A tryptic digest revealed a dimedone adduct on the Cys20 peptide, resulting in a 138-Da mass increase of this peptide compared with the parent peptide (Fig. 3C), confirming that Cys20 is sulfenylated. Cys20 is also partially overoxidized to sulfonic acid (48-Da larger than the parent peptides). In contrast, in the absence of dimedone but in the presence of GSH, MS data clearly show S-glutathionylation at Cys20 (Fig. S4).
To test the 1-mM GSH protection on the DHAR2 activity, we added increasing concentrations of H2O2 to DHAR2 in the presence of 1 mM GSH and determined the initial velocities (Vi) of the progress curves (Fig. 3A). The Vi of the GSH-pretreated DHAR2 sample is slightly higher than that of the nontreated sample, which is, at least, partially sulfenylated after purification (Fig. 3B). At low H2O2 concentrations a disulfide is formed between C6 and C20 together with partial overoxidation of C20 (Table S3), without affecting the DHAR2 activity (Fig. 3A). However, at higher H2O2 concentrations (1 and 5 mM), sulfonic acid is more probably formed (Table S3) and associated with decreased activity (Fig. 3A). This progressive oxidation is further supported by the fact that GSH (1 mM) rescues the 1-mM H2O2 sulfenylation of DHAR2 from overoxidation, whereas at 5 mM H2O2, the DHAR2 activity could only partially be rescued by GSH (Fig. 3A). Possibly, the overoxidation rate is too fast for 1 mM GSH to react with the sulfenic acid to recover all activity (Fig. 3A). Thus, in the absence of substrate, S-glutathionylation occurs after sulfenylation of Cys20 as a reversible protection mechanism, recovering DHAR2 activity, which results in increased initial velocities. Taking these data together, we show that the nucleophilic Cys20 in DHAR2 is vulnerable to oxidation and becomes sulfenylated under H2O2 stress. The formation of a Cys20–Cys6 disulfide bond or the Cys20 S-glutathionylation might protect DHAR2 against irreversible overoxidation.
Materials and Methods
A full discussion of materials and methods can be found in SI Materials and Methods.
Arabidopsis Suspension Cultures.
The Saccharomyces cerevisiae YAP1-coding region corresponding to the Asn565-to-Asn650 region was codon optimized for expression in A. thaliana (L.) Heynh., synthesized, and mutated to create the YAP1C and YAP1A probes, as described previously (12).
Stress Treatments.
Mid-log phase cell cultures expressing the YAP1C/YAP1A N-terminal GS-tag fusions were treated with 0.1, 1, 5, 10, and 20 mM H2O2. Cells were harvested after 1 h.
Protein Extractions, Western Blot Analysis, and TAP.
Plant material was ground on ice in the presence of sand and TAP extraction buffer (39). For the Western blot analysis, soluble proteins were separated on SDS-PAGE gradient gel (Bio-Rad), blotted, and hybridized with a 1:5,000 dilution of PAP-soluble complex (Sigma-Aldrich) to detect the GS tag. The TAP protocol was adapted to allow the use of the redox-active baits YAP1A-GS and YAP1C-GS.
For details on design and cloning of YAP1A-GS and YAP1C-GS and on cloning and purification of recombinant DHAR2, DHAR2 activity/inhibition assays and in vitro sulfenic acid labeling, and MS on DHAR2, see SI Materials and Methods. PCR primers used for cloning of probes are listed in Table S4.
Supplementary Material
Acknowledgments
We thank Eveline Van De Slijke, Nancy De Winne, Brigitte van de Cotte, and Debbie Rombaut for excellent technical assistance; and Martine De Cock for help in preparing the manuscript. This work was supported by grants from Ghent University Multidisciplinary Research Partnership [“Ghent BioEconomy” (Project 01MRB 510W)] and Bijzondere Onderzoeksfonds (BOF 01J11311); the Interuniversity Attraction Poles Program (Grant IUAP VII/29), initiated by the Belgian State, Science Policy Office; Research Foundation-Flanders (Projects G.0D.79.14N and G.0038.09N); the European Cooperation in Science and Research (COST Action BM1203/EU-ROS); a VIB International PhD Program predoctoral fellowship (to C.W.); and an Erasmus Mundus External Cooperation Window predoctoral fellowship (to S.A.); D.V. is “collaborateur logistique” from the Fonds National de la Recherche Scientifique-Fonds de la Recherche Scientifique Belgium. B.D.S. is a predoctoral fellow of the Research Foundation-Flanders. I.V.M. is the recipient of an Omics@vib Marie Curie COFUND fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411607111/-/DCSupplemental.
References
- 1.Garavelli JS. The RESID Database of Protein Modifications as a resource and annotation tool. Proteomics. 2004;4(6):1527–1533. doi: 10.1002/pmic.200300777. [DOI] [PubMed] [Google Scholar]
- 2.Roos G, Messens J. Protein sulfenic acid formation: From cellular damage to redox regulation. Free Radic Biol Med. 2011;51(2):314–326. doi: 10.1016/j.freeradbiomed.2011.04.031. [DOI] [PubMed] [Google Scholar]
- 3.Ma L-H, Takanishi CL, Wood MJ. Molecular mechanism of oxidative stress perception by the Orp1 protein. J Biol Chem. 2007;282(43):31429–31436. doi: 10.1074/jbc.M705953200. [DOI] [PubMed] [Google Scholar]
- 4.Balsera M, Uberegui E, Schürmann P, Buchanan BB. Evolutionary development of redox regulation in chloroplasts. Antioxid Redox Signal. 2014 doi: 10.1089/ars.2013.5817. [DOI] [PubMed] [Google Scholar]
- 5.Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113(7):935–944. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- 6.Tavakoli N, Kluge C, Golldack D, Mimura T, Dietz K-J. Reversible redox control of plant vacuolar H+-ATPase activity is related to disulfide bridge formation in subunit E as well as subunit A. Plant J. 2001;28(1):51–59. doi: 10.1046/j.1365-313x.2001.01130.x. [DOI] [PubMed] [Google Scholar]
- 7.Shaikhali J, et al. The redox-sensitive transcription factor Rap2.4a controls nuclear expression of 2-Cys peroxiredoxin A and other chloroplast antioxidant enzymes. BMC Plant Biol. 2008;8:48. doi: 10.1186/1471-2229-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaikhali J, et al. Redox-mediated mechanisms regulate DNA binding activity of the G-group of basic region leucine zipper (bZIP) transcription factors in Arabidopsis. J Biol Chem. 2012;287(33):27510–27525. doi: 10.1074/jbc.M112.361394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viola IL, Güttlein LN, Gonzalez DH. Redox modulation of plant developmental regulators from the class I TCP transcription factor family. Plant Physiol. 2013;162(3):1434–1447. doi: 10.1104/pp.113.216416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fomenko DE, et al. Thiol peroxidases mediate specific genome-wide regulation of gene expression in response to hydrogen peroxide. Proc Natl Acad Sci USA. 2011;108(7):2729–2734. doi: 10.1073/pnas.1010721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delaunay A, Pflieger D, Barrault M-B, Vinh J, Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111(4):471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- 12.Takanishi CL, Ma L-H, Wood MJ. A genetically encoded probe for cysteine sulfenic acid protein modification in vivo. Biochemistry. 2007;46(50):14725–14732. doi: 10.1021/bi701625s. [DOI] [PubMed] [Google Scholar]
- 13.Van Leene J, Witters E, Inzé D, De Jaeger G. Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci. 2008;13(10):517–520. doi: 10.1016/j.tplants.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Desikan R, A-H-Mackerness S, Hancock JT, Neill SJ. Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 2001;127(1):159–172. doi: 10.1104/pp.127.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovtun Y, Chiu W-L, Tena G, Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA. 2000;97(6):2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, et al. RBOH1-dependent H2O2 production and subsequent activation of MPK1/2 play an important role in acclimation-induced cross-tolerance in tomato. J Exp Bot. 2014;65(2):595–607. doi: 10.1093/jxb/ert404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Truong TH, Carroll KS. Redox regulation of protein kinases. Crit Rev Biochem Mol Biol. 2013;48(4):332–356. doi: 10.3109/10409238.2013.790873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Templeton DJ, Aye M-S, Rady J, Xu F, Cross JV. Purification of reversibly oxidized proteins (PROP) reveals a redox switch controlling p38 MAP kinase activity. PLoS ONE. 2010;5(11):e15012. doi: 10.1371/journal.pone.0015012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanner JJ, Parsons ZD, Cummings AH, Zhou H, Gates KS. Redox regulation of protein tyrosine phosphatases: Structural and chemical aspects. Antioxid Redox Signal. 2011;15(1):77–97. doi: 10.1089/ars.2010.3611. [DOI] [PubMed] [Google Scholar]
- 20.Gupta R, Luan S. Redox control of protein tyrosine phosphatases and mitogen-activated protein kinases in plants. Plant Physiol. 2003;132(3):1149–1152. doi: 10.1104/pp.103.020792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartels S, et al. MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell. 2009;21(9):2884–2897. doi: 10.1105/tpc.109.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulik A, Wawer I, Krzywińska E, Bucholc M, Dobrowolska G. SnRK2 protein kinases—Key regulators of plant response to abiotic stresses. OMICS. 2011;15(12):859–872. doi: 10.1089/omi.2011.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu M, et al. Thiol-based redox proteins in abscisic acid and methyl jasmonate signaling in Brassica napus guard cells. Plant J. 2014;78(3):491–515. doi: 10.1111/tpj.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamazaki D, Motohashi K, Kasama T, Hara Y, Hisabori T. Target proteins of the cytosolic thioredoxins in Arabidopsis thaliana. Plant Cell Physiol. 2004;45(1):18–27. doi: 10.1093/pcp/pch019. [DOI] [PubMed] [Google Scholar]
- 25.Ryu MY, Cho SK, Kim WT. RNAi suppression of RPN12a decreases the expression of type-A ARRs, negative regulators of cytokinin signaling pathway, in Arabidopsis. Mol Cells. 2009;28(4):375–382. doi: 10.1007/s10059-009-0132-x. [DOI] [PubMed] [Google Scholar]
- 26.Jung T, Grune T. The proteasome and the degradation of oxidized proteins: Part I-structure of proteasomes. Redox Biol. 2013;1(1):178–182. doi: 10.1016/j.redox.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung T, Höhn A, Grune T. The proteasome and the degradation of oxidized proteins: Part III-Redox regulation of the proteasomal system. Redox Biol. 2014;2:388–394. doi: 10.1016/j.redox.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basset G, Raymond P, Malek L, Brouquisse R. Changes in the expression and the enzymic properties of the 20S proteasome in sugar-starved maize roots. Evidence for an in vivo oxidation of the proteasome. Plant Physiol. 2002;128(3):1149–1162. doi: 10.1104/pp.010612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulathu Y, et al. Regulation of A20 and other OTU deubiquitinases by reversible oxidation. Nat Commun. 2013;4:1569. doi: 10.1038/ncomms2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchand C, Le Maréchal P, Meyer Y, Decottignies P. Comparative proteomic approaches for the isolation of proteins interacting with thioredoxin. Proteomics. 2006;6(24):6528–6537. doi: 10.1002/pmic.200600443. [DOI] [PubMed] [Google Scholar]
- 31.Wang H, et al. Proteomic analysis of early-responsive redox-sensitive proteins in Arabidopsis. J Proteome Res. 2012;11(1):412–424. doi: 10.1021/pr200918f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riondet C, et al. A dicotyledon-specific glutaredoxin GRXC1 family with dimer-dependent redox regulation is functionally redundant with GRXC2. Plant Cell Environ. 2012;35(2):360–373. doi: 10.1111/j.1365-3040.2011.02355.x. [DOI] [PubMed] [Google Scholar]
- 33.Cheng N-H, et al. Arabidopsis monothiol glutaredoxin, AtGRXS17, is critical for temperature-dependent postembryonic growth and development via modulating auxin response. J Biol Chem. 2011;286(23):20398–20406. doi: 10.1074/jbc.M110.201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z, Gallie DR. Dehydroascorbate reductase affects leaf growth, development, and function. Plant Physiol. 2006;142(2):775–787. doi: 10.1104/pp.106.085506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foyer CH, Halliwell B. Purification and properties of dehydroascorbate reductase from spinach leaves. Phytochemistry. 1977;16(9):1347–1350. [Google Scholar]
- 36.Dixon DP, Davis BG, Edwards R. Functional divergence in the glutathione transferase superfamily in plants. Identification of two classes with putative functions in redox homeostasis in Arabidopsis thaliana. J Biol Chem. 2002;277(34):30859–30869. doi: 10.1074/jbc.M202919200. [DOI] [PubMed] [Google Scholar]
- 37.Benitez LV, Allison WS. The inactivation of the acyl phosphatase activity catalyzed by the sulfenic acid form of glyceraldehyde 3-phosphate dehydrogenase by dimedone and olefins. J Biol Chem. 1974;249(19):6234–6243. [PubMed] [Google Scholar]
- 38.Paulsen CE, Carroll KS. Chemical dissection of an essential redox switch in yeast. Chem Biol. 2009;16(2):217–225. doi: 10.1016/j.chembiol.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 39.Van Leene J, et al. A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol Cell Proteomics. 2007;6(7):1226–1238. doi: 10.1074/mcp.M700078-MCP200. [DOI] [PubMed] [Google Scholar]