Summary
Tumor suppressor genes (TSGs) are often concomitantly lost or mutated in human cancers and have been shown to act synergistically to promote tumorigenesis. In addition to genomic alterations, post-transcriptional regulation by microRNAs (miRNAs) represents another mechanism by which TSG expression is dysregulated in cancers. Although miRNAs which target critical TSGs such as PTEN or p53 have been identified, little is known about miRNAs that concomitantly regulate both these key TSGs. In this study, we characterize miR-518c* and miR-638 as dual PTEN and p53-targeting miRNAs which are upregulated in multiple human cancers. We focus on miR-638 and show that it associates independently with these two tumor suppressor transcripts as well as BRCA1, a known miR-638 target. We find that miR-638 overexpression promotes tumorigenesis and demonstrate cooperativity between miR-638 and its host gene Dnm2, suggesting that the Dnm2 locus encodes two distinct oncogenic components which play important roles in tumorigenesis.
Introduction
PTEN (phosphatase and tensin homolog deleted on chromosome 10) is one of the most frequently lost and mutated tumor suppressor genes (TSGs) in a wide spectrum of human cancers. PTEN encodes a plasma-membrane lipid phosphatase that functions predominantly as an inhibitor of the proto-oncogenic PI3K-AKT signaling pathway which regulates cell growth and survival (Song et al., 2012). In vivo models which demonstrate that heterozygous PTEN loss and even subtler reduction in its expression lead to increased incidence of spontaneous tumors in multiple tissue types provide further support for the key tumor suppressive function of PTEN (Berger et al., 2011; Di Cristofano et al., 1998).
P53, epitomized as the ‘guardian of the genome’, is one of the most studied and frequently mutated or deleted TSGs. It encodes a transcription factor which regulates the expression of target genes involved in DNA repair, cell cycle arrest, apoptosis, senescence and other cellular processes (Vousden and Lane, 2007). Additional evidence for the importance of p53 in tumorigenesis was provided by mouse models which demonstrated that both the heterozygous or homozygous loss of p53 leads to an increased risk of cancer in a variety of tissues (Donehower et al., 1992).
The functions of these two TSGs are intricately intertwined. PTEN has been shown to physically associate with p53, regulate its DNA binding activity and protect it from Mdm2-mediated degradation (Freeman et al., 2003; Mayo et al., 2002). Conversely, p53 has been reported to regulate PTEN transcription (Stambolic et al., 2001). Several studies have examined the functional relevance of this reciprocal cooperation between PTEN and p53: Chen et al found that although Pten inactivation leads to non-lethal invasive cancer in the prostate after long latency and p53 inactivation did not lead to cancer, combined Pten and p53 inactivation resulted in invasive prostate cancer in mice with lethality by 7 months (Chen et al., 2005). Puzio-Kuter et al observed that Pten or p53 single mutant mice did not develop bladder tumors up to a year of age, but inactivation of both genes led to large tumors with 100% penetrance by 6 months (Puzio-Kuter et al., 2009). Andjelkovic et al found that the loss of both these genes occurred frequently in non-small cell lung cancer patients, was associated with increased lymph node invasion and reduced survival (Andjelkovic et al., 2011).
Collectively, these reports suggest that these two TSGs may cross-talk at multiple levels and that their compound inactivation is an event that is strongly selected for in human cancers. This in turn led us to hypothesize that mechanisms which induce the concomitant downregulation of both these tumor suppressors may play key roles in cancer development. Aside from genetic deletions or mutations which result in the heterozygous or homozygous loss of PTEN or p53 function, other mechanisms which contribute to the dysregulation of PTEN or p53 expression in cancers include epigenetic silencing, transcriptional repression, post-translational modifications and post-transcriptional regulation by microRNAs (miRNAs) (Song et al., 2012).
MiRNAs are small non-coding RNAs which modulate gene expression by binding to specific recognition sites on target transcripts (Bartel and Chen, 2004). Several studies have identified miRNAs that target either the PTEN or p53 transcripts (Hermeking, 2012; Tay et al., 2013). However, little is known about miRNAs that concomitantly regulate both these key TSGs. In this study, we characterize miR-518c* and miR-638 as dual PTEN and p53-targeting miRNAs. We find that overexpression of miR-638, which is encoded in the oncogenic Dnm2 locus, promotes tumorigenic properties including cell proliferation, migration and invasion. These data suggest that the Dnm2 gene comprises a two-hit oncogenic locus which plays an important role in cancer progression.
Results
Identification of potential dual PTEN and p53-targeting miRNAs
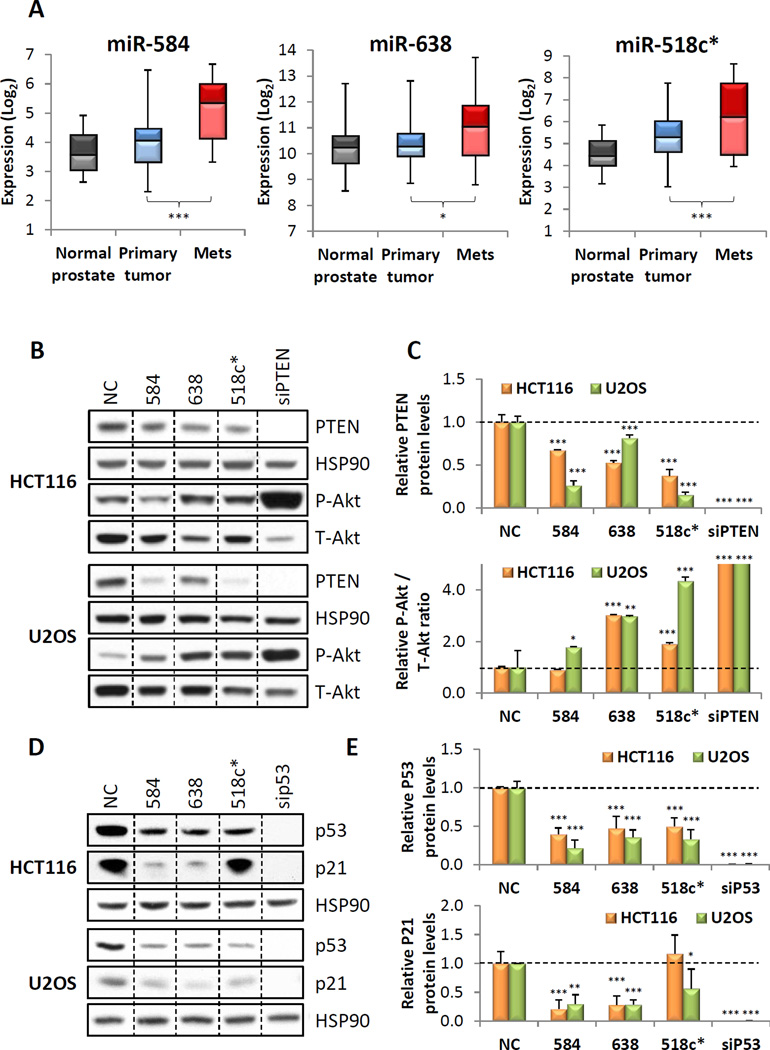
We first performed miRNA target prediction analyses to identify miRNAs which may target both full-length PTEN and p53 transcripts (Table S1). As the combined loss of PTEN and p53 has been shown to promote prostate cancer metastasis (Abou-Kheir et al., 2011), we next examined the expression profiles of these miRNAs in prostate cancer. Of the 51 predicted dual-targeting miRNAs, 14 were upregulated in human metastatic prostate cancer samples (Figure 1A and S1A). We selected miRs-518c*, 584 and 638 for experimental validation as their expression levels were also increased in colon and kidney cancers (Figure S1B and C), additional cancers with frequent loss of PTEN and p53.
Figure 1. Identification of potential dual PTEN and p53-targeting miRNAs.
(A) Expression of miRs-584, 638 and 518c* in primary and metastatic prostate cancer compared to normal prostate. (B,C) Effect of miR-584, 638 or 518c* overexpression on endogenous PTEN and P-Akt expression. Representative western blots are shown in (B), and quantitation of all blots is shown in (C). (D,E) Effect of miR-584, 638 or 518c* overexpression on endogenous p53 and p21 expression. Representative western blots are shown in (D), and quantitation of all blots is shown in (E). (A,C,E) *p < 0.05; **p , 0.01; ***p < 0.001. See also Figure S1 and Table S1.
Next, we investigated their effect on endogenous PTEN and p53 levels. Overexpression of miRs-518c*, 584 and 638 resulted in a significant decrease in PTEN protein levels in both HCT116 colon cancer and U2OS osteosarcoma cell lines which express wild-type PTEN and p53 (Figure 1B and C). This was accompanied by a concomitant increase in Akt activation. Similarly, miRs-518c*, 584 and 638 reduced p53 protein levels and decreased expression of p21, a downstream effector of p53 (Figure 1D and E).
Direct regulation of the PTEN and p53 transcripts by miRs-518c* and 638
As PTEN and p53 have been shown to reciprocally co-regulate each other as discussed above, we next investigated whether these observed effects were due to bona fide miRNA targeting or were alternatively a result of protein-mediated crosstalk. PC3 prostate cancer cells were selected as an ideal model system for this purpose as they are PTEN-null and p53-null, and do not express detectable levels of either transcript or protein (Isaacs et al., 1991; Vlietstra et al., 1998).
miRs-518c*, 584 and 638 have predicted target sites in the 5’UTR and/or coding region of PTEN and p53 (Figure S2A). To examine whether these predictions represented functional binding sites, we first constructed vectors expressing the 5’UTR+coding region of PTEN and p53, and co-transfected them into PC3 cells with miRs-518c*, 584 or 638. Overexpression of miRs-518c* and 638 significantly downregulated the expression of both 5’UTR+CDS constructs, suggesting that their effect on PTEN and p53 is partly 3’UTR-independent (Figure 2A and B). Although miR-584 also downregulated expression of exogenous PTEN, it did not have a significant effect on exogenous p53 expression. This suggests that the predicted miR-584 MREs in p53 are not bona fide MREs, and that its effect on endogenous p53 is most likely indirect.
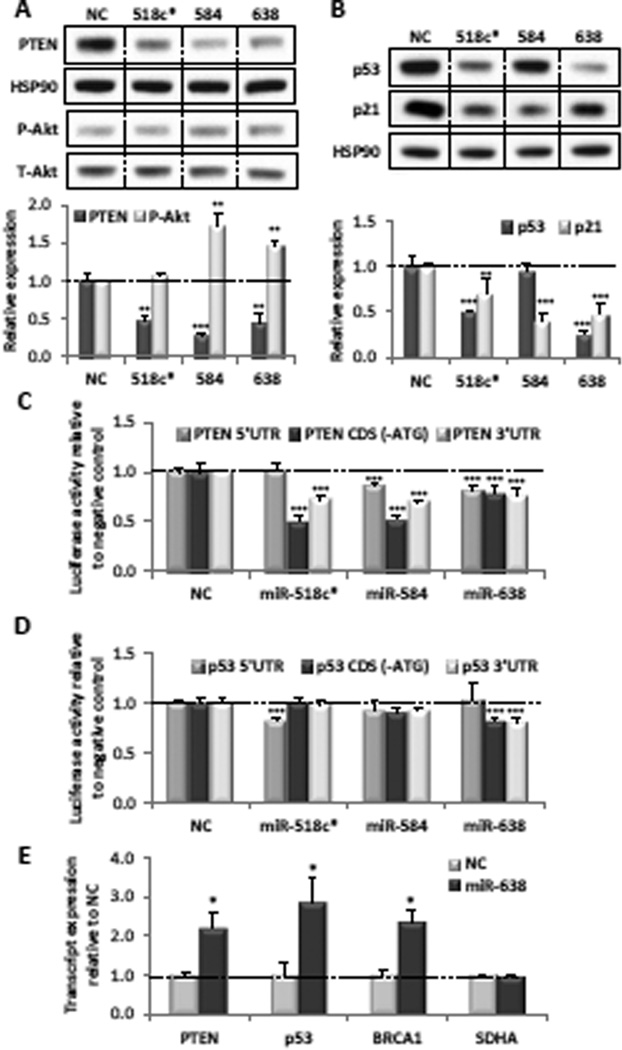
Figure 2. miRs-638 and 518c* directly target PTEN and p53.
(A) Expression of exogenous PTEN and P-Akt in PC3 cells after co-transfection of the negative control (NC), miR-518c*, miR-584 or miR-638 with a plasmid construct expressing only the PTEN 5’UTR+CDS. (B) Expression of exogenous p53 and p21 in PC3 cells after co-transfection of the NC, miR-518c*, miR-584 or miR-638 with a plasmid construct expressing only the p53 5’UTR+CDS. (C,D) Luciferase validation assays demonstrating the effect of miR-518c*, miR-584 or miR-638 overexpression on the 5’UTR, 3’UTR and ATG-less CDS fragments of (C) PTEN and (D) p53 relative to the NC in PC3 cells. (E) Expression of PTEN, p53 and BRCA1 mRNAs in biotinylated miR-638 pulldowns from U2OS cells. (A–E) Mean ± SD; n ≥ 4; *p < 0.05; **p , 0.01; ***p < 0.001. See also Figure S2.
Next, we generated luciferase reporter constructs harbouring the various fragments (5’UTR, CDS [-ATG], 3’UTR) of the PTEN and p53 transcripts. Overexpression of miR-638 resulted in a modest but significant decrease in expression of the PTEN-5’UTR, CDS (-ATG) and 3’UTR reporters, as well as the p53-CDS (-ATG) and 3’UTR reporters (Figure 2C and D). It did not affect the p53-5’UTR reporter. These results are consistent with the location of predicted MREs on these transcripts.
miR-518c* overexpression reduced expression of the PTEN-CDS(-ATG) and p53-5’UTR constructs, consistent with the location of several predicted MREs (Figure 2C and D). However, it did not affect expression of the PTEN-5’UTR or p53-CDS(-ATG) reporters, suggesting that these predictions were not bona fide MREs. Furthermore, it significantly reduced expression of the PTEN-3’UTR reporter, suggesting the existence of additional MREs.
miR-584 overexpression reduced expression of the PTEN-5’UTR and PTEN-CDS(-ATG) constructs, consistent with the location of its predicted PTEN MREs (Figure 2C). Its ability to reduce expression of the PTEN-3’UTR reporter suggests that it may also have additional target sites there. Consistent with our earlier observations, it did not have a significant effect on any of the p53 reporters.
Taken together, these data provide evidence that PTEN is a bona fide target of miR-584 and suggest that both miRs-638 and 518c* are dual PTEN and p53 targeting miRNAs.
miR-638 independently targets PTEN, p53 and BRCA1
Intriguingly, BRCA1 has been identified as a target of miR-638 (Nicoloso et al., 2010). BRCA1 is an important TSG involved in DNA damage repair and the maintenance of genomic stability, and the BRCA1 protein has been shown to reciprocally co-regulate p53 and Akt signalling (Altiok et al., 1999; Zhang et al., 1998). To disentangle potential miRNA-mediated regulation from protein-dependent crosstalk, we performed biotinylated miRNA pulldowns to examine the direct association of miR-638 with each of these tumor suppressor transcripts. We found that PTEN, p53 and BRCA1 transcripts were enriched in miR-638 pulldowns, suggesting that they all independently associate with miR-638 (Figure 2E).
miR-638 was shown to regulate BRCA1 by binding to a single MRE in its coding region. To identify the specific MREs responsible for its regulation of PTEN and p53, we cloned the predicted miR-638 MREs downstream of a luciferase reporter. Overexpression of miR-638 resulted in a modest but significant reduction in the levels of all these reporter constructs (Figure S2B), except for the PTEN-C2 MRE which appeared to have a cell-type-dependent effect.
Although miRNA binding to MREs in the coding regions and 3’UTRs of target genes generally results in the repression of gene expression, miRNA binding to 5’UTRs may enhance translation (Orom et al., 2008). To investigate the potential context-dependent effect of the miR-638 5’UTR and CDS MREs, we generated reporter constructs with the 5’UTR MREs upstream of the luciferase reporter, and the CDS MREs just upstream and in-frame with the luciferase stop codon. Overexpression of miR-638 resulted in a modest but significant reduction in the levels of these PTEN 5’1, 5’3, C1 and p53 C1 MRE reporter constructs in PC3 cells (Figure S2C). These data are consistent with our previous observations when they were cloned into the luciferase 3’UTR and suggest that these MREs do not function in a context-dependent manner. However, we no longer saw a significant reduction in luciferase activity with the PTEN 5’2 MRE cloned into the luciferase 5’UTR or the PTEN C2 MRE cloned into the luciferase coding region, suggesting that these two MREs may not be functional in their endogenous contexts.
We next performed site-directed mutagenesis of the seed regions of the CDS and 3’UTR MREs in the luciferase reporter constructs harbouring the corresponding full-length transcript fragments. Mutation of the PTEN C1 and p53 3’1 MREs resulted in complete rescue of the miR-638 mediated suppression of the respective luciferase constructs, while mutation of the PTEN 3’1 and p53 C1 MREs resulted in a partial but significant rescue of the respective luciferase constructs (Figure S2D). These results provide further support for the functional role of these MREs in miR-638 mediated PTEN and p53 regulation.
Overexpression of miRs-638 and 518c* promotes tumorigenesis
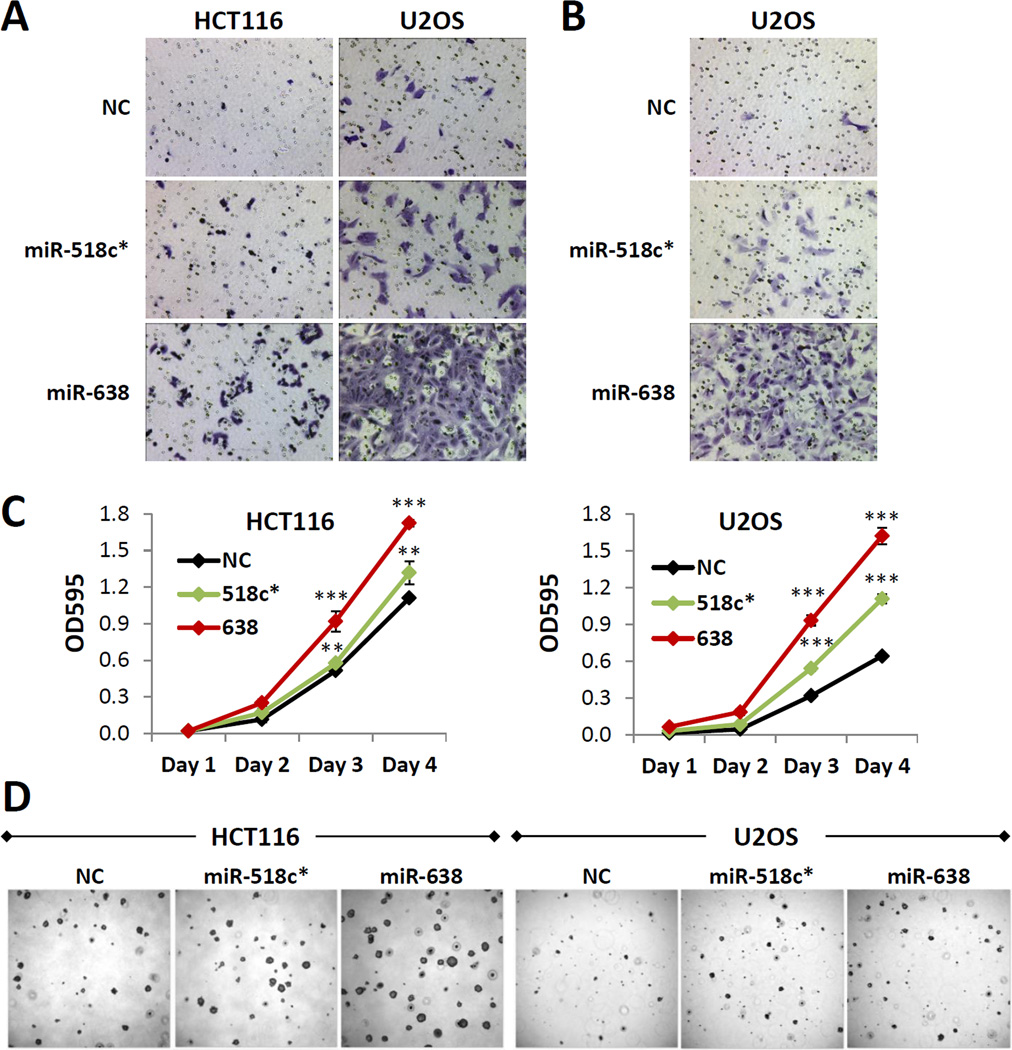
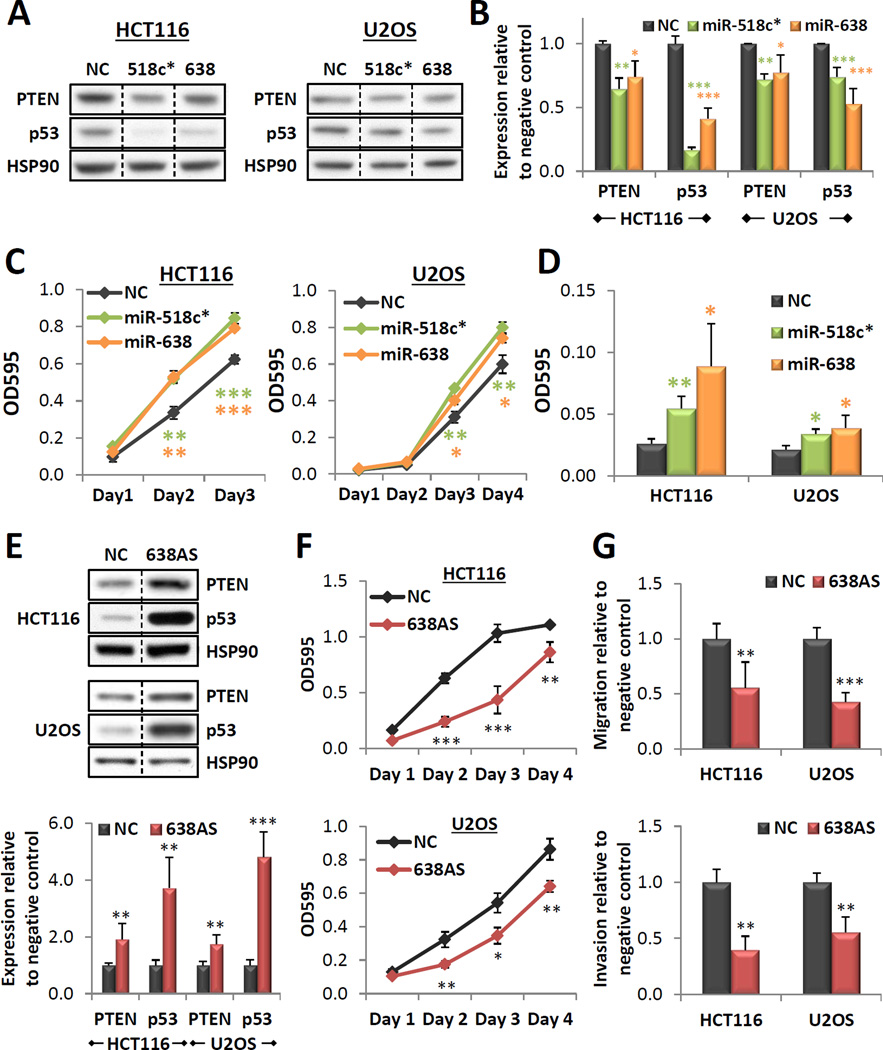
PTEN and p53 have been shown to function as important regulators of cell migration, invasion and proliferation. We thus hypothesized that miRs-518c* and 638 may be critical modulators of these hallmarks of tumorigenesis. In vitro wound healing and Boyden chamber assays demonstrated that overexpression of miR-518c* or miR-638 resulted in a significant increase in both cell migration (Figure 3A, S3A and B) and invasion (Figure 3B and S3C). Additionally, miR-518c* or miR-638 overexpression increased cell proliferation (Figure 3C) and enhanced anchorage-independent growth in soft agar (Figure 3D, S3D and E). Similar effects on target gene expression, cell proliferation and migration were observed with 2nM of transfected mimic (Figures 4A–C). As this amount of transfected mimic is more representative of the fold induction observed in cancers (Figure S3F), these data provide support for the functional relevance of the upregulation of these miRNAs in various pathophysiological conditions.
Figure 3. Overexpression of miRs-638 and 518c* promotes tumorigenesis.
(A–D) Effect of miR-638 or 518c* overexpression on cell migration (A) and invasion (B), proliferation (C) and anchorage-independent growth (D) in vitro. (E) Effect of miR-638 overexpression on HCT116 xenograft tumor formation in nude mice. (C) Mean ± SD; n ≥ 4; *p < 0.05; **p , 0.01; ***p < 0.001. See also Figure S3.
Figure 4. Functional relevance of physiological levels of miRs-638 and 518c*.
(A,B) Effect of miR-518c* or 638 overexpression (transfected at 2nM) on endogenous PTEN and p53 expression. Representative western blots are shown in (A) and quantitation of all blots is shown in (B). (C,D) Effect of miR-518c* or 638 overexpression (transfected at 2nM) on cell proliferation (C) and migration (D). (E) Effect of miR-638 inhibition (638AS) on endogenous PTEN and p53 expression. Representative western blots are shown in the top panel and quantitation of all blots is shown below. (F,G) Effect of miR-638 inhibition on cell proliferation (F), migration (G, top panel) and invasion (G, botton panel). Mean ± SD; n ≥ 4; *p < 0.05; **p , 0.01; ***p < 0.001.
We subsequently focused our attention on miR-638 due to its ability to target PTEN, p53 and BRCA1 and as it had a consistently stronger effect on enhancing tumorigenic potential in vitro. Notably, we found that miR-638 overexpression increased the growth of colon cancer xenograft tumors in nude mice (Figure S3G). Next, we determined endogenous miR-638 expression levels in a panel of cell lines with wild-type PTEN, p53 and BRCA1 (Figure S4A). All cell lines tested expressed detectable levels of miR-638, with the highest levels observed in U2OS and HCT116 cells. Inhibition of endogenous miR-638 resulted in a significant upregulation of PTEN and p53 protein levels (Figure 4E), with a concomitant decrease in cell proliferation, migration and invasion (Figure 5F and G). These data are complementary to those observed in the overexpression experiments and provide further evidence for the functional role of physiological levels of miR-638 in regulating PTEN and p53 levels.
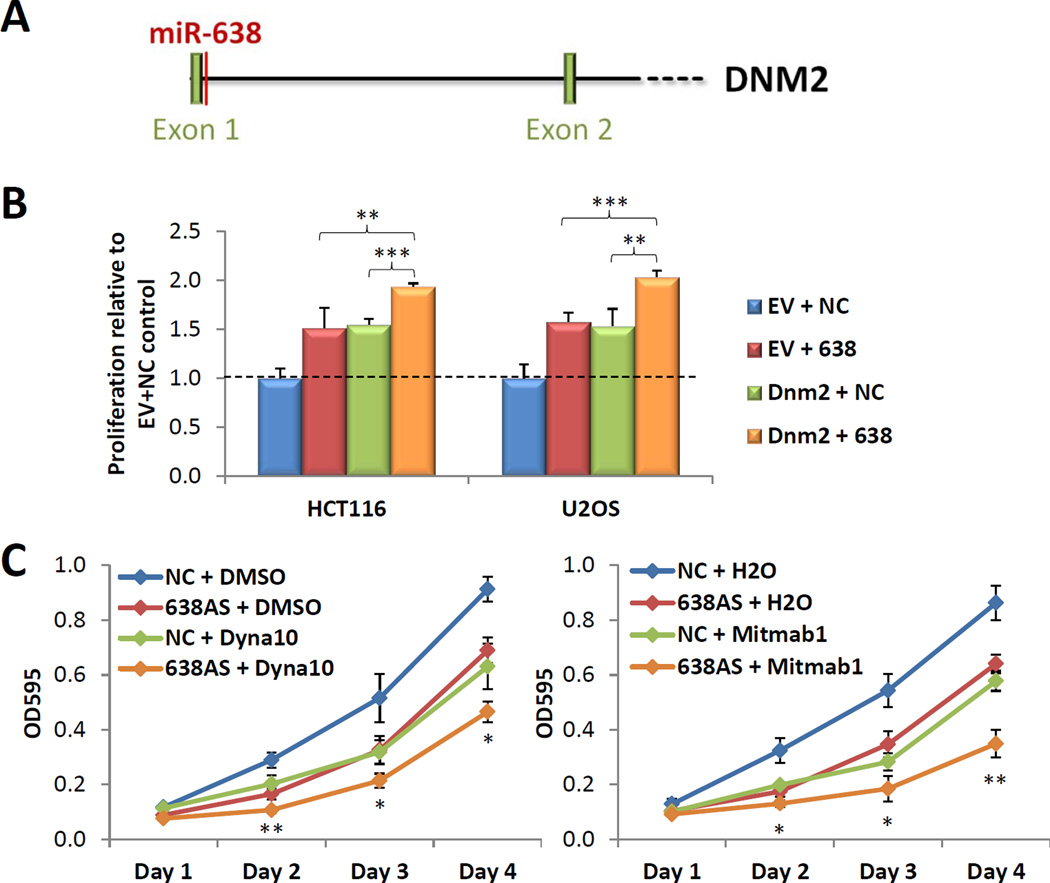
Figure 5. miR-638 is part of the oncogenic DNM2 locus.
(A) Schematic depicting the location of the miR-638 precursor in the DNM2 gene. (B) Effect of the combinatorial overexpression of miR-638 and Dnm2 on cell proliferation. (C) Combined effect of miR-638 knockdown and the Dynamin inhibitors Dynasore (left panel) and Mitmab (right panel) on cell proliferation in U2OS cells. (B,C) Mean ± SD; n ≥ 4; *p < 0.05; **p , 0.01; ***p < 0.001. See also Figure S4.
miR-638 is a functionally relevant component of the oncogenic DNM2 locus
The miR-638 precursor is located in the first intron of the DNM2 gene (Figure 5A). DNM2 is a microtubule-associated, GTP-binding protein and recent reports have demonstrated that its overexpression promotes growth, migration and invasion in various cancers (Eppinga et al., 2012; Feng et al., 2012). On this basis, we hypothesized that the DNM2 locus may represent a two-hit oncogenic locus which produces both the DNM2 protein and miR-638.
To test this hypothesis, we first overexpressed miR-638 and DNM2 independently and in combination in both HCT116 and U2OS cells. As previously reported, DNM2 overexpression led to an increase in cell proliferation (Figure 5B) (Feng et al., 2012). The concurrent overexpression of both miR-638 and DNM2 significantly augmented cell growth above that observed with either miR-638 or DNM2 alone (Figure 5B). Next, we investigated potential cooperativity between miR-638 knockdown and DNM inhibition. U2OS cells were selected for proof-of-principle studies as they expressed the highest levels of endogenous miR-638. Growth curve analyses demonstrated that the combined effect of miR-638 knockdown and either Dynasore or MiTMAB, two small molecule inhibitors of DNM function, led to a reduction in cell proliferation which was significantly greater than that observed with miR-638 knockdown or Dynasore or MiTMAB treatment alone (Figure 5C). These data suggest that miR-638 and the DNM2 protein represent two distinct oncogenic components encoded by the DNM2 locus.
Discussion
In this study, we examine miRNA-mediated post-transcriptional regulation of the full-length PTEN and p53 transcripts and study their role in promoting tumorigenesis. We characterize miR-518c* and miR-638 as PTEN and p53-targeting miRNAs, and confirm previous reports which identify BRCA1 as an additional miR-638 target (Li et al., 2012; Nicoloso et al., 2010). These miRNAs may represent a powerful mechanism to simultaneously modulate levels of these key TSGs which are known to cooperate reciprocally to inhibit tumorigenesis.
Existing studies have focused exclusively on the 3’UTRs of PTEN and p53 as sites for miRNA regulation. It has been shown in both cancer cell lines and primary tumors that transcripts often have shorter 3’UTRs in cancer cells compared to untransformed cells and thus may be subject to less 3’UTR-dependent miRNA regulation (Lembo et al., 2012; Mayr and Bartel, 2009). For example, PTEN, p53 and BRCA1 have 3 (Lianoglou et al., 2013), 3 and 7 validated alternative polyadenylation sites respectively. It is thus conceivable that the ability of miR-638 and 518c* to bind and regulate these transcripts in a partly 3’UTR-independent fashion may enable them to modulate the expression of a larger subset of transcript variants. This may be of particular relevance in cancer cells with reduced 3’UTR-dependent miRNA regulation. Interestingly, the miR-638 binding site in BRCA1 identified by Nicoloso et al was in its coding region.
The role of these miRNAs in tumorigenesis remains largely uncharacterized. miR-518c* is located in the chromosome 19 miRNA cluster (C19MC), which is the largest human miRNA gene cluster discovered to date (Bortolin-Cavaille et al., 2009). This miRNA cluster has been reported to be amplified in aggressive embryonal brain and parathyroid tumors and epigenetically reactivated through DNA demethylation in gastric cancer (Li et al., 2009; Tsai et al., 2009; Vaira et al., 2012). Specifically, elevated levels of miR-518c* were reported to be significantly associated with reduced progression-free survival in bladder carcinoma (Dyrskjot et al., 2009).
The miR-638 precursor is located in the first intron of the Dnm2 gene. Our data suggest that the Dnm2 locus encodes two distinct proto-oncogenic components which play important roles in tumorigenesis. Copy number amplifications at this locus were observed in ovarian and uterine cancers (Figure S4B). In accordance with this, DNM2 transcript expression was found to be significantly elevated in multiple ovarian cancer datasets (Figure S4C). Critically, miR-638 amplification was found to be significantly associated with decreased patient survival in ovarian cancer (Figure S4D). Additional studies reported that high expression levels of miR-638 were significantly associated with shorter overall survival in adrenocortical carcinoma (Ozata et al., 2011) and squamous cell lung carcinoma (Landi et al., 2010). These data provide further support for the functional relevance of miR-638 in human cancers.
Interestingly, it has been reported that miR-638 is one of several miRNAs that is selectively packaged in membrane vesicles released from cancer cells (Jaiswal et al., 2012). A very recent study found that miR-638 was one of only three serum miRNAs inversely associated with overall survival in nasopharyngeal carcinoma patients (Liu et al., 2014). Aside from its function in a cell of origin, miR-638 may be able to regulate gene expression in recipient cells and thus play an important role in the intercellular regulation of cancer development.
Related work from our group and others has demonstrated that transcripts which contain binding sites for shared miRNAs can co-regulate each other by titrating miRNA availability, thus functioning as natural miRNA sponges or competing endogenous RNAs (ceRNAs) (Tay et al., 2014). In particular, much effort has focused on characterization of the PTEN ceRNA network. Our study suggests that in certain conditions whereby the abundance of PTEN, p53 and BRCA1 falls within the range for optimal ceRNA crosstalk, their ability to sequester miR-518c* and miR-638 may represent an additional mechanism of reciprocal co-regulation between these TSGs. Furthermore, existing studies which examine ceRNA crosstalk between protein-coding transcripts have been limited exclusively to 3’UTR-dependent regulation. We suggest that expanding ceRNA crosstalk analysis to include 5’UTRs and coding regions of protein-coding transcripts may lead to new insights into this network of competitive miRNA-RNA interactions and to the identification of novel TSGs or oncogenes based on their ceRNA function.
Experimental Procedures
MiRNA target prediction was performed using Rna22 (Miranda et al., 2006). MREs and transcript fragments were cloned into psicheck2 and pcDNA3.1 according to standard protocols. For the psicheck2 constructs, the 5’UTR fragments were cloned upstream of the luciferase coding region while the CDS and 3’UTR fragments were cloned in the luciferase 3’UTR. Site-directed mutagenesis was performed according to manufacturer’s protocols using the QuikChange Lightning Site-Directed Mutagenesis Kit from Agilent Technologies. HCT116, U2OS and PC3 cells were cultured under standard conditions. MiRNA mimics and inhibitors were purchased from Dharmacon. Transient transfections were performed with 2nM or 100nM of miRNA mimic or inhibitor with Dharmafect 1 according to manufacturer’s protocols. Western blot, real-time PCR and luciferase assays were performed as previously described (Tay et al., 2008). Biotinylated miRNA pulldowns were performed as previously described (Lal et al., 2011). Growth curves and soft agar assays were performed as previously described (Tay et al., 2011). Wound healing, transwell migration and xenograft assays were performed according to standard protocols. Invasion assays were performed using Matrigel Invasion Chambers from BD Biosciences according to manufacturer’s protocols.
Supplementary Material
Highlights.
Identification of microRNAs targeting multiple tumor suppressor genes
These microRNAs are upregulated in multiple human cancers
Their overexpression increases cell migration, invasion and proliferation
miR-638 cooperates with its host gene Dnm2 to promote tumorigenesis
Acknowledgments
We thank Pandolfi laboratory members for critical discussions, and Ivano Legnini and Francesca Orso for technical assistance. Y.T. was supported by a Special Fellow Award from The Leukemia & Lymphoma Society. S.M.T. was supported by a Department of Defense Breast Cancer Research Program (BCRP) postdoctoral fellowship award. F.A.K. was supported by a Department of Defense Prostate Cancer Research Program fellowship award. This work was supported in part by NIH grant R01 CA-82328 to P.P.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abou-Kheir W, Hynes PG, Martin P, Yin JJ, Liu YN, Seng V, Lake R, Spurrier J, Kelly K. Self-renewing Pten-/- TP53-/- protospheres produce metastatic adenocarcinoma cell lines with multipotent progenitor activity. PLoS One. 2011;6:e26112. doi: 10.1371/journal.pone.0026112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altiok S, Batt D, Altiok N, Papautsky A, Downward J, Roberts TM, Avraham H. Heregulin induces phosphorylation of BRCA1 through phosphatidylinositol 3-Kinase/AKT in breast cancer cells. J Biol Chem. 1999;274:32274–32278. doi: 10.1074/jbc.274.45.32274. [DOI] [PubMed] [Google Scholar]
- Andjelkovic T, Bankovic J, Stojsic J, Milinkovic V, Podolski-Renic A, Ruzdijic S, Tanic N. Coalterations of p53 and PTEN tumor suppressor genes in non-small cell lung carcinoma patients. Transl Res. 2011;157:19–28. doi: 10.1016/j.trsl.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan miRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolin-Cavaille ML, Dance M, Weber M, Cavaille J. C19MC miRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009;37:3464–3473. doi: 10.1093/nar/gkp205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. Crucial role of p53-dependent cellular senescence in suppression of Ptendeficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr., Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Dyrskjot L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, Fristrup N, Jensen JL, Andersen CL, Zieger K, et al. Genomic profiling of miRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- Eppinga RD, Krueger EW, Weller SG, Zhang L, Cao H, McNiven MA. Increased expression of the large GTPase dynamin 2 potentiates metastatic migration and invasion of pancreatic ductal carcinoma. Oncogene. 2012;31:1228–1241. doi: 10.1038/onc.2011.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Liu KW, Guo P, Zhang P, Cheng T, McNiven MA, Johnson GR, Hu B, Cheng SY. Dynamin 2 mediates PDGFRalpha-SHP-2-promoted glioblastoma growth and invasion. Oncogene. 2012;31:2691–2702. doi: 10.1038/onc.2011.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman DJ, Li AG, Wei G, Li HH, Kertesz N, Lesche R, Whale AD, Martinez-Diaz H, Rozengurt N, Cardiff RD, et al. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell. 2003;3:117–130. doi: 10.1016/s1535-6108(03)00021-7. [DOI] [PubMed] [Google Scholar]
- Hermeking H. MiRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- Isaacs WB, Carter BS, Ewing CM. Wild-type p53 suppresses growth of human prostate cancer cells containing mutant p53 alleles. Cancer Res. 1991;51:4716–4720. [PubMed] [Google Scholar]
- Jaiswal R, Luk F, Gong J, Mathys JM, Grau GE, Bebawy M. Microparticle conferred miRNA profiles--implications in the transfer and dominance of cancer traits. Mol Cancer. 2012;11:37. doi: 10.1186/1476-4598-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Thomas MP, Altschuler G, Navarro F, O'Day E, Li XL, Concepcion C, Han YC, Thiery J, Rajani DK, et al. Capture of miRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 2011;7:e1002363. doi: 10.1371/journal.pgen.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, Rubagotti M, Goldstein AM, Lembo A, Di Cunto F, Provero P. Shortening of 3'UTRs correlates with poor prognosis in breast and lung cancer. PLoS One. 2012;7:e31129. doi: 10.1371/journal.pone.0031129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wang Q, Liu C, Duan H, Zeng X, Zhang B, Li X, Zhao J, Tang S, Li Z, et al. Aberrant expression of miR-638 contributes to benzo(a)pyrene-induced human cell transformation. Toxicol Sci. 2012;125:382–391. doi: 10.1093/toxsci/kfr299. [DOI] [PubMed] [Google Scholar]
- Li M, Lee KF, Lu Y, Clarke I, Shih D, Eberhart C, Collins VP, Van Meter T, Picard D, Zhou L, et al. Frequent amplification of a chr19q13.41 miRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell. 2009;16:533–546. doi: 10.1016/j.ccr.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013;27:2380–2396. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnoila I, Marincola FM, et al. MiRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16:430–441. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Cui RX, Sun Y, Guo R, Mao YP, Tang LL, Jiang W, Liu X, Cheng YK, He QM, et al. A four-miRNA signature identified from genome-wide serum miRNA profiling predicts survival in patients with nasopharyngeal carcinoma. Int J Cancer. 2014;134:1359–1368. doi: 10.1002/ijc.28468. [DOI] [PubMed] [Google Scholar]
- Mayo LD, Dixon JE, Durden DL, Tonks NK, Donner DB. PTEN protects p53 from Mdm2 and sensitizes cancer cells to chemotherapy. J Biol Chem. 2002;277:5484–5489. doi: 10.1074/jbc.M108302200. [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MiRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Nicoloso MS, Sun H, Spizzo R, Kim H, Wickramasinghe P, Shimizu M, Wojcik SE, Ferdin J, Kunej T, Xiao L, et al. Single-nucleotide polymorphisms inside miRNA target sites influence tumor susceptibility. Cancer Res. 2010;70:2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH. MiRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Ozata DM, Caramuta S, Velazquez-Fernandez D, Akcakaya P, Xie H, Hoog A, Zedenius J, Backdahl M, Larsson C, Lui WO. The role of miRNA deregulation in the pathogenesis of adrenocortical carcinoma. Endocr Relat Cancer. 2011;18:643–655. doi: 10.1530/ERC-11-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzio-Kuter AM, Castillo-Martin M, Kinkade CW, Wang X, Shen TH, Matos T, Shen MM, Cordon-Cardo C, Abate-Shen C. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes Dev. 2009;23:675–680. doi: 10.1101/gad.1772909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak TW. Regulation of PTEN transcription by p53. Mol Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Song SJ, Pandolfi PP. The Lilliputians and the Giant: An emerging oncogenic miRNA network that suppresses the PTEN tumor suppressor in vivo. MicroRNA. 2013;2(2):127–136. doi: 10.2174/22115366113029990017. [DOI] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MiRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Tsai KW, Kao HW, Chen HC, Chen SJ, Lin WC. Epigenetic control of the expression of a primate-specific miRNA cluster in human cancer cells. Epigenetics. 2009;4:587–592. doi: 10.4161/epi.4.8.10230. [DOI] [PubMed] [Google Scholar]
- Vaira V, Elli F, Forno I, Guarnieri V, Verdelli C, Ferrero S, Scillitani A, Vicentini L, Cetani F, Mantovani G, et al. The miRNA cluster C19MC is deregulated in parathyroid tumours. J Mol Endocrinol. 2012;49:115–124. doi: 10.1530/JME-11-0189. [DOI] [PubMed] [Google Scholar]
- Vlietstra RJ, van Alewijk DC, Hermans KG, van Steenbrugge GJ, Trapman J. Frequent inactivation of PTEN in prostate cancer cell lines and xenografts. Cancer Res. 1998;58:2720–2723. [PubMed] [Google Scholar]
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Zhang H, Somasundaram K, Peng Y, Tian H, Bi D, Weber BL, El-Deiry WS. BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene. 1998;16:1713–1721. doi: 10.1038/sj.onc.1201932. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.