Abstract
Aim
Application of quantitative stable-isotope-labeling chemistries and mass spectrometry (MS) to determine alterations in gingival crevicular fluid (GCF) proteome in periodontal disease.
Materials and Methods
Quantitative proteome of GCF from 40 healthy individuals versus 40 patients with periodontal disease was established using 320 GCF samples and stable-isotope-labeling reagents, ICAT and mTRAQ, with MS technology and validated by enzyme-linked immunosorbent methods.
Results
We have identified 238 distinct proteins of which 180 were quantified in GCF of both healthy and periodontal patients with additional 26 and 32 distinct proteins that were found only in GCF of healthy or periodontal patients. In addition, 42 pathogenic bacterial proteins and 11 yeast proteins were quantified. The data highlighted a series of proteins not quantified previously by large-scale MS approaches in GCF with relevance to periodontal disease, such as host derived Ig alpha-2 chain C, Kallikrein-4, S100-A9, transmembrane proteinase 13, peptidase S1 domain, several collagen types and pathogenic bacterial proteins e.g., formamidase, leucine amidopeptidase and virulence factor OMP85.
Conclusions
The innovative analytical approaches provided detailed novel changes in both host and microbial derived GCF proteomes of periodontal patients. The study defined 50 host and 16 pathogenic bacterial proteins significantly elevated in periodontal disease most of which were novel with significant potential for application in the clinical arena of periodontal disease.
Keywords: Gingival crevicular fluid, mass spectrometry, quantitative proteomics, periodontal disease, stable-isotope labeling, biomarkers, diagnostics
INTRODUCTION
Gingival crevicular fluid (GCF) is composed of serum and locally generated materials such as tissue breakdown products, locally produced extracellular proteins, inflammatory mediators, host inflammatory cells, microbial plaque and antibodies directed against dental plaque bacteria (Curtis et al., 1989; Champagne et al., 2003; Delima & Van Dyke, 2003; Uitto, 2003; Armitage, 2004; Rose et al., 2004; Lamster & Ahlo, 2007). GCF can be easily and non-invasively collected and its constituents have the capacity to reflect both locally and systemically derived factors. Due to those properties GCF was considered as a valuable body fluid that may serve as an important source of biomarkers for both periodontal and systemic diseases [Lamster & Ahlo, 2007; Teles et al., 2010; Mantyla et al, 2006; Fitzsimmons, et al., 2010; Kojima et al., 2000; Offenbacher, et al., 2010; Loos and Tjoa, 2000). In those respects numerous GCF derived inflammatory factors such as cytokines, proteins, proteinases, phosphatases and local tissue degradation products have been evaluated as possible diagnostic markers for periodontitis. In such endeavors various methods ranging from traditional enzyme-linked-immunosorbent-assay targeting a single analyte or multianalyte bead-based multiplexing have been used and have been summarized by Loos and Tjoa (Loos and Tjoa 2005). However, high rates of false positive results have been found in tests that evaluate enzymes as diagnostic markers in GCF for periodontal disease progression (Armitage 2004). To date, accurate predictive or diagnostic periodontal disease indicators based on GCF have not been established. In more recent times the advent of mass spectrometry (MS) led to a large-scale proteome documentation of body fluids such as plasma (Schenk et al., 2008), urine (Li et al., 2010; Cardiano et al., 2010), and within the dental field proteome of whole saliva (Xie et al., 2005; Hu et al., 2005), parotid secretion Hardt et al., 2005; Denny et al., 2008; Yan et al. 2009), minor gland secretion (Denny et al., 2008; Yan et al., 2009; Siqueira et al., 2008), acquired enamel pellicle (AEP) (Siqueira et al., 2009), and large-scale phosphoproteome of whole saliva (Salih et al., 2010; Stone et al., 2011). It is of no surprise therefore that MS technology has been also applied to studies of GCF, however, these have been at much limited level because of the sample size 0.2–1.0 μl per site as well as lack of effective methods for sample preparation and the accuracy of approaches used for large-scale MS analysis. It should be noted that the small quantity of GCF volume may not be the only limitation for the highly sensitive contemporary MS technology. For instance, there are other limitations such as dynamic protein range and presence of highly abundant proteins. Such limitations are clearly applicable to GCF which contains abundant serum derived proteins. These become very significant when protein contributions of serum change from 30% to ~70% for GCF from periodontally healthy and diseased sites, respectively. Hence, the serum related protein contributions to GCF become accentuated during gingivitis and periodontitis including additional proteins from local inflammatory response. In this case the well known dynamic-protein range becomes a hindrance whereby presence of highly abundant serum proteins such as albumin, constituting ~50% of the total serum proteins, and immunoglobulins can and do restrict the identification of lower level proteins of both systemic and local origin. These are well known major issues and difficulties encountered in the establishment of serum/plasma proteome by MS technology (Anderson 2005) and directly applicable to GCF analysis by MS technology as we have highlighted recently (Carneiro et al., 2012).
Mass spectrometry (MS) technology permits the identification of a large-scale proteome at a qualitative level with relative ease in complex biological samples without purification of the individual proteins. However, because of the significant contribution of serum which adds further complexity to GCF composition any “biomarker discovery” approach using GCF requires an additional specialized MS approach, namely quantitative or relative quantitative analysis which is far more challenging and should be approached with clear understanding of the sample analysis and complexity imposed by the disease state and the inherent limitations of the analytical tool/technology. To date there has been only a single study that has been reported in an attempt to perform quantitative MS-based protein comparison of GCF from healthy individuals and patients with periodontal disease. That study used “label-free MS” analysis approach with GCF samples collected from 5 healthy and 5 patients with periodontal disease which led to relative quantification of 18 distinct proteins in both groups and an additional 22 proteins that were keratins (Bostanci et al., 2010). Another study carried out using more accurate quantitative analysis using stable-isotope labeling technique, however, this latter study was performed on GCF samples from experimentally “induced gingivitis and not periodontal disease patients” (Grant et al., 2010). More recent study using GCF samples from 12 healthy and 12 periodontal patients attempted to highlight possible biomarkers by grouping what is found only in GCF from healthy or periodontal group at a qualitative level without the use of quantitative analytical methods (Baliban et al., 2012). We have reported a large-scale proteome data set of GCF from periodontally healthy sites by approaches that overcome several limitations of such proteomic analysis of GCF (Carneiro et al., 2012). It is clear that in-depth and more comprehensive documentation of the alterations of GCF proteome during the transition from gingival health to periodontal disease is of major scientific and clinical interest.
The aims of this study were to utilize robust and gold-standard stable-isotope labeling chemistries, isotope-coded affinity tag (ICAT) and amine-specific tag for relative and absolute quantitation (mTRAQ)] coupled with multidimensional protein separation MS analytical approaches to define on a large-scale changes in quantitative proteome of GCF in health and periodontal disease. The aim to document new quantitative changes of both host and pathogenic bacteria derived GCF proteins will advance our limited knowledge of the GCF protein composition and highlight specific alterations that take place in disease state which can be used as biomarker discovery platform.
MATERIALS AND METHODS
This study was approved by the Institutional Review Board for human study with protocol number H-26454. There were no conflict of interest in this work and volunteers of different ethnicities, gender and adult age were included. They were all systemically healthy and were divided in two groups according to their periodontal status. One group consisted of volunteers with healthy periodontium and a second group consisted of patients with moderate to severe periodontal disease. Individuals were excluded from this study if they were known alcohol or illicit drug abusers, pregnant or lactating woman or volunteers that were on or had antibiotic therapy in the 6 months previous to the collection. Additional criteria for exclusion were those with clinically significant illness including unstable heart disease, kidney disease, liver disease and clinically significant mental illness. The healthy group consisted of 16 males and 24 females and ages ranging from 22 to 54 with a mean age of 34.68±9.08. Clinical criteria for selecting periodontally healthy subjects were based on gingival index score of 0, pocket depth ≤ 3 mm, clinical attachment level ≤ 1.5 mm and no bleeding on probing. These criteria were applied to specific sites from which collections were made, as well as full mouth. The moderate to severe group consisted of 19 males and 21 females with ages ranging from 23 to 61 with a mean age of 40.88±10.47. The distance from the cementoenamel junction to the base of crevice, known as clinical attachment level (CAL), was measured in all patients and only patients with 5 mm or more of CAL were selected for the periodontitis group. The clinical bases for the moderate to severe chronic periodontal disease group was based on full mouth examination and patient having at least 4 teeth with pocket depth >5 mm, in at least two different quadrants and bleeding on probing scores of 0.6 –1.0. The pre-selected specific sites with moderate and severe chronic periodontal disease were defined by pocket depth of 5–7 mm (24 patients) and > 7 mm (16 patients), respectively, were then used to collect 5–7 GCF samples using periopapers. Differences between genders and ages were not analyzed since the study was not aimed to define such differences.
The subjects with periodontal disease were recruited from the Postdoctoral Periodontology Clinic at the Boston University Henry M. Goldman School of Dental Medicine and the healthy subjects were recruited through an advertisement about the study that was placed at the Medical Campus of the Boston University. An IRB approved consent form explaining the procedures in detail including the risks and benefits involved was presented to all subjects before the study began. The purpose of this study and its risks and benefits were also explained verbally to all subjects. The medical and dental histories were obtained and reviewed and all subjects were clinically screened at the General Clinical Research Center (GCRC) located at the Boston University Henry M. Goldman School of Dental Medicine. To increase the reliability, all clinical measurements were performed by the same periodontist who was calibrated before the onset of this study.
Gingival crevicular fluid (GCF) collection
GCF was collected from 9:00 am to 12:00 pm in order to minimize the effect of circadian rhythmic variation on the composition of the fluids. All subjects were asked not to eat, drink, brush their teeth or use any type of mouth wash 2 hours prior to fluid collection. The sites selected for collection were subjected to washing by the dental unit’s air-water syringe. The areas were isolated from salivary contamination with cotton rolls, air dried, and GCF was collected with Periopaper strips (Oraflow, Plainview, NY). A sterile Periopaper strip was gently inserted into the entrance of the sulcus/periodontal pocket and left in place for 30 seconds. Mechanical irritation was avoided, and the strips contaminated with blood were discarded. The GCF sample volumes were measured with Periotron 8000 (Periotron 8000, Proflow, Inc., Amityville, NY), and then the readings were converted to an actual volume (μl) by reference to the standard calibration curve using different volumes of water. After the measurement of the volume collected, the paper strips were placed in Eppendorf tubes and either processed immediately for analysis or kept frozen at −80°C until needed. GCF was collected from 40 healthy individuals and 40 patients with periodontal disease. From each subject 4 different sites were used to collect 1 GCF periopaper from each site for MS studies (providing 160 periopaper collections for each group) and additional 1–3 periopapers were also collected from each individual for ELISA and as spares in case of need.
Sample processing and elution of GCF proteins from periopapers
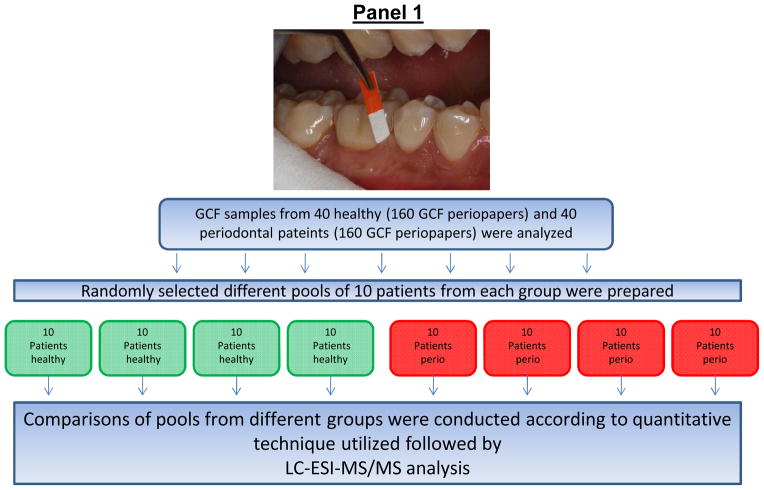
In order to eliminate the individual variation, to reduce the site specific variation, and have a uniform and better knowledge of the differences from two distinct groups, controls and disease, pools of samples from the same group were made. Pools of 10 different GCF samples from each group i.e., health individuals or periodontal disease patients were used for MS analysis. Figure 1 summarizes the above steps of pooling and sample preparation for MS analysis. Furthermore, normally LC-ESI-MS/MS uses liquid chromatography to separate peptides according to hydrophobicity utilizing a micro-reversed phase C-18 column. However, due to limitations introduced by dynamic protein range issues of complex biological protein samples such as GCF during analysis, additional fractionation methods were introduced prior to MS analysis. These included two additional steps one based on affinity capture enrichment and the other separate proteins using SDS-PAGE according to their relative molecular weight (Mr) and analysis of different Mr sections.
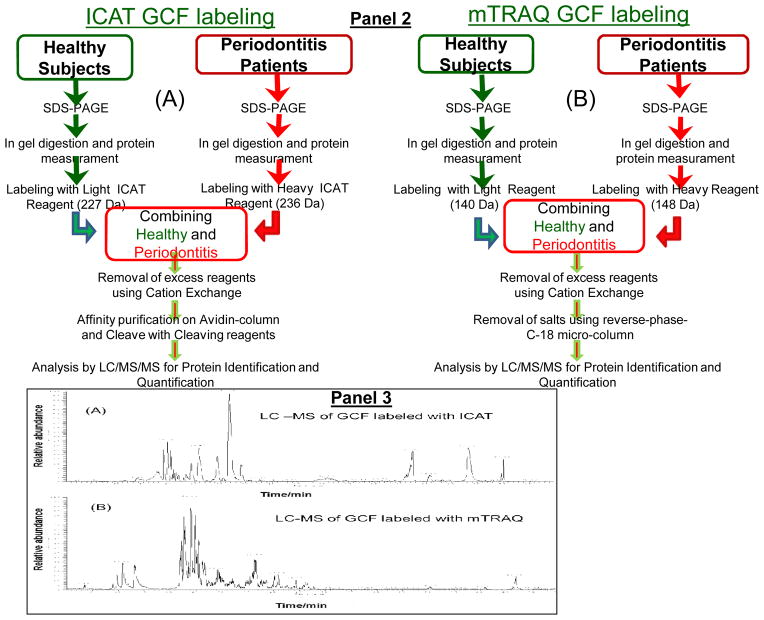
Figure 1. Schematic representation of sample pooling and stable-isotope labeling of GCF samples from healthy and periodontal subjects with ICAT or mTRAQ reagents and processing for LC-ESI-MS/MS analysis.
Panel 1: Different pools of GCF from 10 random patients from each group were prepared for stable-isotope-labeling followed by relative quantitative proteome analysis using LC-ESI-MS/MS. Panel 2: Sequential steps used for stable-isotope labeling of GCF samples for LC-ESI-MS/MS analysis, (A), ICAT and (B), mTRAQ. Panel 3: LC-ESI-MS/MS base-peak ion chromatogram of GCF samples labeled with ICAT and mTRAQ. (A), GCF samples labeled with ICAT, note the simplicity of the LC-MS/MS of ICAT labeled GCF sample illustrating the reduced number of peptides observed since not all of the peptides contain cysteine residues and those peptides without cysteine residues were eliminated during affinity enrichment, and (B), GCF samples labeled with mTRAQ where all of the trypsin peptides are labeled and present.
(a) Electroelution by SDS-PAGE of GCF proteins and in gel digestion
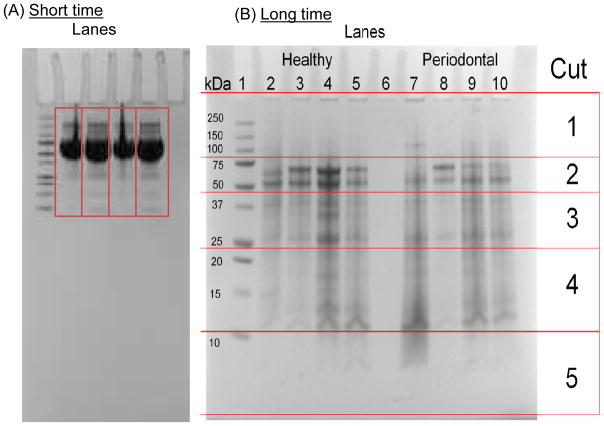
The collection of GCF using periopaper strips (Oraflow, Plainview, NY) requires effective elution and recovery of the GCF proteins prior to MS analysis. We have established the best method with excellent recovery of the GCF proteins from periopapers to be via electroelution. This approach also enabled either to recover all of the proteins as a mixture or fractionated based on molecular weight and use sections of the gel for quantitative analysis. Periopaper strips containing GCF collection were placed into the SDS-PAGE wells, one periopaper per well, (NuPage 12% Bis-Tris Gel 1.0X10mm wells) and subjected to electrophoresis. For extraction of all of the proteins as a mixture without significant separation, electrophoresis was carried out for a short time, 30 minutes at 120 V, during which all of the proteins were mobilized into the SDS-gel and the whole gel lane containing all of the proteins, eluted from 1 periopaper from each of 40 healthy subjects and 40 periodontal patients, were excised (keeping each individuals GCF separate) for in gel trypsin digestion and processes for MS analysis, Figure 2A. This provided trypsin digested samples of each of 40 healthy and 40 periodontal derived GCF and aliquots from each were then used to generate pooled samples (containing 10 different individuals GCF). For electroelution and separation based on molecular weight electrophoresis was carried out at 120 V until the dye front reached to the bottom of the gel, ~2–3 hrs. After eletrophoresis the gels were stained with Coomassie blue and destained with a 40% methanol, 10% acetic acid solution. The de-stained gel was sectioned into 5 different molecular weight ranges by excising these regions with a sharp straight edge razor. The sections comprised Cut 1 (above 75 kDa), Cut 2 (between 50 to 75 kDa), Cut 3 (between 35 to 50 kDa), Cut 4 (between 10 to 25 kDa), and Cut 5 (10 kDa and below), see Figure 2B. Similar to above short gel runs, 1 periopaper was run from each individual at a given time using multiple gels to use periopapers from each of the 40 healthy and 40 periodontal individuals. In this case however, because of the sectioning of the gels, such runs were repeated for 2 periopapers from each individual. The same sections, for example cuts 1 above, generated from the same individual were combined after trypsin digestion. Hence, for each individual from 2 different periopapers, a total of 5 different trypsin digest aliquots were generated i.e., cut 1, cut 2, cut 3 etc. For generating pooled samples an equal aliquot, for example of cut 1 from each of 10 different healthy individuals were combined and similarly a corresponding cut 1 pool for 10 different periodontal patients was generated to be subjected to relative quantitative MS analysis. This process was repeated for cuts 2, 3, 4, and 5.
Figure 2. SDS-PAGE electroelution of GCF proteins from periopaper strips from healthy and periodontal subjects for stable-isotope labeling and quantitative proteome analysis by MS.
(A) Short time SDS-PAGE run: a single periopaper containing ~0.2–0.7 μl GCF were placed in each well containing sample buffer. The SDS-PAGE was run until the dyefront was approximately half-way the gel length. This was sufficient to electroelute proteins from the periopaper into the gel visualized by Coomassie blue staining. Molecular weight standards can be seen on the left side of the gel. The electroeluted GCF proteins are not visible at this point since the gel has not been yet stained with coomassie blue. (B) Long time SDS-PAGE run: 4 periopaper strips derived from healthy individuals and 4 periopapers derived from periodontal patients, respectively, were run in a single SDS-PAGE separated by an empty lane. After comassie staining the gel was sectioned into different molecular weight regions as indicated. Lane 1, standard molecular weight proteins; Lanes 2–5, 4 individual periopapers from healthy and lanes 7–10 individual periopapers from periodontal patients run separately. On the right hand side under “CUT” are the sections of different molecular weight regions excised across and processed for MS analysis, cut 1: Mr range ~100 kDa and above, cut 2: Mr range ~40–80 kDa, cut 3: Mr range ~25–38 kDa, cut 4 ~11–24 kDa and cut 5: Mr range ~2–10 kDa.
(b) In Gel Digestion
After electroelution each separate molecular weight gel section was then cut into smaller pieces (1–2 mm) and placed in an Eppendorf tube. SDS and Coomassie blue stain were removed by washing with buffer 1 (50 mM ammonium bicarbonate pH 8.0) and buffer 2 (50 mM ammonium bicarbonate pH 8.0 + 50% acetonitrile) alternatively with brief agitation (5 min) at each stage. This step was repeated 3 times. After the last buffer treatment the gel pieces were dried under vacuum using Speed-Vac and suspended in buffer 1 containing trypsin (0.5 μg of trypsin per each 25 μl of buffer) and the samples were incubated in Eppendorf tubes for 24 hrs at 37°C. Following in-gel digestion, buffer containing trypsin and the released peptides were removed and placed in a separate Eppendorf tube. The gel pieces were further extracted by washing with buffer 1 and buffer 2 alternatively with brief agitation (5 min) at each stage and all the extracts were pooled in a single Eppendorf tube. This process was repeated 3 times. The peptide extracts from each SDS-PAGE section were then freeze dried, dissolved in 200 μl of buffer A (0,1% trifluoroacetic acid) and cleaned using a C-18 reverse-phase MicroSpin column (The Nest Group, Inc. 45 Valley Road, Southborough, MA) for removal of SDS and salts. Folowing this clean-up step the protein/peptide concentrations were determined using bicinchoninic acid (BCA) protein assay and bovine serum albumin as standard.
Quantitative mass spectrometric analyses of GCF samples from healthy individuals versus periodontal patients using stable-isotope labeling chemistries
For the current study we have utilized the post-extraction stable-isotope labeling relative quantitative proteomic approaches using two complementary labeling chemistries, namely isotope-coded-affinity-tag (ICAT) and amine-specific-tag for relative and absolute quantitation (mTRAQ).
(i) Isotope-Coded-Affinity-Tag (ICAT) labeling
The ICAT reagents are stable isotope variants and were designed to affinity isolate and quantify the relative concentrations of cysteine-containing proteins in control versus experimental samples. Equal amounts of proteins for healthy and periodontal samples were used for relative quantitative MS analysis. 10 μg of protein equivalence from each of the 10 different subjects in each group were pooled, viz., total 100 μg per group (healthy and/or periodontitis). Since we have GCF from 40 healthy and 40 periodontitis patients, 4 such different pools (each containing 10 different individual’s GCF) for healthy and 4 different pools for periodontal patients were made at any given time for isotope labeling and LC-ESI-MS/MS analysis. For GCF samples generated from the short SDS-PAGE elution such 4 pools generated and subjected to the LC-ESI-MS/MS analysis, total of 4 LC-MS/MS runs performed. The trypsin digested (5% w/w in 50 mM NH4HCO3 at 37°C overnight) and speed-vac dried samples from the healthy patients and periodontitis group were labeled with the light reagent (12C-ICAT, 227 Da mass addition after biotin cleavage) and heavy reagent (13C-ICAT, 236 Da mass addition after biotin cleavage), respectively, using the manufacturer’s protocol (Applied Biosystems, Inc, Foster City, CA). Following this step the two samples were combined and excess reagents removed by cation-exchange cartridge (Applied Biosystems, Inc., Foster City CA) and ICAT labeled peptides were affinity purified using avidin-affinity column (Applied Biosystems, Inc., Foster City CA). Following affinity purification step the biotin tag of ICAT was removed by TFA acid treatment prior to LC-MS/MS analysis. The above described steps are summaries in Figure 1 Panel 2A. ICAT-labeled peptides were then subjected to LC-ESI-MS/MS analysis to identify ICAT peptide pairs and to determine the relative [12C]/[13C] ratios. For GCF samples generated from long SDS-PAGE elution similar to above 4 different pools were made for each of the 5 cuts (1 – 5), which led to total of 20 LC-MS/MS runs.
(ii) mTRAQ labeling
Complementing ICAT quantitative method and to expand the scope of identified and quantified proteins, a more universal labeling reagent which labels all the tryptic peptides at the N-terminal amine groups and the side chain of lysine residues was also used. (a) As detailed above for ICAT experiments, equal amounts of proteins from 10 different subjects from each group, i.e., healthy and periodontal patients were used to generate 4 pools of healthy samples utilizing all 40 healthy and 4 pools of periodontal samples. 100 μg protein of each of healthy and periodontal patient pooled GCF were labeled with light mTRAQ for healthy samples (140 Da mass addition) and heavy mTRAQ for the periodontal samples (148 Da mass addition), respectively, using the manufacturer’s protocol (Applied Biosystems, Inc., Foster City CA). Following the labeling step the two differentially labeled samples were combined and excess reagents and salts were removed using cation-exchange cartridge (Applied Biosystems, Inc., Foster City CA) and C-18 reversed-phase micro-spin columns (Nest Group, Inc.). The above steps used are summarized in Figure 1 Panel 2B. The samples were then freeze dried and subjected to LC-ESI-MS/MS analysis for protein identification and quantification. For GCF samples generated from the short SDS-PAGE elution such 4 pools generated and subjected to the LC-ESI-MS/MS analysis, total of 4 LC-MS/MS runs performed. For GCF samples generated from long SDS-PAGE elution similar to above 4 different pools were made for each of the 5 cuts (1 – 5). These overall experimental approaches led to a total of 20 LC-ESI-MS/MS analysis.
Nano-Flow Liquid-Chromatography and Electrospray-Ionization-Tandem Mass Spectrometric (LC-ESI-MS/MS) Analysis
LC-MS/MS analyses were carried out using LTQ-linear ion trap mass spectrometer (Thermo Electron, San Jose, CA). Samples were suspended in 97.4% H2O: 2.5%CH3CN:0.1% formic acid and LC-ESI-MS/MS analyses were carried out using an on-line autosampler (Micro AS, ThermoFinnigan, CA) with auto-injections of 3 μl onto an in-line fused silica microcapillary column, (75 μm X 10 cm), packed in-house with C18 resin (Micron Bioresource, Inc. Auburn, CA) at a flow rate of 250 nl/min. The LC-MS and MS/MS conditions and procedures were as described in detail previously (Salih et al., 2010, Carneiro et al., 2012; Czernick et al., 2013). Figure 1 Panel 3A & 3B show typical LC-ESI-MS/MS relative base-peak ion abundance for ICAT and mTRAQ labeled GCF proteins.
Database search and protein/peptide identification and quantification
All MS/MS spectra from LC-ESI-MS/MS were searched against the human database, Uniprot (Universal Protein Resource, Version 9.0), which combines the data from Swiss-Prot (Version 51), TreMBL (Version 34) and PIR using Bioworks 3.3.1 software and SEQUEST search engine (Eng et al., 1994). The data were searched against 241,242 entries. Search parameters (modifications) used for ICAT labeled samples were 227 Da static modification on cysteine residues labeled with ICAT (light), and a dynamic modification of + 9 Da for cysteine residues labeled with ICAT (heavy). Search parameters used for mTRAQ were 140 Da static modification on lysine residues and N-termini for peptides labeled with mTRAQ (light) and + 8 Da for dynamic modification on lysine residues and N-termini for peptides labeled with mTRAQ (heavy). One important additional mass addition/modification (16 Da, for hydroxyprolines) that was used for the first time in proteomic analysis of GCF was based on the fact that collagens have repeat sequences of almost every third residue is proline and these are frequently hydroxylated. The relative quantifications of mTRAQ (light and heavy), and ICAT (light and heavy) were carried out using the pepQuan option in the Bioworks Rev.3.3.1 software and comparing peak areas of isotope-encoded peptide pairs observed in the corresponding MS spectra. It is noteworthy that this software which calculates automatically the ratios should also be manually evaluated as it can use incorrect peak areas within a given scan time for pairs of peptides. For the mTRAQ labeled samples, only proteins for which two or more unique peptides for a given protein were identified are reported. For the ICAT labeled samples, proteins with single peptide identifications were included for two reasons: (i) The number of cysteine residues present in a given protein is often limited and in some cases may contain only a single cysteine residue, and (ii) ICAT-labeled peptides were affinity purified which eliminates all the other peptides generated from the same protein with no ICAT-label. In order to have a more comprehensive documentation of the GCF proteome, all of the raw LV-MS/MS data files used to document the human proteins were also searched against both bacterial and yeast databases with combined 122,000 entries.
To determine the “false positive rate” the data were searched against a concatenated human sequence database containing both the forward and the reverse sequence version. The false positive rate was calculated as described previously (Peng et al., 2003; Salih et al., 2010). The DTA generation was with a precursor-ion tolerance of 1.5 amu, a fragment ion tolerance of 1.0 amu, and automated calculated charged states +1, +2, and +3. The searches were carried out using partial trypsin specification and 2 miscleavages. The use of partial trypsin searches was to avoid the exclusion of any peptides generated by the unknown proteinases within the GCF samples. The full tryptic and half tryptic peptides were used to create the GCF proteome list in Table 2. The database search results were filtered using the criteria: ΔCn ≥ 0.1; probability ≤ 0.1; for fully tryptic peptides, XCorr ≥ 1.6, 1.8, 3.5 for Z= +1, +2, +3 ; and for partial tryptic peptides, XCorr ≥ 1.8, 2.1, 3.75 for Z= +1, +2, +3.
Table 2. Relative quantitative proteome analysis by mass spectrometry of human GCF from healthy (controls) and periodontal disease subjects using stable-isotope-labeling chemistries, mTRAQ and ICAT.
(A); Relative quantitative proteome analysis of human GCF from healthy versus disease subjects led to the identification and quantification of 180 proteins. The proteins are grouped according to their functional categories with relative quantitative values reflected as ratio (perio/healthy) = mean ± confidence interval. The values of confidence interval (Cl) were calculated as described under methods using the standard deviation (STD), sample number and a 95% level of confidence. S = also found in serum proteome (Schenk et al., 2008). (B); Identification of proteins only in healthy or periodontal GCF derived samples during quantitative proteome analysis using stable-isotope-labeling chemistries. 58 proteins were identified without relative quantification, of which 26 and 32 distinct proteins were found only in GCF of healthy or periodontal samples, respectively.
| Table 2A. Relative quantitative proteome analysis of GCF from healthy and periodontal disease subjects using stable-isotope-labeling chemistries. S.D. = standard deviation, # runs = number of times a given protein was identified, # peptides = number of distinct peptides identified in each run for a given protein and used for quantification, SNS = statistically not significant. | |||||||
|---|---|---|---|---|---|---|---|
| Accession # | GCF Proteins | mTRAQMean ±S.D | P value | ICATMean ±S.D. | P value | # Runs | # Peptides |
| Apoptosis and Signal Transduction | |||||||
| SP02647 | Apolipoprotein A-I | 1.25 ± 0.20 | <0.03 | 9 | 4 | ||
| SP04114 | Apolipoprotein B-100 | 1.44 ± 0.46 | SNS | 5 | 3 | ||
| P53355 | Death-associated protein kinase | 2.5 ± 1.14 | SNS | 4 | 2 | ||
| Q9BTC0 | Death-inducer obliterator 1 | 3.75 ± 2.40 | SNS | 3 | 2 | ||
| P98164 | Low-density lipoprotein receptor-related protein 2 | 0.89 ± 0.19 | SNS | 3 | 2 | ||
| Q8IZF6 | Probable G-protein coupled receptor 112 | 0.95 ± 0.48 | SNS | 3 | 2 | ||
| Q13972 | Ras-specific guanine nucleotide-releasing factor 1 | 0.82 ± 0.58 | SNS | 3 | 2 | ||
| P21817 | Ryanodine receptor 1 | 0.63 ± 0.34 | SNS | 4 | 2 | ||
| Q92736 | Ryanodine receptor 3 | 1.09 ± 0.97 | SNS | 3 | 2 | ||
| O60292 | Signal-induced proliferation-associated 1-like protein 3 | 0.55 ± 0.52 | SNS | 3 | 2 | ||
| Q6ZSZ6 | Teashirt homolog 1 | 0.97 ± 0.83 | SNS | 4 | 2 | ||
| P62736 | Actin, aortic smooth muscle | 1.39 ± 0.32 | SNS | 10 | 15 | ||
| SP60709 | Actin, cytoplasmic 1 | 1.62 ± 0.72 | <0.01 | 7 | 4 | ||
| Q562M3 | Actin-like protein | 1.69 ± 0.45 | <0.03 | 5 | 2 | ||
| O94833 | Bullous pemphigoid antigen 1 | 1.13 ± 0.93 | SNS | 3 | 3 | ||
| SP06396 | Gelsolin | 1.25 ± 0.18 | SNS | 3 | 2 | ||
| SP02533 | Keratin, type I cytoskeletal 14 | 1.04 ± 0.30 | SNS | 13 | 13 | ||
| SP08779 | Keratin, type I cytoskeletal 16 | 0.88 ± 0.11 | SNS | 4 | 2 | ||
| P04264 | Keratin, type II cytoskeletal 1 | 1.20 ± 0.32 | SNS | 8 | 2 | ||
| Q01546 | Keratin, type II cytoskeletal 2 oral | 0.95 ± 0.20 | SNS | 4 | 2 | ||
| SP35908 | Keratin, type II cytoskeletal 2 epidermal | 1.47 ± 0.15 | <0.001 | 17 | 3 | ||
| P12035 | Keratin, type II cytoskeletal 3 | 1.30 ± 0.35 | SNS | 7 | 4 | ||
| P19013 | Keratin, type II cytoskeletal 4 | 1.01 ± 0.31 | SNS | 9 | 6 | ||
| SP13647 | Keratin, type II cytoskeletal 5 | 1.09 ± 0.11 | <0.02 | 8 | 6 | ||
| SP02538 | Keratin, type II cytoskeletal 6A | 1.11 ± 0.15 | SNS | 16 | 10 | ||
| P08729 | Keratin, type II cytoskeletal 7 | 1.31 ± 0.03 | <0.05 | 4 | 2 | ||
| SP13646 | Keratin, type I cytoskeletal 13 | 1.42 ± 0.27 | <0.001 | 16 | 15 | ||
| P20929 | Nebulin | 0.77 ± 0.35 | SNS | 4 | 3 | ||
| Q8WXH0 | Nesprin-2 | 2.04 ± 0.86 | <0.03 | 6 | 2 | ||
| Extracellular Structural Proteins | |||||||
| O95996 | Adenomatous polyposis coli protein 2 | 0.60 ± 0.18 | SNS | 3 | 2 | ||
| SP39060 | Collagen alpha-1(XVIII) chain | 0.71 ± 0.16 | SNS | 3 | 2 | ||
| Q9ULU4 | Protein kinase C-binding protein 1 | 1.18 ± 0.39 | SNS | 3 | 3 | ||
| P12107 | Collagen alpha-1(XI) chain | 1.75 ± 0.72 | SNS | 6 | 2 | ||
| Q05707 | Collagen alpha-1(XIV) chain | 0.43 ± 0.07 | <0.02 | 3 | 2 | ||
| P02462 | Collagen alpha-1(IV) chain | 0.87 ± 0.41 | SNS | 7 | 2 | ||
| P12107 | Collagen alpha-1(XI) chain | 1.75 ± .96 | SNS | 6 | 2 | ||
| Q05707 | Collagen alpha-1(XIV) chain | 0.43 ± 0.07 | <0.004 | 3 | 2 | ||
| Q8NFW1 | Collagen alpha-1(XXII) chain | 1.46 ± 0.75 | SNS | 3 | 4 | ||
| Q8IZC6 | Collagen alpha-1(XXVII) chain | 0.72 ± 0.36 | SNS | 5 | 2 | ||
| P02462 | Collagen alpha-4(IV) chain | 0.9 ± 0.21 | SNS | 4 | 3 | ||
| S08123 | Collagen alpha-2(I) chain | 0.91 ± 0.05 | SNS | 7 | 3 | ||
| P29400 | Collagen alpha-5(IV) chain | 1.14 ± 0.42 | SNS | 8 | 10 | ||
| SP00738 | Haptoglobin | 1.45 ± 0.08 | <0.02 | 1.12 ± 0.22 | SNS | 5 | 3 |
| P24158 | Myeloblastin | 1.17 ± 0.16 | SNS | 7 | 2 | ||
| Q5SZK8 | FRAS1-related extracellular matrix protein 2 | 0.34 ± 0.01 | <0.01 | 3 | 3 | ||
| P13645 | Keratin, type I cytoskeletal 10 | 0.79 ± 0.38 | SNS | 6 | 2 | ||
| Q8WXI7 | Mucin-16 | 1.14 ± 0.59 | SNS | 5 | 7 | ||
| Q9HC84 | Mucin-5B | 1.63 ± 0.83 | SNS | 1.26 ± 0.35 | SNS | 7 | 8 |
| Q14517 | Protocadherin Fat 1 | 1.38 ± 0.59 | SNS | 5 | 2 | ||
| Q96MS0 | Roundabout homolog 3 | 0.72 ± 0.43 | SNS | 5 | 2 | ||
| Q96QU1 | Photocadherin 15 | 1.35 ± 0.15 | SNS | 4 | 3 | ||
| SP02768 | Serum albumin | 2.40 ± 0.67 | <0.0001 | 2.21 ± 0.68 | <0.01 | 19 | 41 |
| Q86UP0 | Cadherin-24 | 1.62 ± 0.12 | <0.003 | 1.68 ± 0.23 | <0.02 | 4 | 2 |
| Hydrolytic Enzymes and Proteinases | |||||||
| Q9UDR5 | Alpha-aminoadipic semialdehyde synthase, | 1.62 ± 0.38 | SNS | 3 | 2 | ||
| P16050 | Arachidonate 15-lipoxygenase | 0.94 ± 0.14 | SNS | 3 | 2 | ||
| Q86UQ4 | ATP-binding cassette sub-family A member 13 | 1.37 ± 0.28 | SNS | 7 | 4 | ||
| O00408 | cGMP-dependent 3′,5′-cyclic phosphodiesterase | 1.17 ± 0.32 | SNS | 3 | 2 | ||
| Q9Y5K2 | Kallikrein-4 (enamel matrix serine proteinase 1) | 2.16 ± 0.45 | <0.002 | 9 | 3 | ||
| SP14618 | Pyruvate kinase isozymes M1/M2 | 3.15 ± 2.14 | SNS | 5 | 3 | ||
| SP07477 | Trypsin-1 | 1.37 ± 0.16 | SNS | 11 | 4 | ||
| P35030 | Trypsin-3 | 1.00 ± 0.53 | SNS | 1.18 ± 0.15 | SNS | 10 | 4 |
| A4D1T9 | Peptidase S1 domain-containing protein LOC136242 | 2.67 ± 0.73 | <0.001 | 5 | 2 | ||
| A2RTX5 | Probable threonyl-tRNA synthetase 2, cytoplasmic | 1.25 ± 0.59 | SNS | 3 | 2 | ||
| O95714 | Probable E3 ubiquitin-protein ligase HERC2 | 1.03 ± 0.70 | SNS | 3 | 3 | ||
| Q5S007 | Leucine-rich repeat serine/threonine-protein kinase 2 | 0.74 ± 0.21 | SNS | 4 | 2 | ||
| Q6P4F7 | Rho GTPase-activating protein 11A | 0.99 ±1.02 | SNS | 3 | 2 | ||
| Q8IVL0 | Neuron navigator 3 | 1.17 ± 0.61 | SNS | 6 | 3 | ||
| P36871 | Phosphoglucomutase-1 | 0.69 ± 0.22 | SNS | 3 | 2 | ||
| Q9P212 | 1-Phosphatidylinositol-phosphodiesterase ε-1 | 1.65 ± 0.31 | <0.03 | 6 | 2 | ||
| Q9BYE2 | Transmembrane protease, serine 13 | 1.53 ± 0.35 | SNS | 8 | 2 | ||
| P30048 | Thioredoxin-dependent peroxide reductase | 1.69 ± 0.25 | <0.04 | 7 | 2 | ||
| SP04406 | Glyceraldehyde-3-phosphate dehydrogenase | 2.35 ± 0.45 | <0.02 | 7 | 3 | ||
| P08246 | Leukocyte elastase | 1.71 ± 0.18 | <0.002 | 4 | 2 | ||
| Q9UIQ6 | Leucyl-cystinyl aminopeptidase | 0.94 ± 0.28 | SNS | 3 | 2 | ||
| Inflammatory and Immune Response | |||||||
| P04083 | Annexin A1 | 1.41 ± 0.17 | <0.001 | 13 | 8 | ||
| SP01024 | Complement C3 | 1.44 ± 0.68 | SNS | 5 | 4 | ||
| SP01857 | Ig gamma-1 chain C region | 1.85 ± 0.58 | <0.0001 | 1.98 ±0.61 | <0.003 | 19 | 11 |
| SP01859 | Ig gamma-2 chain C region | 1.63 ± 0.30 | SNS | 1.64 ±0.38 | <0.01 | 6 | 3 |
| SP01861 | Ig gamma-4 chain C region | 2.34 ± 1.72 | SNS | 4 | 2 | ||
| SP01834 | Ig kappa chain C region | 1.34 ± 0.43 | SNS | 2.19 ± 0.83 | SNS | 8 | 4 |
| SP01842 | Ig lambda chain C regions | 1.57 ± 0.48 | SNS | 1.7 ± 0.37 | <0.05 | 5 | 5 |
| SP02788 | Lactotransferrin | 1.65 ± 0.24 | <0.04 | 1.61 ± 0.33 | <0.05 | 8 | 3 |
| P01833 | Polymeric immunoglobulin receptor | 1.4 ± 0.23 | SNS | 3 | 3 | ||
| P05164 | Myeloperoxidase | 2.12 ± 0.51 | <0.01 | 1.13 ± 0.27 | SNS | 6 | 4 |
| SP59665 | Neutrophil defensin 1 | 1.67 ± 0.37 | <0.03 | 1.26 ± 0.12 | <0.04 | 7 | 3 |
| SP05109 | Protein S100-A8 | 2.34 ± 0.65 | <0.0001 | 1.25 ± 0.30 | SNS | 13 | 13 |
| SP06702 | Protein S100-A9 | 2.41 ± 0.51 | <0.0001 | 1.89 ± 0.26 | <0.01 | 14 | 8 |
| Intracellular Protein/Nucleotide Binding | |||||||
| Q9UPY3 | Endoribonuclease Dicer | 2.42 ± 0.55 | SNS | 4 | 2 | ||
| Q8IVF2 | AHNK2 HUMAN Protein AHNAK2 | 1.24 ± 0.33 | SNS | 3 | 3 | ||
| Q6S8J3 | ANKRD26-like family C member 1A | 1.36 ± 0.32 | <0.02 | 11 | 6 | ||
| P13010 | ATP-dependent DNA helicase 2 subunit 2 | 0.5 ± 0.29 | SNS | 3 | 2 | ||
| Q9NVP1 | ATP-dependent RNA helicase DDX18 | 2.1 ± 2.60 | SNS | 3 | 2 | ||
| Q9NRL2 | Bromodomain adjacent to zinc finger domainprotein 1A | 1.03 ± 0.26 | SNS | 3 | 2 | ||
| Q2M1Z3 | Cdc42 GTPase-activating protein | 2.39 ± 0.48 | <0.004 | 7 | 2 | ||
| Q7Z7A1 | Centriolin | 1.09 ± 0.36 | SNS | 5 | 3 | ||
| Q8TD26 | Chromodomain-helicase-DNA-binding protein 6 | 1.48 ± 0.04 | <0.01 | 3 | 2 | ||
| Q5H945 | Chromosome 1 open reading frame 9 | 0.63 ± 0.12 | SNS | 3 | 2 | ||
| Q14999 | Cullin-7 (CUL-7) | 3.05 ± 0.4 | <0.01 | 3 | 2 | ||
| P16333 | Cytoplasmic protein NCK1 | 2.05 ± 2.44 | SNS | 3 | 2 | ||
| Q9H1X3 | DnaJ homolog subfamily C member 25 | 1.28 ± 0.85 | SNS | 3 | 2 | ||
| Q92621 | Nuclear pore complex protein Nup205 | 1.32 ± 0.09 | SNS | 9 | 2 | ||
| P52948 | Nuclear pore complex protein Nup98-Nup96 | 2.82 ± 1.84 | SNS | 3 | 2 | ||
| Q12830 | Nucleosome-remodeling factor subunit BPTF | 2.92 ± 0.22 | <0.001 | 3 | 2 | ||
| Q8NG31 | Protein CASC5 | 1.06 ± 0.45 | SNS | 4 | 2 | ||
| Q8IVF4 | Dynein heavy chain 10 | 1.29 ± 0.08 | SNS | 3 | 2 | ||
| Q9UFH2 | Dynein heavy chain 17, axonemal | 0.80 ± 0.14 | SNS | 4 | 3 | ||
| Q8TE73 | Dynein heavy chain 5 | 0.98 ± 0.22 | SNS | 3 | 2 | ||
| Q8WXX0 | Dynein heavy chain 7, axonemal | 0.95 ± 0.23 | SNS | 3 | 2 | ||
| O60447 | Ecotropic viral integration site 5 protein homolog | 1.38 ± 1.85 | SNS | 3 | 2 | ||
| SP13639 | Elongation factor 2 | 0.63 ± 0.17 | SNS | 5 | 2 | ||
| SP21333 | Filamin-A | 0.86 ± 0.15 | SNS | 7 | 2 | ||
| Q3V6T2 | Girdin | 2.69 ± 0.12 | <0.03 | 3 | 2 | ||
| Q9BXL5 | Hemogen | 2.35 ± 0.09 | <0.03 | 3 | 2 | ||
| SP69905 | Hemoglobin subunit alpha | 1.42 ± 0.13 | <0.01 | 7 | 4 | ||
| SP68871 | Hemoglobin subunit beta | 1.29 ± 0.11 | <0.01 | 8 | 6 | ||
| P0C0S8 | Histone H2A type 1 | 1.92 ± 0.53 | <0.004 | 7 | 2 | ||
| Q96A08 | Histone H2B type 1-A | 1.34 ± 0.55 | SNS | 7 | 2 | ||
| P62807 | Histone H2B type 1-C/E/F/G/I | 1.18 ± 0.12 | SNS | 3 | 2 | ||
| P68431 | Histone H3.1 | 1.07 ± 0.15 | SNS | 6 | 4 | ||
| Q09666 | Neuroblast differentiation-associated protein AHNAK | 1.01 ± 0.03 | SNS | 5 | 4 | ||
| Q7Z2Y8 | Interferon-induced very large GTPase 1 | 1.05 ± 0.11 | SNS | 7 | 3 | ||
| Q9UKX3 | Myosin-13 | 1.67 ± 0.14 | <0.03 | 3 | 2 | ||
| SP62805 | Histone H4 | 1.58 ± 0.33 | <0.01 | 9 | 4 | ||
| Q149N8 | E3 ubiquitin-protein ligase SHPRH | 1.39 ± 0.27 | SNS | 5 | 2 | ||
| Q5THR3 | EF-hand calcium-binding domain-containing protein 6 | 0.51 ± 0.10 | SNS | 3 | 2 | ||
| Q9BXW9 | Fanconi anemia group D2 protein subunit BPTF | 2.16 ± 0.18 | <0.03 | 3 | 2 | ||
| Q96ST3 | Paired amphipathic helix protein | 1.28 ± 0.18 | SNS | 3 | 2 | ||
| Q9C0D5 | Protein TANC1 | 1.45 ± 0.18 | SNS | 5 | 3 | ||
| O00360 | Putative p150 | 2.37 ± 0.36 | <0.05 | 3 | 2 | ||
| Q9C0B0 | RING finger protein unkempt | 3.67 ± 1.32 | SNS | 4 | 2 | ||
| Q9NSC2 | Sal-like protein 1 | 0.85 ±0.70 | SNS | 3 | 2 | ||
| O75691 | Small subunit processome component 20 homolog | 1.03 ± 0.18 | SNS | 3 | 2 | ||
| P53804 | Tetratricopeptide repeat protein 3 | 1.49 ± 0.61 | SNS | 3 | 2 | ||
| O95359 | Transforming acidic coiled-coil-containing protein 2 | 0.96 ± 0.13 | SNS | 6 | 2 | ||
| Q9HCJ0 | Trinucleotide repeat-containing gene 6C protein | 0.74 ± 0.31 | SNS | 4 | 3 | ||
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | 1.35 ± 0.57 | SNS | 3 | 2 | ||
| Q96RL7 | Vacuolar protein sorting-associated protein 13A | 2.03 ± 0.75 | <0.01 | 5 | 3 | ||
| Q9UNX4 | WD repeat-containing protein 3 | 0.78 ± 0.22 | SNS | 4 | 2 | ||
| Q5JSH3 | WD repeat-containing protein 44 | 1.23 ± 0.48 | SNS | 3 | 2 | ||
| Q15911 | Zinc finger homeobox protein 3 | 0.49 ± 0.19 | SNS | 3 | 2 | ||
| Q86UP3 | Zinc finger homeobox protein 4 | 1.14 ± 0.92 | SNS | 3 | 3 | ||
| Q86YH2 | Zinc finger protein 280B | 0.66 ± 0.45 | SNS | 3 | 2 | ||
| O94822 | Zinc finger protein 294 | 1.23 ± 0.53 | SNS | 3 | 3 | ||
| Q5T7W0 | Zinc finger protein 618 | 1.16 ± 0.12 | SNS | 4 | 3 | ||
| Proteins Falling Into Other Categories | |||||||
| P40145 | Adenylate cyclase type 8 | 1.11 ± 0.43 | SNS | 5 | 2 | ||
| Q9ULX6 | A-kinase anchor protein 8-like | 1.57 ± 0.86 | SNS | 3 | 2 | ||
| P46013 | Antigen KI-67 | 1.03 ± 0.71 | SNS | 4 | 4 | ||
| Q5W041 | Armadillo repeat-containing protein | 4.32 ± 0.36 | <0.001 | 3 | 2 | ||
| P31513 | Dimethylaniline monooxygenase [N-oxide-forming] 3 | 0.26 ± 0.17 | <0.03 | 5 | 3 | ||
| Q8WY64 | E3 ubiquitin-protein ligase MYLIP | 1.62 ± 0.83 | SNS | 4 | 2 | ||
| Q9BUH6 | Uncharacterized protein C9orf142 | 1.67 ± 0.23 | <0.03 | 3 | 2 | ||
| SQ8WZ4 | Titin | 2.37 ± 0.93 | <0.001 | 8 | 10 | ||
| A0AVI2 | Fer-1-like protein 5 | 1.04 ± 0.12 | SNS | 3 | 2 | ||
| Q5CZC0 | Fibrous sheath-interacting protein 2 | 2.16 ± 1.15 | SNS | 4 | 2 | ||
| Q17R60 | Interphotoreceptor matrix proteoglycan 1 | 0.97 ± 0.04 | SNS | 5 | 2 | ||
| Q9P041 | HSPC-109 | 1.12 ± 0.12 | SNS | 0.91 ± 0.21 | SNS | 7 | 3 |
| Q8WVZ9 | Kelch repeat and BTB domain-containing protein 7 | 0.82 ± 0.51 | SNS | 4 | 2 | ||
| Q9H825 | Methyltransferase-like protein 8 | 1.24 ± 0.12 | SNS | 5 | 2 | ||
| Q9NU22 | Midasin | 0.67 ± 0.17 | SNS | 3 | 2 | ||
| Q5VTT5 | Myomesin-3 | 0.14 ± 0.04 | <0.001 | 3 | 2 | ||
| Q8IVL1 | Neuron navigator 2 | 0.51 ± 0.31 | SNS | 3 | 2 | ||
| Q9P2E3 | NFX1-type zinc finger-containing protein 1 | 0.54 ± 0.65 | SNS | 4 | 2 | ||
| A8MV47 | Uncharacterized protein ENSP00000380627 | 0.93 ± 0.11 | SNS | 6 | 2 | ||
| Q8NG94 | Olfactory receptor 11H1 | 1.08 ± 0.26 | SNS | 3 | 2 | ||
| SP13796 | Plastin-2 | 1.37 ± 0.34 | SNS | 4 | 2 | ||
| Q96KK3 | Potassium voltage-gated channel subfamily S member | 0.73 ± 0.22 | SNS | 3 | 2 | ||
| SP07737 | Profilin-1 | 0.92 ± 0.24 | SNS | 7 | 3 | ||
| Q658L4 | Putative uncharacterized protein DKFZp666E157 | 1.13 ± 0.17 | SNS | 3 | 2 | ||
| Q5VUG0 | Scm-like with four MBT domains protein 2 | 0.44 ± 0.15 | <0.04 | 3 | 2 | ||
| SP02787 | Serotransferrin | 1.07 ± 0.12 | SNS | 1.59 ± 0.33 | SNS | 9 | 9 |
| Q6IQ55 | Tau-tubulin kinase 2 | 0.95 ± 0.33 | SNS | 5 | 3 | ||
| Q5T6L9 | Transmembrane protein C6orf70 | 1.55 ± 0.65 | SNS | 3 | 2 | ||
| Q6ZXV5 | Transmembrane and TPR repeat-containing protein 3 | 8.68 ± 6.6 | SNS | 3 | 2 | ||
| Q0VAA2 | Uncharacterized protein C14orf166B | 1.24 ± 0.06 | SNS | 3 | 2 | ||
| Q15878 | Voltage-dependent R-type Ca+2 channel subunit α-1E | 1.28 ± 0.91 | SNS | 6 | 2 | ||
| Q8N4N8 | Kinesin-like protein KIF2B | 2.69 ± 0.37 | <0.001 | 2.88 ± 0.26 | <0.001 | 7 | 5 |
| Protease and enzyme inhibitors | |||||||
| SP01009 | Alpha-1-antitrypsin | 1.88 ± 0.84 | <0.01 | 12 | 9 | ||
| SP04080 | Cystatin-B | 1.42 ± 0.56 | SNS | 8 | 2 | ||
| SP01023 | Alpha-2-macroglobulin | 1.51 ± 0.60 | SNS | 9 | 7 | ||
| Table 2B. Proteins found only in GCF derived from healthy periodontal sites labeled by light stable-isotope or periodontal sites labeled by heavy stable-isotope. | |||
|---|---|---|---|
| Proteins found only in GCF of healthy sites | |||
| Accession # | Protein name | Accession # | Protein name |
| P49641 | Alpha-mannosidase IIx | P26045 | Tyrosine-protein phosphatase non-receptor type 3 |
| Q9NR09 | Baculoviral IAP repeat-containing protein 6 | P08579 | U2 small nuclear ribonucleoprotein B″ |
| Q14CN2 | Calcium-activated chloride channel regulator 4 | Q6NSZ9 | Zinc finger protein 498 |
| Q14031 | Collagen alpha-6(IV) | Q8WXB4 | Zinc finger protein 606 |
| O15061 | Desmuslin | P33908 | Mannosyl-oligosaccharide-1,2-α-mannosidase IA |
| Q5T4S7 | E3 ubiquitin-protein ligase UBR4 | Q8TCU4 | Alstrom syndrome protein 1 |
| P29322 | Ephrin type-A receptor 8 | Q9UQ05 | Potassium voltage-gated channel subfamily H 4 |
| Q86XX4 | Extracellular matrix protein FRAS1 | P20648 | Potassium-transporting ATPase alpha chain 1 |
| Q0JRZ9 | FCH domain only protein 2 | Q6ZRV2 | Protein FAM83H |
| Q13233 | Mitogen-activated protein kinase kinase kinase 1 | Q5TBA9 | Protein furry homolog |
| P41218 | Myeloid cell nuclear differentiation antigen | Q4ZG55 | Protein GREB1 |
| Q01804 | OTU domain-containing protein 4 | Q9NSE7 | Putative ATP-binding cassette transporter sub-family C member 13 |
| P56645 | Period circadian protein homolog 3 | P23471 | Receptor-type tyrosine-protein phosphatase zeta |
| Proteins found only in GCF of periodontal sites | |||
| Q65ZQ1 | Anti-colorectal carcinoma heavy chain | A7KAX9 | Rho GTPase-activating protein 32 |
| P12956 | ATP-dependent DNA helicase 2 subunit 1 | Q96AG3 | Solute carrier family 25 member 46 |
| P98160 | Basement membrane-specific heparan sulfate proteoglycan core protein | Q9HCB6 | Spondin-1 |
| P55285 | Cadherin-6 | Q9H2G2 | STE20-like serine/threonine-protein kinase |
| Q9P2D1 | Chromodomain-helicase-DNA-binding protein 7 | Q5T1R4 | Transcription factor HIVEP3 |
| P06681 | Complement C2 | Q6UXZ0 | Transmembrane and immunoglobulin domain-containing protein 1 |
| Q9NY74 | Ewing’s tumor-associated antigen 1 | Q5T3F8 | Transmembrane protein 63B |
| Q9UKT4 | F-box only protein 5 OS=Homo sapiens | O75382 | Tripartite motif-containing protein 3 |
| Q9BUJ2 | Heterogeneous nuclear ribonucleoprotein U-like protein 1 | A6NGQ3 | Uncharacterized protein OBSCN |
| P01877 | Ig alpha-2 chain C region | Q9Y487 | V-type proton ATPase 116 kDa subunit a isoform 2 |
| Q8NFY9 | Kelch repeat/BTB domain-containing protein 8 | Q9UBW7 | Zinc finger MYM-type protein 2 |
| A4D0S4 | Laminin subunit beta-4 | Q5VU65 | Nuclear pore membrane glycoprotein 210-like |
| Q9NS15 | Latent-transforming growth factor β-binding protein 3 | Q9H7F0 | Cation-transporting ATPase 13A3 |
| O00562 | Membrane-associated phosphatidylinositol transfer protein 1 | Q9UPA5 | Protein bassoon |
| Q7Z5P9 | Mucin-19 | P46940 | Ras GTPase-activating-like protein IQGAP1 |
| P21439 | Multidrug resistance protein 3 | Q15303 | Receptor tyrosine-protein kinase erbB-4 |
| Table 2C. Relative quantitative bacterial and yeast proteome analysis of GCF from healthy and periodontal disease subjects using stable-isotope-labeling chemistries. S.D. = standard deviation, # runs identified = number of times a given protein was identified, # peptides identified = number of distinct peptides identified for a given protein and used for quantification, SNS = statistically not significant. | |||||
|---|---|---|---|---|---|
| Accession # | Bacterial proteins | mTRAQMean ±S.D. | P value | # Runs Identified | # Peptides Identified |
| K0ZHN4 | Uncharacterized protein (Streptococcus sp) | 3.87 ± 1.2 | <0.02 | 2 | 4 |
| U2JW31 | Helicase protein (Porphyromonas gingivalis) | 2.36 ± 1.4 | SNS | 2 | 4 |
| M9VKY7 | Sec-independent translocase (Propionbacterium acnes) | 4.33 ± 3.45 | SNS | 2 | 2 |
| B7GQL2 | Uncharacterized protein ( Bifidobacterium longum ) | 1.80 ± 0.75 | SNS | 2 | 3 |
| U2JW31 | Oxidoreductase (Porphyromonas gingivalis) | 2.86 ± 0.91 | <0.04 | 2 | 3 |
| G6FFC9 | Chromosmal partition protein Smc (Lactococcus lactis) | 14.11 ±9.22 | SNS | 2 | 4 |
| U2IB04 | RelA/SpoT family protein (Porphyromonas gingivalis) | 3.30 ± 1.12 | <0.02 | 3 | 4 |
| F1YR10 | Glycine dehydrogenase (decarboxylating) (Acetobacter pomorum) | 2.38 ± 0.44 | <0.03 | 2 | 2 |
| U2JN88 | Outer membrane protein (Porphyromonas gingivalis) | 2.34 ± 0.66 | <0.03 | 2 | 3 |
| U2JKX9 | Uncharacterized protein (Porphyromonas gingivalis) | 5.33 ± 4.2 | SNS | 2 | 4 |
| G6FCL3 | DNA topoisomerase (Lactococcus lactis) | 1.10 ± 0.67 | SNS | 2 | 4 |
| N6X320 | Glutamate-tRNA ligase (Actinomyces cardiffensis) | 2.38 ± 0.11 | <0.04 | 2 | 2 |
| G6FBX5 | DNA-directed DNA polymerase (Lactococcus lactis) | 1.83 ± 1.66 | SNS | 2 | 4 |
| G6FAW1 | GMP synthase (glutamine-hydrolyzing) (Lactococcus lactis) | 4.25 ± 5.36 | SNS | 2 | 3 |
| M9VHN8 | Uncharacterized protein (Propionibacterium acnes) | 1.29 ± 1.33 | SNS | 2 | 2 |
| B7GN98 | Uncharacterized protein (Bifidobacterium longum) | 5.63 ± 0.94 | <0.04 | 3 | 2 |
| G6FFE2 | Putative uncharacterized protein (Lactococcus lactis) | 2.28 ± 0.61 | <0.03 | 2 | 4 |
| F9E9H1 | aminopeptidase (Streptococcus sanguinis) | 14.36 ±12.25 | SNS | 2 | 2 |
| U2K2Z6 | GTP-binding protein typeA (Porphyromonas gingivalis) | 2.66 ± 1.8 | SNS | 2 | 2 |
| Q6MQE9 | Serine proteinase (peptidase) (Bdellovibrio bacteriovorus) | 4.50 ± 4.10 | SNS | 2 | 2 |
| Q4YIL3 | Leucine amidopeptidase PfLAP (Plasmodium chabaudi) | 2.50 ± 0.85 | <0.04 | 2 | 3 |
| K1A3W6 | PTS system, Lactose/cellobiose-specific IICB (Enterococcus sp) | 1.50 ± 0.33 | SNS | 2 | 2 |
| M9VQL8 | Uncharacterized protein (Propionibacterium acnes) | 0.87 ± 0.12 | <0.02 | 2 | 2 |
| D7W9K3 | Glycosyl hydrolase (Corynebacterium genitalium) | 3.50 ± 1.2 | <0.05 | 2 | 2 |
| D9ZEW0 | Putative carbohydrate-active enzyme (uncultured) | 2.42 ± 0.60 | <0.05 | 2 | 3 |
| L8A4U1 | PTS system, sucrose subfamily IIABC (Enterococcus faecium) | 1.18 ± 0.13 | SNS | 2 | 2 |
| F1YWU8 | Glutamate synthase large chain (Acetobacter pomorum) | 1.37 ± 0.34 | SNS | 2 | 3 |
| U2IF16 | Putative glycerate kinase (Porphyromonas gingivalis) | 0.95 ± 0.31 | SNS | 2 | 2 |
| F1YUN4 | Formamidase (Acetobacter pomorum) | 5.64 ± 0.2 | <0.01 | 2 | 2 |
| F1YSW6 | Glutamate-ammonia-ligase adenyltransferase (Acetobacter pomorum) | 4.88 ±2.85 | SNS | 2 | 2 |
| Q8KHM3 | Bifunctional protein GlmU (Fusobacterium nucleatum) | 1.78 ± 0.45 | <0.02 | 2 | 5 |
| P31224 | Multidrug efflux pump subunit AcrB (Escherichia coli) | 2.34 ± 2.12 | SNS | 2 | 2 |
| P49331 | Glucosyltransferase-S (Streptococcus mutans) | 1.21 ± 0.51 | SNS | 2 | 4 |
| B7GTQ1 | ABC transporter permease component (Bifidobacterium longum) | 3.61 ± 4.75 | SNS | 2 | 2 |
| G6EUF8 | Putative uncharacterized protein (Lactobacillus lactis bulgaricus) | 0.53 ± 0.22 | <0.03 | 2 | 2 |
| K1A813 | Glycosyl transferase family protein (Enterococcus sp) | 5.7 ± 1.7 | <0.01 | 2 | 4 |
| K1AX94 | Cell wall surface anchor family protein (Enterococcus sp) | 3.78 ± 3.69 | SNS | 2 | 2 |
| B7GUQ5 | Phosphatase (Bifidobacterium longum-infants) | 14.1 +9.4 | SNS | 2 | 2 |
| K1A149 | Surface associated protein (Streptococcus sp) | 1.81 ± 1.30 | SNS | 2 | 3 |
| K0ZUJ1 | Cell wall surface anchor family protein (Streptococcus sp) | 4.40 ± 3.20 | SNS | 2 | 3 |
| M9VPN0 | Cysteine synthase (Propionibacterium acnes) | 1.10 ± 0.42 | SNS | 2 | 3 |
| K1A3L8 | Nicotinate phosphorybosyltransferase (Enterococcus sp) | 1.43 ± 1.33 | SNS | 2 | 2 |
| G6FCY9 | Putative uncharacterized protein (Lactococcus lactis) | 2 | 4 | ||
| Yeast Proteins | |||||
| V5N887 | Beta-actin (Chiloscylium puntatum) | 16.50 ± 9.80 | SNS | 2 | 3 |
| E7Q8H9 | Poly (A) polymerase Trf5p (Saccharomyces cerevisiae) | 1.06 ± 0.17 | SNS | 4 | 3 |
| E3UAT6 | Actin (Cladosporium cladosporioides) | 1.80 ± 0.82 | SNS | 3 | 7 |
| P14741 | Translation initiation factor eIF2B (Saccharomyces cerevisiae) | 3.39 ± 1.04 | <0.02 | 2 | 2 |
| N1P505 | DNA polymerase (Saccharomyces cerevisiae) | 1.20 ± 0.74 | SNS | 2 | 3 |
| Q06593 | Oligopeptide transporter (Saccharomyces cerevisiae) | 1.17 ± 0.75 | SNS | 3 | 4 |
| P14235 | Actin (Candida albicans) | 6.7 ± 2.67 | SNS | 2 | 4 |
| N1NWJ3 | Predicted protein Tcb1p(Saccharomyces cerevisiae) | 0.94 ± 0.55 | SNS | 2 | 3 |
| Q6FNY7 | DNA polymerase (Candida glabrata) | 1.20 ± 0.74 | SNS | 2 | 3 |
| T1WEZ8 | Actin (Collectotrichum truncotum) | 1.6 + 0.58 | <0.003 | 2 | 4 |
| C7BFY6 | Histone H3 protein (Tristoma integrum) | 1.34 + 0.19 | <0.0001 | 3 | 4 |
In addition to the search parameters and criteria used, the identified peptide sequences were stringently evaluated and assessed manually by examining each of the identified peptide MS/MS data for the quality and the confidence through the b and y ion fragment series. The MS quantitative data was analyzed and in cases where no ratio was determined the data was checked whether there was any peptide identified and labeled only by light or only by heavy reagent. If a peptide identified was only labeled with light reagent, there will be no ratio calculated and hence it was absent in the control group. Conversely, if the identified peptides were only labeled with the heavy reagent, this reflects that the protein was present only in the disease sample.
Protein annotations
The identified proteins were classified and assigned by molecular function, biological process and cellular component using three web-based applications: Babelomics database http://babelomics.bioinfo.cipf.es/index.html, AmiGO database (http://amigo.geneontology.org/cgi-bin/amigo/go.cgi advanced_query=yes) and Swiss protein database (http://ca.expasy.org/).
Validation of MS data by classical Enzyme-Linked Immunosorbent Assay (ELISA)
To validate the large-scale LC-ESI-MS/MS quantitative analytical approaches protein S100A9 and human serum albumin were selected for confirmation of the results with a different method. S100A9 and human serum albumin levels were measured in the samples by commercially available ELISA kits, according to the manufacturer’s instructions. S100A9 was used for validation because its level was significantly increased in the GCF of patients with periodontal disease as well as its biological significance. The latter relates to its inflammatory origin and expression by macrophages and neutrophils. These properties reflect potentially important biomarker value of this protein for periodontal disease. Albumin was selected because it is well known to be serum derived and its levels increase in patients with periodontal disease due to increased serum protein contributions into the GCF microenvironment during local inflammation/periodontitis. One periopaper from each of 40 healthy subjects and periodontal patients were used to elute proteins for ELISA studies. To optimize the protein elution the periopaper margin containing the GCF was immersed in 20 μl of 50 mM NH4HCO3, pH ~8, (containing 6 M guanidinium HCL) in an Eppendorf tube. After 5 min at room temperature the periopaper was raised above the liquid level and clipped with the Eppendorf cap and centrifuged at 10,000 rpm using bench top centrifuge to elute the residual buffer from the periopaper. The periopaper was subjected to 2x repeat of the above step using each time 20 μl of 50 mM NH4HCO3 (containing no guanidinium HCL) and the eluted GCF proteins were combined. Such samples were generated for all 40 healthy subjects and 40 periodontal patients with each individuals GCF kept separately.
(i) Human Albumin ELISA Kit
The concentrations of human albumin in GCF samples were determined by ELISA Kit (Bethy Laboratories, Inc, Montgomery, TX) as detailed in the manufacturer’s protocol was used for identification and quantitation of human albumin in GCF. The absorbance was measured at 450 nm and the human albumin concentrations in the samples were determined from the standard albumin calibration curve. These analyses were carried out on the 4 different pooled samples (each containing GCF from 10 different individuals) for healthy subjects and similarly for periodontal patients.
(ii) S100-A9 protein quantification by enzyme-Immunoassay
The concentrations of S100-A9 protein in GCF samples were determined by ELISA kit (Peninsula Laboratories, Inc, San Carlos, CA) as detailed in the manufacturer’s protocol. The absorbance at 450 nm was recorded and the S100-A9 protein concentrations in the samples were determined from the standard S100-A9 calibration curve. These analyses were carried out on both the pooled samples used as indicated above for albumin assay, however, in this case additional ELISA was performed using each of the 40 healthy and 40 periodontal GCF samples individually to gain insights to the individual variations.
RESULTS
The results showed ~84% protein recovery with electroelution by SDS-PAGE method which was higher than the 70% obtained when proteins were eluted by simple NH4HCO3 washing technique. We have recently detailed the significance of the type of elution methods used for periopaper collected GCF on the identified proteins for proteome analysis by MS technology (Carneiro et al., 2012). These become even more critical and important when MS-based quantitative approaches are used. The results from the current study showed an average GCF volume of 0.49±0.12 μl (~47 μg protein) and 0.73±0.19 μl (~56 μg protein) per site per paper strip from healthy and periodontal sites with total protein concentrations of 96.38±36.51 μg/μl and 79.1±22.0, respectively, consistent with previous reports (Hattingh & Ho, 1980). ELISA for serum albumin in GCF samples indicated ~15 μg/μl albumin, and assuming albumin represents ~50% (w/w) (50–60% w/w, Burtis & Ashwood, 1999; McPherson & Pincus, 2011) of the total serum proteins an estimate of other serum proteins in GCF is ~15 μg/μl of GCF. This provides ~30 μg/μl (w/w) serum proteins in GCF of periodontally healthy sites. Hence, of the total protein composition in GCF from healthy sites, only small portion ~30 μg/μl (~30%) was serum derived and the remaining 66 μg/μl was local GCF microenvironment specific. These results very closely correlate with those in the literature where GCF of healthy sites have low-albumin content which reflects overall low serum content (Alfano 1974; Pashley 1976; Bickel et al., 1985). Bickel et al showed that GCF collected from healthy and inflamed sites and analysis of albumin concentrations indicated 16.8±9.2 μg/μl, 23.1±7.1 μg/μl and 36.6±8.0 μg/μl after not brushing for 12 hrs, 36 hrs and chronically inflamed sites, respectively. This is consistent with the notion that GCF in healthy sites is not a free flowing serum but rather contains small percentage (30%) of serum proteins and may have > 120 locally generated proteins not serum related as we have shown recently (Carneiro et al., 2012). Furthermore, the level of serum contribution to GCF more than doubles, 73% w/w, as compared with 27% w/w total proteins derived from the GCF microenvironment, Table 1.
Table 1.
Gingival crevicular fluid volume, total protein and albumin concentrations, and their relative distributions in health and periodontal disease.
| Gingival Crevicular Fluid | Serum | Reference | ||
|---|---|---|---|---|
| Healthy | Periodontal | |||
| Mean GCF volume (μl) | 0.49 ± 0.12 | 0.73 ± 0.19 | Present study | |
| Mean GCF protein μg/periopaper | ~ 47 | ~56 | Present study | |
| Total protein (μg/μl) | 96.38 ± 36.51 | 79.1 ± 22.0 | Present study | |
| Literature | 93.1 ± 14.7 | 69.0 ± 2.5 | 70–92 |
Hattingh & HO, 1980. Burtis & Ashwood, 1999; McPherson & Pincus, 2011. |
| Total albumin (μg/μl) | 15.4 ± 1.4 | 29.1 ± 5.1 | Present study | |
| Literature | 16.8 ± 9.2 | 36.6 ± 8.0 | 39.7 ± 4.9 | Bickel et al., 1985. |
| Total serum proteins in GCF (μg/μl) | ~30 (30% w/w) | ~58 (73% w/w) | Present study | |
| Total GCF microenviroment specific proteins (μg/μl) | ~66 (70% w/w) | ~21 (27% w/w) | Present study | |
Quantitative Proteome of GCF from Healthy versus Periodontitis Patients Using LC-ESI-MS/MS Technology Coupled with Stable-Isotope Labeling Chemistry Approaches
While qualitative identification of proteins in biological samples has its own merit and biological importance, in general, studies aiming to define novel biomarkers useful for monitoring disease states require relative quantitative MS approaches. The GCF proteome reported in Table 2 from mTRAQ labeling was constructed using only proteins identified by two or more peptides which are the accepted criteria for general proteomic studies whereas those derived from ICAT labeling included also proteins identified by a single peptide. Using the filtering criteria chosen the results were associated with a false-positive-rate of ~1% at protein level for the human proteins, and 2% and <1% at protein level for the bacterial and yeast proteins, respectively. The GCF proteins identified reflect a wide range of components with a variety of different biological functions and origins. Apart from the expected serum derived proteins, the data revealed the presence of macromolecules belonging to the early inflammation, immune response, cellular and extracellular matrix components, a variety of enzymes including proteinases, esterases and their inhibitors. The data presented in Table 2A are arranged and categorized according to their biological groups. For the current work peptides labeled and identified in both groups were used for relative quantitation whereas those found only in healthy or periodontal group are listed separately in Table 2B. Since the volume of GCF collected was measured for each paper strip using the periotron apparatus during collection procedure it was possible to calculate the protein concentration of GCF after recovery of proteins from periopaper. After extraction of the proteins through electroelution by SDS-PAGE and in gel digestion of the full lanes as well as those sets of experiments using sections of the gels corresponding to different Mr, a total of 238 proteins were identified and 180 of those were quantified in GCF using ICAT and mTRAQ quantitative approaches. Table 2A shows the list of these proteins with their corresponding relative quantifications (as ratios of healthy versus periodontal samples) and their associated statistical significance, “p values”. Of these 212 proteins were identified by mTRAQ and 26 were identified using ICAT consistent with the LC-ESI-MS ion abundance profiles depicted in Figure 1 Panel 3A & 3B. As expected because of the universal nature of the chemistry of mTRAQ, the total number of proteins identified was far greater than those using ICAT. Also relative quantification of 26 proteins by two different reagents with data closely overlapping provided further confidence to the overall MS data set. The statistical significance of the ratios of each protein from healthy versus periodontal disease were derived by using the raw LC-MS/MS peak area data of each identified peptide pairs within a given run and performing two-tailed student’s paired t-test utilizing the combined data generated for a given protein in all runs. Although GCF from periodontally healthy sites has only ~30% (w/w) protein contributions from serum it was of interest to establish what possible proportion of all the GCF proteins identified in the present work could be derived from serum. This was possible since the plasma proteome has been published using large-scale MS analytical methods, (Schenk et al., 2008), permitting us to establish cross-correlation with our quantitative GCF proteome. This analysis led to 47 proteins (~20%) of our GCF proteome that were also identified in plasma by MS and surprisingly a larger proportion, 180 proteins (~80%), were not identified in plasma/serum proteome by MS technology, Table 2. Hence, we refer to these latter set of proteins as the local GCF microenvironment specific proteins. Within the interest of this study we have also identified and quantified 42 bacterial proteins and 11 yeast proteins, Table 2C. The bacterial proteins were derived from 14 different species some of which are well known pathogens that participate in the induction and progression of periodontal disease. The proteins listed in Table 2C are those that have been identified at least in 2 runs or more which is the same criteria used for the human proteins so that comparison can be made in terms of protein abundance and bacterial load. Interestingly, the dominant bacterial proteins were derived from both gram +ve and gram −ve bacteria such as Porphyromonas gingivalis (7 proteins), Streptococcus species (5 proteins), Lactococcus lactis (6 proteins), Propionibacterium acnes (4 proteins), Bifidobacterium longum (4 proteins), Acetobacter pomorum (4 proteins), Enterococcus sp (5 proteins). Importantly, of the 42 bacterial proteins 16 were found to be elevated in periodontal disease and these were statistically significant, Table 2C. Of the latter elevated proteins, 3 were derived from Porphyromonas gingivalis, 2 from each of Lactococcus lactis and Acetobacter pomorum, and 1 from each of Streptococcus species, Bifidobacterium longum, Enterococcus sp, Fusobacterium nucleatum, Plasmodium chabaudi, and Actinomyces cardiffensis.
Validation of MS data by classical Enzyme-Linked Immunosorbent Assay (ELISA)
One of the advantages of quantitative MS approach is the ability to obtain relative quantitation data for a large number of proteins simultaneously and rapidly followed by targeted approaches using selected small number of proteins to evaluate their potential as biomarkers. ELISA of specific highlighted proteins can be used to validate the MS-data and confirming the results by an additional classical method. To validate our LC-ESI-MS/MS-data and confirm the results we have chosen two proteins, namely S100A9 and human serum albumin, and performed ELISA. These two proteins were chosen based on the MS data, their properties and existing knowledge of their biology. Protein S100A9 was selected because it showed a significant increase in GCF of subjects with periodontal disease, and its association with inflammation and specific expression by macrophages and neutrophils. Serum albumin was selected to test the reliability and accuracy of MS-based approach since this is a well know abundant protein in GCF and its quantitative levels in periodontal disease relative to healthy have been well established.
(i) Serum Albumin
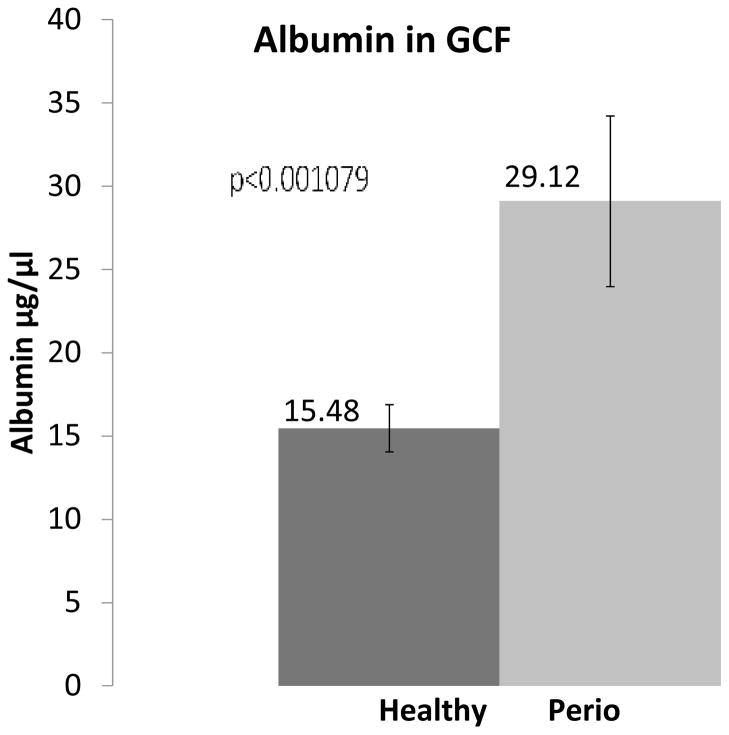
The MS relative quantitation data showed ~2.55 and 2.4 fold increase in human serum albumin in GCF with mTRAQ and ICAT approaches, respectively. Similarly, ELISA for albumin of pooled sets of samples corresponding to those used for MS analysis of GCF from 40 healthy and 40 periodontal patients showed concentrations to be 15.5±1.4 and 29.1±5.1 μg/μl of GCF (~2 fold increase), respectively, Figure 3. The Student paired t-test showed that this difference between the controls and periodontal samples were statistically significant, p<0.001.
Figure 3. ELISA for serum albumin in GCF of patients with periodontal disease and control patients.
4 different pools of GCF samples (each containing equal amounts of NH4HCO3 eluted GCF proteins from 10 different healthy individuals) were generated using GCF from all 40 healthy subjects and 4 different pools of GCF samples (each containing equal amounts of NH4HCO3 eluted GCF proteins from 10 different periodontal patients) were generated using GCF from all 40 periodontal patients, as illustrated in Figure 1, Panel 1. ELISA was carried out in triplicates on aliquots of pooled 4 healthy and 4 periodontal GCF samples representing GCF derived from 40 individuals of each respective group. The data plotted as the mean and variation within each group is shown with standard deviation. The student t-test showed that the two sets of data, disease versus control, were different and statistically significant p<0.001.
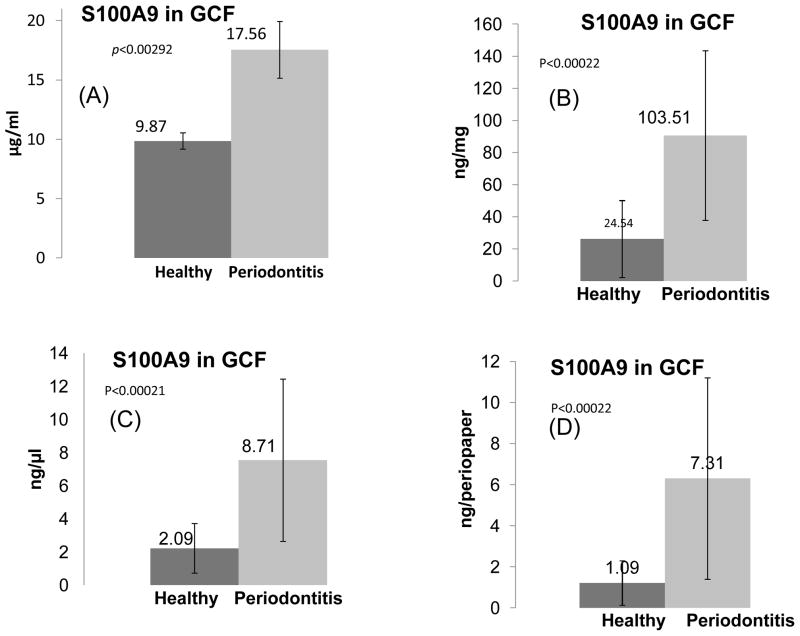
(ii) Protein S100A9
Figure 4 show results of ELISA for S100A9 protein using pooled samples as well as analysis of 40 GCF samples from healthy and 40 GCF samples from periodontal patients individually validating the MS data. The results from the ELISA of pooled samples showed the concentrations of S100A9 to be 9.9 μg/ml and 17.6 μg/ml in the GCF of healthy and periodontal individuals, respectively, Figure 4A. These data reflected ~ 2 fold increase which was statistically significant with a p<0.003, and consistent with the results from the MS relative quantitative data in Table 2. To further evaluate effectiveness of S100A9 as a biomarker and its potential utility under chair-side diagnostic approach, we have also analyzed individual samples which demonstrated clear major increase of this protein in periodontal patients. Although as expected the standard deviation for such analysis was much higher indicative of significant individual variations, nevertheless the results for healthy versus periodontal samples were still different and statistically significant, p<0.0002. It was of interest to evaluate the data further to establish whether the observed differences between the healthy and periodontal GCF samples were statistically significant and can be potentially useful as a “chair-side” biomarker. We have expressed/normalized the observed results from analysis of all of the samples individually in terms of: per μl of GCF volume, (ii) per mg GCF protein, and (iii) per periopaper. Interestingly irrespective as to how the data were normalized, the differences remained statistically significant, p=0.0002, Figure 4B, 4C & 4D.
Figure 4. ELISA of S100A9 protein in GCF of healthy individuals and those with periodontal disease.
(A), ELISA of S100A9 in pools of GCF from 40 healthy subjects and 40 periodontal patients were used as described for albumin in Figure 3 and demonstrated in Figure 1, Panel 1. (B), ELISA of S100A9 in each of the 40 GCF of healthy and 40 periodontal disease, all individually analyzed and normalized per mg of protein. (C), ELISA of S100A9 in each of the 40 GCF of healthy and 40 periodontal disease all individually analyzed and normalized per μl of GCF volume. (D), ELISA of S100A9 in each of the 40 GCF of healthy and 40 periodontal disease, all individually analyzed and normalized per GCF periopaper. The variation within each group is shown with standard deviation. The student t-test showed that the two sets of data, disease versus control, are different and these were statistically significant with p values as noted within the Figures.
DISCUSSION
In the past decade or so there has been significant interest in the development of non-invasive oral and systemic diagnostic biomarkers by large-scale protein analysis which was fueled by the advances made in state-of-the-art mass spectrometry (MS) (Xie et al., 2005; Hu et al., 2005; Hardt et al., 2005; Denny et al., 2008; Yan et al., 2009; Wong, 2006; Carneiro et al., 2012, Salih et al., 2010). These extensive studies have been carried out using whole saliva or parotid secretions from healthy individuals with no systemic or periodontal disease in order to establish proteome baseline in health which can then be used to compare with diseased states for diagnostic biomarkers discovery. Another oral cavity specific fluid is GCF which represents a special protein composition with constitutes ranging from local microenvironment proteins, degradation products as well as serum derived proteins. While MS-based proteome studies of GCF from healthy and periodontal origin have been carried out, these were predominantly at a “qualitative level” (Ngo et al., 2010, Heo et al 2011; Carneiro et al., 2012; Baliban et al., 2012; Tsuchida 2012) or at a quantitative level but using GCF from experimentally induced 21 day gingivitis model (Grant et al., 2010; Bostanci et al., 2013). The only attempt to perform relative quantitative analysis using GCF from healthy and periodontal patients by direct-large scale MS-based technology used a “label-free” approach. Hence, to date the quantitative proteome of GCF from periodontally healthy individuals versus those with periodontal disease by large-scale MS technology remains at its infancy with minimal application of “gold standard analytical methods”. There are multiple reasons for the limited application and effective analysis of GCF proteome on a large-scale ranging from inherent small volume availability, effective elution of total protein from collection strips/glass for accurate quantitation, dynamic protein range problems which become substantially accentuated during inflammation and periodontal disease. These issues are of major importance when quantitative approaches are used to define what changes in healthy versus periodontal disease because of the shift in serum contribution from ~30% in health to ~70% in periodontal disease. This reflects increased level of abundant proteins such as albumin, immunogrobulins and other serum related proteins in GCF of periodontal samples which would automatically change the number and types of proteins identifiable in healthy versus periodontal samples. This leads to lack of quantitation of many proteins due to identified proteins being either found only in healthy or periodontal sample and not in both, hence, no relative comparison can be made. This latter case was clearly exemplified by the study of Bostanci et al., (2010), where only ~40 distinct proteins were found in both healthy and periodontal samples, albeit in that study there was additional limitation because of the low total protein level due to analysis of individual samples without pooling. Similarly, although the study by Ballian et al., (2012) was only a qualitative one, nevertheless, there were only 94 proteins identified in both healthy and periodontal samples based on acceptable criteria for MS-based proteomics where two or more peptides must be identified for a given protein.
In the present work we have utilized multiple strategies to clearly overcome many of the existing limitations that could be encountered in attempts to establish large-scale relative quantitative proteome analysis of GCF from healthy periodontium versus periodontal sites using MS technology. It is noteworthy that use of two distinct stable-isotope labelling chemistries revealed that the universal amine-labelling reagent mTRAQ which enables identification and quantification of every peptide showed major advantages over ICAT. Although the capacity of ICAT to affinity enrich cysteine containing peptides may have been expected to provide identification and quantification of lower abundant peptides, our results showed that this was not the case at least with GCF samples. The limitation of ICAT to only label cysteine containing peptides combined with the presence of abundant proteins in GCF such as albumin and immunoglobulins containing many cysteine residues appear to be one of the major factors that led to identification and quantification of a very small number of proteins. This is clearly exemplified in Table 2 where such proteins dominate the quantified proteins by ICAT. The second issue related to the limitation of ICAT relates to many proteins lacking cysteine residues and those proteins which do contain such residues, there are often only potential one or two peptides with cysteine residue. This latter case leads to not only less number of proteins being identified and quantified but also to the differences that can be observed in the quantitative levels of some of the proteins defined by mTRAQ versus ICAT such as with Ig kappa chain C, myeloperoxidase, protein S100-A8 and serotransferrin. The differences in absolute numeric values of the ratios defined between the two chemistries while have no consequence if there is no true biochemical difference between healthy and disease samples and both chemistries show the level of change being statistically not significant, e.g., serotransferrin and Ig kappa C. However, in other situations there can be clear deficiencies reflected by ICAT whereby a given protein is truly different in two samples and mTRAQ shows statistically significant elevated levels but not ICAT, e.g., with proteins haptoglobulin, S100A8 and myeloperoxidase. Indeed, myeloperoxidase is an excellent example where ICAT falls short of defining its elevation which has been shown to increase in GCF from patients with periodontal disease and gingivitis by both enzyme activity and antibody ELISA (Marcaccini et al. 2010; Leppilahti et al. 2014). On the other hand ICAT in most cases showed consistency with the mTRAQ in quantitation of proteins such as albumin, a number of immunoglobulins and neutrphil defensin 1. While there is continual interest in performing label-free quantitative proteomics, the complexity of proteomic samples poses a special challenge for the label-free MS quantitation methods (Andreev et al. 2007). There are clearly also drawbacks of “label-based” proteomics such as use of expensive stable isotope reagents as well as involving several time consuming steps in labeling and clean up with potential for loss of sample amount which ultimately can limit the number of proteins that can be identified and quantified. Similarly, random sample pooling can have its own negative impact in terms of potential for not all of the available samples being used and repeat use of the same sample that may bias the results in a negative or positive manner. In such cases use of more than one pool sampling approach as we have utilized help to remove or reduce such occurrences.
Interestingly, within the context of the identified proteins in GCF of healthy and diseased periodontal sites further evaluation of the data revealed important occurrences. Figure 5A shows Venn diagram of a comparison between qualitative healthy site proteome, (Carneiro et al. 2012), and the current quantitative study with only a small overlap suggesting significant alterations in the proteome composition during periodontal disease. This is further supported by the comparison of the distribution of functional categories of the GCF proteins highlight a shift from almost equal distribution (~11–13% of each of the categories) for proteins that may be derived from serum to those distinct major categories for proteins specific to GCF microenvironment or increased number of protein in a specific category, Fig. 5B–C. It is possible, however, that our analysis in both sets of data, viz., present study and Carneiro et al. (2012), which were performed under the same LC-MS/MS conditions through evaluation of top 5 peptides per scan can affect the depth of proteins discovered and hence, the degree of overlap observed. Apart from expected serum derived proteins, the data revealed the presence of sets of macromolecules more specific to local GCF microenvironment that have the potential to act as biomarkers during disease progression and not identified in plasma proteome by MS technology. These ranged from the early inflammation, immune response, innate immune response proteins, specific bactericidal proteins-defense response to bacteria, proteins that regulate production of cytokines such as IL-6 & 8 and regulate macrophage activity and TNF production, cellular and extracellular matrix components, a variety of enzymes including proteinases (cathepsin G, leuckocyte elastase, leukitriene A4 and amidase), and an array of proteinase inhibitors. Of the 180 proteins present both in GCF of healthy and periodontal patients which were quantified, ~50 were found to be elevated in periodontal patients with statistical significance. In comparison, total number of bacterial proteins identified and quantified was substantially smaller, 42 proteins, and 16 of these were elevated in the GCF of periodontal patients with statistical significance. These results reflect clear low abundance of the bacterial proteins, at least an order of magnitude less than the host protein concentrations in GCF, as indicated by the limited number of runs and peptides they were identified by. Despite this however, the significance of bacterial proteins can be appreciated as many are derived from pathologic bacteria. Of particular note is the 7 proteins derived from the classic pathogen that plays a major role during the induction and progression of periodontal disease, Porphyromonas gingivalis. Of those, 3 were elevated by 2–3 fold with statistical significance namely oxidoreductase, Rel/SpoT protein and outer membrane protein 85 (OMP85). Importantly, OMP85 proteins are highly conserved outer membrane proteins in all Gram −ve bacteria. While only a few “glycoproteins are known for Gram −ve bacteria, most glycoproteins found in bacteria are localized on bacterial surface and are involved in host-bacteria interactions, immunogenic response and play critical role in virulence. Hence, the present findings of the virulence factor OMP85 specific to Porphyromonas gingivalis in combination with several other host and bacterial derived elevated proteins in periodontal disease contribute important new sets of data with clinical relevance. The yeast data appears to be even more limited with 11 proteins quantified only and of these only three proteins elevated levels with statistical significance. These were Histone H3 protein from Tristoma integrum, actin from Collectotrichum truncotum, and translational initiating factor eIF2B from Saccharomyces cerevisiae, whose relevance and impact on periodontal disease not easy to decipher.
Figure 5. Venn diagram summarizing the quantitative studies for GCF with ICAT and mTRAQ and pie charts representing comparative distributions of functional categories between qualitative, quantitative and plasma MS data.
(A), Venn diagram showing a total of 238 proteins identified in the GCF of healthy versus periodontal sites by stable-isotope labeling multi-dimensional quantitative MS analysis relative to those 199 identified by qualitative MS analysis of GCF from healthy individuals only. The number of proteins found only in quantitative or qualitative experimental approach (non-overlapped regions) and those that are common (overlapped regions) are highlighted. (B), Pie chart of the molecular and functional categories of the GCF proteome from quantitative MS analysis using stable-isotope labeling; (C) Pie chart of the molecular and functional categories of the GCF proteome from qualitative MS analysis; (D) Pie chart of the molecular and functional categories of the plasma proteome by qualitative MS analysis (Schenk et al., 2008).
One area of MS-based proteome analysis of GCF has been the critical lack of identifying any collagens in all of the studies carried out to date. This surprising deficiency is of major significance since collagen as a major and dominant ECM component with a number of different types present in specific microenvironments such as collagen type IV in basement membrane and types I & III in bone. In periodontal disease both the soft tissue gingiva and bone degradation are inherent components of the developing pathophysiology. Hence, the identification and quantification of collagens are intimately related to disease state whereby any increased tissue degradation would be reflected by increased collagen fragmentation. We have for the first time in the present study identified a series of collagen types by knowledge-based approach to MS data search using mass addition of 16 Da for hydroxyproline residues in collagens. The lack of identification of collagens in previous studies is due to presence of proline residues almost every third amino-acid and frequently these are hydroxylated within the collagen sequences which required use of the search algorithm with inclusion of hydroxyproline modification (16 Da mass addition for –OH on prolines, Zhou et al., 2010; Liu et al., 2013).
The notion to use GCF as a source of diagnostic biomarker for periodontal disease is of course not new, however, the ability to identify a large number of GCF proteins simultaneously and highlighting numerous new components is clearly new. For several decades many potential periodontal disease biomarkers have been screened but all of these have been based on targeting a single analyte at a given time (Loos & Tjoa, 2005). However, in more recent times there have been paradigm shift towards possibility of using a panel of independent disease-related proteins that may serve better and reflect more accurately the disease correlation. In this respect the ability to highlight a large number of proteins with local tissue/cell specificity and defining their relative levels in health versus disease become major interests. Amongst a large number of proteins identified and quantified by MS analysis of GCF, this study targeted two distinct proteins, namely serum albumin and S100A9 protein, for validation by ELISA. The validation interest of albumin in our study was to serve as an internal control to evaluate accuracy of the experimental and technical methods with added confidence to the MS-based results. The protein S100A9 was chosen for reasons that its levels were significantly increased in periodontal disease and it is produced specifically by neutrophils and macrophages which are immune response cells. Importantly, macrophages are the precursors for osteoclasts which are responsible for the bone resorption, a process that ultimately defines advanced periodontitis. We have clearly defined the elevated levels of S100A9 protein in the GCF of patients with periodontal disease by MS analysis of multiple pooled samples of 40 patients and by ELISA using same pooled samples as well as individually all of the 40 healthy and 40 periodontal GCF samples. The results summarized in Figure 4 highlight importance of the data analysis within which we also identified that the difference remain irrespective of whether normalization was based upon per volume of GCF or per total protein amount or purely per periopaper. These are desired characteristics and of major interest for any potential “chair-side” biomarker. Protein S100A9 has been identified in other studies but none of those studies actually analyzed the possibility of this protein being a periodontal disease biomarker, whereas studies have suggested correlation of protein S100A8 to periodontal disease.
The precise roles of protein S100A9 are still not clear and need to be further investigated however, our findings by MS analysis and validation by ELISA suggests that S100A9 protein plays an important role and could be a potential predictive/diagnostic biomarker for periodontal disease. Furthermore, its origin as an inflammatory response product produced by neutrophils and macrophages, of which the latter differentiate to bone resorbing osteoclasts, all culminate to signify its capacity to act as a predictive biomarker for the onset and progression of periodontal disease. Support for S100A9 protein as a predictive/diagnostic biomarker for inflammatory disease in general has been demonstrated in other inflammatory associated disorders. The elevated levels of this protein have been used to suggest as candidate for serological biomarker for colorectal cancer (CRC) in combination with other serum markers for CRC diagnosis (Kim et al. 2009). S100A9 protein was also suggested as a biomarker for acute appendicitis and indicated diagnostic potential in conjunction with other established approaches (Bealer and Colgin 2010). In other cases S100A9 in conjunction with S100A8 have been used not as a biomarker to diagnose a disease but rather as a marker for monitoring the progression or regression of irritable bowel disease, S100-A8/S-100A9 (calprotectin) (Fagerhol 2000; Carroccio et al., 2003; Loos and Tjoa 2005). Despite these studies, to date there has been no direct study reported providing experimental evidence for the increase of S100A9 level in clinical samples of GCF from healthy versus periodontal disease sites. However, on the bases of qualitative comparison of serum and GCF from healthy and periodontal patients using 2D-PAGE, Kojima et al., (2000) identified low Mr bands related to S100-A8/S-100A9 that were absent in serum and suggested that these may have an important implications in periodontal disease. The first immune cells to reach infection sites are the neutrophils and they constitute the first line of defense against microbial invaders. In order to reach the inflammatory site, neutrophils migrate across endothelial cells and surrounding tissues. For the migration, the secretion of tissue-degrading enzymes such as gelatinase and elastase is necessary. Protein S100A9 and not S100A8 or S100A12 induces neutrophil degranulation and the release of secretory vesicles (Simard et al., 2010). These data in combination with up regulation of S100A9 by Porphyromonas gingivalis, (Inaba et al., 2009), which is a bacteria associated with periodontal disease, all point towards the significance of S100A9 protein both at biologic level and as potential biomarker. In connection with the inflammation and immune response the identified Ig alpha-2 chain C (IgA2) in the present study only in the GCF of periodontal patients is consistent with its major role as a locally secreted first line of defense against pathogenic bacteria in preventing access of foreign antigens to the general systemic immune system. Similarly, current study also defined elevated levels of neutrophil defensin 1 and lactotransferrin both of which have been implicated as markers of chronic periodontal disease (Puklo et al. 2008, Glimvall et al. 2012).
Conclusions
This study developed and utilized technical methodologies to maximize the number of proteins that can be identified and quantified using gold-standard stable-isotope labeling chemistries in the study of GCF samples from healthy periodontium sites relative to those from patients with periodontal disease for large-scale quantitative LC-ESI-MS/MS-based analysis for biomarker discovery. We have also introduced and used technical and practical innovations in order to overcome many of the problems associated in the studies of GCF by large-scale MS-based technology to provide gold-standards and highlight shortfalls and acceptable criteria when such analysis are carried out/reported. Within this study we have concluded that the universal stable-isotope labeling reagent mTRAQ has major advantages over ICAT in such studies involving large-scale quantitative proteomics using GCF samples. The data highlighted enumerate number of proteins that have not been known to exist in GCF and the MS-data were validated by classical ELISA of selected proteins, e.g., S100A9 and serum albumin. The use of serum albumin which is classically well known to increase in GCF of periodontal patients provided additional confidence with respect to the accuracy and precision of the MS results. Furthermore, several proteins that have been defined in the present study to be elevated in GCF of periodontal patients such as IgA2, Lactotransferrin, neutrophil defensing 1, myeloperoxidase, and S100 A8 protein have been also highlighted previously using a single analyte targeted approaches by activity or ELISA as important markers for periodontal disease. While these are testament to the robustness and power of our large-scale MS approaches, they represent only a very small portion of our defined list of elevated proteins with statistical significance in periodontal disease. Hence, 50 host proteins and 16 bacterial proteins that have been elevated in GCF of periodontal patients including specific virulence factor OMP85 of Porphyromonas gingivalis, altogether provide a wealth of new information and large number of novel potential candidates that have never been probed yet. Those combined with the identification of 187 proteins that are specific to GCF microenvironment, are deemed to provide major opportunities to utilize the data towards clinical application for evaluating risk factors, predictive biomarkers and monitoring treatment effectiveness of periodontal disease as well as add to our understanding of the interplay between oral disease and systemic physiology.
Supplementary Material
CLINICAL RELEVANCE.
Scientific rationale for the study
Develop and utilize quantitative proteome analysis by multidimensional protein separation and MS technology of GCF from healthy and periodontal patients using “gold standard” stable-isotope labeling chemistries to establish with high accuracy and confidence potential predictive/diagnostic biomarkers.
Principal findings
Identification of GCF microenvironment specific novel groups of proteins derived from host and pathogenic bacteria whose levels are altered in periodontal disease which have not being defined previously. Highlight potential chair-side utility of several host proteins including S100A9 protein, identification/quantification of collagens for the first time in GCF proteomics and pathogenic bacterial proteins including virulence factor OMP 85 from Porphyromonas gingivalis.
Practical implications
The results culminated in quantification of unique and disease related host and pathogenic bacterial proteins whose alterations in GCF of periodontal disease patients can be utilized as biomarkers towards chair-side application in clinical dentistry/chemistry.
Acknowledgments
Source of funding: This work was partially supported by a grant from NIDCR; DE 018448 (E.S.) and personal/private funds of Dr. Salih.
The authors thank Dr. Hatice Hasturk for her help and guidance during the collection of GCF from patients at Henry M. Goldman School of Dental Medicine, Boston University.
Abbreviations used
- GCF
gingival crevicular fluid, ICAT, isotope-coded-affinity-tag
- mTRAQ
amine-specific tag for relative and absolute quantitation
- OMP85
outer membrane protein 85
- WS
whole saliva
- ECM
extracellular matrix
- MS
mass spectrometry
- CID
collision-induced dissociation
- DTT
dithiothreitol
- LC-ESI-MS/MS
liquid-chromatography electrospray-ionization-tandem-mass spectrometry
- NH4HCO3
ammonium bicarbonate
- ELISA
enzyme-linked immunosorbent assay
- CH3CN
acetonitrile
Footnotes
Conflict of interest :The authors declare that there is no conflict of interest in this study.
This work was presented in part at the American Association for Dental Research (AADR) 39th Meeting and Exhibition, 2010, Washington DC, USA, Abstract 992.
References
- Alfano MC. The origin of gingival fluid. J Theor Biol. 1974;47:127–136. doi: 10.1016/0022-5193(74)90103-9. [DOI] [PubMed] [Google Scholar]
- Anderson L. Candidate-based proteomics in the search for biomarkers of cardiovascular disease. J Physiol. 2005;563:23–60. doi: 10.1113/jphysiol.2004.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev VP, Li L, Cao L, Gu Y, Rejtar T, Wu SL, Karger BL. A new algorithm using cross-assignment for label-free quantitation with LC-LTQ-FT MS. J Proteome Res. 2007;6(6):2186–2194. doi: 10.1021/pr0606880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage GC. Analysis of gingival crevice fluid and risk of progression of periodontitis. Periodontol 2000. 2004;34:109–119. doi: 10.1046/j.0906-6713.2002.003427.x. [DOI] [PubMed] [Google Scholar]
- Baliban RC, Sakellari D, Li Z, DiMaggio PA, Garcia BA, Floudas CA. Novel protein identification methods for biomarker discovery via a proteomic analysis of periodontally healthy and diseased gingival crevicular fluid samples. J Clin Periodontol. 2012;39:203–212. doi: 10.1111/j.1600-051X.2011.01805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bealer JF, Colgin M. S100A8/A9: a potential new diagnostic aid for acute appendicitis. Academic emergency medicine : official journal of the Society for Academic Emergency Medicine. 2010;17(3):333–336. doi: 10.1111/j.1553-2712.2010.00663.x. [DOI] [PubMed] [Google Scholar]
- Bickel M, Cimasoni G, Andersen E. Flow and albumin content of early (pre-inflammatory) gingival crevicular fluid from human subjects. Archives of Oral Biology. 1985;30:599–602. doi: 10.1016/0003-9969(85)90079-2. [DOI] [PubMed] [Google Scholar]
- Bostanci N, Heywood W, Mills K, Parkar M, Nibali L, Donos N. Application of Label-Free Absolute Quantitative Proteomics in Human Gingival Crevicular Fluid by LC/MS(E) (Gingival Exudatome) J Proteome Res. 2010;9:2191–2199. doi: 10.1021/pr900941z. [DOI] [PubMed] [Google Scholar]
- Bostanci N, Ramberg P, Wahlander A, Grossman J, Jonsson D, Barnes VM, Papapanou PN. Label-free quantitative proteomics revealed differentially regulated proteins in experimental gingivatis. J Proteome Res. 2013;12:657–678. doi: 10.1021/pr300761e. [DOI] [PubMed] [Google Scholar]
- Burtis CA, Ashwood MD. Tietz Textbook of Clinical Chemistry. 3. Saunders; 1999. [Google Scholar]
- Candiano G, Santucci L, Petretto A, Bruschi M, Dimuccio V, Urbani A, Bagnasco S, Ghiggeri GM. 2D-electrophoresis and the urine proteome map: where do we stand? J Proteomics. 2010;73:829–844. doi: 10.1016/j.jprot.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Carneiro LG, Venuleo C, Oppenheim FG, Salih E. Proteome data set of human gingival crevicular fluid from healthy periodontium sites by multidimensional protein separation and mass spectrometry. J Periodont Res. 2012;47:248–262. doi: 10.1111/j.1600-0765.2011.01429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroccio A, Iacono G, Cottone M, Di Prima L, Cartabellotta F, Cavataio F, Scalici C, Montalto G, Di Fede G, Rini G, Notarbartolo A, Averna MR. Diagnostic accuracy of fecal calprotectin assay in distinguishing organic causes of chronic diarrhea from irritable bowel syndrome: a prospective study in adults and children. Clin Chem. 2003;49 (6 Pt 1):861–867. doi: 10.1373/49.6.861. [DOI] [PubMed] [Google Scholar]
- Champagne CM, Buchanan W, Reddy MS, Preisser JS, Beck JD, Offenbacher S. Potential for gingival crevice fluid measures as predictors of risk for periodontal diseases. Periodontol 2000. 2003;31:167–180. doi: 10.1034/j.1600-0757.2003.03110.x. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Heo SH, Lee JM, Cho JY. Identification of azurocidin as a potential periodontitis biomarker by a proteomic analysis of gingival crevicular fluid. Proteome Sci. 2011;9 doi: 10.1186/1477-5956-9-42. 477-5956-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MA, Gillett IR, Griffiths GS, Maiden MFJ, Sterne JAC, Wilson DT, Wilton JMA, Johnson NW. Detection of high-risk groups and individuals for periodontal-diseases - laboratory markers from analysis of gingival crevicular fluid. Journal of Clinical Periodontology. 1989;16:1–11. doi: 10.1111/j.1600-051x.1989.tb01604.x. [DOI] [PubMed] [Google Scholar]
- Czernick D, Liu J, Dibart S, Salih E. Topographical Distribution of Phosphorylation Sites of Phosvitins by Mass Spectrometry: Discovery of a novel phosvitin expressed by vitellogenin I gene. J Proteomics. 2013;83:76–98. doi: 10.1016/j.jprot.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, Bassilian S, Bedi GS, Boontheung P, Cociorva D, Delahunty CM, Denny T, Dunsmore J, Faull KF, Gilligan J, Gonzalez-Begne M, Halgand F, Hall SC, Han X, Henson B, Hewel J, Hu S, Jeffrey S, Jiang J, Loo JA, Ogorzalek Loo RR, Malamud D, Melvin JE, Miroshnychenko O, Navazesh M, Niles R, Park SK, Prakobphol A, Ramachandran P, Richert M, Robinson S, Sondej M, Souda P, Sullivan MA, Takashima J, Than S, Wang J, Whitelegge JP, Witkowska HE, Wolinsky L, Xie Y, Xu T, Yu W, Ytterberg J, Wong DT, Yates JR, 3rd, Fisher SJ. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J Proteome Res. 2008;7:1994–2006. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delima AJ, Van Dyke TE. Origin and function of the cellular components in gingival crevice fluid. Periodontolgy 2000. 2003;31:55–76. doi: 10.1034/j.1600-0757.2003.03105.x. [DOI] [PubMed] [Google Scholar]
- Eng JK, Mccormack AL, Yates JR. An Approach to Correlate Tandem Mass-Spectral Data of Peptides with Amino-Acid-Sequences in a Protein Database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Fagerhol MK. Calprotectin, a faecal marker of organic gastrointestinal abnormality. Lancet. 2000;356(9244):1783–1784. doi: 10.1016/S0140-6736(00)03224-4. [DOI] [PubMed] [Google Scholar]
- Fitzsimmons TR, Sanders AE, Bartold PM, Slade GD. Local and systemic biomarkers in gingival crevicular fluid increase odds of periodontitis. Journal of Clinical Periodontology. 2010;37:30–36. doi: 10.1111/j.1600-051X.2009.01506.x. [DOI] [PubMed] [Google Scholar]
- Hardt M, Thomas LR, Dixon SE, Newport G, Agabian N, Prakobphol A, Hall SC, Witkowska HE, Fisher SJ. Toward defining the human parotid gland salivary proteome and peptidome: identification and characterization using 2D SDS-PAGE, ultrafiltration, HPLC, and mass spectrometry. Biochemistry. 2005;44:2885–2899. doi: 10.1021/bi048176r. [DOI] [PubMed] [Google Scholar]
- Hattingh J, Ho E. The concentration of proteins in human gingival crevicular fluid. J Periodontal Res. 1980;15:90–95. doi: 10.1111/j.1600-0765.1980.tb00262.x. [DOI] [PubMed] [Google Scholar]
- Hu S, Xie Y, Ramachandran P, Ogorzalek Loo RR, Li Y, Loo JA, Wong DT. Large-scale identification of proteins in human salivary proteome by liquid chromatography/mass spectrometry and two-dimensional gel electrophoresis-mass spectrometry. Proteomics. 2005;5:1714–1728. doi: 10.1002/pmic.200401037. [DOI] [PubMed] [Google Scholar]
- Glimvall P, Wickstrom C, Jansson H. Elevated levels of salivary lactoferrin, a marker for chronic periodontitis? J Periodont Res. 2012;47:655–660. doi: 10.1111/j.1600-0765.2012.01479.x. [DOI] [PubMed] [Google Scholar]
- Grant MM, Creese AJ, Barr G, Ling MR, Scott AE, Matthews JB, Griffiths HR, Cooper HJ, Chapple IL. Proteomic analysis of a noninvasive human model of acute inflammation and its resolution: the twenty-one day gingivitis model. J Proteome Res. 2010;9:4732–4744. doi: 10.1021/pr100446f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba H, Hokamura K, Nakano K, Nomura R, Katayama K, Nakajima A, Yoshioka H, Taniguchi K, Kamisaki Y, Ooshima T, Umemura K, Murad F, Wada K, Amano A. Upregulation of S100 calcium-binding protein A9 is required for induction of smooth muscle cell proliferation by a periodontal pathogen. FEBS Lett. 2009;583(1):128–134. doi: 10.1016/j.febslet.2008.11.036. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kang HJ, Lee H, Lee ST, Yu MH, Kim H, Lee C. Identification of S100A8 and S100A9 as serological markers for colorectal cancer. Journal of proteome research. 2009;8(3):1368–1379. doi: 10.1021/pr8007573. [DOI] [PubMed] [Google Scholar]
- Kojima T, Andersen E, Sanchez JC, Wilkins MR, Hochstrasser DF, Pralong WF, Cimasoni G. Human gingival crevicular fluid contains MRP8 (S100A8) and MRP14 (S100A9), two calcium-binding proteins of the S100 family. J Dent Res. 2000;79:740–747. doi: 10.1177/00220345000790020701. [DOI] [PubMed] [Google Scholar]
- Lamster IB, Ahlo JK. Analysis of gingival crevicular fluid as applied to the diagnosis of oral and systemic diseases. Ann N Y Acad Sci. 2007;1098:216–229. doi: 10.1196/annals.1384.027. [DOI] [PubMed] [Google Scholar]
- Leppilahti JM, Hernández-Ríos PA, Gamonal JA, Tervahartiala T, Brignardello-Petersen R, Mantyla P, Sorsa T, Hernández M. Matrix metalloproteinases and myeloperoxidase in gingival crevicular fluid provide site-specific diagnostic value for chronic periodontitis. J Clin Periodontol. 2014;41:348–356. doi: 10.1111/jcpe.12223. [DOI] [PubMed] [Google Scholar]
- Li QR, Fan KX, Li RX, Dai J, Wu CC, Zhao SL, Wu JR, Shieh CH, Zeng R. A comprehensive and non-prefractionation on the protein level approach for the human urinary proteome: touching phosphorylation in urine. Rapid communications in mass spectrometry : RCM. 2010;24:823–832. doi: 10.1002/rcm.4441. [DOI] [PubMed] [Google Scholar]
- Liu J, Czernick D, Lin S-C, Alasmani A, Dibart S, Salih E. Novel Bioactivity of Phosvitin in Connective Tissue and Bone Organogenesis Revealed by Live Calvarial Bone Organ Culture Models. Dev Biol. 2013;381:256–275. doi: 10.1016/j.ydbio.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Loos BG, Tjoa S. Host-derived diagnostic markers for periodontitis: do they exist in gingival crevice fluid? Periodontol 2000. 2005;39:53–72. doi: 10.1111/j.1600-0757.2005.00129.x. [DOI] [PubMed] [Google Scholar]
- Mantyla P, Stenman M, Kinane D, Salo T, Suomalainen K, Tikanoja S, Sorsa T. Monitoring periodontal disease status in smokers and nonsmokers using a gingival crevicular fluid matrix metalloproteinase-8-specific chair-side test. J Periodontal Res. 2006;41:503–512. doi: 10.1111/j.1600-0765.2006.00897.x. [DOI] [PubMed] [Google Scholar]
- Marcaccini AM, Meschiari CA, Zuardi LR, de Sousa TS, Taba M, Jr, Teofilo JM, Jacob-Ferreira AL, Tanus-Santos JE, Novaes AB, Jr, Gerlach RF. Gingival crevicular fluid levels of MMP-8, MMP-9, TIMP-2, and MPO decrease after periodontal therapy. J Clin Periodontol. 2010;37:180–190. doi: 10.1111/j.1600-051X.2009.01512.x. [DOI] [PubMed] [Google Scholar]
- McPherson RA, Pincus MR. Henry’s Clinical Diagnosis and Management by Laboratory Methods: Expert Consult - Online and Print. 22. Saunders; 2011. [Google Scholar]
- Ngo LH, Veith PD, Chen YY, Chen D, Darby IB, Reynolds EC. Mass spectrometric analyses of peptides and proteins in human gingival crevicular fluid. J Proteome Res. 2010;9:1683–1693. doi: 10.1021/pr900775s. [DOI] [PubMed] [Google Scholar]
- Offenbacher S, Barros S, Mendoza L, Mauriello S, Preisser J, Moss K, de Jager M, Aspiras M. Changes in gingival crevicular fluid inflammatory mediator levels during the induction and resolution of experimental gingivitis in humans. Journal of Clinical Periodontology. 2010;37:324–333. doi: 10.1111/j.1600-051X.2010.01543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashley DH. A mechanistic analysis of gingival fluid production. J Periodontal Res. 1976;11:121–134. doi: 10.1111/j.1600-0765.1976.tb00060.x. [DOI] [PubMed] [Google Scholar]
- Peng J, Elias, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- Puklo M, Guentsch A, Hiemstra PS, Eick S, Potempa J. Analysis of neutrophil-derived antimicrobial peptides in gingival crevicular fluid suggests importance of cathelicidin LL-37 in the innate immune response against periodontogenic bacteria. Oral Microbiol Immunol. 2008;23:328–335. doi: 10.1111/j.1399-302X.2008.00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Molecular and Cellular Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Salih E, Siqueira WL, Helmerhorst EJ, Oppenheim FG. Large-scale phosphoproteome of human whole saliva using disulfide-thiol interchange covalent chromatography and mass spectrometry. Anal Biochem. 2010;407:19–33. doi: 10.1016/j.ab.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk S, Schoenhals GJ, de Souza G, Mann M. A high confidence, manually validated human blood plasma protein reference set. BMC medical genomics. 2008;1:41. doi: 10.1186/1755-8794-1-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard JC, Girard D, Tessier PA. Induction of neutrophil degranulation by S100A9 via a MAPK-dependent mechanism. J Leukoc Biol. 2010;87(5):905–914. doi: 10.1189/jlb.1009676. [DOI] [PubMed] [Google Scholar]
- Siqueira WL, Salih E, Wan DL, Helmerhorst EJ, Oppenheim FG. Proteome of human minor salivary gland secretion. J Dent Res. 2008;87:445–450. doi: 10.1177/154405910808700508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira WL, Zhang, Helmerhorst EJ, Gygi SP, Oppenheim FG. Identification of protein components in in vivo human acquired enamel pellicle using LC-ESI-MS/MS. J Proteome Res. 2007;6:2152–2160. doi: 10.1021/pr060580k. [DOI] [PubMed] [Google Scholar]
- Stone MD, Chen X, McGowan T, Bandhakavi S, Cheng B, Rhodus NL, Griffin TJ. Large-scale phosphoproteomics analysis of whole saliva reveals a distinct phosphorylation pattern. J Proteome Res. 2011;10:1728–1736. doi: 10.1021/pr1010247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles RP, Gursky LC, Faveri M, Rosa EA, Teles FRF, Feres M, Socransky SS, Haffajee AD. Relationships between subgingival microbiota and GCF biomarkers in generalized aggressive periodontitis. Journal of Clinical Periodontology. 2010;37:313–323. doi: 10.1111/j.1600-051X.2010.01534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida S, Satoh M, Umemura H, Sogawa K, Kawashima Y, Kado S, Sawai S, Nishimura M, Kodera Y, Matsushita K, Nomura F. Proteomic analysis of gingival crevicular fluid for discovery of novel periodontal disease markers. Proteomics. 2012;12:2190–2202. doi: 10.1002/pmic.201100655. [DOI] [PubMed] [Google Scholar]
- Wong DT. Salivary diagnostics powered by nanotechnologies, proteomics and genomics. J Am Dent Assoc. 2006;137:313–321. doi: 10.14219/jada.archive.2006.0180. [DOI] [PubMed] [Google Scholar]
- Xie H, Rhodus NL, Griffin RJ, Carlis JV, Griffin TJ. A catalogue of human saliva proteins identified by free flow electrophoresis-based peptide separation and tandem mass spectrometry. Mol Cell Proteomics. 2005;4:1826–1830. doi: 10.1074/mcp.D500008-MCP200. [DOI] [PubMed] [Google Scholar]
- Yan W, Apweiler R, Balgley BM, Boontheung P, Bundy JL, Cargile BJ, Cole S, Fang X, Gonzalez-Begne M, Griffin TJ, Hagen F, Hu S, Wolinsky LE, Lee CS, Malamud D, Melvin JE, Menon R, Mueller M, Qiao R, Rhodus NL, Sevinsky JR, States D, Stephenson JL, Than S, Yates JR, Yu W, Xie H, Xie Y, Omenn GS, Loo JA, Wong DT. Systematic comparison of the human saliva and plasma proteomes. Proteomics Clin Appl. 2009;3:116–134. doi: 10.1002/prca.200800140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H-Y, Salih E, Glimcher MJ. Isolation and characterization of glycosylated phosphoproteins from herring bones. J Biol Chem. 2010;285:36170–36178. doi: 10.1074/jbc.M110.146910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.