Abstract
Ricin is a Select Agent Toxin and a member of the RNA N-glycosidase family of medically important plant and bacterial ribosome-inactivating proteins (RIPs). In this study, we determined x-ray crystal structures of the enzymatic subunit of ricin (RTA) in complex with the antigen binding domains (VHH) of five unique single-chain monoclonal antibodies that differ in their respective toxin-neutralizing activities. None of the VHHs made direct contact with residues involved in RTA’s RNA N-glycosidase activity or induced notable allosteric changes in the toxin’s subunit. Rather, the five VHHs had overlapping structural epitopes on the surface of the toxin and differed in the degree to which they made contact with prominent structural elements in two folding domains of the RTA. In general, RTA interactions were influenced most by the VHH CDR3 elements, with the most potent neutralizing antibody having the shortest and most conformationally constrained CDR3. These structures provide unique insights into the mechanisms underlying toxin neutralization and provide critically important information required for the rational design of ricin toxin subunit vaccines.
INTRODUCTION
Ricin is a member of the type II ribosome-inactivating protein (RIP) family that includes abrin and Shiga toxins1. It is also classified as a Select Toxin due to its potential use as a biothreat agent2; 3. Ricin’s catalytic A subunit (RTA) is an RNA N-glycosidase whose sole target is the highly conserved adenosine residue (A2662) within the sarcin-ricin loop (SRL) of eukaryotic 28S ribosomal RNA4; 5. Ricin’s binding subunit (RTB) is a lectin specific for galactose- and N-acetylgalactosamine (Gal/GalNAc)6. RTB promotes ricin attachment and entry into mammalian host cells. Following uptake, RTB facilitates receptor-mediated endocytosis and retrograde trafficking of ricin from early endosomes to the trans-Golgi network and endoplasmic reticulum. Within the lumen of the ER, RTA is liberated from RTB and retrotranslocated into the host cell cytosol where is gains access to its ribosomal RNA substrate7. Cleavage of the SRL results in protein synthesis arrest and concomitant activation of the ribotoxic-stress response8.
Structurally, RTA consists of three distinct folding domains (Figure S1)9; 10. Folding domain I (residues 1–117) is dominated by a six-stranded β sheet that terminates in a solvent exposed loop-helix-loop11. The solvent exposed α-helix, known as α-helix B (residues N97-F108) is conserved among RIPs and is a target of three well-characterized toxin-neutralizing monoclonal antibodies (mAbs), R70, PB10 and 6C211; 12; 13; 14; 15. Folding domain II (residues 118–210) is dominated by five α-helices (C-G) that run through the center of RTA, while folding domain III (residues 211–267) forms a protruding element that slides into the cleft between RTB’s two domains9. RTA’s active site constitutes a shallow pocket in the central portion of the protein. Five residues (i.e., Tyr80, Tyr123, Glu177, Arg180, and Trp211) situated within or near the active site cleft are essential for RTA’s enzymatic activity10; 16.
RTA is the focus of ongoing efforts to develop a recombinant ricin toxin subunit vaccine for use by military personnel and at-risk civilians3; 17. However, improving the safety, solubility and immunogenicity of RTA through protein engineering is being done in the absence of a complete understanding of the linear and conformational B cell epitopes that are important in eliciting toxin-neutralizing antibodies18; 19. Thus, at the present time, it is not possible to predict whether a particular mutation, deletion or truncation will adversely affect the potency of RTA as a vaccine antigen. In an effort to generate a more comprehensive B cell epitope map of RTA, a number of groups, including ours, have characterized and localized the epitopes recognized by more than a dozen RTA-specific, toxin-neutralizing and non-neutralizing mouse mAbs20. Consequently, a partial, albeit largely low resolution, B cell epitope map is now available13.
In order to address the lack of detailed molecular information concerning RTA epitopes, we recently produced a collection of toxin-neutralizing and non-neutralizing ricin-specific, monomeric single-chain VH domain (VHH) antibodies from two alpacas that were immunized with ricin toxin subunit antigens and ricin toxoid21. This collection included 11 unique RTA-specific VHHs that ranged in their affinities and in their toxinneutralizing activities (i.e, strong, moderate, weak or no activity). Pepscan analysis indicated that all 11 RTA-specific VHHs bound non-linear epitopes (D. Vance and N. Mantis, unpublished results). As VHHs are relatively small (~15 kDa) and highly stable22, they afforded us an excellent opportunity to generate co-crystals with ricin’s enzymatic subunit, as a means to generate a high-resolution B cell epitope map of RTA and to better understand the mechanisms by which antibodies neutralize (or fail to neutralize) ricin. In this study, we have determined X-ray crystal structures of RTA in complex with five different VHHs that have overlapping structural epitopes on the surface of ricin but differ in the degree to which they engage with three of RTA’s prominent secondary structural elements. These five structures highlight specific molecular features that promote ricin-toxin neutralization while underscoring critical differences between ricin-neutralizing and non-neutralizing antibodies. This knowledge will contribute to a more rational design of ricin toxin vaccines.
RESULTS
Among the 11 unique RTA-specific VHHs we recently characterized21, the complexes for five of them with recombinant RTA produced X-ray crystal structures (Table 1). The five VHHs fall into four categories with respect to their IC50 values as previously determined in a Vero cell cytotoxicity assay: strong (E5), moderate (D10 and G11), weak (G12) and non-neutralizing (A7) (Table 2). The VHHs with strong, moderate and weak toxin-neutralizing activities had similar equilibrium dissociation constants (Kd) for ricin, ranging from 0.11–0.56 × 10−9 M whereas the non-neutralizing VHH, A7, had a Kd of 4.4 × 10−9 M (Table 1; Figure S2). Sequence analysis previously revealed that the lengths of the complementarity determining regions 1 (CDR1) and 2 (CDR2) were similar among these five VHHs, but that the lengths of the CDR3s varied considerably, ranging from 10 to 22 residues (Table 2; Figure 1A). Moreover, E5, G12, and G11, as well as the non-neutralizing VHH A7, were competitively inhibited from binding ricin by mAb PB10, which recognizes an epitope within solvent exposed α-helix B (residues 97–107)15. The binding of D10, on the other hand, was not affected by PB10 or any of the other RTA-specific neutralizing mAbs tested, which led us to speculate that it may recognize an undocumented neutralizing epitope on RTA.
Table 1.
Summary of RTA-VHH structures
| RTA-VHHa | |||||
|---|---|---|---|---|---|
| E5 | D10 | G12 | G11 | A7 | |
| dmin (Å) | 2.4 | 1.8 | 2.7 | 1.8 | 2.3 |
| Space group | P212121 | P212121 | C2 | P21212 | P41 |
| Rc/Rfreed (%) | 22.5/28.0 | 18.0/20.4 | 20.0/24.8 | 19.8/24.9 | 19.3/24.8 |
| PDB code | 4LGP | 4LGR | 4LGS | 4LHJ | 4LHQ |
, all complexes formed by co-crystallization.
R = ∑||Fo| − |Fc||/∑||Fo||, where Fo and Fc denote observe and calculated structure factors, respectively.
Rfree was calculated using 5% of data excluded from refinement.
Table 2.
Summary of RTA-VHH binding data and interface information
| H-bondsd | Buried Surface Area (Å2) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| VHH | Kda | IC50b | CDR3c | Total | CDR1/2/3 | Total | α-helix B | β-strand H | α-helix D |
| E5 | 0.23 | 5 | 10 | 16 | 4/3/6 | 1755 | 472 | 892 | 140 |
| D10 | 0.11 | 25 | 13 | 13 | 5/6/2 | 1400 | - | 500 | 590 |
| G11 | 0.56 | 90 | 15 | 10 | 0/0/8 | 1280 | - | 890 | 140 |
| G12 | 0.15 | >330 | 22 | 9 | 0/0/9 | 1100 | - | 870 | 70 |
| A7 | 4.4 | -- | 22 | 6 | 0/0/6 | 1060 | - | 715 | 85 |
, Kd, equilibrium dissociation constants (× 10−9 M);
, determined in Vero cell cytotoxicity assay;
, amino acid length;
, H-bonds between RTA and VHH (total) and CDR1, CDR2 and CDR3, respectively.
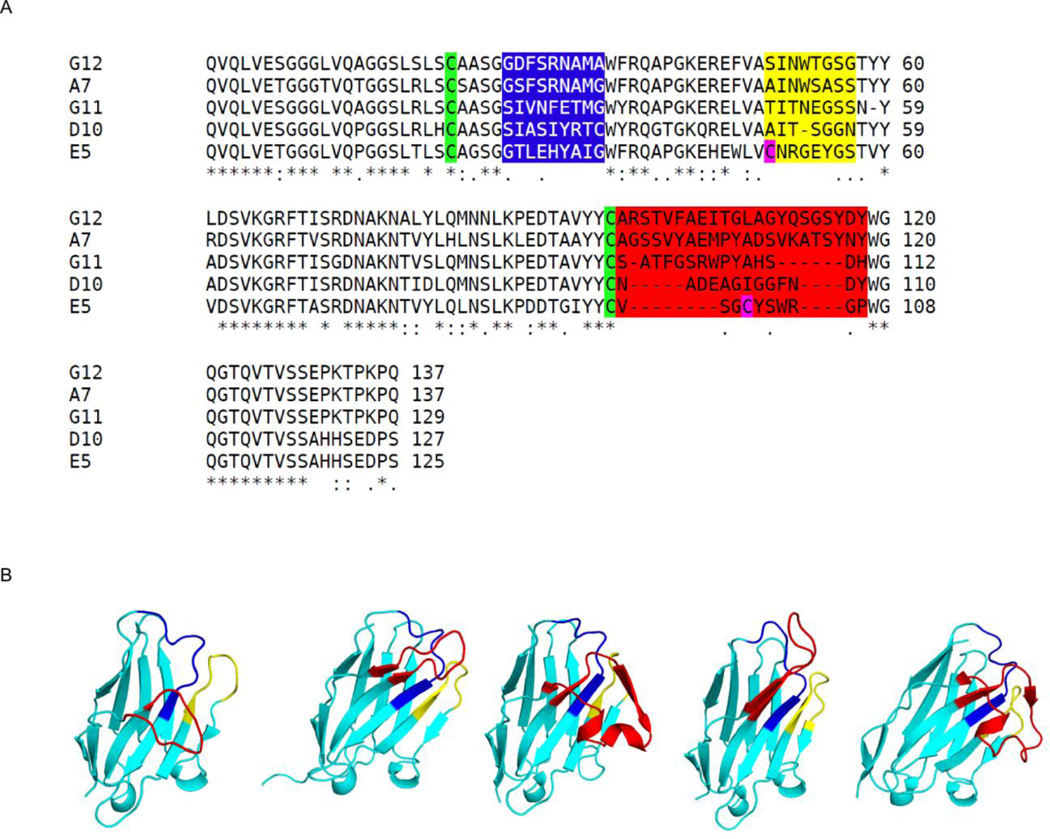
Figure 1. Amino acid sequences and X-ray crystal structures of five RTA-specific VHHs described in this study.
(A) Sequence alignment of all five VHHs (E5, D10, G12, G11, and A7) with the CDRs highlighted with blue, yellow, and red, respectively. Cysteines in the first disulfide are green and the second disulfide in magenta. Figure made with ClustalW51. (B) The structures of VHH (left to right) E5, D10, G12, G11, and A7, drawn as ribbon diagrams with CDR 1, 2, and 3 colored blue, yellow, and red, respectively. All VHHs are similarly oriented with CDR3 in front.
The crystal structures of each of the five VHHs in complex with RTA were solved at resolutions ranging from 1.8 to 2.7 Å (Table 1; Table S1). All five VHH molecules assumed a classical immunoglobulin fold consisting of nine β-strands arranged in two β-sheets with CDRs 1–3 on one face of the molecule along with either two or three 310 helices (Figure 1B). The neutralizing VHHs (E5, D10, G12, G11) formed a disulfide bridge between Cys-22 and Cys-96 (e.g., E5, G12) or Cys-22 and Cys-95 (e.g., D10, G11), effectively forming a stabilizing intramolecular bond between CDR1 and CRD3. The most potent toxin-neutralizing VHH, E5, possessed a second disulfide bridge between residue Cys-50 of CDR2 and Cys-100 of CDR3 (Figure 1B).
The non-neutralizing VHH A7 lacked the conserved disulfide bond between Cys-22 and Cys-96. While this is unusual within single-chain VH domain antibodies23, the local structure of A7 was not perturbed relative to the other VHHs we examined, as evidenced by the low RMSD of 0.5 to 0.6 Å for all superimposed Cα atoms within this region of all five VHH's. Furthermore, the absence of the intramolecular disulfide bond between residues Cys-22 and Cys-96 in A7 cannot be explained by crystallization conditions or synchrotron radiation. For example, while the crystallization conditions for the RTA-A7 complex were at pH 4.6 (a condition which has the propensity to reduce disulfide bonds) RTA-D10 and RTA-G11 were crystallized at a slightly lower pH (i.e., pH 4.5) and yet the disulfide bond between Cys-22 and Cys-96 was intact in both cases. Moreover, we also considered the possibility that the disulfide bond between Cys-22 and Cys-96 in A7 was reduced by synchrotron (X29 at NSLS) radiation. This is unlikely considering that as VHH G11 had greater X-ray exposure time (i.e., 360 seconds) than did A7 (i.e., 180 second) yet the disulfide bond between Cys-22 and Cys-96 was intact in G11 but not A7.
RTA consists of seven α-helices (A-G), three 310 helices (310 a, 310 b, and 310 c), and 10 β-strands (a–j) (Figure S1). These secondary structural elements were evident in each of the five RTA-VHH complexes (Figure 2) and superimposing the Cα atoms of the five RTA structures revealed an RMSD range from 0.4 to 0.6 Å. The largest contribution to the structural differences were caused by crystal contacts and variations within N and C-terminal residues, demonstrating that the structure of RTA was largely unaltered and did not undergo any large conformational changes when bound to the antibodies. This structure is very similar to the unliganded form of recombinant RTA (PDB: 1RTC)24 with an overall RMSD of 0.7 Å.
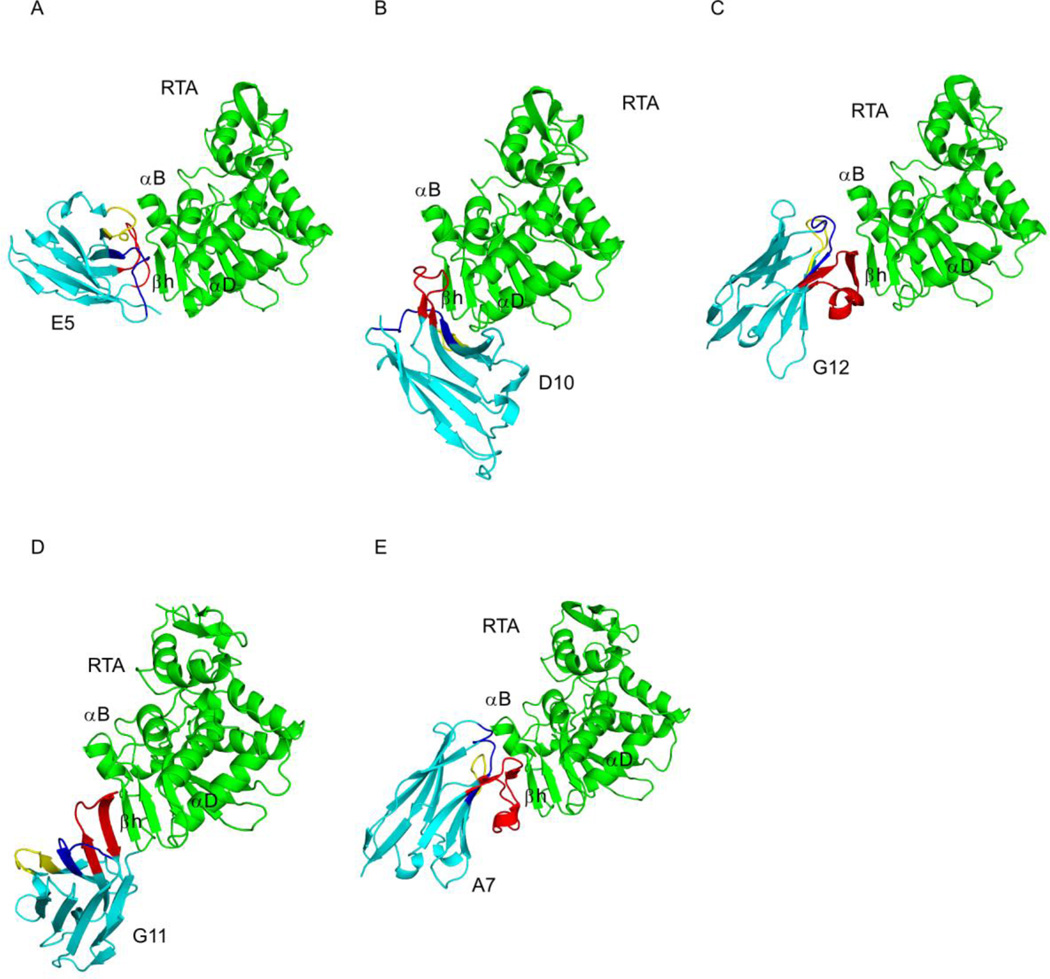
Figure 2. Structures of RTA-VHH complexes.
Structures of RTA (green) in complex with neutralizing VHHs (A) E5, (B) D10, (C) G12, (D) G11, and the non-neutralizing VHH (E) A7, depicted as ribbon diagrams. The VHHs are colored cyan, with CDRs 1, 2, and 3 colored blue, yellow, and red, respectively. RTA secondary structural elements β-strand h (residues 112–118), α-helix B (residues 97–107), and α-helix D (residues 150–157) are indicated, as necessary. RTA is similarly oriented in each panel, highlighting the different VHH binding modes.
VHHs differentially contact three secondary structural elements within RTA’s folding domains 1 and 2
Analysis of the VHH-RTA X-ray crystal structures revealed that all five VHHs had overlapping structural epitopes on RTA (Figure 3). The VHH recognition of RTA involved a common epitope with varying degrees of contact with three different secondary structural elements: β-strand h (residues 112–118), α-helix B (residues 97–107), and α-helix D (residues 150–157) (Figure 4; Table 2). We find that toxin-neutralizing activity correlates with the strength of VHH-RTA interaction. As will be detailed below, VHHs with the highest toxin-neutralizing activity (i.e., lowest IC50) generally had the greatest combined association (e.g., buried surface area, Hbonding, etc.) with these secondary elements signifying that specificity and affinity play a considerable role in RTA neutralization. In addition, we also looked at the correlation between affinity (Kd) and shape complementarity within the RTA-VHH interface using the shape correlation parameter Sc25. Although shape complementarity was high in all five RTA-VHH complexes, it was not a strong predictor of RTA-neutralization or binding affinity. For example, the most potent toxin-neutralizing VHH E5 had a Kd of 2.3 × 10−10 and a shape correlation score of 0.765, while the non-neutralizing VHH A7 had a considerably lower Kd of 4.4 × 10−9 but a shape correlation score of 0.771.
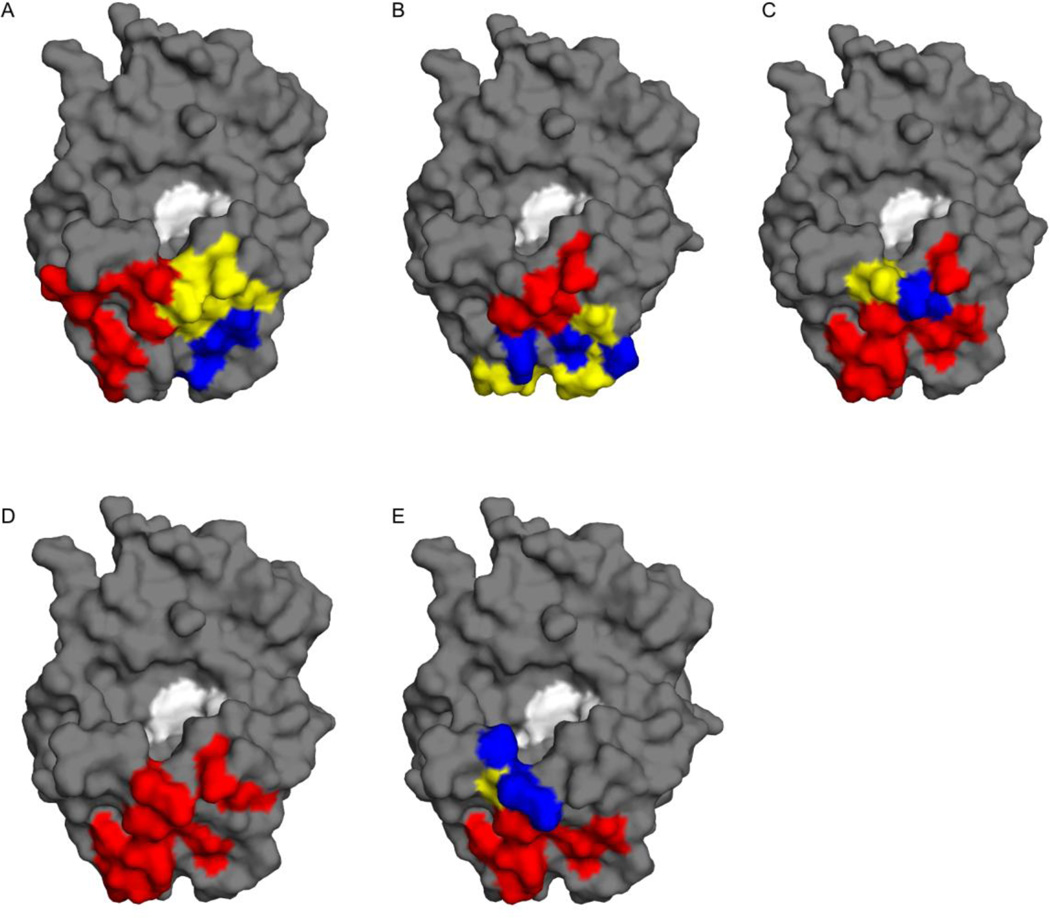
Figure 3. Neutralizing and non-neutralizing VHHs have overlapping structural epitopes on RTA.
Surface representations of RTA (gray) with VHH CDR contact points highlighted: CDR1, blue; CDR2, yellow; CDR3, red. The respective images correspond to VHHs (A) E5, (B) D10, (C) G12, (D) G11, and (E) A7. RTA’s active site, defined by catalytic residues Tyr 80 and Tyr 123 is colored white.
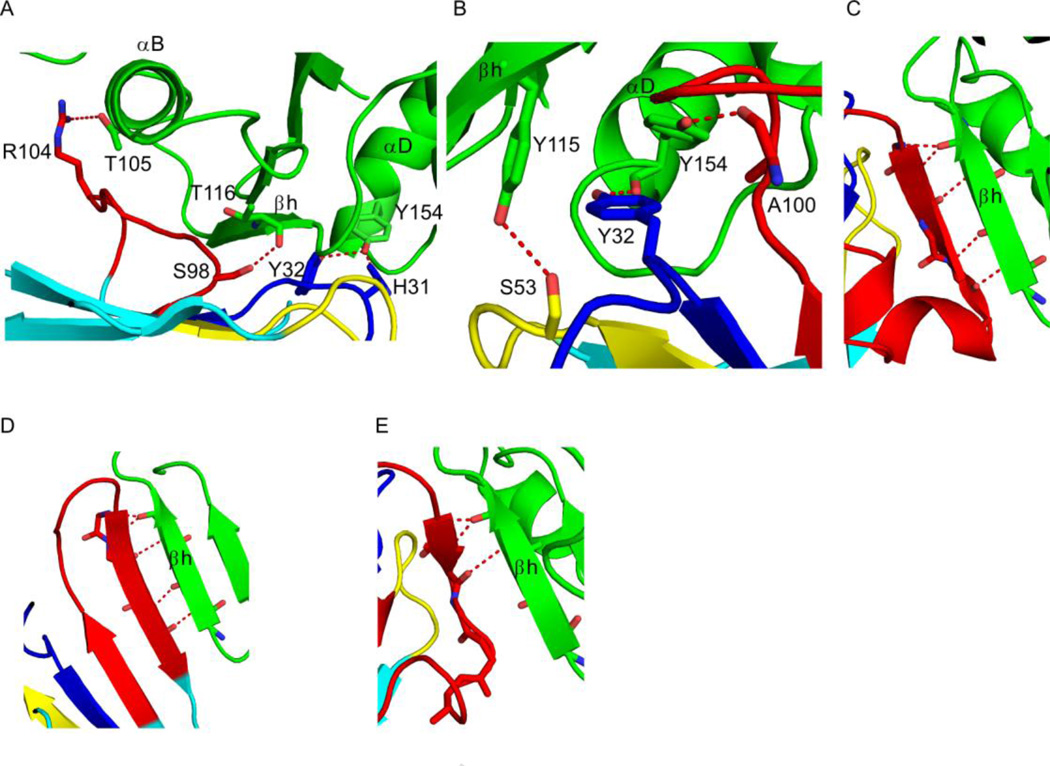
Figure 4. VHH interactions with key RTA secondary structural elements.
RTA (green) and VHHs (cyan) are drawn as ribbon diagrams. VHH CDRs 1, 2, and 3 are colored blue, yellow, and red, respectively. Side chains are drawn as sticks and color coordinated to the main chain color. Hydrogen bonds are represented as red dashes. (Panel A) Close-up of the interaction between VHH E5 and RTA’s secondary structural elements α-helix B (residues 97–107), β-strand h (residues 112–118), and α-helix D (residues 150–157). Only four (of a total of 16) of the hydrogen bonds in the interface are depicted for clarity. (Panel B) Zoom in of the interface between VHH D10 and RTA’s β-strand h (residues 112–118) and α-helix D (residues 150–157) portraying three (of a total of 13) of the hydrogen bonds in the interface. (Panels C–E) Main-chain hydrogen bond interactions between CDR3 from VHHs (C) G12, (D) G11, and (E) A7 with β-strand h (residues 112–118).
The most potent toxin-neutralizing VHH, E5 (IC50, 5 nM) made significant contact with all three epitopic secondary-structural elements: α-helix B (buried surface area (bsa) = 472 Å2), β-strand h (bsa = 892 Å2) and α-helix D (bsa = 140 Å2) (Figure 2A; 4A). Collectively, E5 resulted in the burial of 1755 Å2 of solvent-exposed surface area. There are a total of 16 hydrogen bonds at the interface between RTA and VHH E5, with the three CDRs contributing most of the hydrogen bonds (13) in this interaction (Table 2). Although the CDR1 interaction with RTA is modest, there are four stabilizing hydrogen bonds between the two proteins including the key hydrogen bonds between CDR1 residues His31 and Tyr32 to RTA’s Tyr154 in α-helix D. CDR2 forms three hydrogen bonds with RTA including two important interactions between CDR2 residue Arg52 with RTA residue Ala118. In addition, CDR3 generates six hydrogen bonds to RTA with one crucial hydrogen bond between CDR3 residue Ser 98 and RTAs Thr 116 in β-strand H and the most prominent interaction occurring between CDR3 and α-helix B (residues 97–107) that forms two of the six hydrogen bonds. This interaction results in the burial of 472 Å2 and, importantly, involves hydrogen bonding between CDR3 residue Arg104 and RTA residue Thr105 (Figure 4A), a residue within α-helix B that we have predicted is a common contact point among a number of very potent ricin toxin-neutralizing murine mAbs15; 26.
The second most potent toxin-neutralizing VHH, D10 (IC50, 25 nM), established substantial interactions with α-helix D (bsa = 590 Å2) and β-strand h (bsa = 500 Å2) (Table 2; Figure 2B, 4B), resulting in the complete burial of 1400 Å2 of solvent-exposed surface area. All three CDRs of D10 contribute to RTA recognition, as evidenced by a total of 13 intermolecular hydrogen bonds: five contributed by CDR1, six contributed by CDR2, and two contributed by CDR3 (Table 2). D10’s primary point of contact on RTA is residues Tyr154-Gly157, located in the C-terminus of α-helix D (Figure 4B). This interaction is extensive, as it buries a total surface area of 590 Å2 while establishing five hydrogen bonds between CDR1 residues Tyr32 and Arg33 and RTA residues Tyr154 and Gly157, two more hydrogen bonds between the CDR2 residues Asn56 and Tyr58 and RTA residues Ser155 and Thr156, and one final hydrogen bond between the CDR3 residues Ala100 and RTA residues Tyr154. Another interaction that likely has consequences for D10’s neutralizing activity is the hydrogen bond between CDR2 residue Ser53 and RTA's residue Tyr115 within β-strand h. D10 does not, however, contact α-helix B, demonstrating that the interaction with this secondary structural element is not absolutely essential for potent toxin-neutralizing activity and likely explaining why D10’s binding to ricin was not competitively inhibited by PB1021.
The moderate and weakly toxin-neutralizing VHHs, G12 and G11, respectively, interacted with RTA to a similar extent, as evidenced by comparable degrees of buried surface areas (1100 Å2 and 1280 Å2, respectively). In both instances, the primary contacts with RTA were focused on β-strand h, although each VHH also formed additional, albeit weak, interactions elsewhere on RTA. G12’s CDR3 is responsible for most of the interactions with RTA, as it forms a total of nine intermolecular hydrogen bonds with β-strand h, effectively burying 870 Å2 of solvent-exposed surface area (Table 2). The most notable region within this interaction involves five main chain hydrogen bonds between CDR3’s β-strand (residues 103–107) and RTA’s β–strand h (residues 113–117) (Figure 4C). G12’s CDR3-derived β-strand essentially forms the seventh β-strand within RTA’s centrally located β-sheet (Figure 2C). The additional interplay between G12 and RTA primarily involves CDR1 residue Arg31 and CDR2 residue Trp53 with RTA residues His94 and Pro95, burying an additional 230 Å2 without the formation of any hydrogen bonds.
The interaction of G11 with ricin toxin is also dominated by CDR3, which forms a total of eight intermolecular hydrogen bonds with RTA and results in the burial of 1055 Å2 of solvent-exposed surface area (Table 2). The primary interface involves a β-strand of CDR3 (residues 106–109), which forms five main chain hydrogen bonds with RTA’s β-strand h (residues 113–116) (Figure 4D). In this respect G11 and G12 are similar. However, G11’s interaction is more complicated in that CDR3 residues 109–111 establish three intramolecular main chain hydrogen bonds with CDR3 residues 97–99, resulting in a contiguous intermolecular β-sheet between the two polypeptides. Thus, there are a total of 11 β–strands at the interface between G11 and RTA in which five β-strands are donated by the VHH and six from RTA (Figure 4D). G11 also formed weak (bsa = 225 Å2) contacts with RTA’s α-helix D that involves two hydrogen bonds between G11’s residues Gln1 and Tyr105 with RTA’s residue Tyr154.
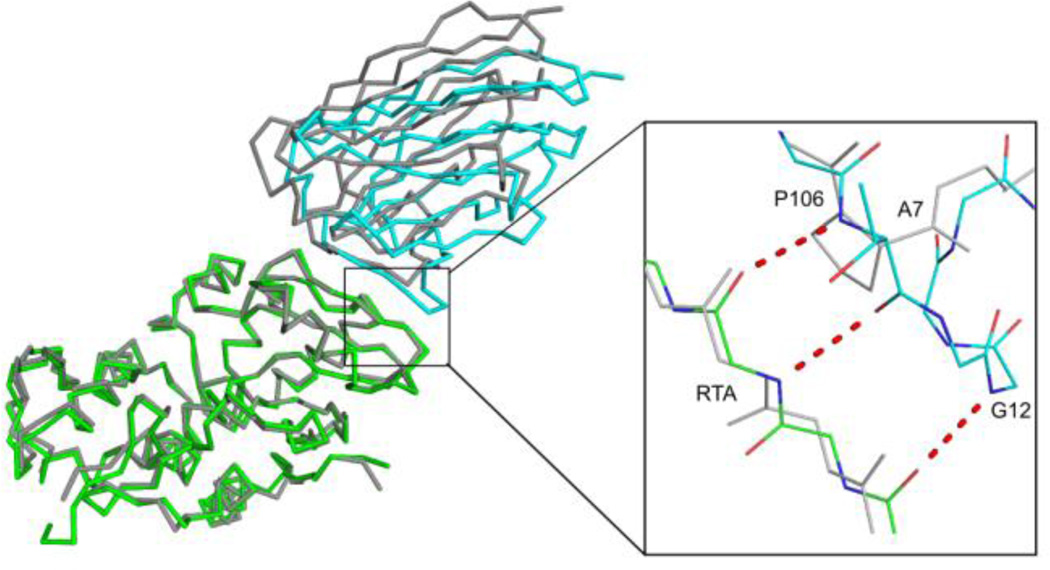
The interaction between the non-neutralizing VHH A7 and RTA consists of six intermolecular hydrogen bonds and results in 1060 Å2 of solvent-exposed surface area being buried (Table 2). Although all three CDRs contribute to RTA recognition, CDR3 dominates the interaction and is highly reminiscent of the interaction of G12 with RTA (Figure 2E; 4E). In fact, G12 and A7 share 74% primary sequence identity and are related phylogenetically suggesting a common clonal origin (C. Shoemaker, unpublished results). The primary difference between the two VHHs is that G12’s CDR3 (residues 103–107) forms a five residue β-strand that interacts with RTA β-strand h (residues 113–117), whereas A7’s CDR3 (residues 103 and 104) forms a truncated β-strand with residues 116 and 117 of RTA β-strand h (Figure 5). The abbreviated interaction between A7 and RTA is due to a proline residue at position 106, which not only lacks the requisite amide hydrogen required for β-sheet formation but also generates a bend within the VHH main chain that causes the ensuing C-terminal residues within the CDR3 to project away from RTA, thereby minimizing the interface between these two molecules. Interestingly, the relatively diminished interaction between A7 and RTA likely contributes to its ~10 fold lower binding affinity (4.4 nM) for RTA as compared to G12 (0.15 nM). The fact that G12 has toxin-neutralizing activity whereas A7 does not, underscores the role that binding affinity likely plays in influencing antibody functionality.
Figure 5. Comparison of RTA-G12 and RTA-A7 interfaces.
The super positioned Cα-traces of RTA in complex with VHH G12 (green-cyan, respectively) and VHH A7 (gray-gray, respectively). The inset illustrates the 3 additional main-chain hydrogen bonds that occur at the interface between RTA and VHH G12 that are absent between RTA and VHH A7. Hydrogen bonds are drawn as red dashes. The key residue in VHH A7, Pro 106, is labeled in gray.
DISCUSSION
Ricin toxin’s enzymatic subunit, RTA, is at the center of efforts to develop a safe and effective ricin toxin vaccine for use by military personnel, laboratory research staff, and emergency first responders tasked with attending to bioterrorism-related incidents3; 17; 27; 28; 29. The two candidate vaccine antigens under development are RiVax and RVEc. RiVax is a recombinant full-length version of RTA with point mutations at residues Tyr80 and Val76: the Tyr80Ala mutation abolishes RTA’s RNA N-glycosidase activity, while the Val76Met mutation eliminates RTA’s ability to elicit vascular leak syndrome30; 31. RVEc is a derivative of RTA that lacks residues 199–267, roughly corresponding to folding domain 3, and residues 34–43, which normally form a small hydrophobic loop32; 33; 34. To improve the long term stability of RiVax and RVEc, which are prone to aggregation, the recombinant proteins have been subjected to computational modeling and site-directed mutagenesis18; 19; 35. However, in the absence of a comprehensive B cell epitope map of RTA, it is unclear whether the introduction of stabilizing deletions, point mutations, and disulfide bonds will perturb potential structural epitopes that are critical in eliciting toxin-neutralizing antibodies. The challenge associated with RTA mutagenesis is even greater considering that neutralizing B cell epitopes on ricin toxin are postulated to not only be rare, but also predominantly (>90%) conformational in nature13; 14.
Preliminary mouse and human B cell epitope localization studies, mainly relying on peptide array-based approaches, have suggested that there are distinct clusters of neutralizing and non-neutralizing epitopes on the surface of RTA13; 14; 36; 37. Of particular interest is a linear epitope(s) within α-helix B (residues N97-F108), which was first identified by Lemley and colleagues as the target of the potent neutralizing murine mAb R70 (UNIVAX 70)11; 38. We subsequently characterized a second mAb, PB10, which recognizes a linear epitope that overlaps, but is distinct from R70’s14; 15. Based on differential peptide reactivity, we postulate that R70 and PB10 contact residues Q98, E99, E102, T105 and H106 (Table S2; Figure S3). Another R70/PB10-like murine mAb, 6C2, was recently described12 and the X-ray crystal structure solved of its Fab fragments in complex with RTA [PDB: 4KUC]26. The crystal structure revealed that 6C2 is virtually identical to R70/PB10 in that it makes key contacts with RTA at residues Gln98, Glu99, Glu102, and Thr105.
In this report we solved the X-ray crystal structures of RTA in complex with the antigen binding domains of five unique VHHs that differ in their respective ricin toxinneutralizing activities. Four of the VHHs, E5, G12, G11 and A7 (but not D10) were previously shown to be competitively inhibited by PB10 from binding to ricin, which led us to speculate that they each recognize epitopes in the vicinity of α-helix B (residues Asn97-Phe108)21. Unlike R70 and PB10, however, none of the VHHs reacted with an RTA peptide array, indicating that they bind discontinuous epitopes. The X-ray crystal structures revealed that in fact all five VHHs, including D10, recognized overlapping structural epitopes, but that they differed in the degree to which they made contact with β-strand h (residues 113–117), α-helix D (residues 150–157), and α-helix B (residues 97–107). None of the VHHs made direct contact with residues known to be involved in RTA’s RNA N-glycosidase activity (i.e., Tyr80, Tyr123, Glu177, Arg180, and Trp211), nor was there any evidence for antibody-induced allosteric changes in RTA that might influence its catalytic activity.
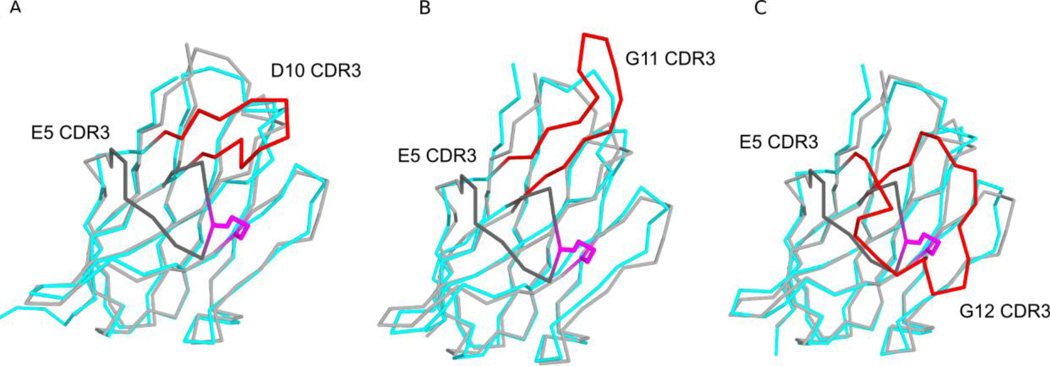
Overall, ricin toxin-neutralizing activity correlated with the degree of interaction between each VHH and RTA, with larger interfaces (i.e., buried surface areas) corresponding to greater neutralizing activity. The interactions between the different VHHs and RTA were, in turn, largely dictated by the different configurations of the VHH CDR3 elements. The different CDR3 configurations were primarily influenced by sequence length and disulfide bond formation. The shortest CDR3 restrained by a disulfide bond in E5 and the second shortest CDR3 without a disulfide bond in D10 were both more spatially constrained in comparison to the longer CDR3s that were unrestrained by disulfide bond formation (e.g., G11, G12, A7) and thus protruded away from the VHH surface (Figure 6). Specifically, the different CDR3 conformations heavily influenced the interaction with RTA’s β-strand h. The more extended CDR3s, exemplified by G11, G12, and A7, which are moderate, weak and non-neutralizing VHHs, respectively, formed β-sheet interactions with RTA’s β-strand h. The formation of these β-sheet interactions positioned CDR1 and CDR2 away from the RTA-VHH interface, thereby minimizing the possible contributions of these hypervariable regions to contact the toxin. A7’s CDR3 alignment with β-strand h accounts for its weak affinity for ricin holotoxin and explains how an antibody that recognizes an apparent “hot spot” on the surface of RTA (based on competition ELISAs with PB10) lacks toxin-neutralizing activity.
Figure 6. Different CDR3 conformations represented by toxin-neutralizing VHHs.
Shown are the super positioned Cα-traces of VHH E5 with (A) VHH D10, (B) VHH G11, and (C) G12. VHH E5’s CDR3 element is colored dark grey, while CDR3 elements of VHHs D10, G11, and A7 are colored red. The disulfide bond between residues Cys50 and Cys100 of VHH E5 is shown in stick representation and colored magenta.
A closer look at the interactions between RTA and G12 and A7 is warranted, as it illustrates the effect that a single residue can have to antibody affinity and toxinneutralizing activity. As noted in the Results section, G12 and A7 are phylogenetically related and are likely derived from a common clonal origin (C. Shoemaker, unpublished results). The two antibodies interact with RTA in a similar fashion, but differ in the extent to which they form β-sheet interactions with β-strand h. G12’s CDR3 (residues 103–107) forms a five-residue β–sheet with β-strand h (residues 113–117), while A7’s CDR3 (residues 103–104) only forms a two-residue β–sheet with β-strand h (116 and 117). This difference is accounted for by the presence of a proline residue in A7’s CDR3 at position 106, which effectively truncates the β-sheet interaction. The shortening of the A7 β–sheet interaction relative to G12 minimizes the size of the interface by 155 Å2, while simultaneously removing three main-chain hydrogen bonds. This difference likely accounts for A7’s 10-fold lower binding affinity for ricin, as compared to G12.
VHHs with more compact CDR3s, exemplified by strong and moderately neutralizing antibodies E5 and D10, associated with β-strand h without forming an RTA-VHH intermolecular β-sheet. In the absence of this β-sheet formation, CDR1 and CDR2 were free to form extensive interactions with α-helix D, and, in the case of E5, with α-helix B. E5 and D10’s CDR1 and CDR2 elements not only contribute to greater total buried surface areas (1755 Å2 and 1400 Å2, respectively), but also improve contact with the key secondary structural elements that are likely to be key determinants in neutralizing ricin. For example, E5 is interesting because its CDR3 forms a hydrogen bond with RTA’s Thr105. That residue, Thr105, figures prominently in the interactions between R70, PB10 and 6C2 with RTA (Table S2; Figure S3)15. While the functional significance of Thr105 within the context of ricin cytotoxicity is not known, our studies would argue that preserving this single amino acid residue within a candidate ricin vaccine antigen is critical for the elicitation of at least one class of toxin-neutralizing antibodies.
Finally, it remains unclear how antibodies against RTA neutralize ricin. Dai and colleagues have proposed that 6C2 affects the flexibility of α-helix B, which in turn influences RTA’s RNA N-glycosidase activity12. While there is in vitro evidence to support this notion, we think it unlikely that antibodies would remain associated with RTA in the cytoplasm of host cells, considering that delivery of RTA into the cytoplasmic compartment involves ERAD-mediated unfolding and retrotranslocation across the ER membrane7. We favor a model in which antibodies work more upstream in the cytotoxic pathway. Song and colleagues reported that the anti-RTA specific mAb affects toxin attachment, uptake and trafficking to the TGN39. Recent work from our laboratory similarly suggests that R70 and PB10 may interfere with retrograde transport of ricin to the TGN and/or block protein disulfide isomerase-mediated reduction of the intermolecular disulfide bond that links RTA and RTB (A. Yermakova, J. O’Hara, TI Klokk, K. Sandvig, and N. Mantis, manuscript in preparation)40. Indeed, there is evidence from systems biology to suggest that intracellular transport of ricin involves interactions with possibly dozens of host proteins41. It is not unreasonable to suggest that α-helix B may be central in one or more of these interactions. Our future studies will be aimed at defining the mechanism(s) by which the five VHHs described in this manuscript are able (or not able) to neutralize ricin.
MATERIALS AND METHODS
Ricin, ELISAs and toxin-neutralization assays
Ricin toxin (RCA-II) was obtained from Vector Laboratories (Burlingame, CA). ELISAs, affinity determinations, and toxin-neutralizing assays were performed as described14; 21; 42.
Cloning, expression and purification of VHHs
PCR amplicons corresponding to E5 (residues 1–125), D10 (residues 1–127), G12 (residues 1–137), G11 (residues 1–129), and A7 (residues 1–126) VHHs were subcloned into the N-terminally deca-histidine maltose binding protein (MBP) tagged MCSG9 expression vector using a standard ligase independent cloning protocol. All five VHH proteins were expressed in E. coli strain BL21(DE3)-pRARE. The transformed bacteria were grown at 37°C in TB medium and induced at 20°C with 0.1 mM (IPTG) at an OD600 of 0.6 for ~16 hours. After induction, cells were harvested and resuspended in 20 mM Tris-Cl pH 7.5 and 150 mM NaCl. The cell suspension was sonicated and centrifuged at 30,000 g for 30 minutes. After centrifugation, the VHH-containing supernatant was purified by nickel-affinity and size-exclusion chromatography on an AKTAxpress system (GE Healthcare), which consisted of a 1mL nickel affinity column followed by a Superdex 75 16/20 gel filtration column. The elution buffer consisted of 0.5M imidazole in binding buffer, and the gel filtration buffer consisted of 20mM HEPES pH 7.6, 150mM NaCl, and 20mM imidazole. Fractions containing VHH were pooled and subject to TEV protease cleavage (1:20 weight ratio) for 3 hours at room temperature in order to remove the decahistidine-maltose binding protein tag. The cleaved protein was passed over a 1mL Ni-NTA agarose (Qiagen) and 1 mL Amylose-agarose gravity column to remove the added TEV protease, cleaved residues, and uncleaved fusion protein. The pUTA-RTA expression construct was supplied by Jon Robertus. RTA was expressed and purified as described previously43; 44. In order to generate RTA-VHH protein complexes, after purification RTA was mixed in a 1:1 stoichiometry with each purified VHH and put over a Superdex 75 10/300 gel filtration column pre-equilibrated in 20 mM Tris pH 7.5 and 150 mM NaCl to isolate the complex from monomeric RTA or VHH. Purified RTA-VHH complex was concentrated to a final total concentration of 10 mg/ml for crystallization experiments
Crystallization and Data Collection
All RTA-VHH complex crystals were grown by sitting drop vapor diffusion at 20°C using a protein to reservoir volume ratio of 1:1 with total drop volumes of 0.4 µL. Crystals of the RTA-E5 complex were grown against crystallization buffer containing 100 mM Bicine (pH 8.5) and 20 % PEG 6K. Crystals of the RTA-E5 complex nucleated within a week and grew slowly to full size of ~60 µm over a period of 10 days. Crystals of the RTA-D10 complex were grown against crystallization buffer containing 100 mM NaAcetate (pH 4.5), 200 mM Zinc Acetate, and 10% PEG 3000. Plate shaped crystals of the RTA-D10 complex nucleated within 24 hours and grew to full size of ~200 µm within 2 days. Crystals of the RTA-G12 complex were grown against crystallization buffer containing 100 mM NaHepes (pH 7.5) and 20% PEG 8000. Clustered rod-shaped crystals (20 × 20 × 200µm) of the RTA-G12 complex nucleated within 24 hours and grew to full size within 5 days. Crystals of the RTA-G11 complex were grown against crystallization buffer containing 100 mM NaAcetate (pH 4.5), 200 mM NaCl, and 40% PEG 300. Crystals of the RTA-G11 complex nucleated within two weeks and grew slowly to full size of ~50 µm in 3.5 weeks. Crystals of the RTA-A7 complex were grown against crystallization buffer containing 100 mM NaAcetate (pH 4.6) and 2 M NaFormate. Crystal clusters of the RTA-A7 complex nucleated within 1 day and grew to full size of 120 µm within 3 days. All crystals were flash frozen in liquid nitrogen after a short soak in the appropriate crystallization buffers supplemented with 20 – 25% ethylene glycol. Data were collected at the X29 beamline at the National Synchrotron Light Source, Brookhaven National Labs. All data was indexed, merged, and scaled using HKL200045 then converted to structure factors using CCP446.
Structure Determination and Refinement
The structures of all RTA-VHH complexes were solved by molecular replacement using the program Phaser47. Molecular replacement calculations were performed using the coordinates of the ricin a chain as a search model for RTA (PDB code 1RTC) in all RTA-VHH complexes. The VHH coordinates used as a search model for D10 in the RTA-D10 structure had the highest sequence identity to D10 (PDB code 3EZJ). All subsequent molecular replacement calculations for the other RTA-VHH complexes used the refined RTA-D10 coordinates as a search model. The resulting phase information was used to autobuild the polypeptide chains for each RTA-VHH complex using the program ARP48, which was able to trace most residues. Further manual model building was performed with COOT49 combined with structural refinement employing the PHENIX package50. Twinned refinement was performed for the RTA-A7 complex using the twin operator h,-k,-l with a twinning fraction of 0.5. During refinement a cross-validation test set was created from a random 5% of the reflections. Data collection and refinement statistics are listed in Table S1. Molecular graphics were prepared using PyMOL(Schrodinger) (DeLano Scientific LLC, Palo Alto, CA).
Surface Plasmon Resonance (SPR) kinetic analysis
Affinity of antibodies for ricin was determined by surface plasmon resonance (SPR) using the ProteOn XPR36 (Bio-Rad Inc. CA, USA). A GLC chip was equilibrated in running buffer PBS-T (phosphate buffered saline plus 0.005% Tween) at a flow rate of 30 µl/min for the ricin immobilization steps. Following 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide HCl (EDAC; 200 mM) N-hydroxysulfosuccinimide (sulfo-NHS; 50 mM) activation (3 min), ricin was diluted in 10 mM sodium acetate at pH 5.0 at two different concentrations, 4 µg/mL and 8 µg/ml and injected for 2 min. A third parallel, vertical channel received acetate buffer only and served as a reference channel. The surfaces were deactivated by injection of 1M Ethanolamine (5 min). Ricin densities of 2509 and 1288 RU were obtained in the respective channels. The ProteOn Multi-channel module was then rotated to the horizontal orientation. Binding of the antibodies was determined using five concentrations ranging from 10–100 nM that were then subjected to 3-fold dilutions. All antibody solutions were made in PBS-T, pH 7.4. Injections were made at 50 µl/min for 480 s and dissociation times varied from 1 hr-2 hr. Chips were regenerated using two injections of glycine, pH 2.0 each at 100 µl/min for 18 s. All experiments were performed at 25°C and run overnight due to the extended dissociation times required for adequate modeling. To calculate the kinetic constants, the data obtained was modeled using the Langmuir fit selecting the grouped settings, unless noted otherwise, using ProteOn Manager Software.
Supplementary Material
HIGHLIGHTS.
Ricin is a Select Toxin and ribosome-inactivating protein.
Solved x-ray crystal structures of ricin’s enzymatic subunit in complex with five nanobodies.
Identified relationships between nanobody contact points and toxinneutralizing activity.
Provide insights into structure-based subunit vaccine design
ACKNOWLEDGEMENTS
We thank Dr. Carmen Mannella and Office of the Director at the Wadsworth Center for facilitating the collaboration between the Wadsworth Center and the NYSBC. We thank Dr. Wayne A. Hendrickson for his advice and comments on the manuscript and Dr. Joanne O’Hara for helpful discussions. We gratefully acknowledge the X29 beamline staff at the National Synchrotron Light Source for their assistance in data collection and Dr. Jon Robertus (University of Texas, Austin) for providing us with a plasmid encoding RTA. This work was supported in part by grants AI097688 (NJM) and U54 AI057159 (CBS) from the National Institutes of Health (USA), the Wadsworth Center and the New York Structural Biology Center. CH is a University at Albany Carson Carr Graduate Diversity Scholar. Use of the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-98CH10886.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers. The structures generated in this study were deposited in the Protein Data Bank (PDB; http://www.rcsb.org/pdb/) under accession numbers 4LGP, 4LGR, 4LGS, 4LHJ, 4LHQ, as described in Table 1.
REFERENCES CITED
- 1.Stirpe F. Ribosome-inactivating proteins. Toxicon. 2004;44:371–383. doi: 10.1016/j.toxicon.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Audi J, Belson M, Patel M, Schier J, Osterloh J. Ricin poisoning: a comprehensive review. JAMA. 2005;294:2342–2351. doi: 10.1001/jama.294.18.2342. [DOI] [PubMed] [Google Scholar]
- 3.Reisler RB, Smith LA. The need for continued development of ricin countermeasures. Adv Prev Med. 2012;2012:149–737. doi: 10.1155/2012/149737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Endo Y, Mitsui K, Motizuki M, Tsurugi K. The mechanism of action of ricin and related toxins on eukaryotic ribosomes. J. Biol. Chem. 1987;262:5908–5912. [PubMed] [Google Scholar]
- 5.Endo Y, Tsurugi K. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J Biol Chem. 1987;262:8128–8130. [PubMed] [Google Scholar]
- 6.Rutenber E, Ready M, Robertus JD. Structure and evolution of ricin B chain. Nature. 1987;326:624–626. doi: 10.1038/326624a0. [DOI] [PubMed] [Google Scholar]
- 7.Spooner RA, Lord JM. How Ricin and Shiga Toxin Reach the Cytosol of Target Cells: Retrotranslocation from the Endoplasmic Reticulum. Curr Top Microbiol Immunol. 2012;357:19–40. doi: 10.1007/82_2011_154. [DOI] [PubMed] [Google Scholar]
- 8.Jandhyala DM, Thorpe CM, Magun B. Ricin and Shiga toxins: effects on host cell signal transduction. Curr Top Microbiol Immunol. 2012;357:41–65. doi: 10.1007/82_2011_181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montfort W, Villafranca JE, Monzingo AF, Ernst SR, Katzin B, Rutenber E, Xuong NH, Hamlin R, Robertus JD. The three-dimensional structure of ricin at 2.8 A. Journal of Biological Chemistry. 1987;262:5398–5403. [PubMed] [Google Scholar]
- 10.Rutenber E, Katzin BJ, Ernst S, Collins EJ, Mlsna D, Ready MP, Robertus JD. Crystallographic refinement of ricin to 2.5 A. Proteins. 1991;10:240–250. doi: 10.1002/prot.340100308. [DOI] [PubMed] [Google Scholar]
- 11.Lebeda FJ, Olson MA. Prediction of a conserved, neutralizing epitope in ribosome-inactivating proteins. Int J Biol Macromol. 1999;24:19–26. doi: 10.1016/s0141-8130(98)00059-2. [DOI] [PubMed] [Google Scholar]
- 12.Dai J, Zhao L, Yang H, Guo H, Fan K, Wang H, Qian W, Zhang D, Li B, Guo Y. Identification of a novel functional domain of ricin responsible for its potent toxicity. J Biol Chem. 2011;286:12166–12171. doi: 10.1074/jbc.M110.196584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Hara JM, Kasten-Jolly JC, Reynolds CE, Mantis NJ. Localization of non-linear neutralizing B cell epitopes on ricin toxin's enzymatic subunit (RTA) Immunol Lett. 2014;158:7–13. doi: 10.1016/j.imlet.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Hara JM, Neal LM, McCarthy EA, Kasten-Jolly JA, Brey RN, 3rd, Mantis NJ. Folding domains within the ricin toxin A subunit as targets of protective antibodies. Vaccine. 2010;28:7035–7046. doi: 10.1016/j.vaccine.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vance DJ, Mantis NJ. Resolution of two overlapping neutralizing B cell epitopes within a solvent exposed, immunodominant alpha-helix in ricin toxin's enzymatic subunit. Toxicon. 2012;60:874–877. doi: 10.1016/j.toxicon.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monzingo AF, Robertus JD. X-ray analysis of substrate analogs in the ricin A-chain active site. J Mol Biol. 1992;227:1136–1145. doi: 10.1016/0022-2836(92)90526-p. [DOI] [PubMed] [Google Scholar]
- 17.Smallshaw JE, Vitetta ES. Ricin Vaccine Development. Curr Top Microbiol Immunol. 2012;357:259–272. doi: 10.1007/82_2011_156. [DOI] [PubMed] [Google Scholar]
- 18.Compton JR, Legler PM, Clingan BV, Olson MA, Millard CB. Introduction of a disulfide bond leads to stabilization and crystallization of a ricin immunogen. Proteins. 2011;79:1048–1060. doi: 10.1002/prot.22933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas JC, O'Hara JM, Hu L, Gao FP, Joshi SB, Volkin DB, Brey RN, Fang J, Karanicolas J, Mantis NJ, Middaugh CR. Effect of single-point mutations on the stability and immunogenicity of a recombinant ricin A chain subunit vaccine antigen. Hum Vaccin Immunother. 2013;9:740–748. doi: 10.4161/hv.22998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Hara JM, Yermakova A, Mantis NJ. Immunity to ricin: fundamental insights into toxin-antibody interactions. Curr Top Microbiol Immunol. 2012;357:209–241. doi: 10.1007/82_2011_193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vance DJ, Tremblay JM, Mantis NJ, Shoemaker CB. Stepwise engineering of heterodimeric single domain camelid VHH antibodies that passively protect mice from ricin toxin. J Biol Chem. 2013;288:36538–36547. doi: 10.1074/jbc.M113.519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 23.Muyldermans S, Cambillau C, Wyns L. Recognition of antigens by single-domain antibody fragments: the superfluous luxury of paired domains. Trends Biochem Sci. 2001;26:230–235. doi: 10.1016/s0968-0004(01)01790-x. [DOI] [PubMed] [Google Scholar]
- 24.Mlsna D, Monzingo AF, Katzin BJ, Ernst S, Robertus JD. Structure of recombinant ricin A chain at 2.3 A. Protein Sci. 1993;2:429–435. doi: 10.1002/pro.5560020315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Y, Dai J, Zhang T, Li X, Fang P, Wang H, Jiang Y, Yu X, Xia T, Niu L, Guo Y, Teng M. Structural insights into the neutralization mechanism of monoclonal antibody 6C2 against ricin. J Biol Chem. 2013;288:25165–25172. doi: 10.1074/jbc.M113.480830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLain DE, Horn TL, Detrisac CJ, Lindsey CY, Smith LA. Progress in Biological Threat Agent Vaccine Development: A Repeat-Dose Toxicity Study of a Recombinant Ricin Toxin A-Chain (rRTA) 1–33/44–198 Vaccine (RVEc) in Male and Female New Zealand White Rabbits. Int J Toxicol. 2011;30:143–152. doi: 10.1177/1091581810396730. [DOI] [PubMed] [Google Scholar]
- 28.McLain DE, Lewis BS, Chapman JL, Wannemacher RW, Lindsey CY, Smith LA. Protective effect of two recombinant ricin subunit vaccines in the New Zealand white rabbit subjected to a lethal aerosolized ricin challenge: survival, immunological response, and histopathological findings. Toxicol Sci. 2012;126:72–83. doi: 10.1093/toxsci/kfr274. [DOI] [PubMed] [Google Scholar]
- 29.Vitetta ES, Smallshaw JE, Schindler J. A Small Phase IB Clinical Trial of an Alhydrogel-Adsorbed Recombinant Ricin Vaccine (RiVax) Clin Vaccine Immunol. 2012 doi: 10.1128/CVI.00381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smallshaw JE, Firan A, Fulmer JR, Ruback SL, Ghetie V, Vitetta ES. A novel recombinant vaccine which protects mice against ricin intoxication. Vaccine. 2002;20:3422–3427. doi: 10.1016/s0264-410x(02)00312-2. [DOI] [PubMed] [Google Scholar]
- 31.Smallshaw JE, Ghetie V, Rizo J, Fulmer JR, Trahan LL, Ghetie MA, Vitetta ES. Genetic engineering of an immunotoxin to eliminate pulmonary vascular leak in mice. Nat Biotechnol. 2003;21:387–391. doi: 10.1038/nbt800. [DOI] [PubMed] [Google Scholar]
- 32.Carra JH, Wannemacher RW, Tammariello RF, Lindsey CY, Dinterman RE, Schokman RD, Smith LA. Improved formulation of a recombinant ricin A-chain vaccine increases its stability and effective antigenicity. Vaccine. 2007;25:4149–4158. doi: 10.1016/j.vaccine.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 33.McHugh CA, Tammariello RF, Millard CB, Carra JH. Improved stability of a protein vaccine through elimination of a partially unfolded state. Protein Sci. 2004;13:2736–2743. doi: 10.1110/ps.04897904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson MA, Carra JH, Roxas-Duncan V, Wannemacher RW, Smith LA, Millard CB. Finding a new vaccine in the ricin protein fold. Protein Eng Des Sel. 2004;17:391–397. doi: 10.1093/protein/gzh043. [DOI] [PubMed] [Google Scholar]
- 35.Peek LJ, Brey RN, Middaugh CR. A rapid, three-step process for the preformulation of a recombinant ricin toxin A-chain vaccine. J Pharm Sci. 2007;96:44–60. doi: 10.1002/jps.20675. [DOI] [PubMed] [Google Scholar]
- 36.Castelletti D, Fracasso G, Righetti S, Tridente G, Schnell R, Engert A, Colombatti M. A dominant linear B-cell epitope of ricin A-chain is the target of a neutralizing antibody response in Hodgkin's lymphoma patients treated with an anti-CD25 immunotoxin. Clin. Exp. Immunol. 2004;136:365–372. doi: 10.1111/j.1365-2249.2004.02442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maddaloni M, Cooke C, Wilkinson R, Stout AV, Eng L, Pincus SH. Immunological characteristics associated with the protective efficacy of antibodies to ricin. J. Immunol. 2004;172:6221–6228. doi: 10.4049/jimmunol.172.10.6221. [DOI] [PubMed] [Google Scholar]
- 38.Lemley PV, Amanatides P, Wright DC. Identification and characterization of a monoclonal antibody that neutralizes ricin toxicity in vitro and in vivo. Hybridoma. 1994;13:417–421. doi: 10.1089/hyb.1994.13.417. [DOI] [PubMed] [Google Scholar]
- 39.Song K, Mize RR, Marrero L, Corti M, Kirk JM, Pincus SH. Antibody to ricin a chain hinders intracellular routing of toxin and protects cells even after toxin has been internalized. PLoS One. 2013;8:e62417. doi: 10.1371/journal.pone.0062417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Hara JM, Mantis NJ. Neutralizing monoclonal antibodies against ricin's enzymatic subunit interfere with protein disulfide isomerase-mediated reduction of ricin holotoxin in vitro. J Immunol Methods. 2013;395:71–78. doi: 10.1016/j.jim.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, Hyman AA, Leproust EM, McManus MT, Weissman JS. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Hara JM, Brey RN, 3rd, Mantis NJ. Comparative efficacy of two leading candidate ricin toxin a subunit vaccines in mice. Clin Vaccine Immunol. 2013;20:789–794. doi: 10.1128/CVI.00098-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis MS, Youle RJ. Ricin subunit association. Thermodynamics and the role of the disulfide bond in toxicity. J Biol Chem. 1986;261:11571–11577. [PubMed] [Google Scholar]
- 44.Villafranca JE, Robertus JD. Crystallographic study of the anti-tumor protein ricin. J Mol Biol. 1977;116:331–335. doi: 10.1016/0022-2836(77)90220-0. [DOI] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.