Summary
Stem cells reside within specialized microenvironments, or niches, that control many aspects of stem cell behaviour. Somatic hub cells in the Drosophila testis regulate the behaviour of cyst stem cells (CySCs) and germline stem cells (GSCs) and are a primary component of the testis stem cell niche. The shutoff (shof) mutation, characterized by premature loss of GSCs and CySCs, was mapped to a locus encoding the evolutionarily conserved transcription factor Escargot (Esg). Hub cells depleted of Esg acquire CySC characteristics and differentiate as cyst cells, resulting in complete loss of hub cells and eventually, CySCs and GSCs, similar to the shof mutant phenotype. We identified Esg-interacting proteins and demonstrate an interaction between Esg and the co-repressor C-terminal binding protein (CtBP), which was also required for maintenance of hub cell fate. Our results indicate that niche cells can acquire stem cell properties upon removal of a single transcription factor in vivo.
Keywords: stem cell, niche, germline, Drosophila, Escargot, CtBP
Introduction
Adult stem cells possess the capacity to self-renew and generate differentiated progeny that contribute to tissue maintenance. The capacity to undergo self-renewing divisions is regulated by intrinsic cellular determinants and requires an instructive local microenvironment, also known as the stem cell ‘niche’ (Schofield, 1978). Stem cell niches are comprised of a variety of components including support cells, soluble signalling factors, adhesion molecules, extracellular matrix, and circulatory or neuronal inputs (reviewed in Jones and Wagers, 2008). A primary role of the niche is to maintain the strict balance between stem and progenitor cells during tissue homeostasis; however, niche components must also coordinate an appropriate stem cell response to acute environmental changes and/or tissue damage.
Drosophila melanogaster has provided a genetically tractable model organism for addressing how communication between stem cells and the niche is regulated in vivo. At the tip of the Drosophila testis, approximately ten somatic hub cells are in direct physical contact with two stem cell populations: germline stem cells (GSCs) and somatic cyst stem cells (CySCs) (Figure 1A) (reviewed in Fuller, 1993). Hub cells secrete factors, such as the ligand Unpaired (Upd), which activates the JAK-STAT pathway in adjacent GSCs and CySCs, to regulate stem cell behaviour (Kiger et al., 2001b; Leatherman and Dinardo, 2008; Tulina and Matunis, 2001). In addition to the JAK-STAT pathway, Hh (Amoyel et al., 2013; Michel et al., 2012; Zhang et al., 2013) and BMP (Kawase et al., 2004; Leatherman and Dinardo, 2010; Michel et al., 2011; Shivdasani and Ingham, 2003; Zheng et al., 2011) signalling also play an important role in regulating stem cell behaviour within the testis stem cell niche.
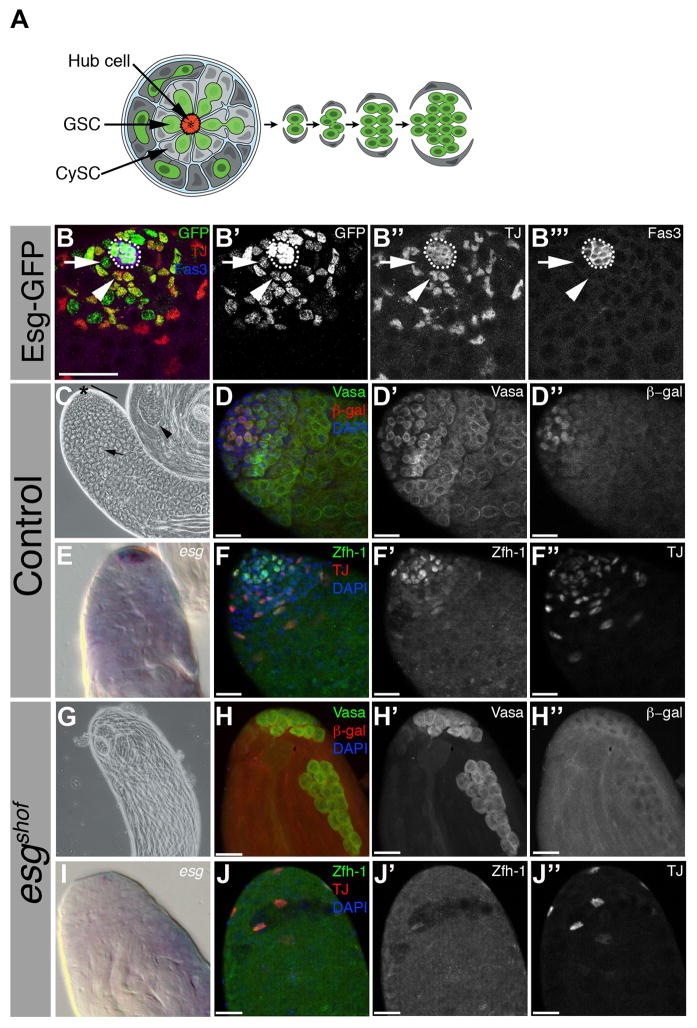
Figure 1. The stem cell niche is lost in adult esgshof males.
(A) Schematic cross-section of testis tip. Germline stem cells (GSC, light green) contact hub cells (red) and somatic cyst stem cells (CySC, light grey). GSCs divide asymmetrically to self-renew and produce goniablasts, which generate spermatogonial cysts (dark green) via transit amplifying (TA) divisions. CySCs generate cyst cells (dark grey) and encapulate spermatogonia. Phase contrast images of testes from (B) 1-day old control and (F) esgshof males. (B) Asterisk: apical tip. Bar: transit amplification zone. Arrow: spermatocytes. Arrowhead: spermatids. (F) Note spermatids in esgshof testis. Wild-type (C) and esgshof (G) flies with S3-46-lacZ enhancer trap immunostained for Vasa (green, C′, G′), β-gal (red, C″, G″), DAPI (DNA, blue). (C) Vasa marks germ cells and S3-46 marks GSCs and early spermatogonia. (G) esgshof testes lack S3-46 expression and contain late stage Vasa+ germ cells. (E, I) Testes immunostained for early somatic cell markers Zfh-1 (green, E′, I′) and Traffic Jam (TJ, red, E″, I″) in control testes (E), and largely absent in esgshof (I). (D, H) DIC images of RNA in situ for esg in control (D) and esgshof (H) adult testes. (J) Adult testis in esg-GFP enhancer trap testes immunostained for GFP (green, J′), TJ (red, J″), and Fasciclin 3 (Fas3, blue, J‴). Hub cells (outline), GSCs (arrow), and CySCs (arrowhead) express GFP. Scale bars, 20 μM (C, E, G, I) and 10 μM (J).
CySCs are anchored at the tip of the testis, adjacent to hub cells, where they divide to self-renew and generate cyst cells that will differentiate in concert with the germ cells they surround (Cheng et al., 2011; Gönczy and DiNardo, 1996; Issigonis et al., 2009). JAK-STAT signalling acts intrinsically within CySCs to regulate CySC self-renewal and maintenance. In addition, activation of Stat92E, the single Stat orthologue in Drosophila, in CySCs is also important for regulating self-renewal of adjacent GSCs in a non-autonomous manner (Leatherman and Dinardo, 2008; Leatherman and Dinardo, 2010). Putative Stat92E targets have been identified in cyst cells, such as zfh-1 and chinmo, that act intrinsically to regulate CySC behaviour and are sufficient to direct GSC proliferation (Flaherty et al., 2010; Leatherman and Dinardo, 2008). Signaling via the BMP pathway is also an important mechanism by which the CySCs can regulate germ cell behaviour (Shivdasani 2003; Kawase 2004; Leatherman and Dinardo, 2010). Results from lineage-tracing analysis also suggested that CySCs are capable of contributing cells to the hub; however, questions remain regarding the frequency of contribution and to what degree it is influenced by genetic variation (Dinardo et al., 2011; Voog et al., 2008).
GSCs are in direct contact with hub cells via adherens junctions and primarily undergo asymmetric divisions, with the mitotic spindle orientated orthogonal to hub cells, as a mechanism to ensure GSC self-renewal and continual production of gametes (Inaba et al., 2010; Yamashita et al., 2003). However, in contrast to CySCs, activation of Stat92E within GSCs appears to be important for regulating hub cell-GSC adhesion, rather than proliferation (Leatherman and Dinardo, 2010). Taken together, these data highlight the intricate relationship between the hub, CySCs, and GSCs and underscore the critical role that CySCs play within the stem cell niche.
Recent work has demonstrated that fully differentiated cells are capable of being “reprogrammed” back to a pluripotent stem cell-like state upon the addition of defined factors (Papp and Plath, 2013). Direct conversion between differentiated cell types, without passing through a pluripotent state, has also been reported, highlighting a previously underappreciated cellular plasticity (reviewed in Graf, 2011). While these advances are promising for regenerative medicine, little is understood about how the stem cell niche can influence reprogramming and de-differentiation or trans-differentiation decisions. Here we show in the Drosophila testis that niche cells can acquire somatic stem cell properties upon removal of a single transcription factor in vivo, underscoring the importance of understanding the mechanisms utilized to balance cell fates within the stem cell niche.
Results
shutoff is an allele of escargot
During the course of a genetic screen, a recessive, loss-of-function allele, named shutoff (shof), was recovered that resulted in premature and progressive loss of early male germ cells in testes evident by phase contrast microscopy (Figure 1B, F). Early germ cell loss was confirmed by examining the expression of an enhancer trap line that marks early germ cells, in combination with the germ cell marker Vasa (Figure 1C–C″, G–G″). Similarly, staining for the early cyst cell markers Zfh-1 and Traffic jam (TJ) revealed loss of early somatic CySCs and cyst cells in the testis (Figure 1E–E″, I–I″) (Leatherman and Dinardo, 2008; Li et al., 2003). Loss of stem cells appeared to be due to direct differentiation, as early somatic and germline cells differentiated at the apical tip of mutant testes (Figure 1F,G,I, Figure S3), and excessive apoptosis during development was not observed (Figure 2K–L).
Figure 2. Loss of hub marker expression during larval development in esgshof males.
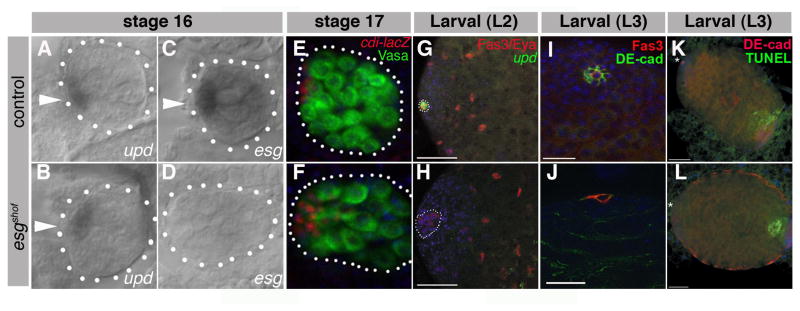
DIC images of RNA in situ for upd (A,B) or esg (C,D) in control (A, C) and esgshof (B, D) stage 16 embryonic gonads (outlined, arrowheads). Gonads from cdi-lacZ (E) and cdi-lacZ; esgshof (F) stage 17 embryos (outlined) stained for Vasa (green), β-galactosidase (red), DAPI (blue). Larval L2 updGAL4, UAS-GFP (G) and updGAL4, UAS-GFP; esgshof (H) gonads stained for Fas3 (red, outline), Eyes Absent (Eya, red), GFP (green), DAPI (blue). Note loss of GFP expression in (H), despite residual Fas3. Larval L3 control (I) and esgshof (J) gonads stained for Fas3 (red), E-cad (green), DAPI (blue). Larval L3 updGal4, UAS-GFP (K) and updGal4, UAS-GFP; esgshof (L) gonads stained for E-cadherin (red, asterisk), DAPI (blue), and TUNEL assay for apoptotic cells (green).
Genetic recombination and mapping with deficiency chromosomes revealed that shof was likely an allele of escargot (esg), a member of the Snail family of transcriptional repressors (Nieto, 2002). Previous studies demonstrated that esg is one of the first, sexually dimorphic markers expressed in Drosophila (Le Bras and Van Doren, 2006; Streit et al., 2002), as it is expressed at the tip of the testis, within hub cells, CySCs and GSCs, but undetectable in ovaries (Figure 1D, J–J‴ and Figure S2) (Gönczy et al., 1992; Kiger et al., 2000; Streit et al., 2002). Characterization of the shof mutation revealed an 18kb insertion approximately 5kb downstream of the esg transcriptional start site (Figure S1F), and testes from flies carrying strong, loss-of-function esg alleles in combination with the shof mutation exhibited phenotypes similar to shof homozygotes, with loss of both GSC and CySC populations (Figure S1B–E), indicating that shof is an allele of esg. Accordingly, RNA in situ hybridization revealed a lack of esg expression in testes from newly eclosed esgshof males (Figure 1H). Furthermore, while esg expression was highly enriched at the anterior end of ~50% (53/93) of control embryonic gonads, esg RNA was absent from ~90% (61/70) of esgshof mutant gonads (Figure 2C–D), indicating that the esgshof mutation results in loss of esg expression at the testis tip from late embryogenesis and into adulthood.
esg is required for maintenance of apical hub cells
The premature loss of early germline and somatic cells in testes from esgshof flies was accompanied by a reduction in hub cells and loss of function of the testis stem cell niche. Hub specification and formation appeared normal during embryonic stages 16 and 17 in esgshof embryos, based on hub cell morphology and marker expression (Figure 2A–F). Similar to esg, RNA in situ analysis revealed upd mRNA was expressed at the anterior tip of wild-type embryonic testes (Le Bras and Van Doren, 2006; Streit et al., 2002): roughly 50% of control embryonic gonads expressed upd (24/44, Figure 2A), as expected for a sexually dimorphic trait. However, contrary to the loss in esg expression (Figure 2C, D), approximately 50% of esgshof embryonic gonads (12/27) maintained upd expression (Figure 2AB). An additional hub marker, center divider (cdi-lacZ), was also detected in testes from esgshof animals during embryogenesis. Furthermore, early germ cells encircled the embryonic hub (Figure 2E, F), indicating that the hub was present and functional as an organizing center at this stage (Le Bras and Van Doren, 2006; Sheng et al., 2009; Wawersik et al., 2005).
Although hub formation and specification appeared unaffected in esgshof embryonic testes, disruption of normal hub morphology was detected during larval stages in mutant animals (Figure 2G–J). This first and most obvious defect in the esgshof mutant gonads preceded the progressive loss of GSCs and CySCs. Expression of Fas3, E-Cadherin (DE-Cad), and cdi (Le Bras and Van Doren, 2006) was markedly reduced or absent in hub cells from ~90% of 1-day old esgshof males (Figure S1A,B (n=18); Figure 3B,D). In addition, upd and Drosophila N-cadherin (DN-Cad) were lost, in most cases, by the second larval instar (L2) (Figure 2G, H and data not shown).
Figure 3. Esg is required autonomously in hub cells to maintain the stem cell niche.
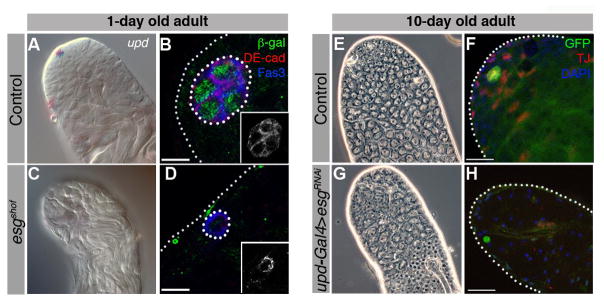
(A, C) DIC images of RNA in situ for upd in 1-day old wild-type (A) or esgshof (C) flies. (B, D) Testes from 1-day old cdi-lacZ (B) or esgshof; cdi-lacZ (D) flies immunostained for Fas3 (blue), DE-cadherin (DE-Cad, red, insets), β-gal (green). Phase contrast image of testes from 10-day old updGal4, UAS-GFP;Gal80ts (E) or updGal4;UAS-esgRNAi/Gal80ts (G) flies shifted to 29°C upon eclosion to induce transgene expression. Immunofluorescence images of testes from 10-day old updGal4, UAS-GFP (F) and updGal4, UAS-GFP; UAS-esgRNAi (H) males for GFP (green, hub), TJ (red) and DAPI shifted to 29°C upon eclosion. Scale bars, 10 μM.
escargot is required autonomously for hub cell maintenance
Loss of hub cells and stem cells in esgshof mutants could be due to cell intrinsic requirements for esg within all of the three cell types (hub, CySC, GSC). However, cell-type specific knock-down and rescue experiments suggested that esg is required in hub cells to maintain CySCs and GSCs, consistent with previous experiments suggesting that esg is required for somatic cells to acquire or maintain hub cell fate (Voog et al., 2008). To probe the function of esg in hub cells, an esg RNAi construct was expressed specifically in hub cells using the bipartite GAL4-UAS expression system (Brand and Perrimon, 1993), in combination with the temperature sensitive Gal80 allele (Gal80ts) (Lee and Luo, 1999). Use of Gal80ts permits tight control of gene expression during development and adulthood, as Gal80ts is active and inhibits GAL4 activity at 18°C but becomes inactive at 29°C. The updGAL4 driver promotes expression of UAS constructs in hub cells but not in CySCs, cyst cells, GSCs, or their differentiating progeny. Flies carrying updGAL4; UAS-esgRNAi/Gal80ts (hereafter referred to as esgRNAi) were raised at 18°C to suppress transgene expression during development. Upon eclosion, flies were shifted to 29°C to induce esgRNAi expression within hub cells. Testes from updGAL4, UAS-GFP;Gal80ts (controls) and esgRNAi flies maintained at 18°C appeared similar to wild-type testes (data not shown). When shifted upon eclosion to 29°C and maintained there for 10 days, controls looked normal by phase contrast microscopy (Figure 3E) and expressed markers for hub cells, as well as early somatic and germ cells (Figure 3F). In contrast, testes from esgRNAi flies shifted to 29°C for 10 days strongly resembled those from 1-day old esgshof homozygotes displaying progressive loss of hub cells and expression of early somatic and germline markers (Figure 3C–D, G–H). In esgRNAi flies, only a few hub cells were detected, with an average hub cell number of 0.2 per testis (n=38) after 10 days at 29°C (Table S1). Control flies did not exhibit loss of hub cells (Table S1), as flies that did not carry the RNAi transgene maintained an average of 8.1 hub cells over 10 days (n=30). Taken together, these experiments suggest that esg is required autonomously within hub cells of adults to maintain hub integrity.
escargot is required autonomously in CySCs, but dispensable for GSC maintenance
The function of esg was also required cell autonomously in CySCs for maintenance of somatic stem cells, as determined by generating positively marked (GFP+) CySC clones homozygous mutant for either of two strong, hypomorphic esg alleles by FRT-mediated recombination. One day post clone induction, 53% of control testes contained at least one marked CySC adjacent to the hub (n=54 testes), and 60% (esgG66, n=43) or 26% (esgL2, n=38) of testes examined contained at least one GFP+ esg mutant CySC (Figure S4 A–E). Marked control CySC clones were observed in 39% (n=66), 33% (n=85), and 30% (n=61) of testes examined at 5, 10, and 15 days after clone induction, indicating that control CySCs are maintained over time (Figure S4 A–B, E). In contrast, CySCs homozygous mutant for esgG66 (16%, n=43) or esgL2 (5%, n=40) were quickly lost by 5 days after clone induction, and no esg mutant CySCs were observed after 10 (esgG66, n= 16) (esgL2, n=36) or 15 days (esgG66, n= 27) (esgL2, n=52) (Figure S4C–E). Mutant cyst cells expressed somatic differentiation markers, encapsulated germline cysts and no increase in CySC apoptosis was detected, indicating that esg is not absolutely required for cyst cell differentiation or survival but is required for CySC maintenance (Figure S4C and data not shown).
In contrast to a direct role for esg in hub cells and CySCs, no effect on GSC maintenance was observed when germline clones were generated that were mutant for either the esgG66 or esgL2 allele (Table S2), which is likely due to functional redundancy provided by the founding family member snail (G. Hime, unpublished data). These data confirm that the requirement of esg for GSC maintenance, as suggested by the shof phenotype, is non-cell autonomous (Streit et al., 2002) and likely due to its autonomous role within the hub and/or CySCs.
Expression of escargot in hub cells rescues hub loss in testes from shof flies
Expression of esg in hub cells was sufficient to rescue the esgshof phenotype. A GFP-tagged Esg construct (UAS-esgNLAP) was expressed in hub cells or cyst cells in the esgshof background (Figure 4A and Figure S5A–D), and rescue of the esgshof phenotype was observed when esgNLAP was expressed in hub cells (Figure 4B–F) but not in cyst cells (Figure S5F). The extent of rescue appeared to be dependent upon the level of esg expression, as shifting to higher temperatures to increase the activity of GAL4 resulted in an average hub cell number closer to wild-type (compare Figures 4G and S5E).
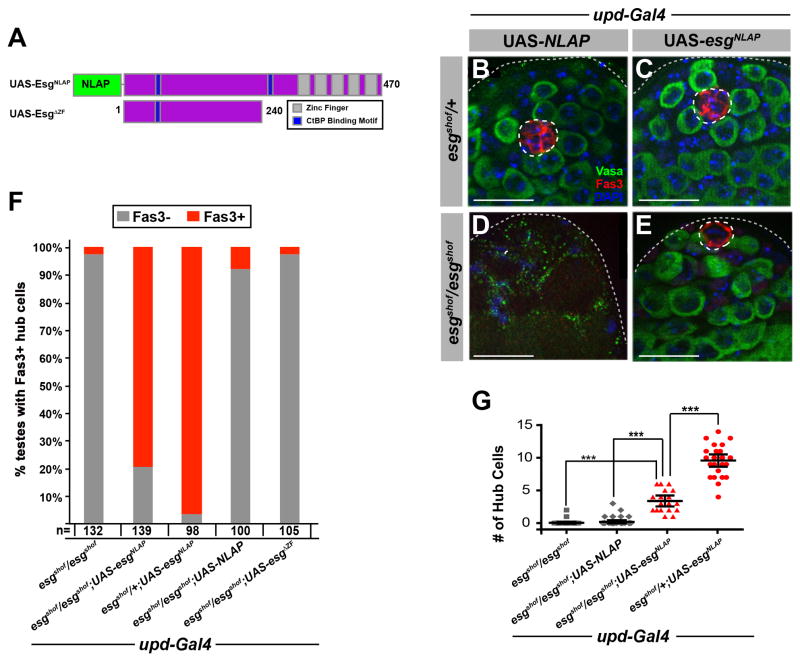
Figure 4. The esgshof phenotype is rescued by hub specific expression of Esg.
(A) Schematic of Esg protein with N-terminal GFP LAP tag. Compare esg construct without zinc fingers (UAS-esgΔZF) (Fuse et al., 1994). Two CtBP binding domains are depicted (aa #40–46, PQDLCVK and aa#259–265, PEDLSLK) (B–E) UAS-esgNLAP (E), but not control transgene (UAS-NLAP) (D) expression under updGAL4 control resulted in rescued hub in esgshof flies. (C) Overexpression of UAS-esgNLAP had no effect on hub in esgshof heterozygotes. (B–E) Testes from 3–5 day-old flies immunostained for Vasa (germline, green), Fas3 (hub, red), DAPI (blue). Scale bars, 20μM. (F). Proportion of testes from 3–5 day old adult flies with Fas3+ hub cells in given genotype. (G) Hub cell number quantification in genotypes from panels (C–E) and esgshof homozygous (DAPI used to count individual cells). UAS-esgNLAP expression rescues hub loss, although the total number of hub cells was less than in the esgshof heterozygote (p<0.001).
The carboxy-terminus of Esg contains five zinc finger domains, which are necessary for DNA binding and transcriptional regulation (Fuse et al., 1994). Expression of a truncated version of esg in which the C-terminal portion of the protein was deleted (UAS-esgΔZF) (Figure 4A) was not sufficient to suppress loss of the hub and stem cells in esgshof males (Figure 4F), suggesting that the ability of Esg to regulate hub maintenance is dependent upon its ability to bind to DNA.
Hub cells become cyst cells upon loss of escargot
No apoptosis was observed in hub cells during development in testes from esgshof males (Figure 2K, L and data not shown), raising the possibility that hub cells are lost due to a change in fate, rather than to cell death. Indeed, lineage-tracing experiments using the G-TRACE system, in combination with esgRNAi in hub cells suggested that hub cells become cyst cells upon loss of esg. The G-TRACE system provides a real-time readout of GAL4 activity (UAS-dsRed expression), as well as permanent labeling (ubi-GFP) of cells that are expressing GAL4 or were derived from GAL4 expressing cells (Evans et al., 2009) (Figure 5A–F, I, J). The G-TRACE cassette was utilized in combination with Gal80ts to suppress activation of RNAi and the lineage-tracing cassette during development.
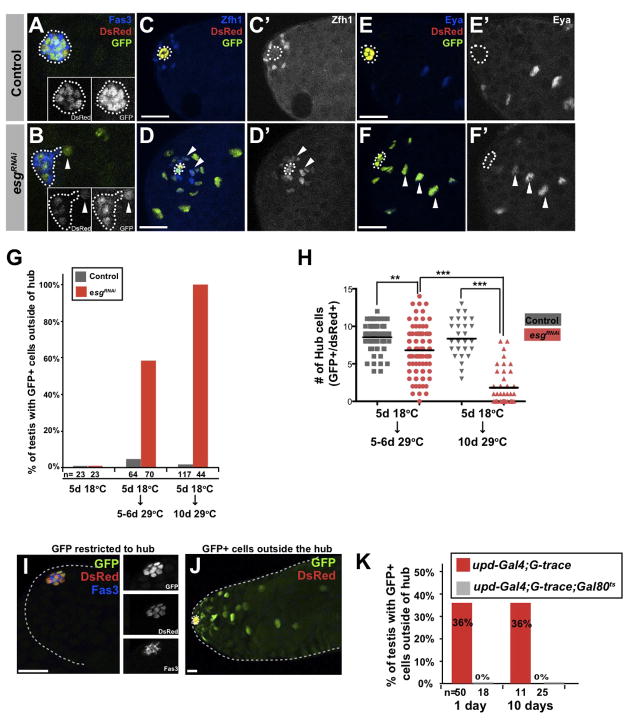
Figure 5. Loss of esg results in hub cell-cyst cell conversion.
(A–F) Immunofluorescence images of testes in control (updGal4, UAS-GFP; G-TRACE) (A,C,E) and updGal4, UAS-GFP; G-TRACE; UAS-esgRNAi (B,D,F) flies raised at 18°C and shifted to 29°C for 5 days to induce transgene expression. Testes immunostained for GFP (green), DsRed (red), Fas3 (blue, outline, A–B), Zfh-1 (blue, arrowheads, C–D′), Eyes absent (Eya, blue, arrowheads, E-F′). Hub outlined in all panels. Scale bars, 20μM. Frequency of hub-cyst cell conversion (G) and quantification of GFP+/DsRed+ hub cells (H) in control (updGal4;G-TRACE;Gal80ts) and esgRNAi (updGal4;G-TRACE;UAS-esgRNAi/Gal80ts) flies raised and maintained at 18°C for 5 days after eclosion, then shifted to 29°C for 5 or 10 days. The mean number of GFP+/DsRed+ cells in testes from esgRNAi flies was significantly lower after 5 days at 29°C (6.8) than in controls (8.5, **p<0.01), which decreased further after 10 days (esgRNAi, 1.8; control, 8.4, ***p<0.001). Statistical significance shown with one-way ANOVA (Kruskal-Wallis test) and Dunn’s multiple comparison test. (I–J) updGal4;G-TRACE flies with restricted hub GFP expression pattern (I), GFP+ cells outside of the hub (J). Testes immunostained for GFP (green, top inset) and Fas3 (blue, bottom inset). DsRed expression detectable without immunostaining (red, middle inset). Scale bars, 20μM. (K) Quantification of testes containing GFP+ cells outside the hub in updGal4; G-TRACE; Gal80ts flies raised at 18°C and shifted to 25°C upon eclosion (OFF during development) or updGal4; G-TRACE flies raised and maintained at 25°C for 10 days (ON during development).
Testes from control flies raised at 18°C and shifted to 29°C upon eclosion exhibited restricted expression of DsRed and GFP within hub cells, which co-stained with Fas3 (95.3%, n=64) (Figure 5A). However, three days after RNAi-mediated knockdown of esg within hub cells, GFP expression was detected in cells that appeared outside of the hub in the majority of testes examined (58.6%, n=70) (Figure 5B). Importantly, the increase in the total number of GFP+ cells appearing outside the hub after 5 days was coincident with a loss of hub cells (Figure 5H; mean for control=8.5 hub cells compared to 6.8 for esgRNAi, p<0.01). After 10 days of RNAi induction, 100% (n=44) of testes from esgRNAi flies contained GFP+ cells that were outside of the hub, in contrast to 1.7% (n=117) of control testes (Figure 5G). After 10 days, the average number of hub cells in esgRNAi flies was 1.8 hub cells/testis, compared to control flies, which was unchanged at 8.4 hub cells/testis (p<0.001, Figure 5H).
The GFP+/Fas3− cells in close proximity to the hub expressed high levels of Zfh-1 (Figure 5D, arrowheads) (Leatherman and Dinardo, 2008), whereas GFP+ cells that were located further away from the apical tip expressed the differentiation marker Eyes Absent (Eya) (Figure 5F, arrowheads), indicating that cells derived from the hub could differentiate along the cyst lineage. These data suggest that Esg is required to maintain hub cell identity by preventing conversion into somatic cyst cells. This is in contrast to other factors, such as Headcase, that appear to maintain hub cells by preventing loss due to programmed cell death (Resende et al., 2013).
Interestingly, analysis of hub cell fate using the G-TRACE system revealed that wild-type hub cells also convert to cyst cells during development (Figure 5I–K). When the system was suppressed prior to eclosion, 0% of testes showed GFP+ cells outside of the hub [Figures 5K (n=18), 5G (n=23), and 6F (n=16)]. In contrast, when the system was active during development, this number increased significantly to 36% (n=50, Figure 5K). This is consistent with previous data demonstrating that hub cells and CySCs are derived from a common progenitor pool during development (Dinardo et al., 2011) and our observation that larval testes contain, on average, approximately twice as many hub cells as adults (Toledano et al., 2012a). Therefore, the conversion of hub cells to the cyst lineage may be a normal, programmed event during development of the male germ line.
Cyst cells derived from hub cells can proliferate
Upon loss of esg activity in hub cells, the majority of GFP+/Fas3− cells expressed differentiation markers specific to cyst cells; however, the number of GFP+ cells was greater than the overall number of hub cells, suggesting that hub cells which convert to the cyst lineage may have progressed through a mitotic CySC stage. Immunostaining for the mitotic marker phospho-histone H3 (pHH3) revealed that hub cell descendants were capable of cell division. GFP+/pHH3+ mitotic, hub-derived cells were observed in 3/16 testes from esgRNAi flies (Figure 6A). In addition, after labelling ex vivo with the thymidine analogue, EdU, to mark cells progressing through S phase, at least one GFP+/EdU+ hub cell was observed in 8/22 (36%) testes in which esgRNAi was expressed in hub cells. GFP+/EdU+ cells were never observed in control flies (n=32) within the hub. Interestingly, we also found rare dsRed+/EdU+ cells within hubs following Esg downregulation (Figure 6B–C), suggesting that some cells in the hub re-entered the cell cycle, consistent with loss of hub cell identity. Accordingly, downregulation in the expression of DEcad and DN-cad, two common markers of hub identity, was also observe after knockdown of esg (Figure S5G–J′).
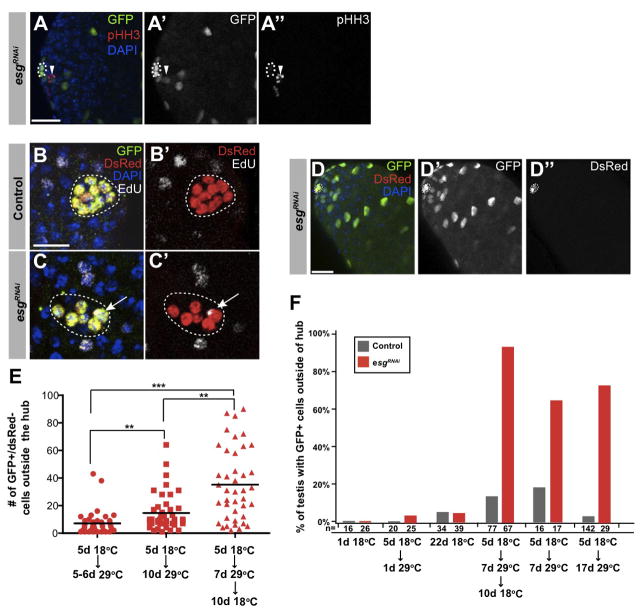
Figure 6. Hub cells that lack Esg convert to functional CySCs.
(A) Testes from updGal4, UAS-GFP; G-TRACE; UAS-esgRNAi flies immunostained for GFP (green), phospho-histone H3 (pHH3, red), DAPI (blue). Scale bars, 20μM. (B–C) Immunofluorescence images from control (updGal4; G-TRACE; Gal80ts) (B–B′) and updGal4; G-TRACE; UAS-esgRNAi/Gal80ts (C–C′) flies raised and maintained at 18°C for 5 days after eclosion, then shifted to 29°C for 5 days. Testes immunostained for GFP (green), DsRed (red), EdU (white) to mark cells in S-phase. Scale bars, 10μM. (D–D″) updGal4; G-TRACE; UAS-esgRNAi/Gal80ts flies shifted to 18°C for 10 days after 7 days at 29°C and immunostained for GFP (green), DsRed (red), DAPI (blue). Scale bar, 20μM (E) Quantification of the number of GFP+/DsRed− cells. Statistical significance shown with one-way ANOVA (Kruskal-Wallis test) and Dunn’s multiple comparison test (**p<0.01, ***p<0.001). (F) Quantification of GFP+ cells outside of the hub for noted experimental paradigms. After 17 days at 29°C, ~ 25% of esgRNAi testes lacked GFP+ cells likely reflecting complete hub loss.
While the overall number of GFP+ cells increased approximately two-fold between 5 and 10 days (p<0.01), most hub cells were lost after 10 days (Figure 5H). Given the critical role that hub cells play in regulating CySC behaviour, this precluded our ability to determine if the converted hub cells were true CySCs or differentiated into cyst cells after one cell cycle. To assess the ability of converted hub cells to maintain CySC function, flies that expressed esgRNAi for 7 days (at 29°C) were shifted back to 18°C for 10 days to suppress transgene expression and further hub cell loss (Figure 6E, F). An approximate 7-fold increase in GFP+ cells was observed over 10 days, when compared to flies that were maintained at 29°C (p<0.001, Figure 6D–F), indicating that a proportion of GFP+/Fas3− hub derived cells were capable of acting as bona fide CySCs, provided that active hub cells remained. Overall these results suggest that esg maintains hub cell fate by blocking conversion to the cyst cell lineage
Escargot interacts with the co-repressor CtBP, which is also required for maintenance of hub cell fate
In addition to expression in the testis, Esg is expressed in numerous other tissues and stem cell populations in Drosophila, including neural stem cells, known as neuroblasts, and the digestive tract (Ashraf et al., 1999b; Cai et al., 2001; Micchelli and Perrimon, 2006; Toledano et al., 2012b). To identify Esg interacting proteins that may be co-factors utilized to regulate cell fate decisions, we developed an unbiased biochemical purification strategy. The UAS-esgNLAP construct used to rescue the esgshof phenotype was adapted for use in cell culture such that stable S2 cell lines were generated that expressed EsgNLAP under an inducible promoter (Figure 7A) (Kyriakakis et al., 2008). Both one-step and two-step immuno-purification (IP) strategies were used to pull down the Esg-GFP fusion using anti-GFP antibodies, and protein interactors were identified by mass spectrometry (MS) (Table S3). Proteins that were also identified in control IPs were considered as background.
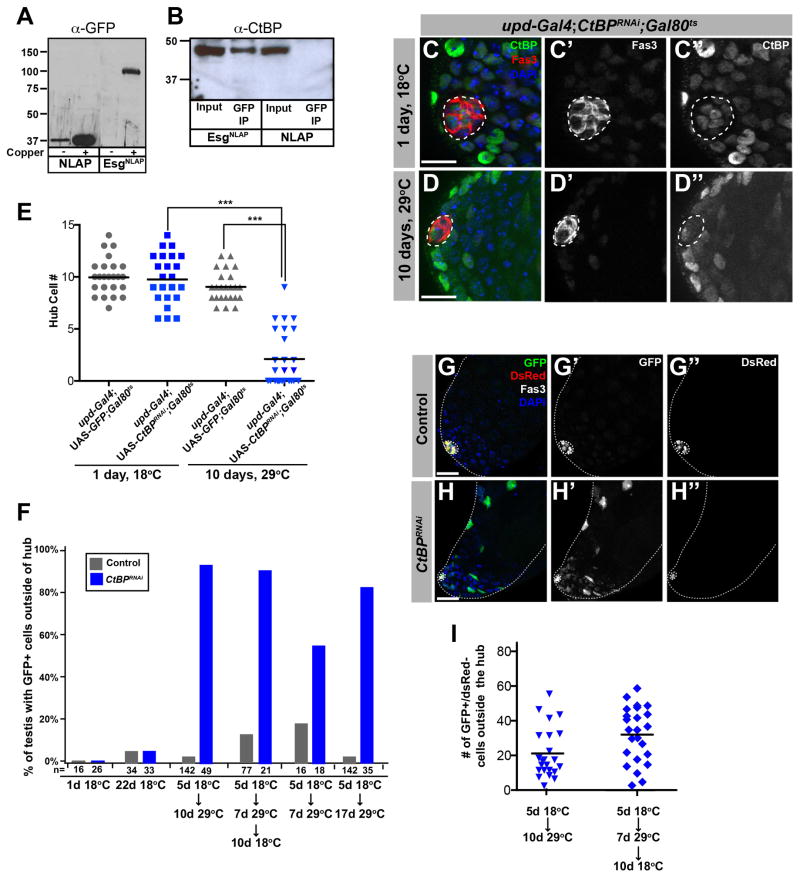
Figure 7. CtBP binds to Esg and is required for hub maintenance.
(A) Western blot analysis from stable line extracts confirmed expression of EsgNLAP and NLAP with copper induction. Expected sizes of NLAP (33.3kDa) and EsgNLAP (85.3kDa). (B) Immunoprecipitation with anti-GFP antibodies and Western blot with anti-CtBP antibodies confirmed Esg and CtBP interaction. Expected size of CtBP (42.3 kDa). (C,D) Testes from 1-day old adult updGal4;UAS-CtBPRNAi;Gal80ts flies raised at 18°C (C) or shifted to 29°C for 10 days (D) immunostained for CtBP (green) and Fas3 (hub, red). Scale bars, 10μM. (E) Quantification of hub cell number in genotypes. Statistical significance shown with one-way ANOVA (Kruskal-Wallis test) and Dunn’s multiple comparison test (***p<0.001). (F) Frequency of hub cell-cyst cell conversion in control (updGal4;;G-TRACE/Gal80ts) or CtBPRNAi (updGal4; UAS-CtBPRNAi; G-TRACE/Gal80ts) flies. Testes with no detectable GFP+ cells at the 17 day-29°C time point were scored as negative. (G–H) Immunofluorescence images from control (G) or UAS-CtBPRNAi (H) flies raised and maintained at 18°C until 5 days after eclosion, shifted to 29°C for 7 days, and shifted back to 18°C. Testes immunostained for GFP (green), DsRed (red), and Fas3 (white). (I) Quantification of GFP+/dsRed− cells for the noted temperature regimes.
In both the one and two-step immunoprecipitation followed by mass spectrometry, the most abundant protein identified as judged by spectra counts was the co-repressor protein, C-terminal binding protein (CtBP) (Table S3). The interaction of Esg and CtBP was independently confirmed by repeating the immuno-precipitation and directly immunoblotting for CtBP (Figure 7B). Although Esg had never been shown to bind CtBP directly, it contains two CtBP binding domains (P-DLS-K), and other Snail family transcription factors are known to interact with CtBP (Ashraf et al., 1999a; Hemavathy et al., 2004; Nieto, 2002; Qi et al., 2008). Therefore, these experiments confirm a predicted interaction and demonstrate the validity of our approach.
CtBP is required cell autonomously for CySC maintenance (Leatherman and Dinardo, 2008), similar to Esg (Figure S4); however, CtBP also appears to play a cell autonomous role in maintaining hub cell fate. Immunofluorescence analysis revealed that CtBP is expressed in the nuclei of all cells at the apical tip of the Drosophila testis: hub cells, CySCs, and GSCs (Figure 7C). Expression of a CtBPRNAi construct specifically in hub cells of adults (genotype: updGal4;UAS-CtBPRNAi; Gal80ts) led to a loss of hub cells similar to that observed upon reduction of esg (Figure 7E, D; compare Figures 7E and 5H). While hub size was normal in adult flies that had been raised at 18°C, flies that had been shifted to 29°C to activate transgene expression displayed hub cell loss by 3 days. After 10 days of transgene expression, the mean number of hub cells per testis had decreased to 2.1 (n=28, p<0.001), with several testes displaying complete loss of the hub (Figure 7E).
Lineage-tracing experiments utilizing the G-TRACE system in combination with CtBPRNAi revealed that loss of CtBP in hub cells resulted in 93.9% of testes displaying GFP+ cells outside of the hub after 10 days of RNAi induction (n= 49; Figure 7F). This was in contrast to control flies of the same genotype maintained at 18°C (6.1%, n=33), as well as control flies expressing the driver alone (genotype: updGal4;Gal80ts) (18°C; 5.9%, n=34) (29°C; 3.5%, n=142) (Figure 7F). Lastly, when CtBPRNAi flies were shifted back to 18°C to suppress transgene expression and further loss of hub cells, the total number of GFP+ cells increased, again suggesting a subset of hub-derived cells were maintained as CySCs (Fig 7F, I). The total number of GFP+ cells at the 10 day time point was slightly higher in the CtBPRNAi flies than in esgRNAi flies, and remaining hub cell numbers were higher at all time points examined. Zfh-1 has been shown to regulate the maintenance of CySCs in a CtBP-dependent manner (Leatherman and Dinardo, 2008). However, in contrast to loss of Esg or CtBP, targeted depletion of Zfh-1 in hub cells did not result in loss of hub cells or conversion of hub cells into CySCs (Figure S6).
Discussion
Given the integral role of the niche in regulating stem cell behaviour, changes in niche size or function could accompany alterations in stem cell activity during development or lead to decreased stem cell activity as a consequence of disease or aging (Jones and Wagers, 2008; Toledano et al., 2012a). Here we identify the Snail class transcriptional repressor Escargot as a factor that regulates the balance of cell fates within the stem cell niche. Our mapping and characterization of the shutoff mutation, in combination with targeted loss of function experiments, identified Esg as an essential factor that is required in hub cells to maintain niche integrity by blocking conversion of hub cells to the cyst cell lineage. Furthermore, immunoprecipitation followed by mass spectrometry identified the co-repressor protein, CtBP, as an interacting partner of Esg (Figure 7). Our data suggest that Esg acts in concert with CtBP in hub cells to regulate maintenance of hub cell identity. In addition to maintaining hub cell fate, Esg acts autonomously to regulate CySC maintenance and self-renewal. We predict that a systematic, functional analysis of other candidates identified through our IP/MS analysis (Table S3) will lead to the characterization of cell-type specific co-factors for Esg. These subsequent studies are likely to identify regulatory networks that provide insight into how Esg can act at nodes to regulate hub cell vs cyst cell identity, as well as stem cell behaviour in distinct tissues (e.g. testis vs intestine).
Although our data suggest that support cells within the testis stem cell niche (hub cells) can assume stem cell function, this is likely not a common event in adults. However, conversion of hub cells to CySCs may be a normal step of maturation of the testis niche during development (Figure 5J), which is restrained by Esg and its partners in adults. An alternative model is that hub cells are comprised of a pool of quiescent CySCs, although given the differences in gene expression observed between the two cell types, the transition between states would require more than cell cycle withdrawal.
Indeed, the close relationship between CySCs and hub cells is quite complex. Previous lineage-tracing and BrdU pulse-chase studies have suggested that CySCs can contribute to the pool of hub cells (Voog et al., 2008). For example, in experiments where 1 labelled CySC was generated, on average, using FLP-mediated genetic recombination, labelled hub cells were observed in 25.8% (49/190) and 24.5% (52/212) of testes examined at 5 and 10 days following clone induction, respectively. In contrast, another group used a similar FLP-mediated lineage-tracing strategy that resulted in 1.5 to 3.3 labelled CySCs per testis and did not observe labelled hub cells after 5 days (n=20) and observed labelled hub cells in 5% (1/20) of testes examined after 10 days (Dinardo et al., 2011). The reasons underlying the discrepancy between our findings and those described by Dinardo et al. remain unclear but could be attributed to a number of factors, including genetic variation. Interestingly, we also reported that BrdU pulse-chase experiments in wild type animals resulted in labelled hub cells in 4% (n=143) and 3% (n=96) of testes examined at 5 and 10 days, respectively (Voog et al., 2008), frequencies that are closer to those reported by Dinardo et al. (Dinardo et al., 2011). In addition, subsequent experiments from our lab using the G-TRACE strategy as an alternative method for labelling all CySCs and their immediate progeny revealed an estimated rate of CySC contribution to the hub of 1.35% (see Materials and Methods for details). Given these data, we are less inclined to think of the CySC to hub cell transition as a homeostatic mechanism to maintain a specific number of hub cells and now favor a model where CySCs can become hub cells as a result of damage or a block in proliferative capacity. Indeed, recent findings from our lab suggest that contribution of CySCs to the hub may be affected by increased replicative stress in CySCs (Landais et al., 2014).
The intimate relationship between somatic stem cells and supporting niche cells observed in the Drosophila testis appears to be conserved in more complex systems. For example, mouse hair follicle stem cells can give rise to K6+ niche cells (Hsu et al., 2011). In addition, in the mouse small intestine, Lgr5+ crypt base columnar cells (CBCs) generate Paneth cells, which are a critical component of the stem cell niche at the base of each crypt (Sato et al., 2011). Interestingly, recent studies in the small intestine demonstrated that quiescent Paneth cell precursors could be stimulated to undergo significant proliferation upon injury, and thus recalled into an active stem cell state (Buczacki et al., 2013).
Although the hub is established normally during embryogenesis in esgshof males, upd expression is lost during development (Figure 2). Subsequent loss of hub cells and decreased Jak-STAT signalling is likely a major mechanism contributing to loss of tissue homeostasis. However, hub cells also express high levels of cell adhesion proteins, such as E-cadherin, N-cadherin, and Fasciclin. Therefore, one explanation for our findings could be that decreased expression of cell-cell adhesion molecules results in loss of hub cells upon loss of esg (Figure S5 H′-J′). However, several lines of evidence suggest that downregulation of conventional cell adhesion molecules is not the major mechanism underlying hub cell loss in shof mutants or upon RNAi-mediated depletion of esg: 1) RNAi-mediated depletion of shotgun, the gene encoding Drosophila E-cadherin (DEcad), in hub cells was not sufficient to induce hub cell loss (Michel et al., 2011; Voog et al., 2008), 2) loss of esg in individual hub cells did not result in downregulation of DE-cad (Voog et al., 2008) and 3) RNAi-mediated depletion of components of the exocyst complex, which traffics adhesion molecules to the cell’s surface, in hub cells, resulted in disintegration of the hub, but not loss of hub cells (Michel et al., 2011). Attempts to overexpress cell adhesion molecules in hub cells to rescue the shof or esgRNAi phenotype resulted in lethality; therefore, we were unable to test this hypothesis directly. Data from chromatin profiling experiments, together with analysis of gene expression changes as a consequence of loss and gain of function of Esg, will provide specific transcriptional targets that could act downstream of Esg in hub cells and cyst cells to maintain cell identity and integrity of the testis niche.
Recent research has uncovered the remarkable ability of highly differentiated cells to become ‘reprogrammed’ into a stem cell-like state upon expression of a limited number of factors (Takahashi and Yamanaka, 2006). However, to date, the process of cellular reprogramming has been studied primarily in vitro using cell cultures. Regeneration of tissues in vertebrates such as zebrafish, axolotl, salamanders, and mice suggest that de-differentiation may be possible in vivo, although it is not evident that differentiated cells in a blastema pass through a stem cell state (King and Newmark, 2012; Tanaka and Reddien, 2011). In contrast, spermatogonia can revert back to spermatogonial stem cells under certain conditions in the male germ line of both Drosophila and mice (Barroca et al., 2009; Brawley and Matunis, 2004; Kai and Spradling, 2004; Nakagawa et al., 2010). In addition, recent reports of de-differentiation of cells within the mammalian small intestine, stomach and lung into stem cells upon injury indicate that somatic tissues also possess this capacity (Buczacki et al., 2013; Stange et al., 2013; Tata et al., 2013), although the molecular mechanisms regulating de-differentiation are not well understood.
Our data reveal that differentiated niche support cells can also acquire stem cell properties upon removal of a single transcription factor in vivo, which underscores the importance of balancing cell fates within a niche and provides a tool to begin probing the mechanism by which this balance is achieved. Importantly, niche components may function more directly than previously appreciated to maintain an adequate number of stem cells available for tissue repair and regeneration. Furthermore, as data suggest that differentiated cells in the body naturally possess the ability to dedifferentiate in vivo, it may be possible to harness this activity to facilitate the repair of tissues without the need for cell transplantation. However, uncontrolled dedifferentiation could result in excess stem cells, which would disrupt tissue homeostasis and may even contribute to tumour initiation and growth. Therefore, characterization of genetic programs that maintain optimal niche function will provide a platform for designing niches to support the faithful derivation and maintenance of tissue stem cells in culture, facilitate the development of strategies to enhance the transplantation of stem cells in the course of regenerative medicine, and may provide novel targets for anti-cancer therapies.
Materials and Methods
Mapping the esgshof allele
The shof mutation spontaneously arose in P571, a stock carrying a P{lacW} insertion. The P{lacW} element in stock P571 was recombined away from the shof locus. shof failed to complement Df(2L)TE116(R)GW21 (Hiller et al., 2004) and Df(2L)osp38 (BDSC #6082) but complemented Df(2L)noc11 (BDSC #6080) and Df(2L)TE116(R)GW2 (Hiller et al., 2004), placing shof near esg. Subsequent complementation tests revealed shof/esgL2, shof/esgG66B and shof/esgP3 males had shof phenotypes.
Primers were designed using Primer 3 (http://frodo.wi.mit.edu/) to regions spanning 3584bp upstream and 6236bp downstream of esg start site. Qiagen Tissue Easy Kit was used to obtain genomic DNA from OreR and S3-46; esgshof/esgshof males. PCR fragments were sequenced by Eton Biosciences Inc., and analyzed using Chromas Pro. Sequences from OreR and S3-46; esgshof/esgshof were identical except for an ~18kb insertion in S3-46; esgshof/esgshof between 5228bp and 5840bp downstream of esg start site.
Clonal analysis
For MARCM analysis adult flies were heat shocked two consecutive days at 37 °C for two hours and collected at indicated times for dissection and immunostaining (described below). Wild-type MARCM genotype: y,w, hsflp122; FRT40A/FRT40A, tubGAL80; tubGAL4/2x-UAS-eGFP. Mutant genotype: esgG66 (y,w, hsflp122; FRT40A esgG66/FRT40A tubGAL80; tubGAL4/2x-UAS-eGFP), and esgL2 (y,w, hsflp122; FRT40A esgL2/FRT40A tubGAL80; tubGAL4/2x-UAS-eGFP). Germline clonal analysis was carried out as described (Kiger et al., 2001a). Control genotypes used were: y,w, hsflp122; FRT40A/FRT40A ubi-GFP. Mutant genotypes used were: esgG66 (y,w, hsflp122; FRT40AesgG66/FRT40A GFP), and esgL2 (y,w, hsflp122; FRT40AesgL2/FRT40A GFP).
Temporal expression of RNAi transgenes and G-TRACE cassette
All updGAL4, UAS-GFP; UAS-esg-RNAi (esg-RNAi) and updGAL4, UAS-GFP; UAS-esg-RNAi/UAS-RedStinger, UAS-FLP, Ubi-p63 FRT>STOP FRT> nEGFP (esg-RNAi/G-TRACE) flies were raised at 18°C unless noted otherwise. Control flies were kept at 18°C during adulthood while experimental flies were shifted to 29°C. This permitted visualization of cell fate changes in esg-RNAi/G-TRACE flies, as DsRed expression reflected hub cells under GAL4 control and descendants of DsRed+ cells are permanently GFP+ (Evans et al., 2009). To further regulate GAL4 activity, the Gal80ts transgene was used. Flies were raised at 18°C for 5 days post-eclosion to restrict Gal4 during final developmental stages. Flies were shifted to 29°C for 5–10 days to inhibit Gal80ts and activate Gal4. For pulse-chase experiments, flies were shifted back to 18°C to suppress GAL4 and track GFP+ cells.
For G-TRACE experiments described in the Discussion, which measured the rate of CySC to hub cell conversion, c587-Gal4; UAS-GFP, UAS-RedStinger, UAS-FLP, Ubi-p63 FRT>STOP FRT> nEGFP; tub-Gal80ts (c587-Gal4ts>G-TRACE) flies were raised at 18°C. Three days after eclosion, the flies were shifted to 29°C for 2 days, then 25°C for 15 days. Control groups were kept at 25°C for 17 days (to control for spurious activation of G-TRACE), or 1 day at 29°C (to estimate baseline G-TRACE activation). Subtracting background control rates from the experimental rate showed an average of 1 additional hub cell in 20% of testis (n=48). Because this paradigm labeled all CySCs (as opposed to approximately one/testis in previous experiments), it is difficult to extrapolate a comparable rate of CySC to hub contribution. However, if we divide the observed rate by an average 15 CySCs per testis, then estimated contribution is 1.35%.
Creation of NLAP stable cell lines and fly strains
The pMK33-NTAP (GS) vector (Kyriakakis et al., 2008) (gift from Alexey Veraksa), was modified with GFP in the IgG domain, resulting in the pMK33-NLAP (N-terminal Localization and Affinity Purification) vector. The coding region of escargot was cloned into the NLAP vector to generate the EsgNLAP fusion protein. pMK33-EsgNLAP and pMK33-NLAP were transfected into S2 cells according to the manufacturer’s recommendations (Fugene HD, Roche). Seventy-two hours after transfection, cells were transferred into media containing 150ug/ml hygromycin (Sigma). Over 3 weeks a stable, hygromycin resistant population was selected.
To generate the UAS-NLAP and UAS-EsgNLAP flies, the pMK33-NLAP pMK33-EsgNLAP plasmids were sub-cloned into the pUAST-attB plasmid and injected into embryos at the att2 site on chromosome 3 by Genetic Services Inc. (Sudbury, MA).
Immunoprecipitations
The expression of EsgNLAP and NLAP was induced adding 0.35mM copper sulfate. After 17–24 hours cells were lysed in TAP buffer (50mM Tris, pH7.5, 5% glycerol, 0.2% IPEGAL, 1.5mM MgCl2, 125mM NaCl, 25mM NaF, 1mM Na3VO4, 1mMDTT, 1mM EDTA, Roche complete protease inhibitors) and the soluble extract incubated with rabbit anti-GFP (Clontech) for 3 hours. IgG dynabeads (Life Technologies) were added for 1.5 hours and samples were washed 3X in TAP buffer. Bound proteins were immunobolotted with either mouse anti-GFP (1:1000, Millipore) or anti-CtBP (1:2000, D. Arnosti). Chemiluminescent detection was performed according to the manufacturer’s protocol (ECL plus, GE Healthcare).
Supplementary Material
Highlights.
Loss of Escargot (Esg) in hub cells results in conversion to the cyst lineage
Hub cells depleted of Esg acquire properties of cyst stem cells (CySCs)
Esg interacts with the co-repressor, C-terminal Binding Protein (CtBP)
CtBP is also required for maintenance of hub cell fate
Niche support cells can acquire stem cell traits
Acknowledgments
The authors thank E. Bach, D. Godt, S. Hayashi, P. Lasko, N. Perrimon, R. Lehman, M. Van Doren, S. DiNardo, D. Wassarman, L. Pile, D. Arnost, D. Montell, the Vienna Drosophila RNAi Center (VDRC), and Bloomington Stock Center for reagents and fly stocks and are grateful to the Jones laboratory and S. DiNardo for helpful discussions and commenting on the manuscript. J.V. was supported by a training grant from the California Institute for Regenerative Medicine to the University of California-San Diego (L. Goldstein), S.L.S. was supported by a postdoctoral fellowship from the Damon Runyon Cancer Research Foundation and the UCSD IRACDA program, and L.P.R is a GABBA fellow funded by the Portuguese Foundation for Science and Technology (FCT; SFRH/BD/33253/2007). This work was funded by the Ellison Medical Foundation, the Emerald Foundation, the G. Harold and Leila Y. Mathers Charitable Foundation, the ACS and the NIH to D.L.J. G.R.H. is supported by a project grant from the NHMRC and the Australian Drosophila Biomedical Research Support Facility. M.T.F. is supported by NIH (GM080501). T.H. is supported by NIH grants CA14915 and CA80100, and holds the Renato Dulbecco Chair in Cancer Research. J.R.Y is supported by the National Institute of General Medical Sciences (8P41GM103533-17) and the National Center for ResearchResources (5P41RR011823-17).
Footnotes
Author Contributions
J.V., S.S, L.P.R., M.F. and L.J. planned experiments. J.V., S.S., L.P.R., A.A. and L.J. performed experiments and analysed data. G.H. identified, mapped and characterized the shof allele. M.L.C. generated reagents used in the study. J.V., S.S., G.H, L.P.R., M.L.C., A.A., T.H., M.F. and L.J. wrote and edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amoyel M, Sanny J, Burel M, Bach EA. Hedgehog is required for CySC self-renewal but does not contribute to the GSC niche in the Drosophila testis. Development. 2013;140:56–65. doi: 10.1242/dev.086413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, Hu X, Roote J, Ip YT. The mesoderm determinant snail collaborates with related zinc-finger proteins to control Drosophila neurogenesis. The EMBO journal. 1999a;18:6426–6438. doi: 10.1093/emboj/18.22.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, Hu X, Roote J, Ip YT. The mesoderm determinant snail collaborates with related zinc-finger proteins to control Drosophila neurogenesis. The EMBO journal. 1999b;18:6426–6438. doi: 10.1093/emboj/18.22.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroca V, Lassalle B, Coureuil M, Louis JP, Le Page F, Testart J, Allemand I, Riou L, Fouchet P. Mouse differentiating spermatogonia can generate germinal stem cells in vivo. Nature cell biology. 2009;11:190–196. doi: 10.1038/ncb1826. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brawley C, Matunis E. Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science. 2004;304:1331–1334. doi: 10.1126/science.1097676. [DOI] [PubMed] [Google Scholar]
- Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, Winton DJ. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- Cai Y, Chia W, Yang X. A family of snail-related zinc finger proteins regulates two distinct and parallel mechanisms that mediate Drosophila neuroblast asymmetric divisions. The EMBO journal. 2001;20:1704–1714. doi: 10.1093/emboj/20.7.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Tiyaboonchai A, Yamashita YM, Hunt AJ. Asymmetric division of cyst stem cells in Drosophila testis is ensured by anaphase spindle repositioning. Development. 2011;138:831–837. doi: 10.1242/dev.057901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinardo S, Okegbe T, Wingert L, Freilich S, Terry N. lines and bowl affect the specification of cyst stem cells and niche cells in the Drosophila testis. Development. 2011;138:1687–1696. doi: 10.1242/dev.057364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CJ, Olson JM, Ngo KT, Kim E, Lee NE, Kuoy E, Patananan AN, Sitz D, Tran P, Do MT, et al. G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat Methods. 2009;6:603–605. doi: 10.1038/nmeth.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty MS, Salis P, Evans CJ, Ekas LA, Marouf A, Zavadil J, Banerjee U, Bach EA. chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation, and stem cell self-renewal in Drosophila. Dev Cell. 2010;18:556–568. doi: 10.1016/j.devcel.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller MT. Spermatogenesis. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Fuse N, Hirose S, Hayashi S. Diploidy of Drosophila imaginal cells is maintained by a transcriptional repressor encoded by escargot. Genes & development. 1994;8:2270–2281. doi: 10.1101/gad.8.19.2270. [DOI] [PubMed] [Google Scholar]
- Gönczy P, DiNardo S. The germ line regulates somatic cyst cell proliferation and fate during Drosophila spermatogenesis. Development. 1996;122:2437–2447. doi: 10.1242/dev.122.8.2437. [DOI] [PubMed] [Google Scholar]
- Gönczy P, Viswanathan S, DiNardo S. Probing spermatogenesis in Drosophila with P-element enhancer detectors. Development. 1992;114:89–98. doi: 10.1242/dev.114.1.89. [DOI] [PubMed] [Google Scholar]
- Graf T. Historical origins of transdifferentiation and reprogramming. Cell Stem Cell. 2011;9:504–516. doi: 10.1016/j.stem.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Hemavathy K, Hu X, Ashraf SI, Small SJ, Ip YT. The repressor function of snail is required for Drosophila gastrulation and is not replaceable by Escargot or Worniu. Developmental biology. 2004;269:411–420. doi: 10.1016/j.ydbio.2004.01.029. [DOI] [PubMed] [Google Scholar]
- Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- Hsu YC, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M, Yuan H, Salzmann V, Fuller MT, Yamashita YM. E-cadherin is required for centrosome and spindle orientation in Drosophila male germline stem cells. PLoS One. 2010;5:e12473. doi: 10.1371/journal.pone.0012473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issigonis M, Tulina N, De Cuevas M, Brawley C, Sandler L, Matunis E. JAK-STAT Signal Inhibition Regulates Competition in the Drosophila Testis Stem Cell Niche. Science. 2009;326:153–156. doi: 10.1126/science.1176817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DL, Wagers AJ. No place like home: anatomy and function of the stem cell niche. Nature reviews. 2008;9:11–21. doi: 10.1038/nrm2319. [DOI] [PubMed] [Google Scholar]
- Kai T, Spradling A. Differentiating germ cells can revert into functional stem cells in Drosophila melanogaster ovaries. Nature. 2004;428:564–569. doi: 10.1038/nature02436. [DOI] [PubMed] [Google Scholar]
- Kawase E, Wong MD, Ding BC, Xie T. Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development. 2004;131:1365–1375. doi: 10.1242/dev.01025. [DOI] [PubMed] [Google Scholar]
- Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- Kiger AA, White-Cooper H, Fuller MT. Somatic support cells restrict germline stem cell self-renewal and promote differentiation. Nature. 2000;407:750–754. doi: 10.1038/35037606. [DOI] [PubMed] [Google Scholar]
- King RS, Newmark PA. The cell biology of regeneration. J Cell Biol. 2012;196:553–562. doi: 10.1083/jcb.201105099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakakis P, Tipping M, Abed L, Veraksa A. Tandem affinity purification in Drosophila: The advantages of the GS-TAP system. Fly. 2008;2 doi: 10.4161/fly.6669. [DOI] [PubMed] [Google Scholar]
- Landais S, D’Alterio C, Jones DL. Persistent replicative stress alters Polycomb phenotypes and tissue homeostasis in Drosophila melanogaster. doi: 10.1016/j.celrep.2014.03.042. In press, online publication date Apr. 17, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bras S, Van Doren M. Development of the male germline stem cell niche in Drosophila. Dev Biol. 2006;294:92–103. doi: 10.1016/j.ydbio.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Leatherman JL, Dinardo S. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell. 2008;3:44–54. doi: 10.1016/j.stem.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherman JL, Dinardo S. Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat Cell Biol. 2010;12:806–811. doi: 10.1038/ncb2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Li MA, Alls JD, Avancini RM, Koo K, Godt D. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nature cell biology. 2003;5:994–1000. doi: 10.1038/ncb1058. [DOI] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Michel M, Kupinski AP, Raabe I, Bokel C. Hh signalling is essential for somatic stem cell maintenance in the Drosophila testis niche. Development. 2012;139:2663–2669. doi: 10.1242/dev.075242. [DOI] [PubMed] [Google Scholar]
- Michel M, Raabe I, Kupinski AP, Perez-Palencia R, Bokel C. Local BMP receptor activation at adherens junctions in the Drosophila germline stem cell niche. Nat Commun. 2011;2:415. doi: 10.1038/ncomms1426. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Sharma M, Nabeshima Y, Braun RE, Yoshida S. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science. 2010;328:62–67. doi: 10.1126/science.1182868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Papp B, Plath K. Epigenetics of reprogramming to induced pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D, Bergman M, Aihara H, Nibu Y, Mannervik M. Drosophila Ebi mediates Snail-dependent transcriptional repression through HDAC3-induced histone deacetylation. EMBO J. 2008;27:898–909. doi: 10.1038/emboj.2008.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende LP, Boyle M, Tran D, Fellner T, Jones DL. Headcase promotes cell survival and niche maintenance in the Drosophila testis. PLoS One. 2013;8:e68026. doi: 10.1371/journal.pone.0068026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 1978;4:7–25. [PubMed] [Google Scholar]
- Sheng XR, Posenau T, Gumulak-Smith JJ, Matunis E, Van Doren M, Wawersik M. Jak-STAT regulation of male germline stem cell establishment during Drosophila embryogenesis. Developmental biology. 2009;334:335–344. doi: 10.1016/j.ydbio.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani AA, Ingham PW. Regulation of stem cell maintenance and transit amplifying cell proliferation by tgf-beta signaling in Drosophila spermatogenesis. Current biology: CB. 2003;13:2065–2072. doi: 10.1016/j.cub.2003.10.063. [DOI] [PubMed] [Google Scholar]
- Stange DE, Koo BK, Huch M, Sibbel G, Basak O, Lyubimova A, Kujala P, Bartfeld S, Koster J, Geahlen JH, et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 2013;155:357–368. doi: 10.1016/j.cell.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A, Bernasconi L, Sergeev P, Cruz A, Steinmann-Zwicky M. mgm 1, the earliest sex-specific germline marker in Drosophila, reflects expression of the gene esg in male stem cells. Int J Dev Biol. 2002;46:159–166. [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tanaka EM, Reddien PW. The cellular basis for animal regeneration. Developmental cell. 2011;21:172–185. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata PR, Mou H, Pardo-Saganta A, Zhao R, Prabhu M, Law BM, Vinarsky V, Cho JL, Breton S, Sahay A, et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledano H, D’Alterio C, Czech B, Levine E, Jones DL. The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature. 2012a;485:605–610. doi: 10.1038/nature11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledano H, D’Alterio C, Loza-Coll M, Jones DL. Dual fluorescence detection of protein and RNA in Drosophila tissues. Nat Protoc. 2012b;7:1808–1817. doi: 10.1038/nprot.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–2549. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- Voog J, D’Alterio C, Jones DL. Multipotent somatic stem cells contribute to the stem cell niche in the Drosophila testis. Nature. 2008;454:1132–1136. doi: 10.1038/nature07173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawersik M, Milutinovich A, Casper AL, Matunis E, Williams B, Van Doren M. Somatic control of germline sexual development is mediated by the JAK/STAT pathway. Nature. 2005;436:563–567. doi: 10.1038/nature03849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C, Jones DL. Efficiency of spermatogonial dedifferentiation during aging. PLoS One. 2012;7:e33635. doi: 10.1371/journal.pone.0033635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Jones DL, Fuller MT. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science (New York, N Y. 2003;301:1547–1550. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lv X, Jiang J, Zhang L, Zhao Y. Dual roles of Hh signaling in the regulation of somatic stem cell self-renewal and germline stem cell maintenance in Drosophila testis. Cell research. 2013;23:573–576. doi: 10.1038/cr.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Wang Y, Vargas E, DiNardo S. magu is required for germline stem cell self-renewal through BMP signaling in the Drosophila testis. Dev Biol. 2011;357:202–210. doi: 10.1016/j.ydbio.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.