Abstract
Glioblastoma multiforme (GBM) tumors are one of the most deadly forms of human cancer and despite improved treatments, median survival time for the majority of patients is a dismal 12–15 months. A hallmark of these aggressive tumors is their unique ability to diffusively infiltrate normal brain tissue. To understand this behavior and successfully target the mechanisms underlying tumor progression, it is crucial to develop robust experimental ex vivo disease models. This review discusses current two-dimensional (2D) experimental models, as well as animal-based models used to examine GBM cell migration, including their advantages and disadvantages. Recent attempts to develop three-dimensional (3D) tissue engineering-inspired models and their utility in unraveling the role of microenvironment on tumor cell behaviors are also highlighted. Further, the use of 3D models to bridge the gap between 2D and animal models is explored. Finally, the broad utility of such models in the context of brain cancer research is examined.
Introduction
Glioblastoma multiforme (GBM), a central nervous system tumor derived from glial or glial-precursor cells, accounts for ∼15% of intracranial tumors and affects over 20,000 individuals annually in the United States.1–4 While their frequency is relatively low, these are among the most malignant of human cancers, and prognoses associated with this lesion are bleak.1,3,5 Despite dramatic improvements in micro-neurosurgical techniques, neuro-imaging, chemotherapy, and radiation therapy, the outcomes for patients with aggressively managed tumors still remains dismal.6 Further, it has been shown that migrating GBM cells at the leading front divide more slowly than those in the core, rendering cytotoxic chemotherapies ineffective.7,8 As a consequence of their highly infiltrative nature, recurrence can occur both locally and distantly within the brain.9 Given these factors, median survival for a patient with optimal care is ∼14 months, with many patients succumbing to their illnesses precipitously.1,3,10
Most therapeutic strategies aimed at GBMs target rapidly proliferating cells through a combination of cytotoxic therapies.11–13 Fewer attempts have been made to target GBM migration, although targeting cell migration could provide significant benefits.11 Understanding the aggressive, invasive behavior of GBMs is therefore, crucial to the development of new, precisely targeted therapeutics.14,15 A major limitation in new anti-invasive treatments is the lack of powerful in vitro experimental models predicting migration in the brain. Current models, specifically two-dimensional (2D) culture on tissue culture polystyrene (TCPS), do not adequately reproduce the complex in vivo tumor microenvironment and therefore, are poor predictors of tumor cell behavior in vivo. To gain detailed insight into GBM dispersion in brain tissue, experimental models that recapitulate both the in vivo niche and provide highly reproducible, tunable microenvironments are needed. These models would allow identification of factors that play a pivotal role in disease progression, eventually leading to novel therapeutic options with implications for cancer treatment in vivo.
This review examines recent developments in models used to study GBM migration, especially those that incorporate elements from the field of tissue engineering to approximate the tumor niche. Changes occurring in tumor versus normal brain extracellular matrix (ECM) compositions are reviewed. Then, state of the art experimental models to study GBM migration in vitro and the limitations of those models in providing reproducible, in vivo-like behaviors are presented. Further, recent attempts to develop three-dimensional (3D) models that mimic several aspects of the in vivo environment are highlighted. Finally, the potential of improved 3D tissue analogs to impact brain cancer research, as well as that of other cancers, is discussed.
The Tumor Niche: Extracellular Matrix in Glioma Versus Normal Brain
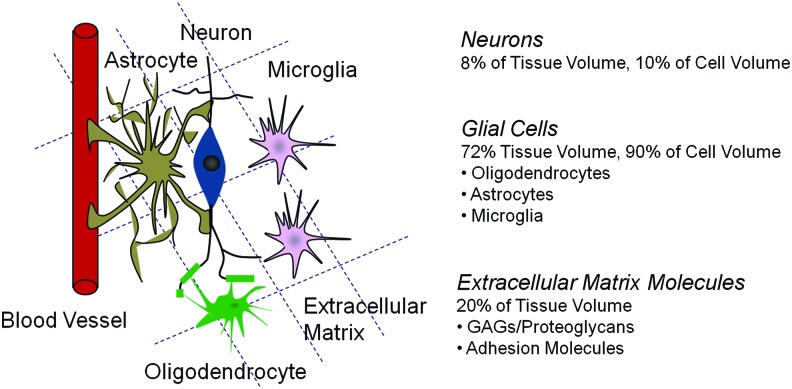
The neural ECM is the macromolecular scaffold surrounding neurons and glial cells, and is comprised of free glycosoaminoglycans (GAG), proteoglycans (PG), and glycoproteins that tether the cells. The ECM has long been recognized as an important contributor in tumorigenesis and tumor cell migration.16,17 The normal central nervous system exhibits a substantially different ECM composition compared to that of other organs. The normal human brain contains ∼20% ECM by volume (Fig. 1), which is comprised predominantly of hyaluronic acid or hyaluronan (HA), a hydrophilic, anionic glycosaminoglycan. HA interacts noncovalently with ECM PGs of the lectican family, HA-binding proteins, and tenascins18 to form the basic ECM scaffold. The primary fibrillar ECM components found in other tissues (e.g., collagens, laminin, and fibronectin) are not found in the brain parenchyma; they are instead restricted to the basal lamina of blood vessels and the subpial surface.19
FIG. 1.
Schematic of the brain microenvironment. Color images available online at www.liebertpub.com/teb
The composition of the ECM changes dramatically in gliomas. Free GAG production increases threefold,20 leading to a significant increase in the volume, tortuosity, and interstitial pressure of the extracellular space, which facilitates cell dispersion and at the same time hinders efficient drug delivery.21 Total PG composition is also altered, with significant up regulation of PGs secreted by glioma cells, such as brevican and neurocan, and marked decrease in the neuronal PG aggrecan resulting from neuronal death during tumor growth.22 Since brevican and versican are HA-binding PGs of the lectican family, their increase in glioma leads to structural changes in the ECM that favor the opening of interstitial spaces for cell motility.19 More importantly, both PGs have been shown to act as signal-triggering molecules, activating EGFR signaling, increasing integrin binding to the ECM, and promoting cell adhesion and motility.23,24 Other ECM molecules in the blood vessels and neuropil are also up- or downregulated compared to normal brain, as summarized in Table 1.18,25 It is worth noting that the mechanisms by which the structure and composition of the ECM change in gliomas compared to normal brain are very poorly understood since the regulation of ECM genes is still poorly defined. However, current evidence suggests that glioma cells themselves introduce most of the changes by secreting large amounts of neural ECM molecules, as well as other molecules that are present in mesenchymal but not in neural ECM;11,19 therefore, creating an environment with novel properties that favors tumor cell adherence, growth, and dispersion.
Table 1.
Composition of the Brain ECM: Major Components Identified in Normal Brain Versus Glioma
| Major proteins | GAGs | PGs | |
|---|---|---|---|
| Blood vessels (tumor associated) | Fibronectin, collagen, laminin, osteopontin, tenascin-C, thrombospondin-1, sparca | CSPGsb | |
| Versican | |||
| Blood vessels (normal) | Fibronectin, collagen, laminin, vitronectin, entactin | HSc | |
| Neuropil (tumor associated) | Vitronectin, osteopontin, tenascin-C, sparc | HAd ↑ | Brevican ↑ |
| CSe, HS ↑ | Versican ↑ | ||
| KSf, DSg ↑ | Aggrecan ↓ | ||
| Neuropil (normal) | No vitronectin, no tenascin-C | HA | Phosphacan |
| Neurocan | |||
| GAGs (three times lower than tumor associated neuropil) | Brevican (most abundant CSPG) | ||
| Versican |
Secreted protein acidic and rich in cysteine.
Chondroitin sulfate PG.
Heparan sulfate.
Hyaluronan/hyaluronic acid.
Chondroitin sulfate.
Keratin sulfate.
Dermatan sulfate.
ECM, extracellular matrix; GAG, glycosoaminoglycans; PG, proteoglycan.
In addition to chemical alterations in the tumor microenvironment, mechanical properties are altered. For example, clinical observations using magnetic resonance elastography have revealed that cancerous brain tissue displays different mechanical properties as compared to normal brain tissue.26–29 However, it has been difficult to conclusively determine whether the tissue becomes stiffer or softer when compared to normal tissue, which likely has repercussions in the ability of cells to metastasize away from the tumor origin. These varied observations result from a number of factors, including the heterogeneity of these tumors, changes in interstitial pressure, and secretion of several ECM components. Nonetheless, recent evidence suggests that the mechanical properties of the microenvironment can strongly influence the migration capabilities of GBM cells.30–32
Finally, the brain has a unique microanatomy that provides “migration highways” that promote tumor dissemination15 (Fig. 2). The most significant of these “highways” are the white matter tracts formed by long axons aligned and organized into myelinated bundles by oligodendrocytes. These fibrillar structures have individual fiber diameters ranging from 0.5 to 3 μm and fiber densities of ∼10,000–30,000 fibers/mm2.33,34 The largest white matter track in the human brain, the corpus callosum, connects the brain hemispheres,35 and constitutes a cellular highway for contralateral tumor dispersion.36 In addition to white matter tracts, GBM cells can also migrate along the surface of brain blood vessels and in the subpial space.9,18,37–39 Recent studies have reinforced the importance of microanatomy in GBM migration, demonstrating that GBM cells respond strongly to topographical cues.40–42 Thus, biochemical, biomechanical, and unique architectural features presented in the brain could potentially contribute to tumor cell migration. Furthermore, certain microanatomies also likely limit invasion into specific regions.
FIG. 2.
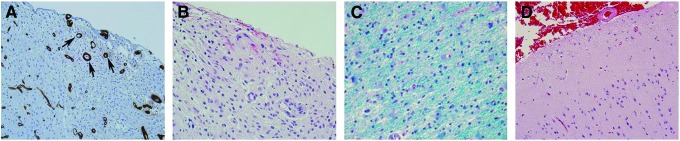
Clinical presentation of glioblastoma multiforme (GBM) tumors. Histology images of brain tissue stained using hematoxylin and eosin: (A) GBM cells seen around blood vessels (labeled brown (collagen IV) via immunoperoxidase staining and indicated by black arrows). (B) GBM cells seen below the surface of pia mater (sub-pial spread). (C) GBM cells migrating along white matter tracts (labeled blue via Luxol fast blue). (D) Histology image of a normal brain cortical tissue showing sparsely populated neurons, preserved cellularity and architectures as opposed to (A–C) that show extensive tumor cell infiltration, disruption of normal architectures, and hypercellularity. Color images available online at www.liebertpub.com/teb
Modeling Cell Migration in 2D
Invasive cell dispersion through the brain parenchyma is the hallmark of malignant gliomas, and one of their most unique properties. Malignant tumors that metastasize to the brain almost never invade soft neural tissue and grow instead as focal metastases.43 Conversely, glioma cells can disperse long distances in the central nervous system without causing clinical symptoms, but almost never metastasize to other tissues and, when implanted peripherally, form compact masses that replicate poorly the phenotype of the original tumor.44 Glioma cell migration is therefore, a complex process that requires a concerted interaction of the tumor cells with their native microenvironment, and is perhaps the most defining phenotypical property of these tumors when compared to other solid cancers. Understanding glioma dispersion provides a critical insight into the biology of these tumors and the key aspects that drive them in the neural microenvironment.
To better understand tumor cell migration, controlled environments that can recapitulate specific physical and chemical features of the tumor microenvironment are required. Unfortunately, the standard model of inquiry—2D culture on TCPS—fails rather dramatically in this regard. Here we provide specific examples of standard 2D culture models used to examine GBM cell migration, and highlight inadequacies in their ability to mimic native tumor cell behavior in vivo.
The monolayer wound-healing assay (scratch assay)
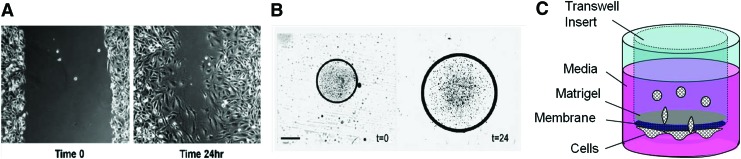
The monolayer wound-healing assay was one of the earliest methods employed to investigate GBM cell migration. In this technique,45 GBM cells are cultured until they form a confluent monolayer (Fig. 3A). A scratch is then made in the monolayer, and the time required for cells to fill the voided gap is measured. The migration rate is calculated by dividing the distance traveled by the cells over time. These assays have been routinely used to investigate the signaling pathways associated with GBM migration (e.g., PTEN, a tumor suppressor protein was shown to inhibit GBM migration through its C2 domain).46 It is important to note that the width of the “wound” often determines the migration rate. For example, cells fill larger wounds more slowly than smaller wounds.45 Although calculation of migration rates is straightforward, this assay examines GBM cell migration on 2D, rigid substrates (e.g., plastic, glass with elastic modulus (E)>∼100 kPa47), culture conditions that do not replicate the in vivo mechanical environment (E ∼ 0.1–1 kPa,47 a 2–3 order of magnitude difference), and are less physiologically relevant. Further, this model provides no topographical cues, similar to those presented by white matter tracts. Chemical cues, however, can be added by coating the TCPS with adherent molecules or by adding soluble factors to the culture medium.
FIG. 3.
State of the art cell culture models. (A) two-dimensional (2D) gap assay. (B) 2D microliter scale migration assay. (C) Transwell™ insert assay or chamber assay. Both (A, B) taken from45 are reprinted with permission from Elsevier, Inc. Copyright © 2005 Elsevier, Inc. Color images available online at www.liebertpub.com/teb
The microliter scale migration assay (radial migration assay)
Microliter-scale migration assays45 examine radial migration of GBM cells, usually in presence of ECM molecule(s) (Fig. 3B) (However, the assay can also be performed without ECM molecules on 2D TCPS.). Selected molecules are deposited on a substrate (e.g., 10-well Teflon printed microscopic slides), and the cell-containing solution is placed at the center of these wells. Radial migration of cells is then monitored by quantifying the increase in area of the circle that encompasses the cells over a stipulated time period. This assay has been used to study tumor cell migration with a variety of ECM molecules found in the brain parenchyma or blood vessel basal lamina, such as collagen type IV,48–50 laminin,48,50 vitronectin,48–51 fibronectin,48–50 merosin,49,51 tenascin,49,52 HA,49 and myelin extracts.53,54 These assays permit investigation of the potential influence of chemical cues and have allowed screening and identification of several ECM components that are permissive or nonpermissive for GBM migration, such as those listed in Table 1. However, quantification of migration area or perimeter may be biased by the presence of outlier cells that make image analysis challenging. Also, similar to the scratch assay, the Microliter scale migration assay examines migration on 2D rigid substrates that do not provide mechanical environments equivalent to those found in vivo.
Boyden chamber assays
The Boyden chamber assay examines migration of cells through a porous insert (Transwell™) in response to specific attractant or repellent cues. Cells are seeded on the top of the insert and the number of cells crossing through the membrane pores is counted. Migration can be promoted with a gradient of culture serum or other chemoattractants. The requirement for myosin II in glioma invasion was demonstrated using these assays utilizing inserts with different pore sizes. In particular, for glioma cells to squeeze through pores smaller than their nuclear diameter, both A and B isoforms of myosin-II were required.55 These results were in contrast to cells migrating on a bare surface, wherein migration proceeded without the involvement of myosin-II. However, it is important to note that the rigidity of the plastic membrane (typically made of poly(ethylene terephthalate) [PET]) may dramatically influence migration capacity.
Matrigel-modified chamber assays overcome some of the shortcomings of the “naked” Boyden chamber assay. Matrigel is an self-gelling suspension of ECM molecules derived from the Engelbreth-Holm-Swarm mouse sarcoma (a connective tissue tumor) and provides a less rigid microenvironment than TCPS.56 In this assay,45,57–61 culture inserts are first coated with Matrigel, forcing the cells to pass through the gellified Matrigel before reaching the underside of the insert (Fig. 3C). Because Matrigel forms a natural gel, it can be employed to explore the role of physiologically relevant ECM molecules (e.g., unmodified HA58,59,61), by incorporating them as an additive or as a chemoattractant in the lower chamber (e.g., type I and IV collagen, laminin, fibronectin60). Although this assay permits some modification of the mechanical environment with the addition of Matrigel and other additives, migrating cells still contact the synthetic membrane, which can influence results substantially. Furthermore, consisting primarily of laminin, entactin, and collagen-IV,62 Matrigel does not adequately recapitulate the composition of brain ECM, which lacks these proteins. In addition, Boyden chamber assays are end point-based, and therefore, fail to provide dynamic information on migration patterns of tumor cells. For example, one has to incorporate additional methods to distinguish cell proliferation from cell migration in static end-point assays that rely on counting cell numbers to calculate migration parameters.
Animal and Animal-Derived Models
Given the deficiencies of 2D culture and the ultimate goal of understanding tumor behavior in vivo, several animal-derived models for studying GBM migration ex vivo have been developed. While these assays are arguably the closest possible approximation to the conditions found in patients, disadvantages may include substantial animal to animal variation, inability to control the tumor microenvironment, and the costs and scalability issues involved with animal studies. Here we discuss the most commonly used models and their applications, as well as their limitations.
Brain slice assays
In this method,37,45,63–65 brain tissue slices (100–300 μm thick) are obtained using a vibratome and cultured on porous inserts. Fluorescent glioma cells are then placed on slices, and their dispersion is observed using wide-field or confocal fluorescence microscopy. Cell invasion is quantified as a function of distance from the initial seeding point over time. Using dynamic time lapse microscopy, brain slice assays have demonstrated that glioma cells migrate along blood vessels within slices in vitro.66 Further, these assays have been applied for identification of genes differentially regulated in glioma invasion.67
Brain slice assays have many positive features, most notably the fact that they model the tissue microenvironment closely and tumor cell invasion in slices largely reproduces the behavior of the cells in vivo.68 However, they also have limitations. These models are time consuming, requiring 3–7 days for completion, and are impractical to scale up as screening assays. In addition, the viability of normal brain cells decays rapidly over time, while some resident cell populations (such as microglia) can become highly proliferative, affecting tumor cell behavior. Additionally, animal to animal variation may necessitate a considerable number of replicates to overcome experimental variability. The major limitation of these assays, however, is that they fail to provide fine investigator control of the local environment for systematic study of the influence of chemistry and mechanics on cell behaviors.
Confrontational tissue assays
Confrontational assays69–73 assess the migration of tumor cells in the interfacial environment between tumor and normal brain by culturing normal and cancerous brain tissue in close proximity (e.g., one tissue on top of the other or side by side). Other versions of this assay employ tumor spheroids in confrontation with normal brain tissue or use normal brain cell aggregates in confrontation with tumor cells. The infiltration of tumor cells into normal tissue is then investigated via microscopy. Confrontational assays have declined in popularity because of their limited utility and present shortcomings similar to those of the brain slice assay.
Tumor xenograft models
Examination of tumor invasion directly in animal models is the most physiologically relevant standard because cells can be embedded in their native microenvironment. In these models, tumor cells are implanted into the brain and their progression over time is monitored by imaging techniques or longitudinal histology.74–78 Although quantification of invasion in tissue sections is usually not straightforward, there are several standards that can be applied in these models, such as quantification of “islets” of cells away from the core mass and measurement of the distance of these islets to the central tumor core.79,80
These models are assumed to reproduce adequately the tumor microenvironment and can be employed to study the spatiotemporal tumor distribution (i.e., the different anatomical structures that favor tumor cell dispersion in vivo) and even dynamics of tumor cell migration (e.g., intracellular cytoskeletal organization81). These models have enabled identification of human brain tumor initiating cells for the first time.82 Moreover, xenograft models are routinely utilized to investigate the in vivo efficacy of individual targeted therapies (e.g., antivascular endothelial growth factor [VEGF] treatment83), chemotherapies (e.g., Carmustine or bis-chloroethylnitrosourea [BCNU]),84 as well as combination therapies (e.g., angiostatin, an angiogenesis inhibitor and ionizing radiation85).
Unfortunately, animal studies present significant downsides, such as animal-to-animal variability, high cost, challenges in live imaging, and impractical scale up for screening assays. Animal studies also fail to provide control of the local environment where the tumor grows and invades, thereby making reliable studies of specific interactions (e.g., tumor cell response to modulus or soluble factors) impossible.
Genetically engineered models
In these models, the tumor is formed spontaneously in the brain of the animal (i.e., mouse).86 This is achieved by inducing genetic alterations seen in human tumors into mice (either via germ-line or somatic modifications87–89). One of first genetically engineered models for glioblastomas involved mutations in two tumor-suppressor genes: Nf1 and Trp53.90 Since then, several models have been developed and are reviewed in detail elsewhere.86–88 Genetically engineered models offer several advantages, including the ability to model key early events in the evolution of the tumor, as well as identification of molecular pathways and oncogenes involved in tumor initiation and progression.86,91 However, the relatively long time period needed for tumors to develop combined with tumor heterogeneity in terms of low tumor penetrance, location, and growth rate complicates the use of such models.92,93
Making the Transition to 3D: Bridging the Gap Between 2D and Animal Models
While most 2D models can be executed rapidly and easily, they fail to recapitulate the highly complex, 3D environment of in vivo niches. Further, it is now widely recognized that 2D cell culture can produce substantially different results in tumor cell behavior and signaling cascades than 3D culture.94–96 Animal models offer one possible solution, but procedures are technically complex, costly, and include additional levels of variability. Thus, researchers have begun to leverage lessons learned from tissue engineering to create well-defined 3D niches for a variety of cell types,97–100 including GBMs. These environments are created using either naturally derived or synthetic biomaterials, can recapitulate several aspects of the in vivo environment, and provide important material cues to tumor cells. Most importantly, engineered materials permit investigator control of the microenvironment, a critical feature for dissecting the multitude of factors—mechanics, chemistry, topography—which govern the biology of tumor cell migration. Not all materials are suitable for such investigations; however, the application of tissue engineering to in vitro models offers the potential to advance these studies. Here we discuss biomaterial development for recreating GBM tumor niches, the application of tissue engineering concepts to these environments, and the degree of engineering control that can be achieved.
Tissue engineering inspired models
Biomaterial platforms inspired from tissue engineering have now begun to be utilized as engineered models of cell behavior to elucidate mechanisms of tumor progression. Hydrogels and electrospun fibers, biomaterials commonly employed in tissue engineering,101,102 are the two most valuable in vitro disease models currently in use. Both models are attractive in terms of their mechanical and chemical tunability and ability to incorporate a number of cell responsive cues into the material (e.g., adhesion molecules, growth factors). Specifically, hydrogels, cross-linked polymeric biomaterials, can be engineered to mimic structural and mechanical features of brain tissue because of their similarity to GAG and PGs found in the native brain ECM. Electrospun fibers mimic fibrous structures (e.g., white matter, blood vessels) that act as highways for GBM dispersion in vivo. These materials can be composed of either natural or synthetic materials designed to closely mimic specific features of the 3D tumor microenvironment. Interactions of GBMs with several biomaterial systems are summarized in Table 2.
Table 2.
Interactions of GBMs with Natural and Synthetic Biomaterials
| Biomaterial | Cell type | Tumor cell Model | Culture type and dimensionality | Assay type | Observations | Refs. |
|---|---|---|---|---|---|---|
| Poly (methylphenyl) siloxane) | SNB-19 | S | H, 2.5D | D | Migration=f (stiffness) | 103 |
| Poly (acryl amide) coated with fibronectin | U-373 | S | H, 2.5D | D | Migration=f (stiffness) | 30 |
| U-87 | ||||||
| U-251 | ||||||
| SNB-19 | ||||||
| C6 (R) | ||||||
| PCL coated with fibronectin | X-12 | S, A | F, 2.5D | S, D | Migration=f (topography) | 40,41 |
| U-251 | ||||||
| U-87 | ||||||
| G-8, | ||||||
| G-9 (M)a | ||||||
| Gelatin-PCL | OSU-2 | S | F, 2.5D | D | Migration=f (nanofiber mechanics) | 42 |
| PCL | ||||||
| PDMS-PCL | HA reduced migration on aligned nanofibers | |||||
| PES-PCL | ||||||
| PCL-collagen | ||||||
| PCL-matrigel | ||||||
| PCL-HA | ||||||
| Matrigel | U-87 | A | H, 3D | D | Tumor exerts mechanical stress and traction on its surrounding | 108 |
| Collagen | U-87 | A | H, 3D | S | Invasion=f (collagen concentration at early time points) | 109 |
| Collagen | U-87 | A | H, 3D | S | Invasion was influenced by tissue cohesion and N-cadherin expression | 111 |
| U-373 | ||||||
| GBM 1 | ||||||
| GBM 2 | ||||||
| GBM 3 | ||||||
| GBM 4 | ||||||
| Collagen | U-87 | S | H, 3D | D | Migration=f (epidermal growth factor stimulation) | 110 |
| Collagen | C6 (R) | S, A | H, 3D | S | Migration=f (pore size) | 114 |
| Collagen-tenascin-C | U-251 | S | H, 3D | S | Presence of tenascin-C increases invasiveness | 112 |
| U-178 | ||||||
| Chitosan-alginate | U-87 | S | H, 2.5D | S | Provided an environment leading to formation of solid tumor-like cells | 115 |
| U-118 | ||||||
| C6 (R) | VEGF and MMP-2 secretion↑for human cell lines | |||||
| Collagen-agarose | U-373 | S, A | H, 3D | S, D | With increasing agarose, migration mechanism is altered and eventually abrogated | 113 |
| HA | U-87 | S | H, 2.5D | S | The extent of invasion was cell type dependent | 119 |
| U-251 | ||||||
| U-343 | ||||||
| U-373 | ||||||
| HA | CB-191 | S | H, 2.5D | S | Invasion=f (hyaluronidase activity) | 120 |
| CB-193 | ||||||
| HA | CB-191 | S | H, 2.5D | S | Invasion was influenced by adhesion molecules (collagens) and growth factors (stromal cell-derived factor-1α and basic fibroblast growth factor) | 116 |
| HA-κE | CB-74 | S | H, 2.5D | S | Invasion increased in the presence of κE, MMP-2 ↑ | 117 |
| CB-109 | ||||||
| CB-191 | ||||||
| HA-RGD | U-373 | S | H, 2.5D | S, D | Migration=f (stiffness and ligand density) | 32 |
| U-87 | A | H, 3D | ||||
| C6 (R) | ||||||
| HA-collagen | OSU-2 | S | H, 3D | D | Migration=f (HA density) | 118,121 |
| Collagen-HA | C6 (R) | A | H, 3D | S | Migration=f (CS concentration) | 122 |
| Collagen-CS | Effect of HA not significant |
Cell source is Hu unless otherwise noted in cell type column in parentheses.
Species: Hu, human; M, mice; R, rat.
Tumor cell model: S, dissociated single cells; A, tumor aggregates/spheroids.
Culture type: H, hydrogel; F, electrospun fiber.
Culture dimensionality: 2D, cells cultured on tissue culture plastic/glass; 2.5D, cells cultured on top of hydrogels/electrospun fibers; 3D, cells encapsulated in hydrogels.
Assay type: S, static, end point based; D, dynamic, time lapse imaging based.
Tumor initiating cells implanted into mice to generate tumors and these tumor explants are cultured on biomaterial surfaces.
f, functional dependence; κE, kappa-elastin; GBM, glioblastoma multiforme; VEGF, vascular endothelial growth factor; PCL, poly(ɛ-caprolactone); PDMS, poly(dimethylsiloxane); PES, poly(ethersulfone); MMP-2, matrix metalloproteinase-2.
Synthetic biomaterials
Several synthetic biomaterials have been used to study GBM cell behavior. For example, sheets of silicone rubber (poly(methylphenyl)siloxane)103 and poly(acrylamide)-based hydrogels30 have been used to elucidate the role of mechanics on glioma cell migration. These studies showed that migration is proportional to the rigidity of the underlying substrate. In particular, with poly(acrylamide)-based hydrogels,30 it was observed that as the rigidity (E ∼ 0.8 kPa) approached that of the brain tissue, migration was drastically reduced compared to that observed on stiff substrates (E ∼ 119 kPa), demonstrating the strong role played by the mechanical environment in guiding migration (e.g., Fig. 4). In these investigations, the primary goal was to modulate mechanical properties, and thus, synthetic materials were used as they offer more flexibility in the range of mechanical properties that can be obtained.
FIG. 4.
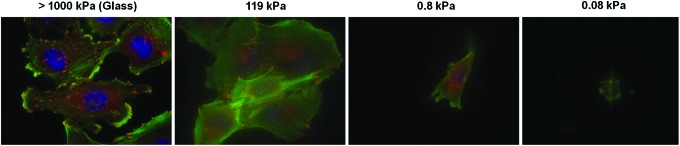
Morphology and cytoskeletal organization of U373-MG human glioblastoma tumor cells on laminin-coated glass and laminin-coated polyacrylamide hydrogels of varying stiffness. Cells were stained for F-actin (green), nuclear DNA (blue), and vinculin (red). Figure courtesy of Sophie Wong, Dr. Theresa Ulrich, and Dr. Sanjay Kumar (University of California, Berkeley; Dr. Ulrich is currently at the Massachusetts Institute of Technology). Color images available online at www.liebertpub.com/teb
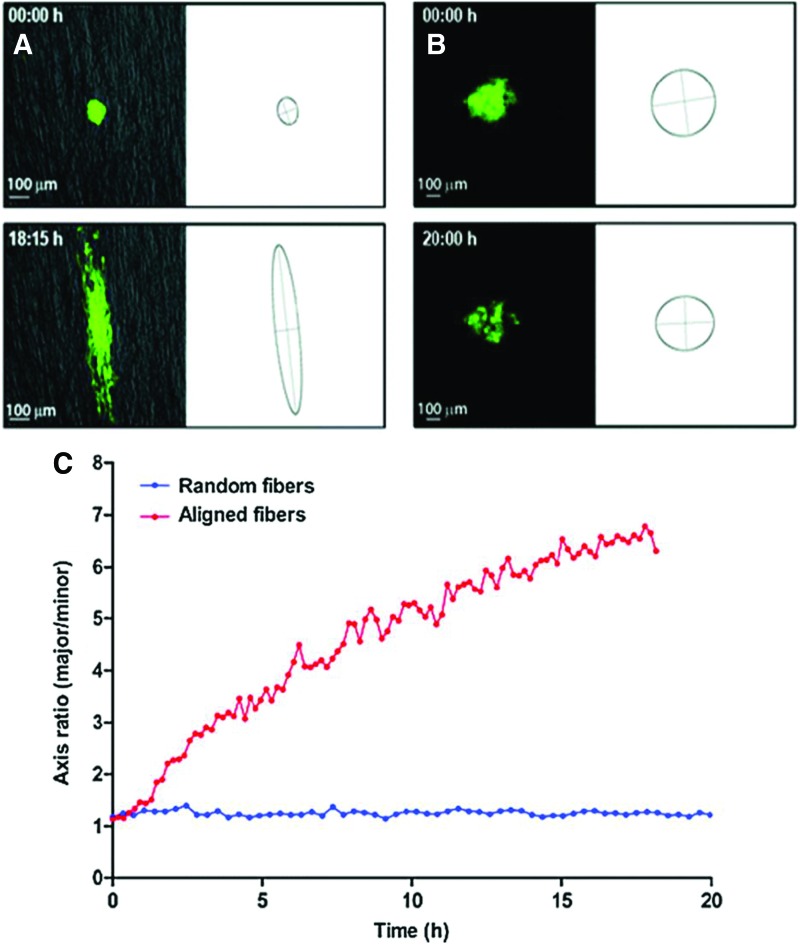
Similarly, to investigate the role of topography on GBM behaviors, synthetic electrospun fibers derived from poly(ɛ-caprolactone) (PCL) have been used. In particular, glioma migration was observed to be a strong function of substrate topography as GBM cells migrated much faster on aligned PCL fibers than on random PCL fibers40 (Fig. 5). In a separate study, the sensitivity of glioma cell migration to low concentrations of signal transducer and activator of transcription-3 (STAT-3) inhibitors was observed in cells dispersing in cultured brain slices and on aligned nanofibers, but not on TCPS,41 demonstrating the unique sensitivity to topography provided by tissue-engineering inspired materials. Furthermore, inhibition of STAT-3 did not reduce cell translocation in a Transwell migration assay at low concentrations of inhibitors, in contrast to observations on both nanofibers and brain slices. This system also recapitulated in vivo-like migratory morphologies with migration correlating to STAT-3 signaling, a known driver of cell migration in vivo.40,41,104 More recently, using aligned core-shell nanofibers (i.e., Gelatin-PCL, poly(dimethylsiloxane) [PDMS]-PCL, poly(ethersulfone) [PES]-PCL) and aligned PCL nanofibers mimicking white matter tract topography, GBM migration was shown to be a strong function of nanofiber mechanics, with a peak in migration speed seen on aligned PCL nanofibers of intermediate modulus (∼8 MPa).42 The few reports that have investigated the mechanical properties of white matter tracts primarily use atomic force microscopy techniques.105 Properties measured using this technique display values that have a similar order of magnitude to the electrospun nanofibers reported in the study. Additional studies to further understand changes in the mechanical modulus of white matter tracts in response to cancer are currently underway and could increase the physiological relevance of electrospun nanofibers studies.
FIG. 5.
GBM tumor cell dispersion from neurospheres on (A) random and (B) aligned poly(ɛ-caprolactone) (PCL) electrospun nanofibers mimicking white matter tract topography. (C) Quantification of cell dispersion as measured by a change in the ratio of elliptic axes over time. Figure taken from40 reprinted with permission from Mary Ann Liebert, Inc. Copyright © Mary Ann Liebert, Inc. Color images available online at www.liebertpub.com/teb
Synthetic materials provide user control over material properties, but unfortunately cannot fully recapitulate the complex, time-varying chemistry, including chemical and mechanical changes, of the in vivo microenvironment. In addition, although cell motility largely reproduces the cellular and molecular parameters of in vivo migration, the cells are usually not challenged by a true, 3D ECM-like system that provides a platform for cell invasion. Therefore, these materials represent an intermediate environment between 3D and 2D cultures (a “2.5D” environment). The lack of a true ECM-like barrier hampers the ability to examine the role of tumor-secreted proteases and glycanases on tumor cell migration. While there have been few studies with these materials, a dramatic increase in their use as 3D tissue analogs with the development of new biomaterial combinations is expected in the coming years primarily because of their simple preparation procedures, tunability, and reproducibility.
Natural biomaterials
Naturally derived scaffold materials are also ideal candidates for developing 3D models for tumor cell migration investigations. Standard models employed include Matrigel31,106–108 and collagen109–112 assays. These assays investigate invasion into the gel by seeding cells on the gel surface or may utilize tumor cell spheroids to examine radial migration of cells away from the tumor core. While these assays are good starting points for studying cell migration in 3D environments and continue to be used, they usually access a limited range of physicochemical properties (e.g., stiffness, ligand density) making certain tumor cell characteristics difficult to capture.
To circumvent these issues, several studies have focused on developing more advanced biomaterial models, including multicomponent and tunable systems, to isolate the role of specific factors on cell behaviors in 3D. For example, using hybrid hydrogels of collagen and agarose, GBM migration was shown to be inversely related to matrix stiffness in 3D. Further, a change in migration pattern from mesenchymal to amoeboid was also observed.113 Similarly, by exploiting the gelation dynamics of collagen gels, GBM migration was examined as a function of pore size in 3D networks.114 Migration was hindered in constructs with a small pore size (i.e., 2 μm); however, the invasion distance was not very sensitive in the pore size range from ∼5 to 12 μm. Using chitosan-alginate composite scaffolds, factors promoting tumor malignancy (i.e., VEGF, matrix metalloproteinase-2 [MMP-2]) were shown to increase in human GBM lines compared to 2D cultures.115
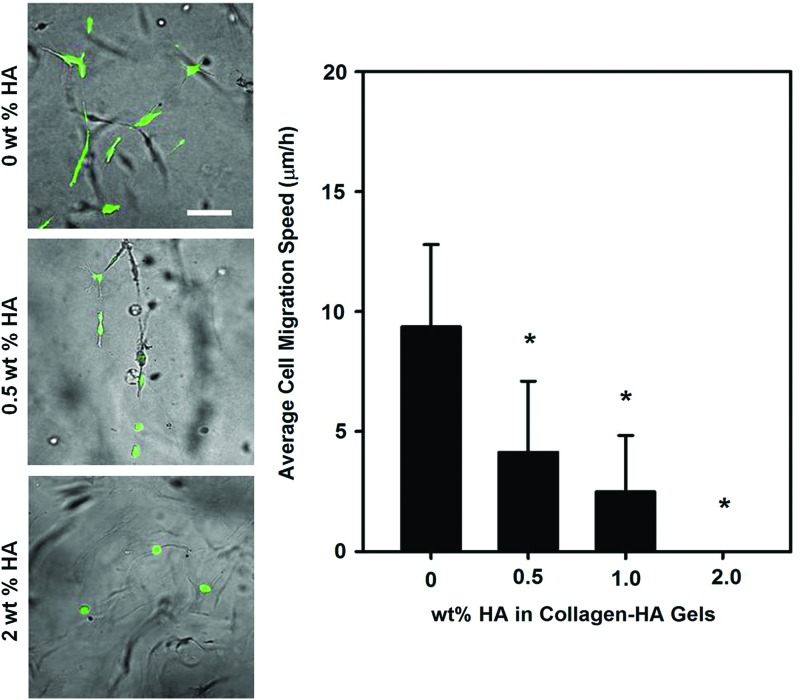
To increase physiological relevance, investigators have also employed HA as a hydrogel biomaterial, singly or with other biomolecules to create multicomponent systems. These biomaterials have been used to investigate migration capacity of GBM cells in both 2.5D and 3D cultures.32,116–121 Specifically, using gels containing HA and the RGD peptide, the migration capacity of GBM cells was shown to be a strong function of stiffness and ligand density (i.e., RGD concentrations of 0–5 mg/mL) in 2.5D culture.32 In contrast, in 3D cultures, migration was permitted only in less dense HA cultures and strongly inhibited by highly dense HA-based hydrogel cultures32,118,121 (Fig. 6). This behavior was also observed with chondroitin sulfate addition to collagen-based gels wherein migration of GBM spheroids was inhibited.122 Similarly, HA also reduced GBM migration when presented as a “shell” on a PCL “core” nanofiber versus bare PCL nanofiber in an aligned white matter topography mimetic setting.42 In addition to matrix parameters, several other factors, such as elastin-derived peptides (i.e., kappa-elastin)117 and growth factors (i.e., stromal cell-derived factor-1α and basic fibroblast growth factor),116 have been shown to increase migratory capacities of GBMs in HA-based hydrogel systems. Studies have pointed toward cell-specific differences in invading HA-based hydrogels, indicating inherent differences in cell type.119 However, it should be noted that HA by itself usually does not support cell migration. Thus, modification with additional materials is required to generate in vivo-like migration behaviors. This most likely results from a number of factors, including the dense nature of HA hydrogel with few open pores to support migration and also chemical factors (i.e., HA hydrogels do not promote cell attachment on their surfaces).
FIG. 6.
Morphology and cell migration speed of patient derived OSU-2 glioblastoma tumor cells within three-dimensional (3D) Collagen-hyaluronic acid or hyaluronan (HA) composite hydrogels of varying HA density. Cells were stained with Cell tracker Green. Scale bar indicates 100 μm. *p<0.0001 compared to 0 wt% HA (i.e., pure collagen gels). Color images available online at www.liebertpub.com/teb
Studies with naturally derived biomaterials offer several advantages.123 For example, with an increase in the development of systems that offer tunability, it is becoming easier to assess the role of competing cues (e.g., chemistry, stiffness) on tumor cell behavior. While this is also possible with synthetic biomaterials, the importance of translating these findings to the clinic using natural biomaterial systems cannot be underscored. Also, naturally derived materials can elicit specific signaling responses that can influence cell behaviors similar to those observed in vivo. However, it is also important to realize that, for natural systems, variations in composition can strongly influence experimental findings and only certain ranges of engineered mechanical and topographical properties may be accessible. In addition to natural and synthetic biomaterials, peptide and protein-based engineered ECMs124 are gaining relevance as they can combine native architectural features with physiologically relevant stiffness and could become a useful tool to investigate GBM behaviors. Similarly, microfluidic devices have begun to be utilized in both 2D and 3D culture microenvironments to investigate GBM behaviors, as they can maintain stable gradients, can incorporate flow, and provide time-sensitive resolution of migration.125–127
Compared to animal tissue-based models, results obtained using 3D in vitro biomaterial assays are more reproducible. Furthermore, these assays are inexpensive, quicker, and provide ease of execution compared to brain slice models or confrontational assays. However, recognizing that GBM behavior is extremely complex, it is likely that a combination of in vitro assays (e.g., brain slice, hydrogel, and/or fiber-based assays) will be required to fully understand tumor cell behavior. For instance, 3D cell culture models could be used to identify specific effects via high-throughput screening, which can be further explored using animal models. Detailed studies of intracellular signaling cascades and other migration regulatory pathways using existing, as well as forthcoming physiological biomaterial platforms should enable development of new therapeutic targets.
A Look to the Future: How Will We Benefit from 3D Brain Tissue Models?
2D cell culture models (e.g., monolayer cultures) are currently used to test efficacy of anticancer drugs and evaluate their potential before clinical trials.128 Despite rapid advances in high-throughput drug screening procedures, 2D assays have largely failed to effectively predict the efficacy of anti-invasive drugs, and several studies have indicated that 2D assays provide only marginal benefit in evaluating anticancer drugs.129,130 This is mainly because cells grown on 2D TCPS adapt to this artificial environment and may no longer display characteristics of the original tumor. Classical human cell lines commonly used to study glioblastoma behavior, such as U-87, U-118, and U-138, were established in the late 1960s and have been subjected to extensive adaptation to conventional culture conditions. Therefore, it is no surprise that models using these cells do not adequately predict tumor in vivo response, creating a huge scientific and financial challenge.
Biomimetic 3D models could become powerful predictors in high-throughput drug screening and drug discovery by uniquely bridging the gap between 2D and animal models in a cost effective manner,96,131 potentially reducing time to market. Further, 3D brain tissue models will also be of immense use to clinicians to investigate tumor migration rate in vitro, thereby guiding patient care and treatment decisions. For example, improved 3D tissue analogs could serve as tailored bioassays for patient-derived tumor biopsies and for testing different drug combinations for personalized medicine approaches. Further, as compared to well established cell lines, GBM tumor stem cells could become a valuable and robust model cell system as they have been demonstrated to closely recapitulate the genotype and phenotype of primary human tumors.132,133
However, there are several significant challenges remaining for 3D cultures. For example, oxygen diffusion in 3D cultures is a substantial concern, especially for long-term cultures. In addition, downstream assays, such as western blots and immunofluorescence assays are difficult to perform in 3D culture. Some of these challenges can be partially overcome. For example, to increase oxygen transport in 3D cultures, bioreactors may be employed; leveraging lessons from the tissue engineering community.134,135 Similarly, downstream assays can be performed on thin 3D slices taken from the samples via histological sectioning.
Future models will increase in complexity. While these early models are mainly designed based on the ECM of the tumor, a variety of cues found in the in vivo microenvironment can be added. For example, cell–cell interactions are also important. Investigating interaction of brain tumor cells with other resident cell types in the brain (e.g., oligodendrocytes, astrocytes, endothelium136,137) through coculture will improve understanding of the influence of these interactions on tumor progression. Further, the role of several growth factors in combination with ECM cues on tumor cell behaviors can be explored. Increasing model complexity is not without challenges; understanding the nutritional requirements and culture conditions for each specific cell type and their cocultures will be crucial.
Tissue engineering-inspired 3D biomaterial models are poised to make substantial contributions to the field of cancer biology and patient care. Brain mimetic 3D models can be used to not only predict tumor cell behaviors, but also provide a powerful tool to understand and evaluate fundamental questions in neuroscience (e.g., migration of other cell types in the brain) that are yet unexplored. For example, crucial information gained from these models may be applicable as a guide in designing scaffolds for neural tissue engineering and creating stable nerve tissue-electrode interfaces; thus, demonstrating the broad applicability of this research.
Acknowledgments
The authors would like to acknowledge Sophie Wong and Dr. Sanjay Kumar (University of California, Berkeley) and Dr. Theresa Ulrich (Massachusetts Institute of Technology) for providing Figure 4. The authors would also like to acknowledge financial support from the National Science Foundation (CBET BME 0854015, to J.O.W. and A.S. and EEC 0425626 to J.J.L.), Women in Philanthropy, OSU (to J.O.W.), the H.C. “Slip” Slider Professorship (to J.O.W.), and a Pelotonia Graduate Fellowship (to S.S.R.). Any opinions, findings, and conclusions expressed in this material are those of the author(s) and do not necessarily reflect those of the Pelotonia Fellowship Program.
Disclosure Statement
No competing financial interests exist.
References
- 1.Sarkar A., and Chiocca E.A.Glioblastoma and malignant astrocytoma. In: Kaye A.H., and Laws E.R., eds. Brain Tumors: An Encyclopedic Approach. 3rd ed. Edinburgh, New York: Churchill Livingstone, 2011, pp. 384 [Google Scholar]
- 2.Nakada M., Nakada S., Demuth T., Tran N.L., Hoelzinger D.B., and Berens M.E.Molecular targets of glioma invasion. Cell Mol Life Sci 64,458, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen P.Y., and Kesari S.Malignant gliomas in adults. N Engl J Med 359,492, 2008 [DOI] [PubMed] [Google Scholar]
- 4.Thoman W.J., Ammirati M., Caragine L.P., Jr., McGregor J.M., Sarkar A., and Chiocca E.A.Brain tumor imaging and surgical management: the neurosurgeon's perspective. Top Magn Reson Imaging 17,121, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Van Meir E.G., Hadjipanayis C.G., Norden A.D., Shu H.K., Wen P.Y., and Olson J.J.Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin 60,166, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldbrunner R.H., Bernstein J.J., and Tonn J.C.Cell-extracellular matrix interaction in glioma invasion. Acta Neurochir (Wien) 141,295, 1999 [DOI] [PubMed] [Google Scholar]
- 7.Demuth T., and Berens M.E.Molecular mechanisms of glioma cell migration and invasion. J Neurooncol 70,217, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Giese A., and Westphal M.Glioma invasion in the central nervous system. Neurosurgery 39,235, 1996 [DOI] [PubMed] [Google Scholar]
- 9.Bellail A.C., Hunter S.B., Brat D.J., Tan C., and Van Meir E.G.Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int J Biochem Cell Biol 36,1046, 2004 [DOI] [PubMed] [Google Scholar]
- 10.Sarkar A., and Chiocca E.A.Multiple craniotomies. J Neurosurg 114,574, 2011 [DOI] [PubMed] [Google Scholar]
- 11.Viapiano M.S., and Lawler S.E.Glioma invasion: mechanisms and therapeutic challenges. In: Van Meir E.G., ed. CNS Cancer: Models, Markers, Prognostic Factors, Targets, and Therapeutic Approaches. 1st ed. New York: Humana Press (Springer), 2009. pp. 1219 [Google Scholar]
- 12.Anderson E., Grant R., Lewis S.C., and Whittle I.R.Randomized Phase III controlled trials of therapy in malignant glioma: where are we after 40 years? Br J Neurosurg 22,339, 2008 [DOI] [PubMed] [Google Scholar]
- 13.Salgaller M.L., and Liau L.M.Current status of clinical trials for glioblastoma. Rev Recent Clin Trials 1,265, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Claes A., Idema A.J., and Wesseling P.Diffuse glioma growth: a guerilla war. Acta Neuropathol 114,443, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louis D.N.Molecular pathology of malignant gliomas. Annu Rev Pathol 1,97, 2006 [DOI] [PubMed] [Google Scholar]
- 16.Rosenblum M.L., Berens M.E., and Rutka J.T.Recent perspectives in brain tumor biology and treatment. Clin Neurosurg 35,314, 1989 [PubMed] [Google Scholar]
- 17.Huang S., and Ingber D.E.A non-genetic basis for cancer progression and metastasis: self-organizing attractors in cell regulatory networks. Breast Dis 26,27, 2006 [DOI] [PubMed] [Google Scholar]
- 18.Gladson C.L.The extracellular matrix of gliomas: modulation of cell function. J Neuropathol Exp Neurol 58,1029, 1999 [DOI] [PubMed] [Google Scholar]
- 19.Viapiano M.S., and Matthews R.T.From barriers to bridges: chondroitin sulfate proteoglycans in neuropathology. Trends Mol Med 12,488, 2006 [DOI] [PubMed] [Google Scholar]
- 20.Bertolotto A., Magrassi M.L., Orsi L., Sitia C., and Schiffer D.Glycosaminoglycan changes in human gliomas. A biochemical study. J Neurooncol 4,43, 1986 [DOI] [PubMed] [Google Scholar]
- 21.Vargova L., Homola A., Zamecnik J., Tichy M., Benes V., and Sykova E.Diffusion parameters of the extracellular space in human gliomas. Glia 42,77, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Sim H., Hu B., and Viapiano M.S.Reduced expression of the hyaluronan and proteoglycan link proteins in malignant gliomas. J Biol Chem 284,26547, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu B., Kong L.L., Matthews R.T., and Viapiano M.S.The proteoglycan brevican binds to fibronectin after proteolytic cleavage and promotes glioma cell motility. J Biol Chem 283,24848, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y.J., La Pierre D.P., Wu J., Yee A.J., and Yang B.B.The interaction of versican with its binding partners. Cell Res 15,483, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Newton H.B.Molecular neuro-oncology and the development of targeted therapeutic strategies for brain tumors. Part 3: brain tumor invasiveness. Expert Rev Anticancer Ther 4,803, 2004 [DOI] [PubMed] [Google Scholar]
- 26.Scholz M., Noack V., Pechlivanis I., Engelhardt M., Fricke B., Linstedt U., et al. Vibrography during tumor neurosurgery. J Ultrasound Med 24,985, 2005 [DOI] [PubMed] [Google Scholar]
- 27.Xu L., Lin Y., Han J.C., Xi Z.N., Shen H., and Gao P.Y.Magnetic resonance elastography of brain tumors: preliminary results. Acta Radiol 48,327, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Unsgaard G., Rygh O.M., Selbekk T., Muller T.B., Kolstad F., Lindseth F., et al. Intra-operative 3D ultrasound in neurosurgery. Acta Neurochir (Wien) 148,235, 2006 [DOI] [PubMed] [Google Scholar]
- 29.Selbekk T., Bang J., and Unsgaard G.Strain processing of intraoperative ultrasound images of brain tumours: initial results. Ultrasound Med Biol 31,45, 2005 [DOI] [PubMed] [Google Scholar]
- 30.Ulrich T.A., de Juan Pardo E.M., and Kumar S.The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res 69,4167, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao S.S., Bentil S., Dejesus J., Larison J., Hissong A., Dupaix R., et al. Inherent interfacial mechanical gradients in 3D hydrogels influence tumor cell behaviors. PLoS One 7,e35852, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ananthanarayanan B., Kim Y., and Kumar S.Elucidating the mechanobiology of malignant brain tumors using a brain matrix-mimetic hyaluronic acid hydrogel platform. Biomaterials 32,7913, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benninger Y., Colognato H., Thurnherr T., Franklin R.J., Leone D.P., Atanasoski S., et al. Beta1-integrin signaling mediates premyelinating oligodendrocyte survival but is not required for CNS myelination and remyelination. J Neurosci 26,7665, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makino M., Mimatsu K., Saito H., Konishi N., and Hashizume Y.Morphometric study of myelinated fibers in human cervical spinal cord white matter. Spine (Phila Pa 1976) 21,1010, 1996 [DOI] [PubMed] [Google Scholar]
- 35.Aboitiz F., Scheibel A.B., Fisher R.S., and Zaidel E.Fiber composition of the human corpus callosum. Brain Res 598,143, 1992 [DOI] [PubMed] [Google Scholar]
- 36.Suzuki S.O., and Iwaki T.Dynamic analysis of glioma cells: looking into “movement phenotypes”. Neuropathology 25,254, 2005 [DOI] [PubMed] [Google Scholar]
- 37.Jung S., Ackerley C., Ivanchuk S., Mondal S., Becker L.E., and Rutka J.T.Tracking the invasiveness of human astrocytoma cells by using green fluorescent protein in an organotypical brain slice model. J Neurosurg 94,80, 2001 [DOI] [PubMed] [Google Scholar]
- 38.Holland E.C.Glioblastoma multiforme: the terminator. Proc Natl Acad Sci USA 97,6242, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao J.S.Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev Cancer 3,489, 2003 [DOI] [PubMed] [Google Scholar]
- 40.Johnson J., Nowicki M.O., Lee C.H., Chiocca E.A., Viapiano M.S., Lawler S.E., et al. Quantitative analysis of complex glioma cell migration on electrospun polycaprolactone using time-lapse microscopy. Tissue Eng Part C Methods 15,531, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agudelo-Garcia P.A., De Jesus J.K., Williams S.P., Nowicki M.O., Chiocca E.A., Liyanarachchi S., et al. Glioma cell migration on three-dimensional nanofiber scaffolds is regulated by substrate topography and abolished by inhibition of STAT3 signaling. Neoplasia 13,831, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao S.S., Nelson M.T., Xue R., Dejesus J.K., Viapiano M.S., Lannutti J.J., et al. Mimicking white matter tract topography using core-shell electrospun nanofibers to examine migration of malignant brain tumors. Biomaterials 34,5181, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian A., Harris A., Piggott K., Shieff C., and Bradford R.Metastasis to and from the central nervous system—the ‘relatively protected site’. Lancet Oncol 3,498, 2002 [DOI] [PubMed] [Google Scholar]
- 44.Pilkington G.J.The paradox of neoplastic glial cell invasion of the brain and apparent metastatic failure. Anticancer Res 17,4103, 1997 [PubMed] [Google Scholar]
- 45.Valster A., Tran N.L., Nakada M., Berens M.E., Chan A.Y., and Symons M.Cell migration and invasion assays. Methods 37,208, 2005 [DOI] [PubMed] [Google Scholar]
- 46.Raftopoulou M., Etienne-Manneville S., Self A., Nicholls S., and Hall A.Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science 303,1179, 2004 [DOI] [PubMed] [Google Scholar]
- 47.Buxboim A., Rajagopal K., Brown A.E., and Discher D.E.How deeply cells feel: methods for thin gels. J Phys Condens Matter 22,194116, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berens M.E., Rief M.D., Loo M.A., and Giese A.The role of extracellular matrix in human astrocytoma migration and proliferation studied in a microliter scale assay. Clin Exp Metastasis 12,405, 1994 [DOI] [PubMed] [Google Scholar]
- 49.Giese A., Loo M.A., Rief M.D., Tran N., and Berens M.E.Substrates for astrocytoma invasion. Neurosurgery 37,294, 1995 [DOI] [PubMed] [Google Scholar]
- 50.Giese A., Rief M.D., Loo M.A., and Berens M.E.Determinants of human astrocytoma migration. Cancer Res 54,3897, 1994 [PubMed] [Google Scholar]
- 51.Giese A., Loo M.A., Tran N., Haskett D., Coons S.W., and Berens M.E.Dichotomy of astrocytoma migration and proliferation. Int J Cancer 67,275, 1996 [DOI] [PubMed] [Google Scholar]
- 52.Giese A., Loo M.A., Norman S.A., Treasurywala S., and Berens M.E.Contrasting migratory response of astrocytoma cells to tenascin mediated by different integrins. J Cell Sci 109(Pt 8),2161, 1996 [DOI] [PubMed] [Google Scholar]
- 53.Giese A., Kluwe L., Laube B., Meissner H., Berens M.E., and Westphal M.Migration of human glioma cells on myelin. Neurosurgery 38,755, 1996 [PubMed] [Google Scholar]
- 54.Amberger V.R., Hensel T., Ogata N., and Schwab M.E.Spreading and migration of human glioma and rat C6 cells on central nervous system myelin in vitro is correlated with tumor malignancy and involves a metalloproteolytic activity. Cancer Res 58,149, 1998 [PubMed] [Google Scholar]
- 55.Beadle C., Assanah M.C., Monzo P., Vallee R., Rosenfeld S.S., and Canoll P.The role of myosin II in glioma invasion of the brain. Mol Biol Cell 19,3357, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weaver V.M., Howlett A.R., Langton-Webster B., Petersen O.W., and Bissell M.J.The development of a functionally relevant cell culture model of progressive human breast cancer. Semin Cancer Biol 6,175, 1995 [DOI] [PubMed] [Google Scholar]
- 57.Amar A.P., DeArmond S.J., Spencer D.R., Coopersmith P.F., Ramos D.M., and Rosenblum M.L.Development of an in vitro extracellular matrix assay for studies of brain tumor cell invasion. J Neurooncol 20,1, 1994 [DOI] [PubMed] [Google Scholar]
- 58.Radotra B., and McCormick D.Glioma invasion in vitro is mediated by CD44-hyaluronan interactions. J Pathol 181,434, 1997 [DOI] [PubMed] [Google Scholar]
- 59.Nakagawa T., Kubota T., Kabuto M., and Kodera T.Hyaluronic acid facilitates glioma cell invasion in vitro. Anticancer Res 16,2917, 1996 [PubMed] [Google Scholar]
- 60.Chintala S.K., Gokaslan Z.L., Go Y., Sawaya R., Nicolson G.L., and Rao J.S.Role of extracellular matrix proteins in regulation of human glioma cell invasion in vitro. Clin Exp Metastasis 14,358, 1996 [DOI] [PubMed] [Google Scholar]
- 61.Koochekpour S., Pilkington G.J., and Merzak A.Hyaluronic acid/CD44H interaction induces cell detachment and stimulates migration and invasion of human glioma cells in vitro. Int J Cancer 63,450, 1995 [DOI] [PubMed] [Google Scholar]
- 62.Vukicevic S., Kleinman H.K., Luyten F.P., Roberts A.B., Roche N.S., and Reddi A.H.Identification of multiple active growth factors in basement membrane Matrigel suggests caution in interpretation of cellular activity related to extracellular matrix components. Exp Cell Res 202,1, 1992 [DOI] [PubMed] [Google Scholar]
- 63.Ohnishi T., Matsumura H., Izumoto S., Hiraga S., and Hayakawa T.A novel model of glioma cell invasion using organotypic brain slice culture. Cancer Res 58,2935, 1998 [PubMed] [Google Scholar]
- 64.Nakada M., Niska J.A., Miyamori H., McDonough W.S., Wu J., Sato H., et al. The phosphorylation of EphB2 receptor regulates migration and invasion of human glioma cells. Cancer Res 64,3179, 2004 [DOI] [PubMed] [Google Scholar]
- 65.Yoshida D., Watanabe K., Noha M., Takahashi H., Teramoto A., and Sugisaki Y.Anti-invasive effect of an anti-matrix metalloproteinase agent in a murine brain slice model using the serial monitoring of green fluorescent protein-labeled glioma cells. Neurosurgery 52,187, 2003 [DOI] [PubMed] [Google Scholar]
- 66.Farin A., Suzuki S.O., Weiker M., Goldman J.E., Bruce J.N., and Canoll P.Transplanted glioma cells migrate and proliferate on host brain vasculature: a dynamic analysis. Glia 53,799, 2006 [DOI] [PubMed] [Google Scholar]
- 67.Holtkamp N., Afanasieva A., Elstner A., van Landeghem F.K., Konneker M., Kuhn S.A., et al. Brain slice invasion model reveals genes differentially regulated in glioma invasion. Biochem Biophys Res Commun 336,1227, 2005 [DOI] [PubMed] [Google Scholar]
- 68.Palfi S., Swanson K.R., De Bouard S., Chretien F., Oliveira R., Gherardi R.K., et al. Correlation of in vitro infiltration with glioma histological type in organotypic brain slices. Br J Cancer 91,745, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Easty G.C., and Easty D.M.An organ culture system for the examination of tumor invasion. Nature 199,1104, 1963 [DOI] [PubMed] [Google Scholar]
- 70.Bjerkvig R., Laerum O.D., and Mella O.Glioma cell interactions with fetal rat brain aggregates in vitro and with brain tissue in vivo. Cancer Res 46,4071, 1986 [PubMed] [Google Scholar]
- 71.Steinsvag S.K., Laerum O.D., and Bjerkvig R.Interaction between rat glioma-cells and normal rat-brain tissue in organ-culture. J Natl Cancer Inst 74,1095, 1985 [PubMed] [Google Scholar]
- 72.Steinsvag S.K., and Laerum O.D.Transmission electron-microscopy of cocultures between normal rat-brain tissue and rat glioma-cells. Anticancer Res 5,137, 1985 [PubMed] [Google Scholar]
- 73.Engebraaten O., Bjerkvig R., Lund-Johansen M., Wester K., Pedersen P.-H., Mork S., et al. Interaction between human brain turnout biopsies and fetal rat brain tissue in vitro. Acta Neuropathol 81,130, 1990 [DOI] [PubMed] [Google Scholar]
- 74.Guillamo J.S., Lisovoski F., Christov C., Le Gu´erinel C., Defer G.L., Peschanski M., et al. Migration pathways of human glioblastoma cells xenografted into the immunosuppressed rat brain. J Neurooncol 52,205, 2001 [DOI] [PubMed] [Google Scholar]
- 75.Laws E.R., Jr., Goldberg W.J., and Bernstein J.J.Migration of human malignant astrocytoma cells in the mammalian brain: Scherer revisited. Int J Dev Neurosci 11,691, 1993 [DOI] [PubMed] [Google Scholar]
- 76.Goldberg W.J., Laws E.R., Jr., and Bernstein J.J.Individual C6 glioma cells migrate in adult rat brain after neural homografting. Int J Dev Neurosci 9,427, 1991 [DOI] [PubMed] [Google Scholar]
- 77.Bernstein J.J., Goldberg W.J., and Laws E.R., Jr.Human malignant astrocytoma xenografts migrate in rat brain: a model for central nervous system cancer research. J Neurosci Res 22,134, 1989 [DOI] [PubMed] [Google Scholar]
- 78.Bernstein J.J., Goldberg W.J., and Laws E.R., Jr.Immunohistochemistry of human malignant astrocytoma cells xenografted to rat brain: apolipoprotein E. Neurosurgery 24,541, 1989 [DOI] [PubMed] [Google Scholar]
- 79.Zhang H., Kelly G., Zerillo C., Jaworski D.M., and Hockfield S.Expression of a cleaved brain-specific extracellular matrix protein mediates glioma cell invasion In vivo. J Neurosci 18,2370, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hu B., Thirtamara-Rajamani K.K., Sim H., and Viapiano M.S.Fibulin-3 is uniquely upregulated in malignant gliomas and promotes tumor cell motility and invasion. Mol Cancer Res 7,1756, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caspani E.M., Echevarria D., Rottner K., and Small J.V.Live imaging of glioblastoma cells in brain tissue shows requirement of actin bundles for migration. Neuron Glia Biol 2,105, 2006 [DOI] [PubMed] [Google Scholar]
- 82.Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., et al. Identification of human brain tumour initiating cells. Nature 432,396, 2004 [DOI] [PubMed] [Google Scholar]
- 83.Kunkel P., Ulbricht U., Bohlen P., Brockmann M.A., Fillbrandt R., Stavrou D., et al. Inhibition of glioma angiogenesis and growth in vivo by systemic treatment with a monoclonal antibody against vascular endothelial growth factor receptor-2. Cancer Res 61,6624, 2001 [PubMed] [Google Scholar]
- 84.Houchens D.P., Ovejera A.A., Riblet S.M., and Slagel D.E.Human-brain tumor xenografts in nude-mice as a chemotherapy model. Eur J Cancer Clin On 19,799, 1983 [DOI] [PubMed] [Google Scholar]
- 85.Mauceri H.J., Hanna N.N., Beckett M.A., Gorski D.H., Staba M.J., Stellato K.A., et al. Combined effects of angiostatin and ionizing radiation in antitumour therapy. Nature 394,287, 1998 [DOI] [PubMed] [Google Scholar]
- 86.Huse J.T., and Holland E.C.Genetically engineered mouse models of brain cancer and the promise of preclinical testing. Brain Pathol 19,132, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Vries N.A., Beijnen J.H., and van Tellingen O.High-grade glioma mouse models and their applicability for preclinical testing. Cancer Treat Rev 35,714, 2009 [DOI] [PubMed] [Google Scholar]
- 88.Schmid R.S., Vitucci M., and Miller C.R.Genetically engineered mouse models of diffuse gliomas. Brain Res Bull 88,72, 2012 [DOI] [PubMed] [Google Scholar]
- 89.Rankin S.L., Zhu G., and Baker S.J.Review: insights gained from modelling high-grade glioma in the mouse. Neuropathol Appl Neurobiol 38,254, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reilly K.M., Loisel D.A., Bronson R.T., McLaughlin M.E., and Jacks T.Nf1;Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nat Genet 26,109, 2000 [DOI] [PubMed] [Google Scholar]
- 91.Hambardzumyan D., Parada L.F., Holland E.C., and Charest A.Genetic modeling of gliomas in mice: new tools to tackle old problems. Glia 59,1155, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fomchenko E.I., and Holland E.C.Mouse models of brain tumors and their applications in preclinical trials. Clin Cancer Res 12,5288, 2006 [DOI] [PubMed] [Google Scholar]
- 93.Becher O.J., and Holland E.C.Genetically engineered models have advantages over xenografts for preclinical studies. Cancer Res 66,3355, 2006 [DOI] [PubMed] [Google Scholar]
- 94.Cukierman E., Pankov R., Stevens D.R., and Yamada K.M.Taking cell-matrix adhesions to the third dimension. Science 294,1708, 2001 [DOI] [PubMed] [Google Scholar]
- 95.Webb D.J., and Horwitz A.F.New dimensions in cell migration. Nat Cell Biol 5,690, 2003 [DOI] [PubMed] [Google Scholar]
- 96.Yamada K.M., and Cukierman E.Modeling tissue morphogenesis and cancer in 3D. Cell 130,601, 2007 [DOI] [PubMed] [Google Scholar]
- 97.Hutmacher D.W., Loessner D., Rizzi S., Kaplan D.L., Mooney D.J., and Clements J.A.Can tissue engineering concepts advance tumor biology research? Trends Biotechnol 28,125, 2010 [DOI] [PubMed] [Google Scholar]
- 98.Burdett E., Kasper F.K., Mikos A.G., and Ludwig J.A.Engineering tumors: a tissue engineering perspective in cancer biology. Tissue Eng Part B Rev 16,351, 2010 [DOI] [PubMed] [Google Scholar]
- 99.Tibbitt M.W., and Anseth K.S.Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol Bioeng 103,655, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Griffith L.G., and Swartz M.A.Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol 7,211, 2006 [DOI] [PubMed] [Google Scholar]
- 101.Peppas N.A., Hilt J.Z., Khademhosseini A., and Langer R.Hydrogels in biology and medicine: from molecular principles to bionanotechnology. Adv Mater 18,1345, 2006 [Google Scholar]
- 102.Pham Q.P., Sharma U., and Mikos A.G.Electrospinning of polymeric nanofibers for tissue engineering applications: a review. Tissue Eng 12,1197, 2006 [DOI] [PubMed] [Google Scholar]
- 103.Thomas T.W., and DiMilla P.A.Spreading and motility of human glioblastoma cells on sheets of silicone rubber depend on substratum compliance. Med Biol Eng Comput 38,360, 2000 [DOI] [PubMed] [Google Scholar]
- 104.Devarajan E., and Huang S.STAT3 as a central regulator of tumor metastases. Curr Mol Med 9,626, 2009 [DOI] [PubMed] [Google Scholar]
- 105.Heredia A., Bui C.C., Suter U., Young P., and Schaffer T.E.AFM combines functional and morphological analysis of peripheral myelinated and demyelinated nerve fibers. Neuroimage 37,1218, 2007 [DOI] [PubMed] [Google Scholar]
- 106.Bernstein J.J., Goldberg W.J., and Laws E.R., Jr.Migration of fresh human malignant astrocytoma cells into hydrated gel wafers in vitro. J Neurooncol 18,151, 1994 [DOI] [PubMed] [Google Scholar]
- 107.Bernstein J.J., Laws E.R., Jr., Levine K.V., Wood L.R., Tadvalkar G., and Goldberg W.J.C6 glioma-astrocytoma cell and fetal astrocyte migration into artificial basement membrane: a permissive substrate for neural tumors but not fetal astrocytes. Neurosurgery 28,652, 1991 [DOI] [PubMed] [Google Scholar]
- 108.Gordon V.D., Valentine M.T., Gardel M.L., Andor-Ardo D., Dennison S., Bogdanov A.A., et al. Measuring the mechanical stress induced by an expanding multicellular tumor system: a case study. Exp Cell Res 289,58, 2003 [DOI] [PubMed] [Google Scholar]
- 109.Kaufman L.J., Brangwynne C.P., Kasza K.E., Filippidi E., Gordon V.D., Deisboeck T.S., et al. Glioma expansion in collagen I matrices: analyzing collagen concentration-dependent growth and motility patterns. Biophys J 89,635, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim H.D., Guo T.W., Wu A.P., Wells A., Gertler F.B., and Lauffenburger D.A.Epidermal growth factor-induced enhancement of glioblastoma cell migration in 3D arises from an intrinsic increase in speed but an extrinsic matrix- and proteolysis-dependent increase in persistence. Mol Biol Cell 19,4249, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hegedus B., Marga F., Jakab K., Sharpe-Timms K.L., and Forgacs G.The interplay of cell-cell and cell-matrix interactions in the invasive properties of brain tumors. Biophys J 91,2708, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sarkar S., Nuttall R.K., Liu S.H., Edwards D.R., and Yong V.W.Tenascin-C stimulates glioma cell invasion through matrix metalloproteinase-12. Cancer Res 66,11771, 2006 [DOI] [PubMed] [Google Scholar]
- 113.Ulrich T.A., Jain A., Tanner K., MacKay J.L., and Kumar S.Probing cellular mechanobiology in three-dimensional culture with collagen-agarose matrices. Biomaterials 31,1875, 2010 [DOI] [PubMed] [Google Scholar]
- 114.Yang Y.L., Motte S., and Kaufman L.J.Pore size variable type I collagen gels and their interaction with glioma cells. Biomaterials 31,5678, 2010 [DOI] [PubMed] [Google Scholar]
- 115.Kievit F.M., Florczyk S.J., Leung M.C., Veiseh O., Park J.O., Disis M.L., et al. Chitosan-alginate 3D scaffolds as a mimic of the glioma tumor microenvironment. Biomaterials 31,5903, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.David L., Dulong V., Coquerel B., Le Cerf D., Cazin L., Lamacz M., et al. Collagens, stromal cell-derived factor-1alpha and basic fibroblast growth factor increase cancer cell invasiveness in a hyaluronan hydrogel. Cell Prolif 41,348, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Coquerel B., Poyer F., Torossian F., Dulong V., Bellon G., Dubus I., et al. Elastin-derived peptides: matrikines critical for glioblastoma cell aggressiveness in a 3-D system. Glia 57,1716, 2009 [DOI] [PubMed] [Google Scholar]
- 118.Rao S.S., Dejesus J., Sarkar A., and Winter J.O.Brain mimetic hydrogels for investigating migration of glioblastoma multiformes in 3D. Transactions of the 35th Annual Meeting of Society for Biomaterials XXXIII, Society for Biomaterials, Mt. Laurel, NJ, 2011, pp. 671 [Google Scholar]
- 119.Jin S.G., Jeong Y.I., Jung S., Ryu H.H., Jin Y.H., and Kim I.Y.The effect of hyaluronic Acid on the invasiveness of malignant glioma cells: comparison of invasion potential at hyaluronic acid hydrogel and matrigel. J Korean Neurosurg Soc 46,472, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.David L., Dulong V., Le Cerf D., Chauzy C., Norris V., Delpech B., et al. Reticulated hyaluronan hydrogels: a model for examining cancer cell invasion in 3D. Matrix Biol 23,183, 2004 [DOI] [PubMed] [Google Scholar]
- 121.Rao S., DeJesus J., Short A.R., Otero J.J., Sarkar A., Winter J.O.Glioblastoma behaviors in 3D collagen-hyaluronan composite hydrogels. ACS Appl Mater Interfaces 5,9276, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang Y.L., Sun C., Wilhelm M.E., Fox L.J., Zhu J.L., and Kaufman L.J.Influence of chondroitin sulfate and hyaluronic acid on structure, mechanical properties, and glioma invasion of collagen I gels. Biomaterials 32,7932, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nyga A., Cheema U., and Loizidou M.3D tumour models: novel in vitro approaches to cancer studies. J Cell Commun Signal 5,239, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Romano N.H., Sengupta D., Chung C., and Heilshorn S.C.Protein-engineered biomaterials: nanoscale mimics of the extracellular matrix. Biochim Biophys Acta 1810,339, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huang Y., Agrawal B., Sun D., Kuo J.S., and Williams J.C.Microfluidics-based devices: new tools for studying cancer and cancer stem cell migration. Biomicrofluidics 5,13412, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chung S., Sudo R., Mack P.J., Wan C.R., Vickerman V., and Kamm R.D.Cell migration into scaffolds under co-culture conditions in a microfluidic platform. Lab Chip 9,269, 2009 [DOI] [PubMed] [Google Scholar]
- 127.Lee K.H., Lee J., Choi H., Lee D., Park Y., and Lee S.H.Integration of microfluidic chip with biomimetic hydrogel for 3D controlling and monitoring of cell alignment and migration. J Biomed Mater Res A, 2013 [DOI] [PubMed] [Google Scholar]
- 128.Shoemaker R.H.The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer 6,813, 2006 [DOI] [PubMed] [Google Scholar]
- 129.Voskoglou-Nomikos T., Pater J.L., and Seymour L.Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin Cancer Res 9,4227, 2003 [PubMed] [Google Scholar]
- 130.Johnson J.I., Decker S., Zaharevitz D., Rubinstein L.V., Venditti J.M., Schepartz S., et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br J Cancer 84,1424, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pampaloni F., Reynaud E.G., and Stelzer E.H.The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol 8,839, 2007 [DOI] [PubMed] [Google Scholar]
- 132.Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N.M., et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9,391, 2006 [DOI] [PubMed] [Google Scholar]
- 133.Hanahan D., and Weinberg R.A.Hallmarks of cancer: the next generation. Cell 144,646, 2011 [DOI] [PubMed] [Google Scholar]
- 134.Martin I., Wendt D., and Heberer M.The role of bioreactors in tissue engineering. Trends Biotechnol 22,80, 2004 [DOI] [PubMed] [Google Scholar]
- 135.Ratcliffe A., and Niklason L.E.Bioreactors and bioprocessing for tissue engineering. Ann N Y Acad Sci 961,210, 2002 [DOI] [PubMed] [Google Scholar]
- 136.Oellers P., Schallenberg M., Stupp T., Charalambous P., Senner V., Paulus W., et al. A coculture assay to visualize and monitor interactions between migrating glioma cells and nerve fibers. Nat Protoc 4,923, 2009 [DOI] [PubMed] [Google Scholar]
- 137.Oellers P., Schroer U., Senner V., Paulus W., and Thanos S.ROCKs are expressed in brain tumors and are required for glioma-cell migration on myelinated axons. Glia 57,499, 2009 [DOI] [PubMed] [Google Scholar]