Summary
In mammals, a cell’s decision to divide is thought to be under the control of the Rb/E2F pathway. We previously found that inactivation of the Rb family of cell cycle inhibitors (Rb, p107, and p130) in quiescent liver progenitors leads to uncontrolled division and cancer initiation. Here we show that, in contrast, deletion of the entire Rb gene family in mature hepatocytes is not sufficient for their long-term proliferation. The cell cycle block in Rb family mutant hepatocytes is independent of the Arf/p53/p21 checkpoint but can be abrogated upon decreasing liver size. At the molecular level, we identify YAP, a transcriptional regulator involved in organ size control, as a factor required for the sustained expression of cell cycle genes in hepatocytes. These experiments identify a novel, higher level of regulation of the cell cycle in vivo, in which signals regulating organ size are dominant regulators of the core cell cycle machinery.
Introduction
A cell’s decision to continue cycling, exit the cell cycle, or start proliferating is paramount to embryonic development, homeostasis in the adult organism, and cancer development. Extra- and intracellular signals that govern the cell cycle are integrated and relayed by the Rb pathway. Under cytostatic conditions, Rb and its family members p107 and p130 block cell cycle progression in G0/G1, in large part by interacting with E2F transcription factors to repress the expression of cell cycle genes. Phosphorylation by Cyclin/Cdk complexes in response to mitogens functionally inactivates Rb family members and results in increased E2F activity and cell cycle progression. Rb pathway members have also been involved in many other biological processes, including differentiation, survival, chromosomal stability, metabolism, and senescence. Alterations in members of the Rb pathway are found in the vast majority of human tumors, underscoring the central role of this pathway in the biology of mammalian cells and the maintenance of homeostasis (reviewed in (Dick and Rubin, 2013; Manning and Dyson, 2012; Nicolay and Dyson, 2013)).
A notable feature of the cell cycle machinery in mammals is a high level of functional redundancy. Accordingly, while overexpression of one Rb family member is often sufficient to promote cell cycle arrest (or cell death), inactivation of only one family protein often results in limited long-term cell cycle defects. For instance, acute loss of Rb function can only partly abrogate quiescence, in part because of a compensatory transcriptional up-regulation of p107 (Sage et al., 2003). In contrast, combined inactivation of Rb family members leads to increased proliferation and, eventually, complete loss of the G1 checkpoint in culture (see for example (Dannenberg et al., 2000; Sage et al., 2000)). Similarly, while Rb mutant mice have a limited tumor spectrum, inactivation of the Rb family results in long-term ectopic proliferation and is often followed by neoplastic transformation in mice (see for example (Dannenberg et al., 2004; McEvoy et al., 2011; Robanus-Maandag et al., 1998; Viatour et al., 2011)).
The liver of mammals has a remarkable regenerative ability compared to other adult organs and tissues. Hepatocytes are capable of proliferation and growth in response to liver damage, including partial hepatectomy. Under certain conditions, liver progenitor cells with the capacity of generating both hepatocytes and bile duct cells are believed to contribute to regeneration in the adult liver (reviewed in (Duncan et al., 2009)). Thus, with a large number of cells having retained the capacity to divide, the adult liver is a system of choice to explore the mechanisms regulating cell cycle progression in vivo.
Here we investigated the consequences of in vivo abrogation of the Rb pathway in the liver of adult mice. We found that the control of cell cycle is different in hepatocytes and their progenitors and identified interactions between the Rb/E2F pathway and organ size sensing mechanisms that are critical for the long-term proliferative potential of hepatocytes in vivo.
Results
Acute loss of the Rb family results in transient proliferation in mature hepatocytes
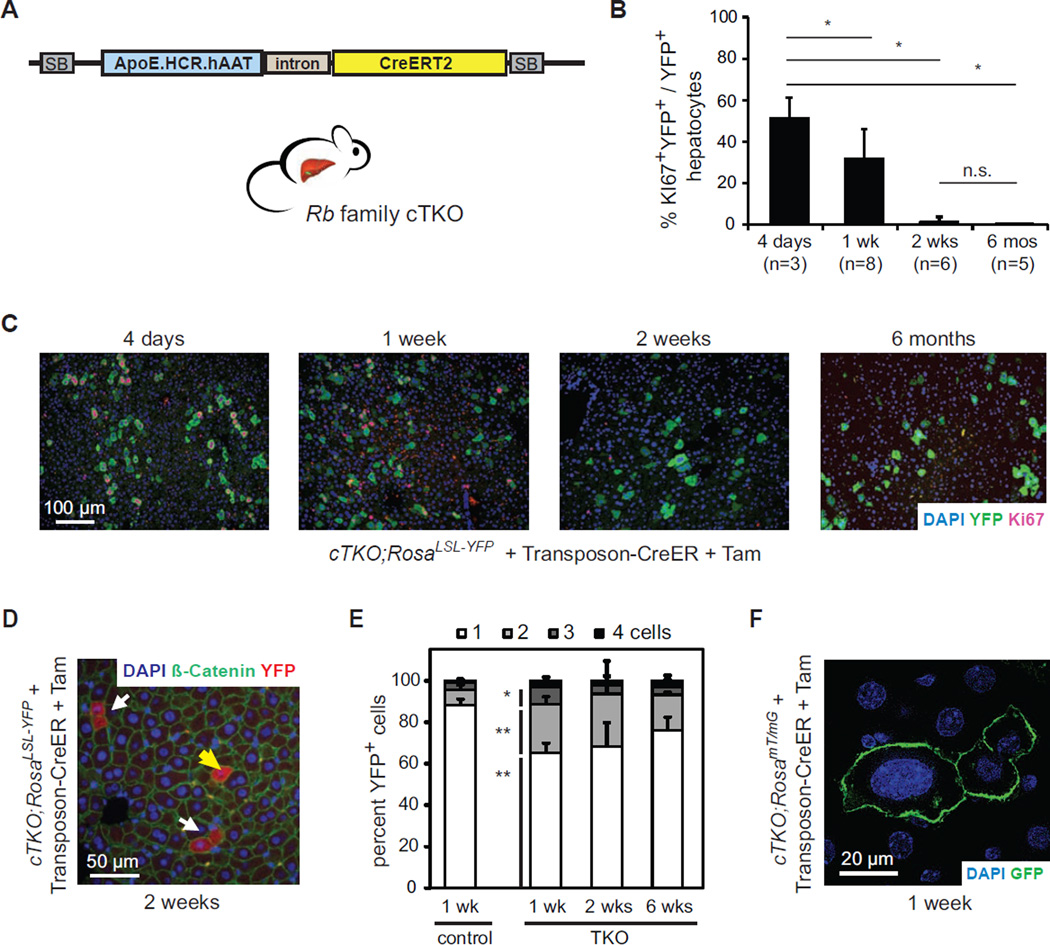
To delete Rb family genes in mature hepatocytes, we constructed a Sleeping Beauty transposon that harbors a tamoxifen (Tam)-inducible Cre recombinase (CreER) under the control of a hepatocyte-specific promoter (Figure 1A). This transposon stably integrates into a low percentage of liver cells following hydrodynamic tail vein injection. We transfected Rb family conditional triple knock-out hepatocytes (cTKO, Rblox/lox; p130lox/lox; p107−/−) with this Transposon-CreER vector. After two weeks of recovery, mice were injected with Tam and TKO hepatocytes were tracked using a Cre-inducible fluorescent reporter. Deletion of Rb family genes initially resulted in cell cycle entry but TKO hepatocytes rapidly and stably exited the cell cycle (Figures 1B and 1C). cTKO hepatocytes remained quiescent after Tam injection (YFP− cells in Figure 1C).
Figure 1. Rb pathway inactivation leads to transient cell cycle entry in hepatocytes.
(A)Schematic representation of the Sleeping Beauty (SB) Transposon-CreER system. ApoE.HCR.hAAT drives the expression of CreERT2 specifically in hepatocytes. (B and C) Acute Cre-mediated Rb gene family deletion in adult hepatocytes leads to cell cycle entry followed by cell cycle arrest. (B) Quantification and (C) representative images of Ki67+ cells (red) after Cre activation in transfected hepatocytes (YFP+, green) in cTKO;RosaLSL-YFP mice. Nuclei are visualized by DNA staining (DAPI, blue). *, p<0.05; n.s., not significant. (D and E) Quantification of single (yellow arrow) and adjacent (white arrows) YFP+ (red) hepatocytes in Transposon-CreER-injected RosaLSL-YFP (control) or cTKO;RosaLSL-YFP (TKO) mice at different time points after Tam. Cell membranes are visualized by immunostaining for β-catenin (green). Significances are shown between control and TKO cells 1 week after Tam. *, p<0.05; **, p<0.01. (F) Representative example of enlarged nuclei (DAPI, blue) in TKO hepatocytes (GFP+, green) in Transposon-CreER-injected cTKO;RosamTmG mice. Data in (B) and (E) are represented as mean +/− SD.
We developed three other systems to delete the Rb gene family in adult hepatocytes: intra-splenic injections of Ad-CMV-Cre (Ad-Cre) in cTKO mice and Tam injections in Rosa26CreER/+ cTKO mice or AlbCreERT2/+ cTKO mice. Figure S1A summarizes the advantages and limitations of the four approaches. Importantly, cell cycle re-entry (7–10 days after Cre induction) and exit (2–4 weeks after Cre induction) were similarly observed in all systems tested (Figures S1B–S1E and data not shown), indicating that these phenotypes are independent of the approach used to delete Rb family genes.
Strikingly, less than one third of TKO hepatocytes underwent one or two cell divisions (Figures 1D and 1E). In all the systems used, a number of TKO hepatocytes had abnormally high ploidy (Figures 1F, S1F, and data not shown). We did not observe apoptotic cell death (data not shown). These experiments indicate that, in response to loss of Rb family function, adult hepatocytes divide once or twice and/or undergo endoreduplication, as has been shown for Rb-only mutant hepatocytes (Mayhew et al., 2005) and is commonly observed in wild-type hepatocytes (Duncan, 2013). Thus, the Rb family is required to maintain cell cycle arrest in adult hepatocytes but loss of Rb family function is not sufficient for their prolonged proliferation.
The cell cycle arrest in Rb family mutant hepatocytes is independent of the p53 pathway
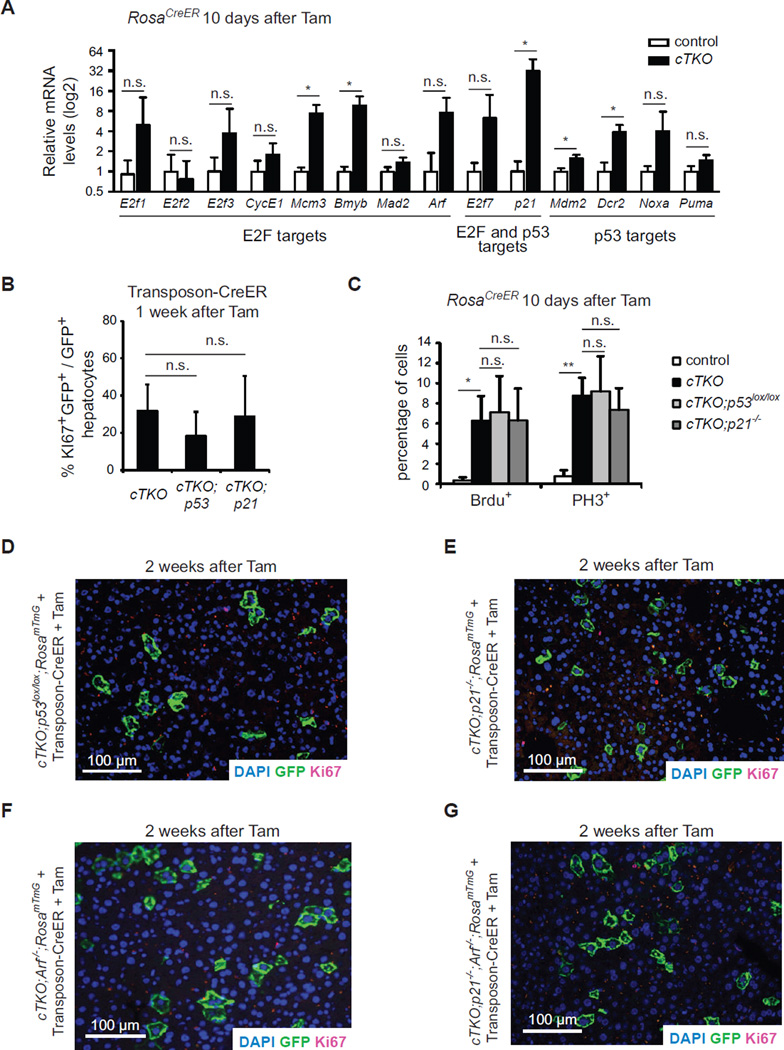
The Rb/E2F and p53 pathways intersect at many levels, including shared targets between E2F and p53, such as the p21Cdkn1a and E2F7 cell cycle inhibitors (Aksoy et al., 2012; Hiyama et al., 1998). Activation of the p53 pathway can serve as a tumor suppressive mechanism in Rb-deficient cells, including in the liver (McClendon et al., 2011; Symonds et al., 1994). We examined the expression of E2F targets and members of the p53 pathway when TKO hepatocytes are exiting the cell cycle 10 days after Cre induction. As expected, loss of Rb family function led to some increased expression of cell cycle genes (Figure 2A and data not shown). We also observed induction of a number of cell cycle inhibitors, including high levels of p21Cdkn1a as well as other known p53 targets (Figures 2A and S2A, and data not shown).
Figure 2. The cell cycle arrest in Rb family mutant hepatocytes is independent of the p53/p21 axis.
(A) RT-PCR analysis of E2F and p53 target genes in control and TKO livers 10 days after Cre activation in cTKO;RosaCreER mice (n≥3) (n.s., not significant; *, p<0.05, paired t-test). (B) Quantification of cell cycle entry in triple and quadruple knockout hepatocytes one week after induction of Cre in Transposon-CreER-injected mice. (n.s., not significant). (C) Quantification of cell proliferation in triple and quadruple KO hepatocytes 10 days after Cre induction (n.s., not significant; *, p<0.05; **, p<0.01). Data in (A–C) represented as mean +/− SD. (D–G) Immunofluorescence analysis of Ki67 (red) expression in mutant (GFP+, green) hepatocytes 2 weeks after Cre induction in the liver of Transposon-CreER-injected mice.
To test whether activation of p53 and/or p21 may be in part responsible for the cell cycle exit in TKO hepatocytes, we crossed cTKO mice to p53 lox/lox mice or p21−/− mice (and to RosamTmG/+ reporter mice). In both cases, the kinetics of cell cycle re-entry were similar in quadruple mutant mice compared to TKO mice (Figures 2B and 2C). We also observed cell cycle exit in quadruple mutant hepatocytes two weeks after Cre (Figures 2D and 2E), despite efficient deletion of p53 (Figure S2B). We detected slightly elevated mRNA levels of the cell cycle inhibitor p19ARF in both TKO and TKO,p21−/− mutant livers compared to controls (Figures 2A and S2C) and generated cTKO;p19ARF−/− mice to investigate p53/p21-independent functions of p19ARF. However, loss of p19ARF did not prevent cell cycle exit (Figures 2G). Similarly, pentuple mutant hepatocytes in cTKO;p21−/−;p19ARF−/− mice still exited the cell cycle (Figure 2H). Therefore, the cell cycle arrest in Rb family mutant hepatocytes is independent of the p19ARF/p53/p21 module.
The cell cycle arrest in Rb family mutant hepatocytes correlates with an inhibition of E2F transcriptional activity
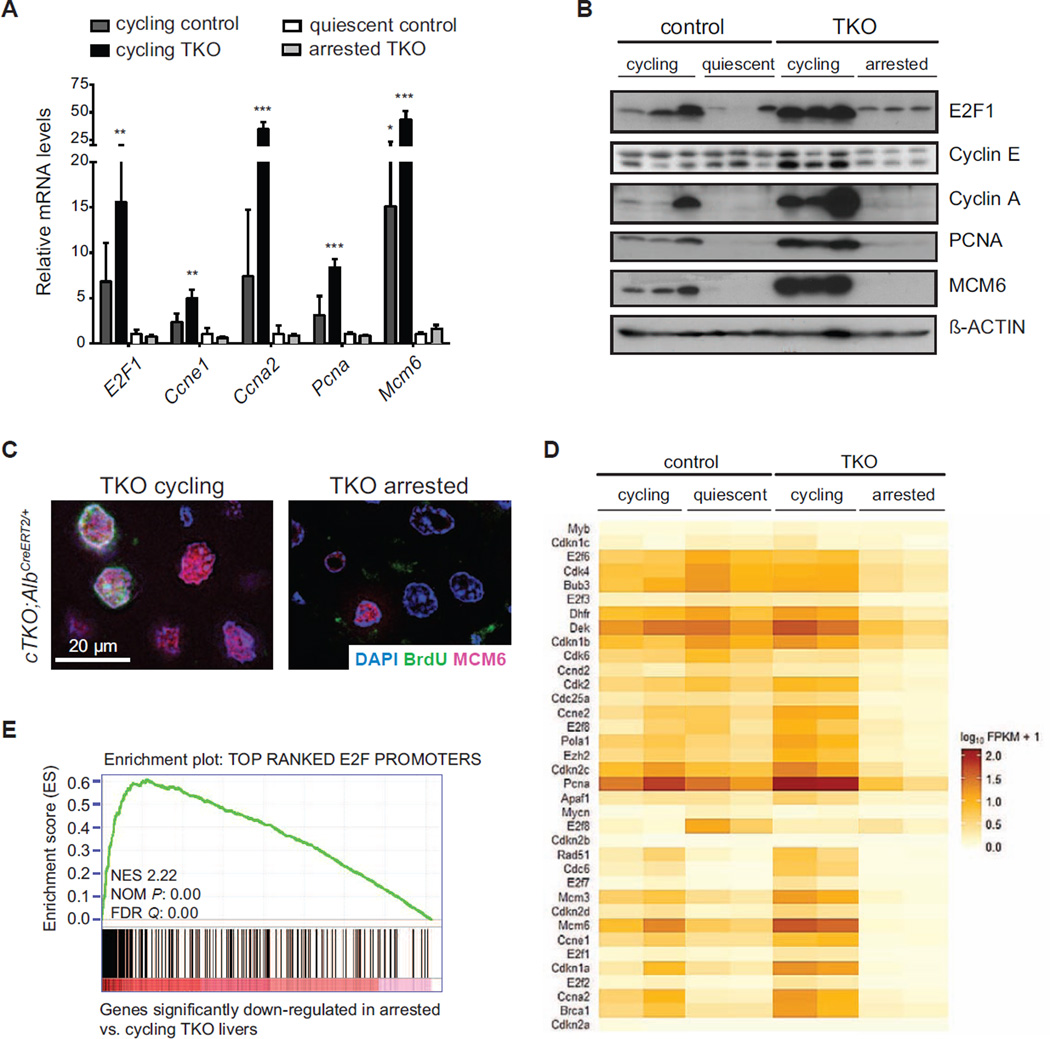
To gain insight into the mechanisms underlying the cell cycle exit in TKO hepatocytes, we extracted RNA and proteins from adult cTKO livers (quiescent controls), cTKO liver 36 hours after partial hepatectomy or 48 hours after treatment with CCl4 (both conditions induce wild-type hepatocytes to cycle), cTKO;AlbCreERT2/+ mice one week after Tam (TKO cycling), and cTKO;AlbCreERT2/+ mice two weeks after Tam (TKO arrested). As expected, Rb/E2F cell cycle targets were expressed at low levels in quiescent control livers and at higher levels in cycling cells. These genes were also down-regulated in arrested TKO hepatocytes (Figures 3A, 3B, and 3C), which was confirmed in an RNA-seq analysis on two samples from each of the four groups (Figures 3D, 3E, S3A, and Table S1). These data suggested that mechanisms exist in adult hepatocytes to inhibit the prolonged transcription of E2F target genes even in absence of the Rb family.
Figure 3. Downregulation of E2F targets in arrested Rb family mutant hepatocytes.
(A) RT-PCR analysis of E2F target genes in control and TKO livers (cycling control: 48 hrs after CCl4 treatment or 36 hrs after partial hepatectomy; quiescent control, Tam-treated cTKO;Alb+/+ mice; cycling TKO, cTKO;AlbCreER/+ mice 1 week after Tam; arrested TKO, cTKO;AlbCreER/+ mice 2 weeks after Tam) (n=3) (*, p<0.05; **, p<0.01; ***, p<0.001). Data represented as mean +/− SD. (B) Immunoblot analysis of control and TKO livers (samples as in 3A). (C) Immunofluorescence analysis of BrdU (green) and the replication factor MCM6 (red), 1 (cycling) and 2 (arrested) weeks after Cre activation in cTKO;AlbCreER/+ mice. (D) CummeRbund heatmap showing differential expression of curated canonical E2F target genes in cycling and non-cycling TKO and control livers (RNA-seq; n=2). (E) Gene Set Enrichment Analysis (GSEA) of genes significantly down-regulated in arrested vs. cycling TKO livers using a gene set generated from top E2F targets (Xu et al., 2007).
Arrested Rb family mutant hepatocytes display low YAP/TEAD levels and activity
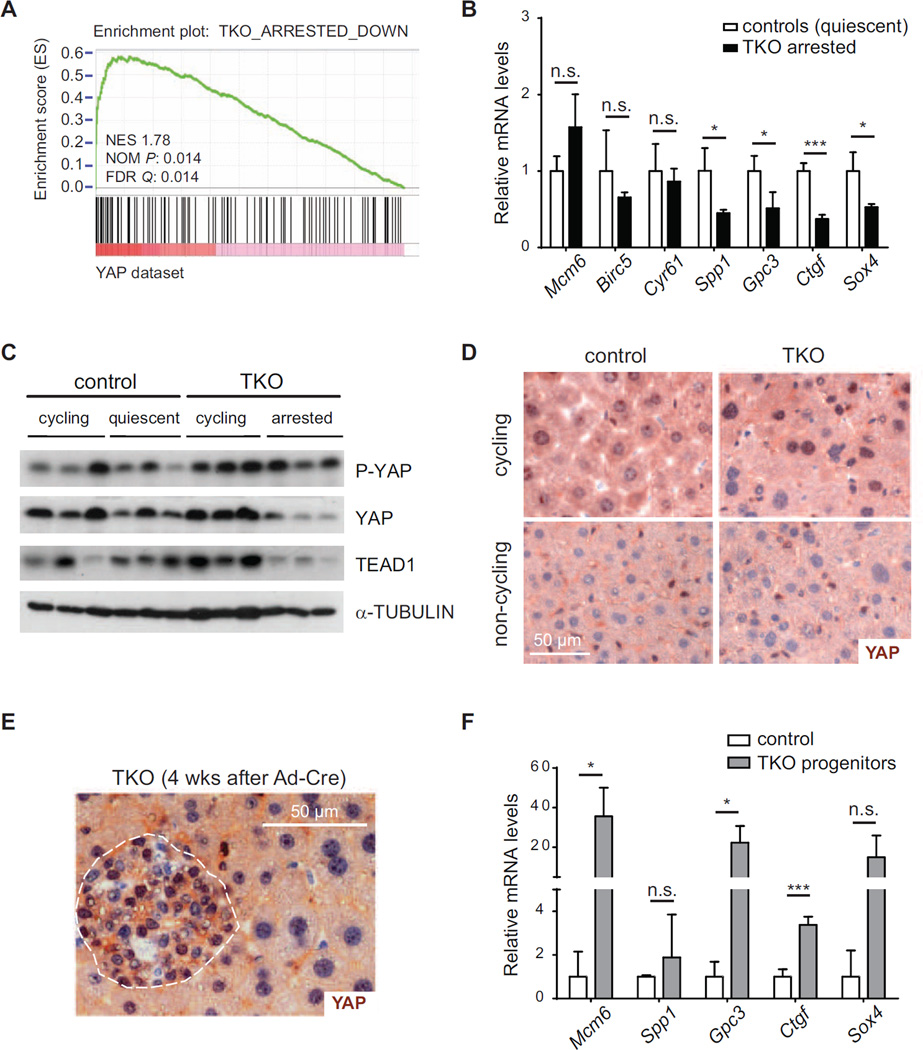
We reasoned that the mechanism enforcing cell cycle exit in TKO hepatocytes may exist in both wild-type and mutant livers but may be even more active in the TKO context to counteract the propensity of the mutant cells to divide. Thus, we sought to identify transcription factors that may regulate the genes whose expression is specifically altered in arrested TKO cells compared to quiescent wild-type cells. A computational analysis of the RNA-seq data identified candidates with known roles in proliferation and tumorigenesis (Figure S3B and Tables S2 and S3), including enrichment for TEF1 binding sites in genes down-regulated in arrested TKO cells. TEF1/TEAD1 is a major partner of the transcriptional co-activators YAP and TAZ downstream of Hippo signaling, one of the key regulators of hepatic growth, liver size control, and liver cancer (Dong et al., 2007; Lu et al., 2010; Zender et al., 2005). Hippo/YAP signaling has also been shown to regulate Wnt pathway activity, another pathway highly enriched in the analysis and that plays a central role in liver biology (Rosenbluh et al., 2012). GSEA showed a strong enrichment for hepatic YAP targets (Dong et al., 2007) in genes significantly down-regulated in arrested TKO livers (Figure 4A), which was confirmed by RT-PCR analysis of known YAP targets (Figure 4B). Furthermore, we observed increased YAP phosphorylation as well as reduced YAP and TEAD1 protein levels in arrested TKO livers (Figures 4C and S4A), suggestive of an active down-regulation of YAP and TEAD1 in TKO arrested cells.
Figure 4. YAP inactivation in arrested TKO livers.
(A) GSEA of genes up-regulated upon hepatic YAP over-expression (Dong et al., 2007) in a gene set of significantly down-regulated genes in arrested TKO livers. (B) mRNA analysis of E2F- (Mcm6, Birc5) and YAP-regulated genes in non-cycling TKO and control livers (n=3; *, p<0.05; ***, p<0.001). (C) Immunoblot analysis of YAP and TEAD1 in livers of control and cTKO;AlbCreER/+ mice. (D) Immunohistochemistry for YAP (brown - counterstain: hematoxylin, blue) in cycling control (36 hrs after partial hepatectomy) and TKO (cTKO;AlbCreERT2/+ 1 week after Tam) livers (top) and non-cycling control and TKO (cTKO;AlbCreERT2/+ 2 weeks after Tam) livers (bottom). (E) YAP staining of early progenitor lesions 4 weeks after intrasplenic injection of Ad-Cre in cTKO mice. (F) mRNA analysis of YAP-regulated genes in TKO progenitor cells (cTKO;AlbCreERT2/+ 4 weeks after Tam) and control livers (n=3; *, p<0.05; **, p<0.01). Data in (B) and (F) are represented as mean +/− SD.
Based on these observations, we wondered whether E2F and YAP may regulate similar sets of genes. We used two independent studies where YAP targets were defined in gene expression profiling analyses (Dong et al., 2007; Zhao et al., 2007). For each study, we performed an enrichment analysis using transcription factor target sites identified by ENCODE ChIP-Seq experiments (Auerbach et al., 2013). In both cases, we found a strong enrichment for E2F binding in YAP targets, with the vast majority of YAP targets being also bound by E2F (Table S4 and Figure S4C). ChIP-PCR assays confirmed binding of YAP to Mcm3 and Mcm6, two canonical E2F targets (Figure S4D). Together, these experiments suggested that decreased YAP/TEAD1 levels and transcriptional activity may affect the long-term expression of key cell cycle E2F targets (Table S5).
YAP was detectable by immunostaining in the nucleus of cycling wild-type and TKO hepatocytes and absent in arrested wild-type and TKO cells (Figure 4D). We recently reported that loss of Rb family function in liver progenitor cells drives these cells out of quiescence and leads to the development of hepatocellular carcinoma (Figure S4E) (Viatour et al., 2011). Cycling TKO progenitors also displayed nuclear YAP (Figure 4E), which correlated with increased expression of YAP target genes (Figure 4F). The expression of nuclear YAP and elevated expression of YAP target genes were conserved in TKO liver tumors (Figures S4F and S4G). Thus, in the adult liver, hepatocyte progenitors and hepatocytes respond differently to loss of Rb family function and YAP/TEAD1 do not become inactivated in progenitor populations.
Reduced liver size and ectopic activation of YAP are sufficient to revert the cell cycle arrest observed in Rb family mutant hepatocytes
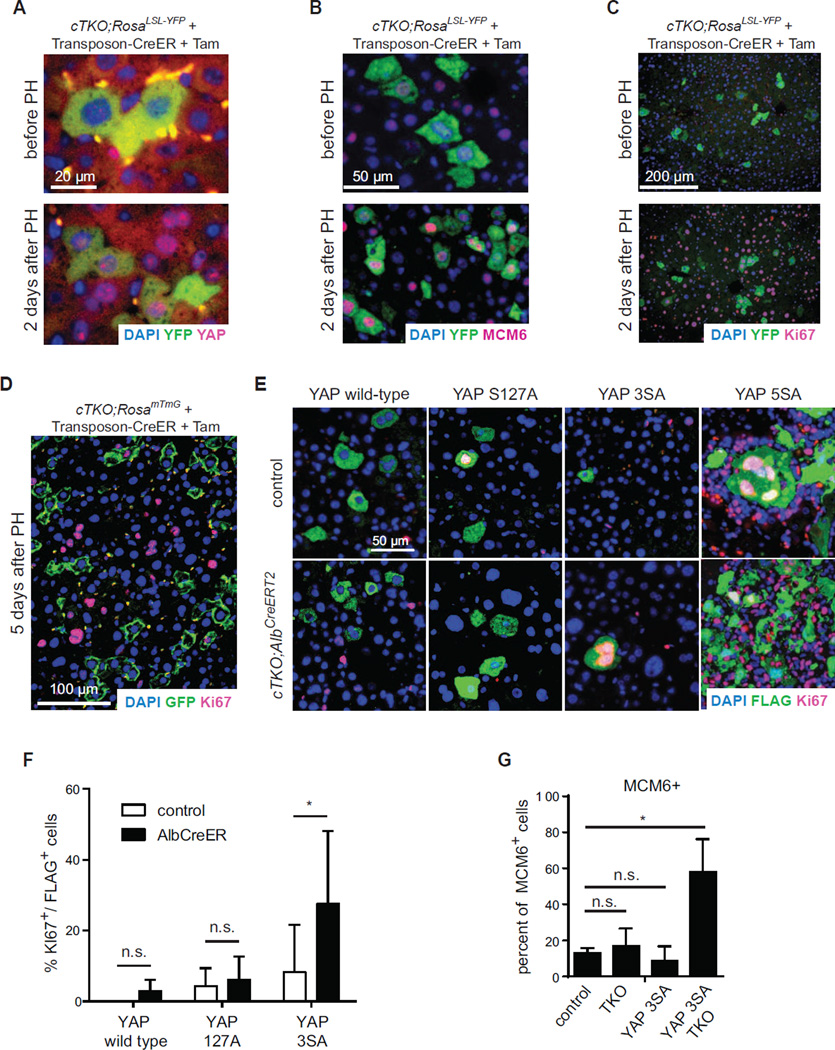
These experiments revealed a striking correlation between E2F activity, proliferation, and YAP activity in liver cells in the hepatocytic lineage, which led us to test the hypothesis that downregulation of YAP/TEAD activity in TKO hepatocytes was limiting for proliferation. YAP activity has been linked to the regulation of organ size (reviewed in (Avruch et al., 2011)). To test if changes in liver size may affect the proliferation of TKO hepatocytes, we performed partial hepatectomy (PH) assays. PH was associated with nuclear translocation of endogenous YAP, expression of E2F targets such as MCM6, and proliferation in both quiescent wild-type and arrested TKO hepatocytes (Figures 5A, 5B, 5C, and S5A). Remarkably, once the livers reached their normal size five days after PH, YAP molecules were again excluded from the nucleus (Figure S5B), and TKO hepatocytes re-exited the cell cycle (Figures 5D). Thus, the cell cycle potential of both wild-type and TKO hepatocytes is under the control of the signals involved in PH response.
Figure 5. YAP activation is sufficient to revert the cell cycle arrest in TKO livers.
(A–C) Immunofluorescence analysis of YFP (green) together with endogenous YAP (A), MCM6 (B), or Ki67 (C), respectively, after 2/3rd partial hepatectomy (PH) 2 weeks after Tam in CreER transposon-injected cTKO;RosaLSL-YFP mice. (D) Immunofluorescence staining of GFP (green) and Ki67 (red) 5 days after 2/3rd PH, 2 weeks after Tam in CreER transposon-injected cTKO;RosamTmG mice. (E) Immunofluorescence analysis of YAP (Flag, green) and Ki67 (red) following hydrodynamic tail vein injection of a transposon expressing either wild type YAP or constitutively active variants of YAP (YAPS127A, YAP3SA, or YAP5SA) into control or arrested TKO livers (transposon injection 1 week after 50 µg Tam, livers analyzed 2 weeks after Tam). (F) Quantification of Ki67 positive YAP-transfected hepatocytes. Data represented as mean +/− SD. (G) Quantification of MCM6 positive hepatocytes following YAP3SA expression in controls or arrested TKO livers (transposon injection 1 week after 50 µg Tam, livers analyzed 2 weeks after Tam). Controls, Flag negative cells in control livers; TKO, Flag negative cells in arrested TKO livers; YAP3SA, Flag positive cells in control livers; YAP3SA TKO, Flag positive cells in arrested TKO livers.
To directly test if low YAP activity was a limiting factor specifically in TKO hepatocytes, we transfected hepatocytes with Transposon vectors expressing wild-type or constitutively active forms of YAP in which phosphorylation sites are mutated to inhibit YAP nuclear exclusion and degradation (YAP S127A, 3SA, or 5SA) (Dong et al., 2007). Wild-type YAP and the partially active S127A mutant were primarily cytoplasmic in both control and TKO cells, which correlated with low cell cycle activity (Figures 5E and 5F). Strong activation of YAP has been shown to force hepatocytes out of quiescence (Dong et al., 2007; Lu et al., 2010; Zhou et al., 2009). Accordingly, expression of the nuclear YAPS5A mutant led to proliferation of both control and TKO hepatocytes (Figure 5E). We also observed the recruitment of immune cells around the transfected cells with this YAP isoform and the elimination of the transfected cells after a few weeks, although the basis of this observation remains unclear (data not shown). Importantly, expression of YAP3SA, a form of YAP with presumed intermediate potency, led to a higher cell cycle activity specifically in TKO cells compared to control cells (Figures 5E and 5F). Additionally, the E2F target MCM6 was significantly expressed only in TKO cells transfected with YAP3SA and not in TKO cells with a control transposon or wild-type hepatocytes transfected with YAP3SA (Figure 5G and S5C). Thus, Rb family mutant hepatocytes are more sensitive to YAP activation than control hepatocytes, indicating that YAP activity is a key limiting factor in the capacity of TKO hepatocytes to proliferate.
Discussion
Experiments in the past three decades have identified the Rb/E2F pathway as an essential regulator of a cell’s decision to enter or exit the cell cycle. Here we show that even in the absence of the Rb family, hepatocytes in the liver of adult mice possess a mechanism that can enforce quiescence in vivo.
Loss of Rb family function in adult stem/progenitor cells often results in the development of pre-neoplastic or neoplastic growth, as was shown for example in the hematopoietic system (Viatour et al., 2008), the mammary gland (Jiang et al., 2010), and the liver (Viatour et al., 2011). In post-mitotic differentiated cells, loss of the Rb family has either no effect or often leads to cell death, which can be triggered by p53 (Symonds et al., 1994; Viatour et al., 2008). Loss of Rb family function can also lead to dedifferentiation, proliferation, and tumor formation (Ajioka et al., 2007) or to prolonged non-cancerous growth (Garfin et al., 2013). In hepatocytes, however, the transient proliferative state triggered by inactivation of the Rb family is followed by a stable cell cycle arrest. Together, these observations indicate that the Rb family is a primary regulator of the cell cycle in stem/progenitor cell populations. In contrast, additional mechanisms may regulate the cell cycle of differentiated cells, even within the same organ or tissue.
The cell cycle entry observed in TKO hepatocytes occurs in the context of a liver of normal size. We propose that this initial proliferative phase, which is associated with an increase in liver size (Figure S5D), triggers a compensatory mechanism connected to organ size sensing in the liver, instructing TKO hepatocytes to stop cycling. At the molecular level, loss of Rb family function initially results in activation of E2F and transcription of cell cycle genes. As this ectopic proliferative phase takes place, compensatory signals eventually lead to the down-regulation of YAP/TEAD transcriptional activity, in part through cytoplasmic relocalization of YAP (see Graphical Abstract). While low YAP levels suggest that kinases in the Hippo pathway may be involved in this response, the lower levels of TEAD, which is not currently known to be regulated by phosphorylation, suggest that additional mechanisms may be at play. One intriguing possibility would be if inhibitors of YAP and TEAD were E2F targets, creating a negative feedback loop. The molecular basis for the differential response to loss of the Rb family in progenitors and mature liver cells remains unknown.
YAP/TEAD complexes bind to many key cell cycle genes and we propose that the long-term transcription of these E2F targets requires YAP/TEAD as co-factors. In support of this idea, evidence in Drosophila has linked E2F and YAP activity at the promoter of target genes. For instance, E2F activity is important for the proliferation of Hippo mutant cells in flies and the concomitant loss of Rb and Hippo signaling leads to proliferation and dedifferentiation in flies (Nicolay et al., 2011; Nicolay et al., 2010). Finally, consensus E2F binding sites are enriched close to YAP binding sites in fly cells (Oh et al., 2013) and human cells (this work). Thus, a decrease in transcriptional YAP activity in response to the ectopic proliferation induced by loss of Rb family function might prevent the activation of cell cycle genes despite initial high E2F activity in TKO hepatocytes. The molecular basis of the functional interaction between E2F and YAP/TEAD may include direct interactions between transcriptional complexes containing these transcription factors at promoters. In addition, our experiments do not rule out the possibility that other pro-proliferative factors may be limiting in TKO cells exiting the cell cycle or may be cooperating with low YAP activity. In particular, we found that Myc levels – but not levels of other putative oncogenes such as activated ERK or AKT – rapidly decreased in TKO hepatocytes entering quiescence (Figure S4B and data not shown). Future experiments will further dissect the regulatory networks implicated in cell cycle decision in adult hepatocytes.
Recent experiments in mammalian cells indicate that low levels of the LATS2 kinase in the Hippo pathway may weaken the ability of Rb to induce cell cycle arrest in tumor cells (Tschop et al., 2011). Furthermore, in breast cancer loss of RB may replace amplification of YAP, further connecting this two pathways (Cheng et al., 2010). The dominant effect of organ size control mechanisms and YAP activity in cells with abrogation of the G1/S checkpoint identifies a novel hierarchy in the regulation of the cell cycle and may have important consequences in regenerative medicine and the development of anti-cancer strategies.
Experimental procedures
Mice
Rb family cTKO mice (Viatour et al., 2011) were maintained in a mixed 129Sv/J;C57/BL6 background. p53lox/lox and p19ARF−/− mice were generous gifts of Drs. Anton Berns and Chuck Sherr, respectively. p21−/−, Rosa26LSL-YFP, and Rosa26mTmG mice were purchased from the Jackson Laboratories. For Ad-Cre injections, 10–12 week-old mice were anesthetized and surgically opened on the upper left quadrant of the abdomen, followed by injection of adenovirus into the spleen. In cTKO; RosaCreERT2/+ mice, Cre was induced by three consecutive injections of 1mg Tam (T-5648; Sigma-Aldrich). In cTKO; AlbCreERT2/+ mice (a kind gift of Pierre Chambon) (Schuler et al., 2004) Cre was activated by injection of 20µg-1mg Tam. CreER was cloned in the transposon vector (Yant et al., 2000) from the MB80 plasmid (Addgene #12168), YAP, YAPS127A, and YAP5SA were cloned from Addgene plasmids #19045, #17794, and # 27371, respectively. YAP3SA (S61A, S109A, S127A) is a swap mutant of YAP and YAP5SA. Samples were 36hrs (for RNA and protein analysis) or 48hrs (for immunostaining and ChIP experiments) after PH. All experiments with mice were approved by Stanford Institutional Animal Care and Use Committee.
Histology, immunostaining, and immunoblot analysis
Antigen retrieval on paraffin sections was performed using Trilogy (Cell Marque). After blocking, sections were incubated with primary antibodies overnight at 4°C, washed in PBS + 0.1% Tween 20 (PBST), and then incubated with secondary antibodies. Quantification was performed using ImageJ software. Primary antibodies used were anti-GFP (Invitrogen), anti-Ki67 (BD), anti-Flag (Sigma), anti-BrdU (BD), anti-YAP, anti-PH3, anti-CC3 (Cell Signaling Technology – data not shown), anti-MCM6, and anti-p21 (Santa Cruz Biotechnology). For immunoblotting, primary antibodies used were anti-YAP, anti-P-ERK, anti-ERK (Cell Signaling), anti-TEF1 (BD Biosciences), anti-p21, anti-E2F1, anti-Cyclin A, anti-PCNA, anti-MCM6 (Santa Cruz Biotechnology), and anti-Cyclin E (eBiosciences).
Flow cytometry
Hepatic nuclei were isolated from frozen liver tissue (Mayhew et al., 2005) and ploidy was analyzed by flow cytometry using staining for propidium iodide (PI).
RT-PCR, RNA-seq, and bioinformatics analysis
RNAs were processed and qPCR reactions were prepared as previously (Viatour et al., 2011). RNA-seq, TopHat and Cufflinks analysis were performed by Centrillion Biosciences. CummeRbund was used for heatmap visualization and clustering of RNA-seq data. Data was submitted to the GEO repository (GSE43631). E2F1/YAP co-target genes were defined as genes with a significant ChIP-seq peak for E2F1 (ENCODE data) with direct overlap or within +/− 2.5 kb from the transcription site of YAP-regulated genes (Dong et al., 2007; Zhao et al., 2007). Curated gene sets for GSEA (version 3.1) (Subramanian et al., 2005) were obtained from http://www.broadinstitute.org/gsea/msigdb/index.jsp. Enrichment was considered significant when the p-value was <0.05 and the FDR was <0.25. Gene Ontology analysis was performed using DAVID (GOTERM_BP_FAT, DAVID Bioinformatics Resources 6.7) and transcription factor target analysis using WebGestalt Gene Set Analysis Toolkit V2 (Zhang et al., 2005).
ChIP analysis
ChIP was performed as described previously (O'Geen et al., 2006). In brief, mouse livers where perfused 5 min with 2% formaldehyde, before homogenization with a Potter-Elvehjem tissue grinder, followed by 10 min crosslinking in 2% formaldehyde. Antibodies used for immunoprecipitation were YAP (Cell Signaling), and p16 (sc-467). Promoter binding was assessed by quantitative PCR using SYBR green (Quanta biosciences).
Statistical analysis
Statistical significance was assayed by two-tailed Student’s t test (*,p<0.05; **, p<0.01;***, p<0.005). Data are represented as mean ± SD if not stated otherwise.
Supplementary Material
Highlights.
Proliferation upon loss of the Rb family is only transient in hepatocytes in vivo
Partial hepatectomy reverts the cell cycle arrest in Rb family mutant hepatocytes
Sustained YAP activity is limiting for prolonged proliferation in hepatocytes
Liver progenitor cells do not obey the same cell cycle regulatory mechanisms
Acknowledgments
We thank Drs. R.L. Johnson, P. Viatour, and members of the Sage lab for helpful discussions. This work was supported by the NIH (grant HL064274 to M.K. and CA114102 to J.S.), the Lucile Packard Foundation for Children’s Health (Ernest and Amelia Gallo Endowed Postdoctoral Fellowship - CTSA grant number UL1 RR025744 to U.E.), and the Dr. Mildred Scheel fellowship from Deutsche Krebshilfe (to U.E.). Dr. Sage is a Leukemia and Lymphoma Society Scholar and the Harriet and Mary Zelencik Scientist in Children’s Cancer and Blood Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: U.E. and A.-F.Z. performed and analyzed the experiments, D.V., R.K.A., and A.J.B. performed the bioinformatics analyses, U.E. and M.A.K. designed the transposon construct, U.E. and J.S. designed the overall research and wrote the manuscript.
References
- Ajioka I, Martins RA, Bayazitov IT, Donovan S, Johnson DA, Frase S, Cicero SA, Boyd K, Zakharenko SS, Dyer MA. Differentiated horizontal interneurons clonally expand to form metastatic retinoblastoma in mice. Cell. 2007;131:378–390. doi: 10.1016/j.cell.2007.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy O, Chicas A, Zeng T, Zhao Z, McCurrach M, Wang X, Lowe SW. The atypical E2F family member E2F7 couples the p53 and RB pathways during cellular senescence. Genes Dev. 2012;26:1546–1557. doi: 10.1101/gad.196238.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach RK, Chen B, Butte AJ. Relating genes to function: identifying enriched transcription factors using the ENCODE ChIP-Seq significance tool. Bioinformatics. 2013;29:1922–1924. doi: 10.1093/bioinformatics/btt316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avruch J, Zhou D, Fitamant J, Bardeesy N. Mst1/2 signalling to Yap: gatekeeper for liver size and tumour development. Br J Cancer. 2011;104:24–32. doi: 10.1038/sj.bjc.6606011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Zhou Z, Flesken-Nikitin A, Toshkov IA, Wang W, Camps J, Ried T, Nikitin AY. Rb inactivation accelerates neoplastic growth and substitutes for recurrent amplification of cIAP1, cIAP2 and Yap1 in sporadic mammary carcinoma associated with p53 deficiency. Oncogene. 2010 doi: 10.1038/onc.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg JH, Schuijff L, Dekker M, van der Valk M, te Riele H. Tissue-specific tumor suppressor activity of retinoblastoma gene homologs p107 and p130. Genes Dev. 2004;18:2952–2962. doi: 10.1101/gad.322004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G (1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 2000;14:3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol. 2013;14:297–306. doi: 10.1038/nrm3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin Cell Dev Biol. 2013;24:347–356. doi: 10.1016/j.semcdb.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Duncan AW, Dorrell C, Grompe M. Stem cells and liver regeneration. Gastroenterology. 2009;137:466–481. doi: 10.1053/j.gastro.2009.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfin PM, Min D, Bryson JL, Serwold T, Edris B, Blackburn CC, Richie ER, Weinberg KI, Manley NR, Sage J, et al. Inactivation of the RB family prevents thymus involution and promotes thymic function by direct control of Foxn1 expression. J Exp Med. 2013 doi: 10.1084/jem.20121716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama H, Iavarone A, Reeves SA. Regulation of the cdk inhibitor p21 gene during cell cycle progression is under the control of the transcription factor E2F. Oncogene. 1998;16:1513–1523. doi: 10.1038/sj.onc.1201667. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Deng T, Jones R, Li H, Herschkowitz JI, Liu JC, Weigman VJ, Tsao MS, Lane TF, Perou CM, et al. Rb deletion in mouse mammary progenitors induces luminal-B or basal-like/EMT tumor subtypes depending on p53 status. J Clin Invest. 2010;120:3296–3309. doi: 10.1172/JCI41490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Halder G, Finegold MJ, Lee JS, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Dyson NJ. RB: mitotic implications of a tumour suppressor. Nat Rev Cancer. 2012 doi: 10.1038/nrc3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayhew CN, Bosco EE, Fox SR, Okaya T, Tarapore P, Schwemberger SJ, Babcock GF, Lentsch AB, Fukasawa K, Knudsen ES. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer Res. 2005;65:4568–4577. doi: 10.1158/0008-5472.CAN-04-4221. [DOI] [PubMed] [Google Scholar]
- McClendon AK, Dean JL, Ertel A, Fu Z, Rivadeneira DB, Reed CA, Bourgo RJ, Witkiewicz A, Addya S, Mayhew CN, et al. RB and p53 cooperate to prevent liver tumorigenesis in response to tissue damage. Gastroenterology. 2011;141:1439–1450. doi: 10.1053/j.gastro.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy J, Flores-Otero J, Zhang J, Nemeth K, Brennan R, Bradley C, Krafcik F, Rodriguez-Galindo C, Wilson M, Xiong S, et al. Coexpression of normally incompatible developmental pathways in retinoblastoma genesis. Cancer Cell. 2011;20:260–275. doi: 10.1016/j.ccr.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay BN, Bayarmagnai B, Islam AB, Lopez-Bigas N, Frolov MV. Cooperation between dE2F1 and Yki/Sd defines a distinct transcriptional program necessary to bypass cell cycle exit. Genes Dev. 2011;25:323–335. doi: 10.1101/gad.1999211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay BN, Bayarmagnai B, Moon NS, Benevolenskaya EV, Frolov MV. Combined inactivation of pRB and hippo pathways induces dedifferentiation in the Drosophila retina. PLoS Genet. 2010;6:e1000918. doi: 10.1371/journal.pgen.1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay BN, Dyson NJ. The multiple connections between pRB and cell metabolism. Curr Opin Cell Biol. 2013 doi: 10.1016/j.ceb.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Geen H, Nicolet CM, Blahnik K, Green R, Farnham PJ. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques. 2006;41:577–580. doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Slattery M, Ma L, Crofts A, White KP, Mann RS, Irvine KD. Genome-wide association of Yorkie with chromatin and chromatin-remodeling complexes. Cell reports. 2013;3:309–318. doi: 10.1016/j.celrep.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robanus-Maandag E, Dekker M, van der Valk M, Carrozza ML, Jeanny JC, Dannenberg JH, Berns A, te Riele H. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev. 1998;12:1599–1609. doi: 10.1101/gad.12.11.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, Zack TI, Wang X, Tsherniak A, Schinzel AC, et al. beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell. 2012;151:1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M, Dierich A, Chambon P, Metzger D. Efficient temporally controlled targeted somatic mutagenesis in hepatocytes of the mouse. Genesis. 2004;39:167–172. doi: 10.1002/gene.20039. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds H, Krall L, Remington L, Saenz-Robles M, Lowe S, Jacks T, Van Dyke T. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell. 1994;78:703–711. doi: 10.1016/0092-8674(94)90534-7. [DOI] [PubMed] [Google Scholar]
- Tschop K, Conery AR, Litovchick L, Decaprio JA, Settleman J, Harlow E, Dyson N. A kinase shRNA screen links LATS2 and the pRB tumor suppressor. Genes Dev. 2011;25:814–830. doi: 10.1101/gad.2000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viatour P, Ehmer U, Saddic LA, Dorrell C, Andersen JB, Lin C, Zmoos AF, Mazur PK, Schaffer BE, Ostermeier A, et al. Notch signaling inhibits hepatocellular carcinoma following inactivation of the RB pathway. J Exp Med. 2011;208:1963–1976. doi: 10.1084/jem.20110198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viatour P, Somervaille TC, Venkatasubrahmanyam S, Kogan S, McLaughlin ME, Weissman IL, Butte AJ, Passegue E, Sage J. Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell. 2008;3:416–428. doi: 10.1016/j.stem.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17:1550–1561. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant SR, Meuse L, Chiu W, Ivics Z, Izsvak Z, Kay MA. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- Zender L, Xue W, Cordon-Cardo C, Hannon GJ, Lucito R, Powers S, Flemming P, Spector MS, Lowe SW. Generation and analysis of genetically defined liver carcinomas derived from bipotential liver progenitors. Cold Spring Harb Symp Quant Biol. 2005;70:251–261. doi: 10.1101/sqb.2005.70.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic acids research. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.