Abstract
The development of therapeutic proteins and peptides is an expensive and time-intensive process. Biologics, which have become a multi-billion dollar industry, are chemically complex products that require constant observation during each stage of development and production. Post-translational modifications along with chemical and physical degradation from oxidation, deamidation, and aggregation, lead to high levels of heterogeneity that affect drug quality and efficacy. The various separation modes of capillary electrophoresis (CE) are commonly utilized to perform quality control and assess protein heterogeneity. This review attempts to highlight the most recent developments and applications of CE separation techniques for the characterization of protein and peptide therapeutics by focusing on papers accepted for publication in the in the two-year period between January 2012 and December 2013. The separation principles and technological advances of CE, capillary gel electrophoresis, capillary isoelectric focusing, capillary electrochromatography and CE-mass spectrometry are discussed, along with exciting new applications of these techniques to relevant pharmaceutical issues. Also included is a small selection of papers on microchip electrophoresis to show the direction this field is moving with regards to the development of inexpensive and portable analysis systems for on-site, high-throughput analysis.
1. Introduction
The characterization of protein therapeutics presents a unique analytical challenge due to the inherent heterogeneity of recombinant protein expression. Even small changes in the manufacturing process can lead to vastly different active pharmaceutical ingredients. Additionally, numerous physical and chemical degradation pathways can occur during manufacturing and storage that compromise protein integrity, leading to a potentially harmful, unstable product [1]. Thorough characterization of protein therapeutics is necessary at every step of the research and development process, from drug discovery to lot release.
Due to the potential complexity of product degradation during preformulation and formulation studies, additional separation techniques are needed to complement the more widely used column liquid chromatography (LC) methods. To address this issue, capillary electrophoresis (CE) has become a popular choice for the separation and analysis of therapeutic proteins and peptides.
CE provides several distinct advantages over LC. First, due to the faster separation times and the use of multi-capillary arrays, hundreds of samples can be processed by CE per day. Second, CE is capable of achieving very high efficiency separations due to the low diffusion coefficients of biomolecules. Lastly, the small dimensions of the capillary and the low sample volume requirements keep reagent and analyte use to a minimum, reducing the cost-per-test. The benefits of CE for the analysis of therapeutic peptides and proteins have been addressed in several excellent reviews to date [2–5].
This review is aimed at highlighting the advances made in the field of CE therapeutic protein analysis during 2012 and 2013 by expanding on a paper that was recently published by Zhao et al. [5]. Following brief descriptions of the working principles of the different CE separation and detection methods, the recent technological improvements and novel applications are discussed. Two additional sections have been included to further explore the use of CE for the determination of protein glycosylation and the comparison of biosimilars. Finally, a brief introduction into microfluidic approaches to protein analysis is given. Microchip electrophoresis (ME) has the additional advantages of increased speed, high-throughput capabilities, and portability for on-site analyses. Tables are presented in each section to highlight the relevant CE and ME application-based citations.
2. Techniques
Historically, capillary zone electrophoresis (CZE) has been the most commonly employed form of CE. Yet, the principles of electrophoretic separations and the benefits of capillary-based techniques are applicable to other CE separation modes as well. Protein analysis based on size can be accomplished by capillary gel electrophoresis (CGE), capillary isoelectric focusing (CIEF) can be used to determine isoelectric points and charge heterogeneity, and capillary electrochromatography (CEC), which combines the high efficiency electrophoretic separation with chromatographic retention, can be used for more selective separations and analysis of neutral species. Depending on the properties of the analyte and requirements of the assay, each of these separation modes can be coupled to a number of detection methods such as UV-Vis absorbance, laser-induced fluorescence (LIF), and mass spectrometry (MS).
2.1 Capillary zone electrophoresis
Of the electrophoresis-based separation techniques, CZE is most frequently used for the analysis of small molecules, carbohydrates, and peptides. It is simple, easy-to-use, and requires minimal reagents compared to chromatographic methods. Additionally, in CZE, the separation of analytes is based on their size-to-charge ratio making it well suited for separations of proteins with post-translational modifications (PTMs) or degradations that affect the charge of the molecule [6, 7] including deamidation, glycosylation, and phosphorylation.
One example of the use of CZE for the investigation of deamidation concerns the stability of oxytocin. Deamidation of Asn and Gln residues is the most common chemical degradation pathway for peptides and proteins [1]. This process leads to the production of an ionizable carboxylic acid from the neutral amide (R-CONH2 → R-COOH), facilitating a separation by CZE. However, if peptides, such as oxytocin, contain several labile Asn and Gln sites, multiple degradation products of the same size-to-charge ratio are produced and a straightforward separation becomes impossible. To distinguish between the seven desamino degradation products of heat-stressed oxytocin, Creamer et al. utilized sulfobutyl ether β-cyclodextrin (SBE β-CD) as a pseudo-stationary phase [8]. The negatively charged SBE β-CD forms an inclusion complex with the hydrophobic Tyr2 residue of oxytocin, affecting the electrophoretic mobility of the peptides. A baseline separation of all eight peptides and a migration time RSD of less than 1.2% was achieved.
Unfortunately, reproducible separations of larger biomolecules using bare fused-silica capillaries are rare due to protein adsorption. Many proteins have large localized regions of positive charge that are electrostatically attracted to the negatively charged silanol groups at the capillary surface. Additional adsorption can be caused by hydrophobic and hydrogen bonding interactions. This adsorption process keeps CZE from obtaining the 106 theoretical plates that should be possible due to the very low diffusion coefficients of large proteins. [9, 10].
One strategy for minimizing protein adsorption is to alter either the charge density of the protein or the capillary wall by changing the pH or ionic strength of the background electrolyte (BGE). Another approach is to simply add a modifier to the BGE to reduce protein-wall interactions. The addition of surfactants, small amines, or anionic salts, such as phytic acid, to the BGE is common [11, 12]. In cases where modification of the run buffer does not obviate protein adsorption, dynamic and static capillary coatings have been used to create a barrier between the ionized silanol groups and the protein of interest.
2.1.1 Dynamic coatings
Dynamic coatings are buffer additives that adsorb to the surface of the capillary, shielding the silanol groups from analyte adsorption [13]. These noncovalent coatings are popular due to their simplicity, versatility, and ease-of-use. However, because of their impermanent nature, the coatings need to be continuously regenerated. This can be accomplished by refreshing the physically adsorbed layer at the capillary with rinses between runs, or adding a small amount to the BGE to prevent coating degradation during electrophoresis. A variety of such coatings have been used for protein separations, ranging from small molecules such as ionic liquids (ILs), to larger molecules such as surfactants and polymers [14].
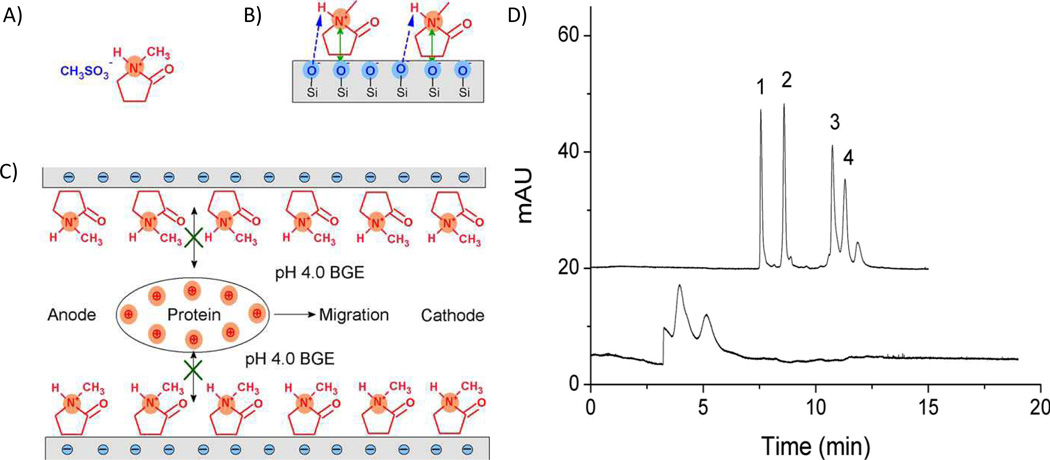
ILs have been previously explored as dynamic coatings for CZE protein separations [15–17]. ILs are salts made up of organic cations and inorganic or organic anions that are liquid at, or around, room temperature. Recently, a new IL, N-methyl-2-pyrrolidonium methyl sulfonate ([NMP]+CH3SO3−), was used to prevent basic protein (pI 9.0–10.7) adsorption to capillary walls during CE separation [18]. The [NMP]+ moiety electrostatically adsorbs to the capillary surface, where it is able to hydrogen bond for additional stability (Fig. 1). Using this coating, the authors were able to achieve a baseline separation of four basic proteins (Table 1) with an interday migration time RSD of less than 1.5%. The improvement in the separation after addition of only 0.02% w/v IL, compared to that obtained with phosphate buffer alone, is easily seen in Fig. 1D.
Fig. 1.
A) The structure of the IL [NMP]+CH3SO3−, B) the interaction between [NMP]+ and the silica capillary inner wall , and C) the mechanism of separation of proteins using [NMP]+ as dynamic coating material, D) Electropherograms of four basic proteins in bare silica capillary (bottom trace) and in the presence of 0.02% w/v IL (top trace). Running buffer: 40 mM pH 4.0 sodium phosphate; voltage: 18 kV; detection: 214 nm; peaks: (1) cytochrome c, (2) lysozyme, (3) ribonuclease A, (4) α-chymotrypsinogen A. Reprinted with permission from ref. 18
Table 1.
CZE and capillary coatings
| Analyte | Coating | Capillary | BGE | Voltage | Detection | Notes | Ref. |
|---|---|---|---|---|---|---|---|
| Therapeutic albumin | Semi-permanent coating with PEO |
57 (50) cm, 50 µm id |
50 mM HEPES, pH 7.5, 0.5 mM SDS |
− 25 kV | UV 214 nm | Separation of human serum albumin isoforms |
[6] |
| In-house IgG1 mAbs | Bare fused silica | 30.2 (20) cm, 50 µm id |
20 nM NaAc, 0.3% PEO, 2 mM triethylenetetraamine, pH 6.0 |
+ 30 kV | UV 214 nm | Rapid method to determine mAb charge variance |
[7] |
| Oxytocin | Bare fused silica | 50 (40) cm, 50 µm id |
50 mM sodium phosphate pH 6.0,12.5 mM SBE β-CD, 10% v/v MeOH, |
+ 22 kV | UV 214 nm | Separation of all oxytocin desamino products |
[8] |
| Cytochrome c, lysozyme, ribonuclease A, and α-chymotrypsinogen A |
Static coating with ionic liquid [NMP]+CH3SO3− |
50 (41.5) cm, 75 µm id |
40 mM sodium phosphate, pH 4.0, 0.3% w/v ionic liquid |
+ 15 kV | UV 214 nm | Minimize protein adsorption | [18] |
| (1,2) Chymotrypsinogen, ribonuclease A, cytochrome c, trypsin inhibitor, lysozyme |
(1) QC (2) HMQC |
47 (40) cm, 75 µm id |
25 mM sodium phosphate over a range of pH 3.0–8.0 |
+ 12 and − 12 kV |
UV 214 nm | The hydrophobic QC provided a more effective for coating |
QC [22] HMQC [23] |
| Purchased mAbs | Various commercial and in-house coatings |
64.5 (56) cm, 50 µm id |
Various BGE composition, pH, and additives |
+ 30 and − 30 kV |
UV 200 nm | Comparison of static capillary coatings |
[26] |
| Enhanced green fluorescent protein and R-phycoerythrin |
Polymerize phospholipid bilayer |
42 (32) cm, various µm i |
Various BGEs over a range of pH 4.0–9.3 |
+ 24 kV | LIF | Best coating stability in capillaries with id of ≤ 50 µm |
[28] |
| (1,2) Lysozyme, cytochrome c, BSA (2) amyloglucosidase, myoglobin |
(1) PVA or (2) PEG and diazoresin |
50 (41) cm, 75 µm id |
40 mM sodium phosphate over a range of pH 3.0–9.0 |
+ 15–18 kV |
UV 214 nm | Easy to form covalently bonded capillary coatings |
PVA [29] PEG [30] |
Capillary: actual length (effective length), inner diameter
Bovine serum albumin (BSA), Hydrophobically modified QC (HMQC), N-methyl-2-pyrrolidonium methyl sulfonate IL ([NMP]+CH3SO3− ), Polyethylene glycol (PEG), Polyethylene oxide (PEO), Polyvinyl alcohol (PVA), Quaternized celluloses (QC), Sulfobutyl ether β-cyclodextrin (SBE β-CD)
Polysaccharides are also attractive candidates for dynamic coatings for protein separations because they are non-toxic, readily abundant, and biocompatible [19–21]. Two novel dynamic coatings based on the chemical substitutions of cellulose have recently been reported [22, 23]. The first, a positively charged quaternarnized cellulose (QC), was synthesized through a reaction of cellulose with 3-chloro-2-hydroxypropyltrimethylammonium chloride. The positive charge of the QC leads to the electrostatic adsorption of the compound to the capillary surface, reversing the electroosmotic flow (EOF). Addition of 5 µg/mL QC to the BGE prevented adsorption of model basic proteins leading to higher separation efficiencies [22]. To increase the reverse EOF by 10%, and further improve separation efficiency, additional substitution of the QC was made using hydrophobic hexadecyl groups [23]. Both QCs were evaluated with a separation of five basic proteins (Table 1). In both cases, the modified capillaries produced a migration time reproducibility with RSD of less than 2.7%.
Despite their simplicity, buffer additives and dynamic coatings are not always the best approach to eliminate protein adsorption. If the modifier is highly charged, band broadening can occur due to high separation currents and Joule heating. Additionally, some buffer modifiers can interfere with protein binding assays [24], disrupt protein stability [25], or be incompatible with downstream detection methods such as MS. In cases where greater stability and reproducibility are needed, static coatings have been used [13].
2.1.2 Static coatings
Static coatings are chemically linked to the capillary wall and do not need to be added to the run buffer to acheive reproducible separations. Therefore they have the potential for large-scale production and can be made commercial available. Several companies are already selling coated capillaries for protein separations including GL Sciences (FunCap®), Target Discovery (UltraTrol™), MicroSOLV (CElixer™), and Beckman Coulter (eCAP™).
Gassner et al. performed a thorough comparison of both commercially available and lab-generated static coatings in 2013 [26]. Eight coatings were selected—four positive: FunCap®-type A, UltraTrol™ HR, Hexadimethrin bromide (polybrene) (PB)-dextran sulfate-PB, and polyethylenimine; and four neutral: FunCap®-type D, UltraTrol™ LN, hydroxypropylcellulose, and polyvinyl alcohol (PVA). The coatings were evaluated for the protein recovery, isoform resolution, and migration time reproducibility of two monoclonal antibodies (mAbs).
For the positively charged coatings, the separation was run in negative polarity. With these capillaries it was determined that the slower the EOF, the better the resolution. Yet, while UltraTrol™ HR had the slowest EOF, it had poor reproducibility (8.9% RSD) and was discarded from the study. For the neutral coatings run in normal polarity, the largest factor for protein adsorption was the presence of residual silanol groups. This was apparent by the fact that some EOF was still generated in the capillary. Of the four neutral coatings in this study, both commercial options, FunCap®-type D and UltraTrol™ LN, generated a small amount of EOF at pH 7.0, indicating that the coating was not uniform and there were still potential sites for protein adsorption. However, it is important to note that the separation performance of each coating was highly dependent on the pH and composition of the BGE. Consequently, care should be taken during method development to fully optimize the BGE for the selected coating.
Due to the varied performance of the commercially available products, new coatings for the separation of basic and hydrophobic proteins are still under development. One particularly attractive choice for static coatings is phospholipid bilayers (PLB) because of the protein resistant nature of the hydrophobic phosphoylcholine polar headgroup. However, the limiting factor for these coatings is their poor long-term chemical and physical stability. This can be remedied by cross-linking the PLB with bis-SorbPC which produces a stabilized phospholipid bilayer (SPB) at the capillary surface [27]. In a recent report, it was shown that the SPB produced a stable coating over a pH range from 4.0–9.3 [28]. Over the course of 45-days dry storage the migration time reproducibility for both model proteins (Table 1) was marginally affected and the overall RSD for the EOF was only changed by 1.1%.
To reduce the preparation time for the preparation of static coated capillaries, self-assembled bilayers and photoinitiated polymerization can be used. An example of such a process was described by Yu et al. using a photosensitive diazoresin (DR) in combination with either PVA [29] or polyethylene glycol (PEG) [30]. After exposure to 365 nm light, both the DR/PVA and DR/PEG coatings were able to prevent protein adsorption and achieve an efficient separation of several model basic proteins (Table 1) with a migration time precision less than 4% RSD.
2.1.3 Evaluation of capillary coating performance
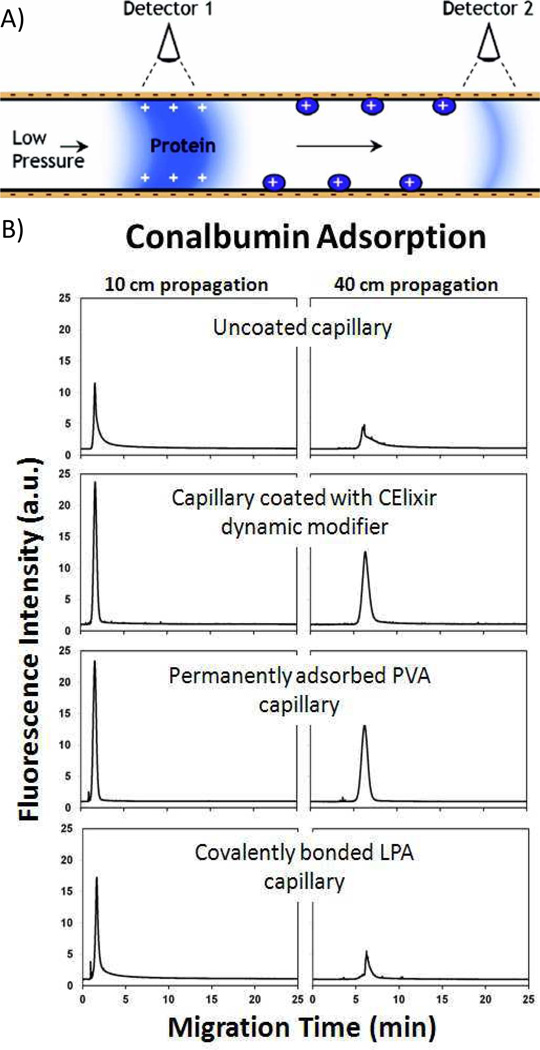
Prior to assay development, the determination of capillary coating performance is extremely important. A previous analytical approach to determine protein adsorption in capillaries involves flushing the capillary with the protein of interest to allow adsorption and then measuring desorption on a subsequent rinse [31–33]. However, with this method, only irreversibly bound proteins are measured. As an alternative, de Jong et al. recently developed a more direct method using pressure-driven flow [34]. Briefly, a plug of sample is pressure injected into a capillary at a low flow rate (0.5 psi) and the Taylor dispersion of the plug is measured at two different detection points along the capillary. Based on these measurements, the magnitude of the protein adsorption can be estimated (Fig. 2).
Fig. 2.
A) Diagram of the set-up for the dual detection pressure-based technique for assessing protein adsoption. B) Pressure-driven propagation of 5.3 µM chromeo-labeled conalbumin detected at 10 and 40 cm. Better protection against adsorption can be seen in both the capillary coated with CElixir dynamic modifier and the capillary with a permanently adsorbed PVA coating. Reprinted with permission from ref. 34
2.2 Capillary gel electrophoresis
The most commonly used analytical method for size determination, purity assessment, and quality control of therapeutic recombinant proteins is sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). SDS is used to coat the proteins, resulting in a uniform negative charge proportional to their size. Under an electric field the proteins are then separated through a sieving gel matrix, allowing for estimation of protein molecular weight (MW). However, conventional SDS-PAGE can be time-consuming, tedious, and yield irreproducible results with limited quantitative abilities [35].
To improve on this important technique, the CGE equivalent, SDS-CGE, has been developed and utilized for the determination of size heterogeneity of therapeutic proteins [36–38]. Here the sieving gel is placed inside the capillary through which the negatively charged SDS-coated proteins are separated. SDS-CGE has many advantages over SDS-PAGE, including high efficiency separations, more accurate MW and concentration determination, and the ability to automate the process for high-throughput analysis.
Shi et al. demonstrated these advantages of SDS-CGE over SDS-PAGE, along with the improved precision of migration time and peak area, for the analysis of the light chain, nonglycosylated heavy chain, and heavy chain fragments of a mAb [39]. Using the capillary format, the authors were able to achieve RSDs of less than 0.5% for migration time and less than 5% for corrected peak area. However, for quality control of biopharmaceuticals, the precision for a quantitative assay needs to be lower than 2% RSD. By switching from hydrodynamic rather than electrokinetic injection, along with increased sample concentration, the precision of a standard SDS-CGE assay was improved to 0.2% RSD for migration time and between 1 and 2% RSD for peak area ratio [40].
Another method to improve assay precision for the SDS-CGE assay is through automation of the sample preparation process. A large number of samples are generated during the development of high-quality biologics. These samples originate from every step of the development process and are presented for analysis in a variety of matrices. The use of an automatic robotic platform for sample preparation can help mitigate user error introduced in the multi-step sample preparation process. The PhyNexus Micro-Extractor Automated Instrument uses a ProA resin column to bind mAb samples prior to separation. Once bound, the instrument performs sample concentration normalization, removal of contaminates, desalting, and mixing with appropriate SDS-CGE buffers. With this method, protein recovery of Fc-fusion proteins, and IgG1 and IgG2 mAbs was increased to 90% [41].
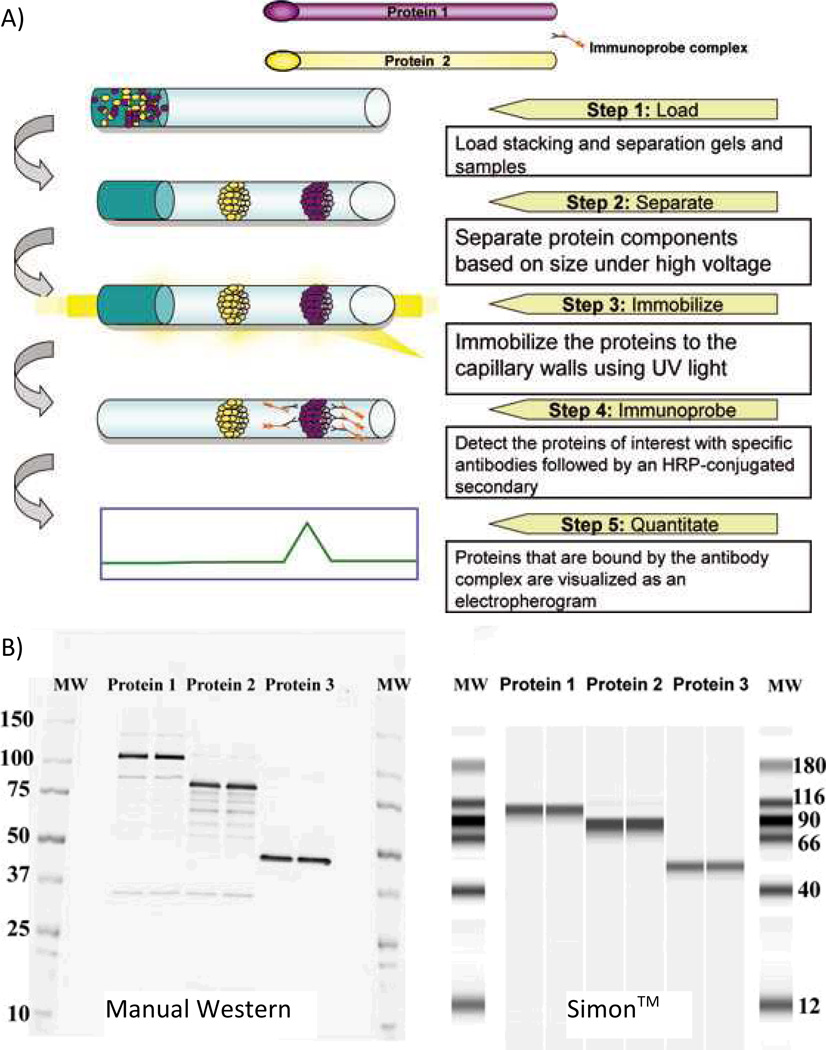
UV absorbance and LIF spectroscopy are the dominant detection methods for SDS-CGE. However, for detection of specific mAbs, Western blot immunoassay detection has also been utilized. The ProteinSimple Simple Western™ (or Simon™) automates the immunoassay detection procedure by performing all separation steps and washes in-capillary. Following a SDS-CGE separation, the proteins are photochemically cross-linked to the capillary wall, where they are exposed to a horseradish peroxidase-conjugated secondary antibody for whole-capillary chemiluminescence imaging (Fig. 3). Simon™ also makes quantitative Western blots possible. Using this instrument, a standard curve was generated for a vaccine candidate protein with linearity from 0.45–7 µg/mL and R2 values of 0.990 or greater for five experiments [42].
Fig. 3.
A) Step-by-step overview of the Simon™ operational procedure. B) Comparison between the manual Western and Simon™ for duplicate runs of three proteins. Experimental details for protein and antibody conditions in ref. 31. Reprinted with permission from ref. 42
Immunoassay detection methods for CGE are useful because coupling CGE with MS by electrospray ionization (ESI) is difficult due to the presence of nonvolatile BGE. However, CGE-SDS has been coupled successfully to matrix-assisted laser desorption ionization (MALDI) MS by moving a poly(tetrafluoroethylene) membrane past the end of the capillary to collect the peaks as they leave the capillary [43]. CGE-MALDI-MS has been utilized for the direct mass measurement of recombinant proteins [44, 45] and neoglycoproteins [46].
2.3 Capillary isoelectric focusing
Another capillary-based technique that was adapted from its original slab-gel format is capillary isoelectric focusing (CIEF). Like SDS-CGE, performing IEF in a capillary exhibits the benefits of faster analysis times, higher resolutions (up to 0.005 pH units [47]), lower limits of detection, and the capacity for high-throughput analysis [48].
CIEF separates proteins based on their isoelectric point (pI) and can be used to determine charge heterogeneity of biogenic products [49]. The assay is typically performed in a coated capillary to eliminate EOF. A pH gradient is self-assembled under an electric field using a mixture of mobile carrier ampholytes with a distribution of pIs. The anodic end of the capillary is then placed in an acidic solution and the cathodic end in a basic solution. Under the applied electric field, the protein will migrate through the ampholyte solution toward the oppositely charged electrode until the pH environment equals its pI.
UV detection at 280 nm is typically used with CIEF because the ampholytes exhibit strong absorbance at wavelengths below 240 nm [50]. Optical detection for CIEF can be accomplished either by a two-step method that requires mobilization after focusing to bring the analyte bands past a small detection window or using whole-capillary imaging CIEF (iCIEF) within a transparent capillary.
An important application of CIEF for the analysis of biologics is for the characterization of charge heterogeneity, as it is possible to identify proteins based on their unique charge profile [51]. Variations in this charge profile are often used to determine protein stability [52, 53] and identify degradation products or PTMs that change the charge of the protein, such as glycosylation and deamidation [54].
As mentioned earlier, deamindation can be a major pathway of protein and peptide degradataion. The rate of deamidation depends on both the primary and secondary structure surrounding the Asn or Gln residue in question [1]. Typically characterization of deamidation sites is accomplished through peptide mapping and MS analysis. However, this process can be complicated, sometimes impossible when a fragment contains multiple desamino sites. Shimura et al. used CIEF and site-directed mutagenesis to determine the rates of deamidation in Fab fragments of mouse IgG1-κ [54]. The rate of disappearance of the parent peak of each mutant was compared to that of the wild type to obtain the single-residue deamidation rates. By monitoring the CIEF charge profile of the six Fab mutants for the additional acidic peaks, a third, previously unknown deamidation hotspot for the mouse IgG1-κ was identified.
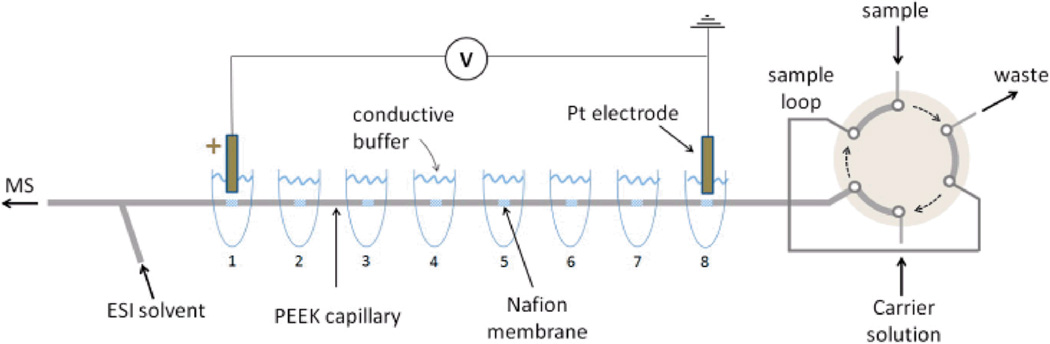
CIEF can be even more powerful when run in combination with an orthogonal separation technique such as SDS-CGE [55] or reversed-phase LC, or in tandem with MS. CIEF has been coupled to MS through both ESI [56, 57] and MALDI interfaces [58, 59]. Due to the presence of the non-volatile ampholytes in the separation buffer, coupling CIEF with ESI can be complicated by ion-suppression and source contamination. To cut down on the intensive sample preparation needed to desalt protein samples from gels, a segmented capillary has been described. In this design, seven segments of PEEK capillary were connected by Nafion joints, each with its own buffer reservoir (Fig. 4) [60]. This allowed analytes in the capillary segments to be selectively mobilized after focusing, creating an online fractionator prior to additional analysis by LC, CE, or MS.
Fig. 4.
Schematic layout of the on-line multiple junction CIEF setup; the six-port injector is shown in the sample-loop loading position. Reprinted with permission from ref. 60
Additional technological advances in CIEF-MS interface development have been reported by Zhong et al. [61] and Wang et al. [62]. Their work is discussed further in the MS detection section of this review. Along with the development of new interfaces, several straightforward BGE buffer modifications have been described in the literature to solve the problems of high backgrounds and ion suppression [63, 64].
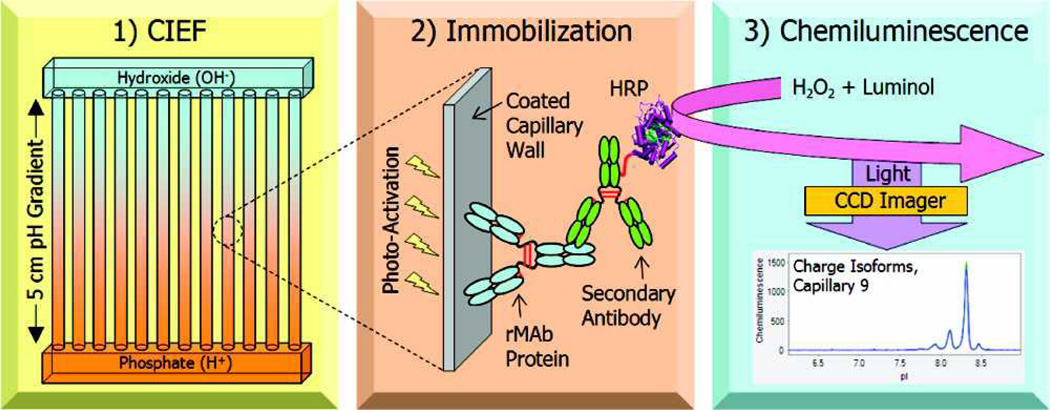
As with SDS-CGE, detection of proteins by immunoassay following separation by CIEF can be used to improve detection limits and specificity without the need for an MS. For example, Michels et al. have described the first multiplexed iCIEF immunoassay for investigation of the charge heterogeneity of mAbs [65]. Once the mAbs were focused, they were then photochemically immobilized to the capillary wall where they were then exposed to a secondary antibody, conjugated with horseradish peroxidase, and detected by chemiluminescence (Fig. 5). The resulting LOD of this assay was 6 ng/mL, which was a 1000-fold increase over UV detection.
Fig. 5.
Schematic of the Nanopro three-step process . 1) Separation by CIEF, 2) immobilization of the antibody to the capillary wall, 3) detection with secondary antibody by chemiluminescence. Reprinted with permission from ref. 65
2.4 Capillary electrochromatography
Capillary electrochromatography (CEC) is a technique that uses both chromatographic retention and electrophoretic migration for the separation of analytes, with bulk fluid flow created by the EOF. This combination enhances the selectivity and efficiency of the separation, drastically lowers the reagent use compared to LC, and enables the separation of neutral species not possible with CZE.
In the first applications of CEC to proteins, capillaries packed with porous particles were utilized because of their similarity to the stationary phase materials used for conventional LC, and the commercial availability of particles with a variety of functionalities. However, the packed CEC columns have significant limitations in terms of stability and fabrication reproducibility and are not yet able to match the robust performance of nano-LC [66]. This limits their usefulness for routine protein assays on a larger industrial scale. In its place, the use of nanoparticles (NP) as a pseudostationary phase (PSP), open-tubular CEC (OTCEC), and monolithic columns have gained momentum.
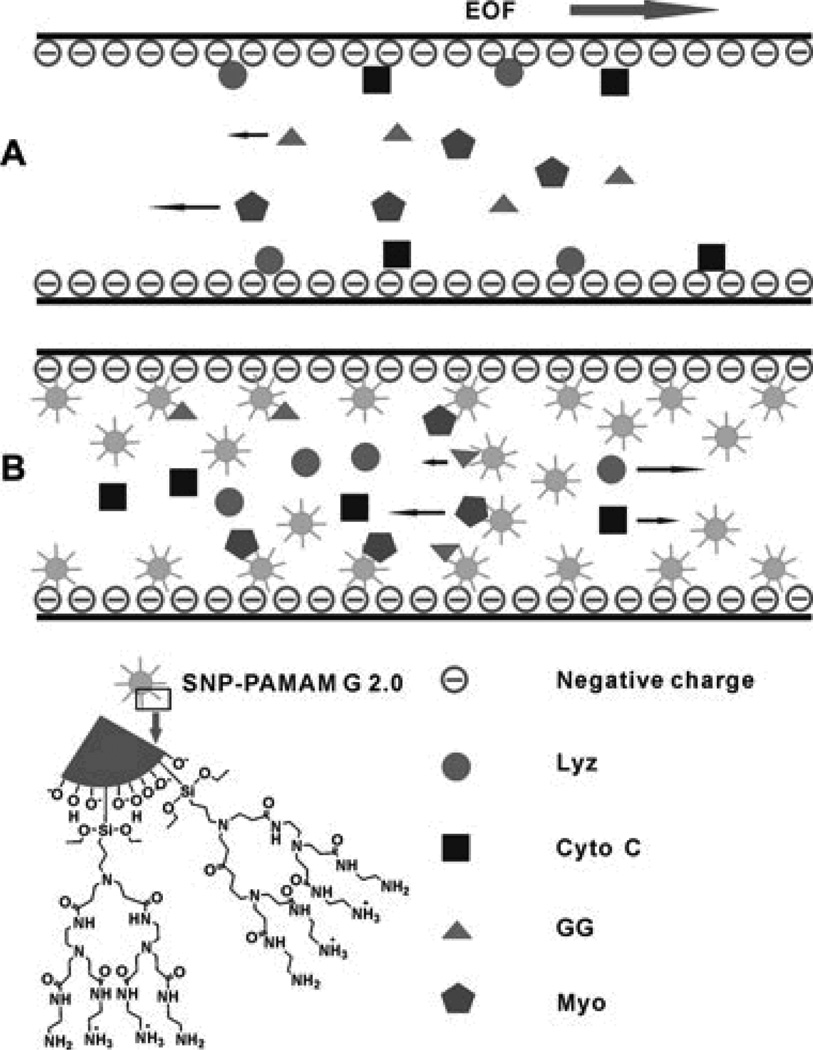
The use of NP as a PSP for CEC has been thoroughly reviewed [67]. In the BGE, the NP can interact with the proteins during the separation, changing their electrophoretic mobility and generating a separation based on the difference in affinity between the analytes for the NP. A wide range of materials have been investigated for PSP-CEC, including polymer NP, carbon nanotubes, gold NP, and silica NP (SNP) [67]. To improve the stability and functionality of SNP, Gao et al. synthesized polyamidoamine-grafted SNP (PAMAM-SNP) and utilized them for a separation of basic and acidic proteins [68] (Fig. 6). With 0.01% PAMAM-SNP in the BGE, a complete separation of all four model proteins (Table 4) was possible. Additionally, the PAMAM-SNP were able to effectively reduce the adsorption of basic proteins to the capillary wall.
Fig. 6.
A) Diagram of the separation of four proteins without and B) with the pseudostationary phase effect of the polyamidoamine (PAMAM)-grafted silica nanoparticles (SNP). Reprinted with permission from ref. 68
Table 4.
CEC
| Analyte | Column type | Column material | Capillary | BGE | Voltage | Detection | Notes | Ref. |
|---|---|---|---|---|---|---|---|---|
| Cytochrome c, myoglobin, gamma globulin, and lysozyme |
Pseudostationary phase |
Polyamidoamine-grafted SNP |
48 (38) cm, 75 µm id |
12.5 mM tetraborate/ phosphate, pH 9.1, 0.01% SNP |
+ 15 kV | UV 214 nm | Improved SNP stability | [68] |
| Tryptic digests of BSA And human transferin |
Open-tubular | Bare AuNP on sol-gel modified surface |
47 (40) cm, 50 µm id |
100 mM sodium phosphate, pH 2.5 |
+ 10 kV | UV 214 nm | AuNP OT column for tryptic digests of native and glycated proteins |
[70] |
| Bradykinin, LHRH, oxytocin, angiotensin I, met-enkephalin, and HAS tryptic digest |
Open-tubular | AuNP on (3-aminopropyl) triethoxysilane modified surface |
41.2 (31) cm, 75 µm id |
20 mM potassium phosphate, pH 8.0 |
+ 12 kV | UV 214 nm | New preparation of AuNP OT column with improved stability |
[71] |
| Egg while proteins | Open-tubular | Graphene oxide and graphene |
60 (50) cm, 75 µm id |
5 mM sodium phosphate, pH 7.0 |
+ 20 kV | UV 214 nm | Separation was only Possible with the GO column |
[72] |
| Egg while proteins | Open-tubular | Ionic adsorption of GO to surface modified with PDDA |
60 (50) cm, 75 µm id |
5mM sodium phosphate, pH 7.5 |
+ 20 kV | UV 214 nm | Improved assembly and stability of GO OT columns |
[73] |
| Cytochrome c and BSA | Open-tubular | Mixture of four monomers in the presence of 1-propanol as sole porogen |
35 (25) cm, 75 µm id |
5 mM sodium borate, 45% ACN, pH 9.04 and 10 mM Tris-HCl, 0.4% PVP, pH 8.86 |
– 10 kV | UV 214 nm | Improve retention of Proteins in OTCEC |
[74] |
| Many model proteins and tryptic digests of cytochrome c |
Monolith | C−8, −12, and −16 methacrylate with pentaerythritol triacrylate |
27 (20) cm, 100 µm id |
1–10 mM sodium phosphate, ACN, pH 7.0 |
+ 15 kV | UV 214 nm | Neutral monoliths to reduce adsorption |
[75] |
| Cytochrome c equine myoglobin, lysozyme, and BSA |
Monolith | Cationic ionic liquid ViOcIm+ with various anions: Br−, BF4−, PF6−, and NTf2− |
32 (20) cm, 100 µm id |
20% ACN, 30 mM sodium phosphate/citric acid uffer, pH 2.5–4.0 |
− 10 kV | UV 210 nm | Only the anion NTf2− was Able to achieve separation of proteins |
[77] |
Capillary: Actual length (effective length), inner diameter
1-vinyl-3-octylimidazolium (ViOcIm+), Bovine serum albumin (BSA), Gold nanoparticles (AuNP), Graphene oxide (GO), Human serum albumin (HSA), Ionic liquid anions (bromide, Br−; tetrafluoroborate, BF4−; hexafluorophosphate, PF6−; and bis-trifluoromethanesulfonylimide, NTf2−) Open-tubular (OT), Luteinizing-hormone-releasing hormone (LHRH), Poly(diallydimethylammonium chloride) (PDDA), Polyvinylpyrrolidone (PVP), Silica nanoparticles (SNP)
OTCEC columns are a popular alternative to packed columns because of their ease of fabrication and excellent separation efficiency [69]. These OTCEC columns can be made by either physically bonding the stationary phase to the capillary wall or several layered coatings. In one report, OTCEC columns were fabricated through the immobilization of gold NP (AuNP) on the surface of the capillary that had been pretreated with a sol-gel. The gold immobilized in the sol-gel participates in noncovalent interactions with thiol and amino groups of proteins, increasing their capacity factor. Using this technique, Miksik et al. were able to separate the peptides generated by the tryptic digestion of native and glycosylated bovine serum albumin (BSA) and human transferrin [70]. Unfortunately, preparation of the AuNP modified columns required several days and many reaction steps, which limited its utility. To alleviate this problem, a new method for AuNP immobilization to the capillary wall through covalent binding using (3-aminopropyl) triethoxysilane has been described [71]. This procedure creates a stable coating that could be reused over 900 times with migration time RSDs less than 1.7% for model proteins (Table 4).
Another novel OTCEC column was described by Qu et al. and was produced by immobilizing graphene (G) and graphene oxide (GO) sheets to the capillary wall to act as the stationary phase. It was found that between the two coatings, only the GO exhibited a reproducible EOF over the pH range of 3–9 and separate a mixture of egg white proteins [72]. The separation was achieved due to the reverse-phase-like interaction between the graphene coated surface and the proteins. To improve the stacking of GO at the capillary wall, a layer-by-layer technique to produce the GO-modified OTCEC column was reported. In this case, GO nanosheets were adsorbed on a poly(diallydimethylammonium chloride)-treated capillary by electrostatic interaction. This created a stable coating for over 200 runs [73]. Both methods for column fabrication produced excellent run-to-run, day-to-day, and column-to-column reproducibility with less than 3% RSD for the EOF.
Often, OTCEC separations suffer from low capacity factors because of the small active surface area and fewer available functional sites. This also can lead to poor separation efficiency and co-eluting peaks. In an attempt to improve peak capacity, a new porous layer for OTCEC has been described that uses the in situ polymerization of a mixture of monomers in the presence of porogen for higher separation efficiencies [74]. A column generated from the porogen, 1-propanol, was able to generate a high abundance of micropores and mesopores, resulting in a large specific surface area. This generated an efficient separation of the two model proteins, BSA and cytochrome-c.
Another widely explored approach for the implementation of CEC is the use of monolithic columns. Monoliths have high permeability, a fast mass transfer rate, and high loading capacity. Many commercially available monoliths are made from silica leading to a risk of band broadening and sample loss due to protein adsorption. Therefore, to minimize protein adsorption during CEC and improve separation efficiencies, neutral and cationic monoliths have been developed.
A series of neutral nonpolar monolithic columns were manufactured and tested for the separation of both intact proteins and peptides from protein tryptic digest. To produce the monoliths, various ratios of monomers C8-methacrylate, C12-methacrylate, and C16-methacrylate were mixed with the crosslinking polymer pentaerythritol (PETA) [75]. In these experiments, it was determined that when the ratio of monomer to PETA was kept constant, the C8 monolith gave the best separations for intact proteins. The C16 column exhibited the best efficiencies for smaller peptides. In their report, Puangpila et al. claim that, even in the absence of a charged surface, there is EOF generated by adsorption of BGE ions to the monolith and it can be controlled by changes in the pH and ACN content of the mobile phase.
Cationic monolithic columns can also be used to reduce electrostatic interaction of basic proteins to the monolithic and capillary surface. Wang et al. developed a novel monolithic IL column that was made by a simple “one pot” approach using thermal free radical copolymerization [76]. Using this method, several counterions (bromide, tetrafluoroborate, hexafluorophosphate, and bis-trifluoromethanesulfonylimide (NTf2−)) were tested with the cation 1-vinyl-3-octylimidazolium (ViOcIm+) to create an IL monolith capillary columns [77]. Each IL monolith was capable of generating a consistent reverse EOF over the pH range 2.9–12.0. However, only the ViOcIm+NTf2− was able to achieve baseline resolution for all proteins in a standard mix (Table 4).
3. Detection methods
3.1 Spectroscopic detection
Spectroscopy is the most common detection method for proteins and peptides separated by CE. UV absorbance tends to be favored over fluorescence spectroscopy due to a natural absorbance of the amide bonds and aromatic residues in the near UV (214 and 280 nm). However, this approach suffers from poor limits of detection due to the micrometer pathlengths characteristic of CE and high background from the UV source. Additionally, BGE composition, pH, and ionic strength can have a significant effect on background. Approaches such as increasing the pathlength through modification of the detection window using Z-shaped capillaries and bubble-cells have been successful in decreasing the LOD by an order of magnitude or greater [78, 79].
Fluorescence detection of proteins can be accomplished based on the native fluorescence of tryptophan, phenylalanine, and tyrosine residues in proteins using a deep UV light sources [80–82]. However, with native fluorescence based detection, the signal is dependent on the number of excitable residues as well as their accessibility within the tertiary structure of the protein. Therefore, the applicability of this technique varies from protein to protein. To improve the LODs for native fluorescence detection of erythropoietin (EPO), Wang et al. utilized a magnetic bead-based extraction system for pre-concentration. Using this procedure, it was possible to obtain an LOD of 10 nM, two orders of magnitude lower than what was possible with UV absorbance at 214 nm [83].
Low limits of detection achievable by LIF can also be obtained through derivatization of the protein or peptide of interest with a fluorophore [84]. The most common derivatization sites for proteins are the primary amines and cysteine residues. These can be tagged with a variety of agents including Alexa Fluor-based dyes, naphthalene-2,3-dicarboxaldehyde, fluoresceinisothiocyanate, and many others. A major disadvantage of pre-separation derivatization for proteins is the complexity of the derivatization process. This approach requires not only that the tag is specific for the functional group on the analyte of interest but also that it does not interfere with the separation by introducing additional fluorescent by-products. Proteins typically have several reactive sites that can be labeled which leads to multiple peaks for one analyte, complicating data analysis [85].
3.2 Mass spectrometry
CE-MS is a powerful combination of high efficiency separations with selective and sensitive detection. This technique can provide important information on identity, glycoforms, degradation, and impurities of protein therapeutics [86, 87]. It is possible to couple CE to MS using different ionization techniques, as has been described in several excellent reviews [88, 89]. For this review, only the recent advances regarding the development and application of the ESI interfaces will be highlighted. CE was first interfaced with MS by ESI in 1987 [90] and it remains the most popular ionization method due to its broad applicability and commercialization.
ESI is a robust soft-ionization technique that produces multiply charged ions for proteins in the gas phase. However, there are many considerations that must be taken into account when coupling it with CE. Primarily, the use of run buffers containing non-volatile salts and additives can lead to their deposition within the instrument, and subsequent contamination of the source. While formic acid and acetate buffers have been used as BGEs for the separation of proteins by CE, they are not always ideal because of inadequate resolution and possible protein instability at low pH. Additionally, the voltages typically applied to the capillary for separation are 2–3 orders of magnitude higher than what is used for ESI. Toward this end, researchers have developed three general approaches for coupling CE to MS with ESI: sheath-liquid, sheathless, or junction-at-the-tip interfaces.
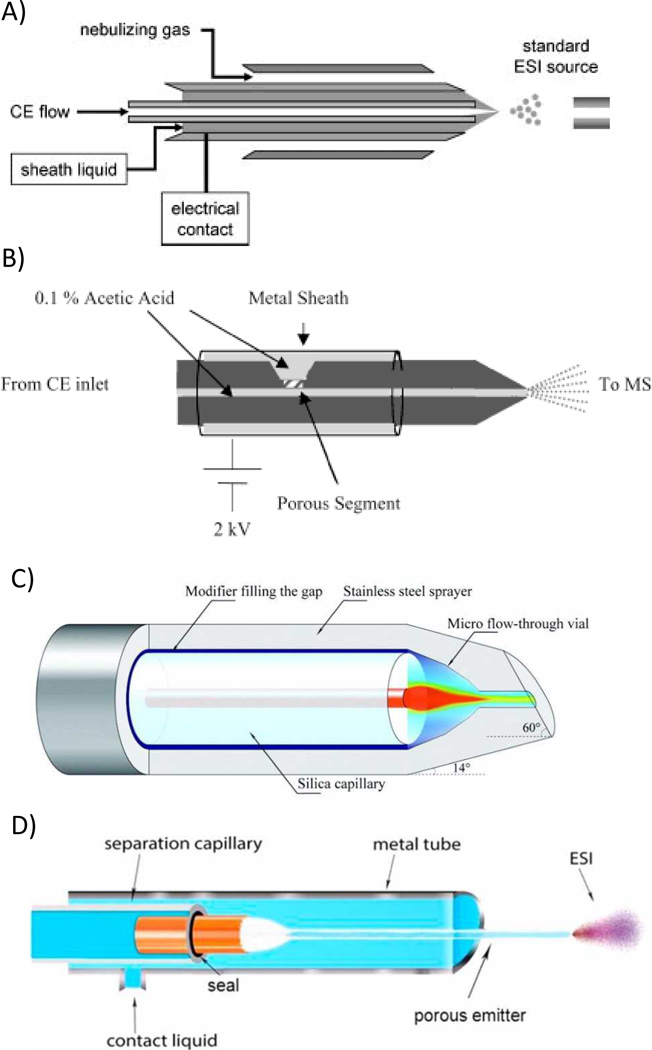
The most widely used and commercially available option is the sheath-liquid interface. This is accomplished by placing the outlet of the CE capillary coaxially within a tube. The tube delivers a MS-compatible sheath liquid (Fig. 7A) that provides easy electrical connections and a flow rate to the ESI of µL/min. This is beneficial because the EOF of the CE is generally much slower (nL/min) than what is compatible for a stable spray.
Fig. 7.
Diagrams of four CE-ESI-MS interfaces. A) Sheath-flow, B) Moini and Whitt sheathless flow, C) Chen junction-at-the-tip, D) sheathless interface for CITP/CZE-nanoESI-MS. Reprinted/adapted with permission from ref. 89, 94, 102, 62 respectively
The compatibility of separation and detection parameters for CE-ESI-MS with a sheath-liquid interface was evaluated for eight model proteins and several EPO isoforms [91]. It was found that the BGE composition and capillary coating play the largest role in the quality of the separation. For all analytes the best signal was obtained with a sheath flow rate between 2–5 µL/min and a sheath flow liquid composed of 1% acetic acid in 1:1 organic:water; in this study, 2-propanol was chosen over MeOH or ACN. The optimal gas pressure was determined to be 0.2 bar, since anything lower lead to a loss of analyte intensity and anything higher was shown to affect the resolution of the separation. As an added benefit, the nebulizer gas pressure can create suction at the capillary outlet, increasing the CE flow rate for separations performed in neutral capillaries.
An obvious disadvantage of the sheath-liquid interface is the loss in detector sensitivity from dilution of the eluting peaks. To improve detection limits, a sheathless interface was developed. The largest downside of this approach is the difficulty in properly completing the electrical circuits for the CE and the ESI. While many attempts have been made, these interfaces were limited by stability and ease of application [89, 92].
Recently, Moini and Whitt developed a sheathless interface based on a porous junction [93, 94]. In this interface, the end of the capillary was made porous to small ions by drilling a well into the polyamide coating and etching the remaining material with hydrofluoric acid. The capillary was then placed within an existing ESI needle filled with BGE, allowing electrical connection to both the CE and ESI (Fig. 7B). The tip of the capillary could then be used for electrospray when voltage is applied. The only drawback to this technique was the difficulty in reproducibly etching the capillary end. To improve the applicability and availability of the Moini and Whitt sheathless interface, Beckman Coulter developed a prototype that has been successfully applied to the analysis of intact proteins [95], protein glycoforms [96], and protein tryptic digests [97, 98].
In a recent report, both CE- and LC-MS were compared for the analysis of a particular therapeutic mAb [98]. With LC-MS, 11 small peptides eluted in the void volume and could not be detected, including two fragments that were critical for the identification of the binding domain of the mAb. The same digest was analyzed by CE-MS employing both a traditional sheath-liquid interface and the Beckman Coulter sheathless interface using a BGE consisting of 10% acetic acid at pH 2.3. Sixty of 61 peptides were detected with the sheath-liquid interface, while all 61 peptides were detected with the sheathless system with higher separation efficiencies and better sensitivity.
Another alternative to the sheath-liquid interface is the junction-at-the-tip design developed by Chen’s group. In this interface, the capillary end is placed within a hollow needle that forms a “flow-through microvial” [99] (Fig. 7C). The hollow needle is filled with a chemical modifier that provides the necessary electrical contacts for the separation and ESI voltages. Similar to a sheath-liquid interface, this modifier increases the CE BGE compatibility with the ESI. However, because the flow rates are much lower (< 1 µL/min), the dilution factor is not significant. Chen’s group has extensively characterized the performance of this interface in several publications [100–102].
Perhaps the most exciting new use of these interfaces is in coupling MS to more complex CE modes, such as CIEF and capillary isotachophoresis (CITP) which require high concentrations of non-volatile components to achieve a separation. In 2011, Zhong et al. described a CIEF-MS approach for the analysis of several model peptides and proteins using the junction-at-the-tip interface with coated and uncoated capillaries [61]. Unfortunately, the ampholytes used for the separation were still able to reach the detector, leading to high backgrounds and ion suppression. To prevent this from occurring, a new sheathless interface was developed by Wang et al. that uses a large bore separation capillary for sample loading and a sheathless interface with a porous emitter for its application with CITP [62] (Fig. 7D). This system was then utilized for the analysis of test peptides spiked into tryptic digests of BSA (Table 5). They were able to obtain a linear range over 4.5 orders of magnitude and a five-fold sensitivity improvement compared to the sheath-liquid interface for two test peptides, kemptide and angiotensin II.
Table 5.
CE-MS and ME-MS
| Analyte | Mode | Interface | Sheath flow | BGE | Capillary coating | MS | Notes | Ref. |
|---|---|---|---|---|---|---|---|---|
| Lysozyme, β-lactoglobulin A, cytochrome c, RNase A, myoglobin, RNase B, trypsin inhibitor, carbonic anhydrase, and EPO glycoforms |
CZE | Agilent sheath-liquid |
2-propanol /water (1:1), 1% acetic acid |
0.5–2 M acetic acid | Various coating solutions and permanently coated capillaries |
TOF | Comparison of coatings. Emphasis on intact protein analysis |
[91] |
| EPO and interferon-β glycoforms |
CZE | Beckman Coulter sheathless |
0.5 mM - 2.0 M acetic acid |
Beckman Coulter neutral bilayer |
TOF | Glycoprofiling of pharmaceutical products |
[96] | |
| Tryptic digest of Trastuzumab | CZE | Beckman Coulter sheathless |
10% acetic acid | Bare fused silica | TOF | Rapid characterization of therapeutic mAbs |
[97] | |
| Tryptic digests of in-house mAbs |
CZE | (1) Agilent sheath- liquid (2) Beckman Coulter sheathless |
(1) 0.1% acetic acid |
10% acetic acid | (1) PVA-coated and bare silica (2) Bare fused silica |
TOF | Improving detection of small peptides from tryptic digrests |
[98] |
| Bradykinin, angiotensin I, neurotensin, fibrinopeptide, substance P, kemptide, leu-enkephalin, angiotensin II, melittin, and renin spiked in tryptic digests of BSA |
CITP | In-house sheathless | 0.1 M acetic acid (90%), MeOH (10%) LE: 25 mM ammonium acetate, pH 4 |
HPMC | TQ | Fivefold sensitivity improvement from the sheath-liquid interface |
[62] | |
| Trypticdigest of in-house IgG2 mAb |
LC-ME | Microchip | 50% ACN, 0.1% formic acid |
APTES | TOF | UPLC followed by MCE and on-chip ESI-MS interface |
[146] |
Capillary: Actual length (effective length), inner diameter
Aminopropyltriethoxysiland (APTES), Bovine serum albumin (BSA), Capillary isotacholphoresis (CITP), Hydroxypropyl cellulose (HPMC), Leading electrolyte (LE), Polyvinyl alcohol (PVA), Triple quadrupole (TQ)
4. Applications
In addition to assessing protein pharmaceutical products based on their size and charge heterogeneity and the presence of impurities, the analysis of biologics poses two additional analytical challenges: 1) how to characterize and better understand the complicated cellular process of glycosylation, and 2) preparing for the onset of biosimilar drugs to the market and how to best prove their similarity to the innovator product.
4.1 Glycosylation
Glycosylation is one of the most prevalent PTMs of therapeutic proteins. In vivo, glycosylation plays several important roles, including protection against degradation and non-specific interactions as well as orientation for the binding domain. The two major types of glycosylation that occur involve N-linked and O-linked carbohydrates. N-linked glycans are attached to the protein backbone at the amine side of Asn and are found in the well-defined amino acid sequence of Asn-X-Ser/Thr, where X is any amino acid but proline. O-linked glycans are not sequence-specific and are found attached to the protein backbone at the OH group of Ser or Thr.
Monoclonal antibody-based therapeutics of the IgG1 sub-type make up a 100 billion dollar annual market [103]. These mAbs consist of 2–3% carbohydrate by mass. Most of the glycosylation occurs as N-linked glycans located on the Asp297 in the CH2 domain of the Fc region of each heavy chain. A number of factors can affect the composition, structure, and frequency of these glycans, posing an interesting challenge for the manufacturing of a homogeneous product. To ensure a homogeneous product and avoid potentially immunogenic glycans, each step of biotherapeutic production from clone selection to lot release needs to be well characterized. This characterization requires fast, high-throughput analytical methods to accurately screen the numerous samples generated per day.
The size and charge characterization of glycoproteins can be accomplished by the various electrophoretic separation techniques mentioned in the previous sections of this review. Several methods and protocols for CZE, SDS-CE, and CIEF separations of glycoproteins have been compiled by Rustandi et al. [104]. A typical downside to CE-based methods is the characteristic migration time irreproducibility. To address this, freely available software, glyXalign, was developed based on a set of rapid algorithms that enables automatic correction of distortions in CGE-LIF data to improve peak identification [105].
For further understanding of the nature, location, and composition of the glycans, methods for the removal and analysis of the sugars themselves are also needed. The majority of these carbohydrate analyses are performed by LC. In particular, hydrophilic interaction chromatography (HILIC) coupled to LIF and MS detection has been useful for the sensitive analysis of glycans [106].
CE-LIF is an excellent orthogonal technique to HILIC-LIF for separation of glycans, and in a comparative study it was shown that they were able to detect an equal number of glycans removed from an IgG [107]. An advantage of CE-LIF for glycan analysis is that it can be used to distinguish both lineage and positional isomers [108, 109]. Using CZE-LIF, carbohydrate sequencing can be performed by both top-down digestion and bottom-up identification using a series of sugar-specific exoglycosidases. Typically, glycans are enzymatically removed, fluorescently labeled, and separated by size or charge. There are several charged fluorescent reagents commercially available for tagging glycans. The most common reagent used in conjunction with CE-LIF is 8-aminopyrene-1,3,6-trisulfonic acid (APTS). However, recently, Kuo et al. published a rapid method for labeling aldoses with 2,3-naphthalenediamine to produce highly fluorescent naphthimidazole derivatives [110]. Using this reagent, it was possible to perform composition analysis and enantioseparation of the glycans using CE with cyclodextrin in the BGE.
An important advantage of CE-LIF over HILIC-LIF is the ability to multiplex 48- and 96-capillary arrays for high-throughput analysis. Callewaert et al. were the first to perform glycan analysis using a commercially available multiplexed CE-based DNA analyzer [111]. Later, this same technique was used along with a 48-capillary array to perform high-throughput analysis of glycans from IgG. In this application, glycans were removed by digestion and labeled with APTS in 96-well plates and then subjected to simultaneous analysis by capillary array. This approach made it possible to run 3000 samples in a single day [112].
In the research and development of mAbs, a particular area of interest is the study of immunogenic non-human glycans. The frequency and type of non-human glycans attached to the therapeutic protein during production differ from cell line-to-cell line [113]. It is well known that the non-human oligosaccharides galactose-α-1,3-galactose (α1,3-Gal) and N-glycolylneuraminic acid (Neu5Gc) can illicit an immune response. In fact, in response to enteric bacteria, approximately 1% of all human antibodies are against the α1,3-Gal epitope [114].
Detection of both α1,3-Gal and Neu5Gc non-human glycans was performed by partial filling affinity CE. In this method, a plug of either anti-Neu5Gc antibody or α-galactosidase (dissolved in BGE) was injected on capillary prior to injection of the APTS-labeled glycans (removed from the target antibody) [115]. Once the electric field was applied, the higher mobility sugars in the sample pass through the antibody or enzyme plug, causing a reaction. This reaction produced additional product peaks upon LIF detection, allowing specific detection and quantification of the two immunogenic sugars.
In another study, six commercially available mAb pharmaceuticals produced in nonhuman mammalian cell lines were analyzed by CZE-LIF, in parallel with LC-ESI-TOF-MS, to determine the presence of nonhuman N-glycans [116]. By CZE, forty-six fluorescently labeled N-glycans were separated using a tris-borate BGE containing 5% PEG to slow the EOF. Of the six mAb pharmaceuticals, three were found to contain nonhuman N-glycan residues. To obtain additional information regarding the attachment of nonhuman N-glycans to therapeutic proteins, CZE-LIF with exoglycosidase digestion and fluorescent tagging was used to achieve LODs of 1 µg allowing characterization of the low-abundance α1,3-Gal epitope [117].
CE-MS can also be used in conjunction with CGE-LIF [118] to obtain additional structural information and identify unknown glycans [2]. For example, Bunz et al. described both alkaline and acidic BGE systems that could be used for the determination of APTS-labeled mAb glycans by CE-TOF-MS [119, 120]. The CE-MS methods were then compared against to two CGE-LIF methods commonly used for routine glycan analysis. While both CE-MS and CGE-LIF were able to resolve and detect the glycans, because of the difference in the separation mechanisms they had different migration orders, making it difficult to directly compare the two electropherograms obtained for a complex sample.
The downside of glycan analysis by MS is the likelihood of unwanted fragmentation of sugars during the ionization process. This can lead to large amounts of difficult-to-interpret data and misidentification [121]. For this reason, it is important not only to insure careful optimization during MS method development but to provide orthogonal analyses such as CZE-LIF or CGE-LIF to validate the findings.
4.2 Biosimilars
Follow-on biologics, also known as biosimilars or biobetters, is the term for the “generic” biopharmaceuticals that have recently entered the market. The European Medicines Agency published regulatory guidelines for biosimilars in 2005, and by 2012 there were 14 products approved for sale in Europe [122]. In 2013 the first mAb biosimilar, Hospira’s Inflectra, hit the European market, and more than a half-dozen prospective biosimilars are in the pipeline. In 2015, as the majority of the leading biologics go off patent, there will be ample opportunity for established and start-up companies to begin producing biosimilars.
While production of biosimilars is an inherently less risky venture, due to the established market and tested safety of the innovator product, proving comparability to regulatory agencies still poses a significant challenge. Unlike chemical synthesis of small molecule generics, the composition of biologics is highly dependent on the manufacturing process. Small changes in production can have significant implications on the quality. In particular, the addition of impurities, aggregation products, and/or PTMs such as glycans can cause the protein to be immunogenic. Without detailed knowledge of how the innovator was produced, it can be very difficult to create an identical product.
Fortunately, dozens of analytical techniques exist to verify the physicochemical and functional comparability of the biosimilar to the innovator [123]. As discussed in the previous sections of this review, electrophoretic techniques are widely used for characterization of size and charge heterogeneity, product degradation, and PTMs. The appropriate method is generally chosen based on protein complexity, which varies from small non-glycosylated proteins like insulin and HGH to large, heterogeneous glycoproteins and mAbs [124].
EPO is a glycoprotein with approved biosimilars making up 12% of its market [122]. EPO has three complex N-glycosylation sites and one O-glycosylation site, which introduce a high level of heterogeneity into the protein. To be able to differentiate between the various formulations of EPO, or prove similarity between innovator and biosimilar, Taichrib et al. evaluated two multivariate statistical approaches for the analysis of CE-MS data [125]. The data were generated using a CE-ESI-TOF-MS method developed previously that exhibited high separation efficiencies and high selectivity for 14 commercially available preparations of EPO [91]. Both statistical approaches proved useful for analyzing the similarity or difference between large sets of glycosylated biologics that were generated under different production conditions, cell lines, and various batch numbers.
With the upcoming mAb biologic patent cliff, much of the biosimilar research has focused on the comparability of antibodies from various sources. Towards this end, CZE [126] and SDS-CGE [127] techniques can be used to determine charge heterogeneity of mAbs. Using CZE, rituximab (Kikuzubam® and Reditux®) and trastuzumab biosimilars were analyzed with respect to existing commercial products, Mabthera® and Herceptin®, respectively [126]. The CZE methods were then compared to existing CIEF and chromatographic methods (HILIC and cation exchange chromatography). They found that, not surprisingly, a single method was not sufficient to resolve and characterize a protein, putting the emphasis on orthogonal techniques. However, they did report that CZE and CIEF gave better resolution of the mAbs than either HILIC and cation exchange chromatography, especially when using coated capillaries, since protein adsorption tends to lead to band broadening.
With the multitude of assays that exist, reproducibility and ruggedness is essential for widespread biosimilar production and regulation. The innovator, the biosimilar manufacturer, and the regulatory agency need to be certain that, despite the various laboratory conditions, the experimental results are comparable. To help facilitate this, Salas-Solano et al. evaluated an iCIEF method in 12 different laboratories across the world using several analysts, a variety of ampholytes, and multiple instruments [128]. The combined precision for the 12 labs was 0.8% RSD for the pI determination and 11% RSD for the percent peak area values for the charge variants of a therapeutic mAb. This study compared these values to those obtained using conventional CIEF, where the RSDs for pI and peak area were of 0.8% and 5.5%, respectively [129].
5. Microchip electrophoresis
Many aspects of CE, such as low sample volume requirements, speed, efficiency, and the ability to use physiologically appropriate BGEs, make it an attractive method for the analysis of biopharmaceuticals. The advantages CE offers over chromatography are a function of the small inner diameter of the capillary. Consequently there has been an effort to further miniaturize bench-top CE instrumentation to a microfluidic format. This has decreased samples sizes needed for analysis from mL to µL, reduced analysis times from minutes to seconds, increased separation efficiencies, decreased costs, and added the ability for portable point-of-care analysis. Additionally, multiplex ME systems can be designed to handle high-throughput analysis on a greater scale than CE systems, making them an attractive technology for drug discovery and analysis [130, 131].
While most CE separation modes can be transferred to ME, the majority of the current published assays have dealt with analysis of biomarkers and small molecule drugs. Recent advances in N-glycan profiling by ME have also been made for clinical chemistry applications [132–134]. However, as the field of protein analysis on-chip grows, so does the possibility that the use of these devices will soon be accepted by the FDA as a validated method, allowing them to be incorporated into industry protocols.
5.1 Microchip gel electrophoresis
The LabChip® GXII, a commercially available microchip gel electrophoresis (MGE) system from PerkinElmer, is used frequently in the pharmaceutical industry [135, 136]. The commercial procedure, which uses indirect fluorescence and a HT Protein Express gel matrix [137], was compared against two new SDS-MGE methods, one for “high-sensitivity” and the other for “high-resolution” [138]. In the “high-sensitivity” method, direct LIF detection of fluorescently labeled proteins was investigated. Two labeling schemes were compared, and it was reported that performing the labeling step prior to protein denaturation improved the signal up to 50-fold for a loading concentration LOD of 1 ng/mL. In the “high-resolution” method, the sieving effect of the commercial gel was increased by the addition of a 6% poly(N,N-dimethylacrylamide) (PDMA) solution. With an optimal ratio of 2:1, gel:PDMA, the assay achieved resolution between Fab heterodimers without increasing the separation time. Additional high-throughput analysis is available to process 96 samples in less than an hour.
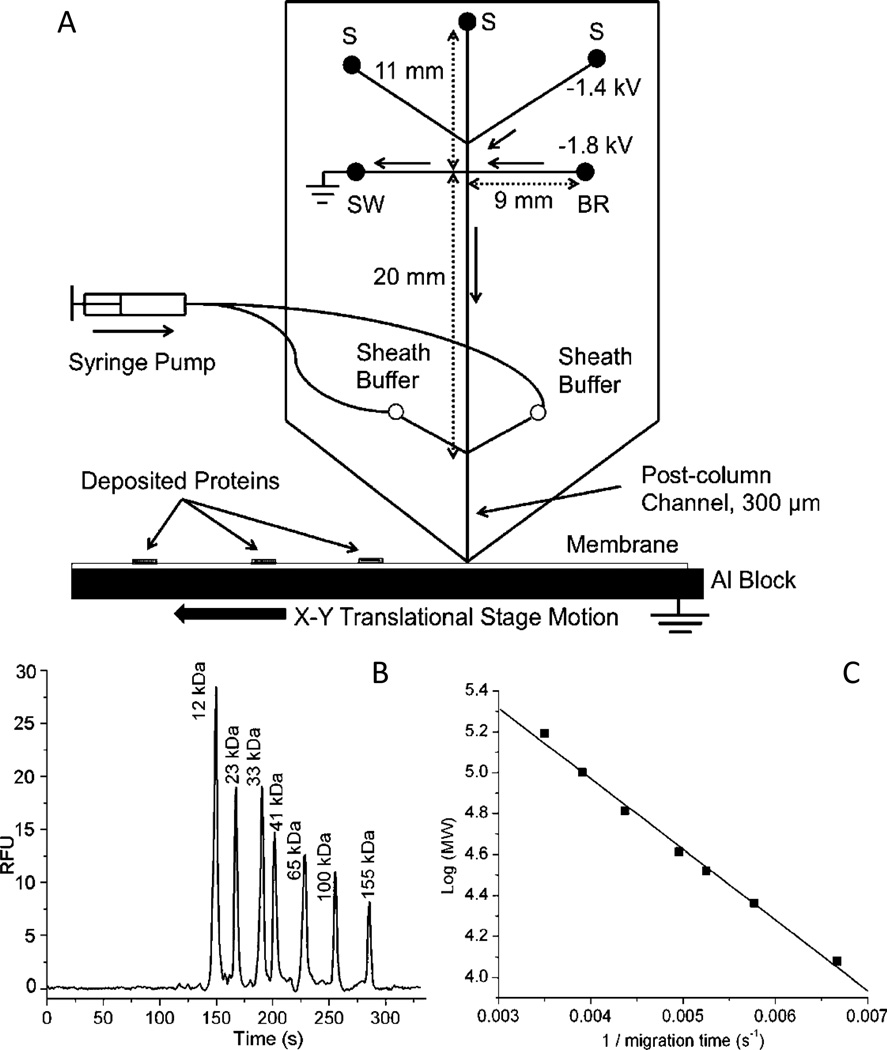
SDS-MGE has also been integrated with Western blot immunoassay detection [139]. The separation of a series of test proteins with a MW range of 11–155 kDa (Table 2) was performed on-chip by Jin et al. [140]. Following the separation, the sample was eluted from the chip onto the Western blot membrane. In order to maintain the discrete zones accomplished during the separation, the chip was held in place vertically while the membrane moved below the outlet on an X–Y stage for spotting (Fig. 8). By carefully controlling the membrane spotting rate and the flow from the SDS-MGE chip, separation efficiencies of 40,000 theoretical plates were possible. With this set-up, the throughput capabilities were improved with a total analysis time of less than 32 min for the separation and immunoassay. This is a dramatic improvement over the traditional Western assay that takes several hours to complete.
Table 2.
CGE and MGE
| Analyte | Mode | Gel | Capillary | Voltage | Detection | Notes | Ref. |
|---|---|---|---|---|---|---|---|
| In-house IgG2λ, IgG2K, and IgG1K mAbs |
Nonreducing | Beckman Coulter SDS-MW gel buffer |
30 (20) cm, 50 µm id |
15 kV | UV 220 nm | Monitor disulfide reduction during production |
[36] |
| In-house IgG1 mAbs |
Reducing and nonreducing |
Agilent High Sensitivity Protein 250 Kit |
Agilent 2100 Bioanalyzer |
NR | LIF | Characterization of size variants |
[37] |
| In-house IgG1 and IgG4 mAbs |
Reducing and nonreducing |
Beckman Coulter SDS-MW gel buffer |
31.2 (20) cm, 50 µm id |
−15 kV | LIF ex. 488 nm / em. 600 nm |
Impurity analysis | [38] |
| IgG 1 mAb | Reducing | Beckman Coulter SDS-MW gel buffer |
30.2 (20) cm, 50 µm id |
−15kV | PDA | Comparison of SDS-CGE To SDS-PAGE |
[39] |
| Myoglobin, carbonic anhydrase I, ovalbumin, BSA |
Nonreducing | Beckman Coulter SDS-MW gel buffer |
33 (24.5) cm, 50 µm id |
−16.5 kV | 220 nm | Demonstration of improved precision |
[40] |
| In-house Fc-fusion proteins, and IgG1 and IgG2 mAbs |
Reducing and nonreducing |
Beckman Coulter SDS-MW gel buffer |
30 (20) cm, 50 µm id |
15 kV | 220 nm | Automated sample preparation |
[41] |
| In-house vaccine proteins |
Reducing | Protein Simple separation matrix |
12-capillary cartridge; 5 cm, 100 µm id |
250 V | Chemiluminescence from secondary antibody |
Automated separation and Western blot |
[42] |
| Ricin A-chain immunotoxins |
Reducing and nonreducing |
Bio-Rad CE-SDS run buffer |
24 (19.5) cm, 50 µm id |
5 or 15 kV | UV 220 nm and MALDI-TOF-MS |
CGE-MALDI-TOF-MS of ricin proteins |
[45] |
| In-house mAbs and proteins |
MGE Reducing and nonreducing |
HT Protein Express gel matrix |
LabChip GXII | NR | Indirect and direct LIF ex. 620 nm / em. 700 nm |
MGE methods for high- resolution and high- sensitivity |
[138] |
| Actin, carbonic, anhydrase II, and, lysozyme |
MGE Nonreducing |
Beckman Coulter SDS-MW gel buffer |
2 cm glass microchip | 480 kV | Western blot | MGE separation with off- chip Western blot detection |
[140] |
Capillary: actual length (effective length), inner diameter
Bovine serum albumin (BSA), Not reported (NR)
Fig. 8.
(A) Microchip overview. Samples are loaded in different sample reservoirs (S). Samples are injected by floating the buffer reservoir (BR) and sample waste (SW) with voltage applied between the desired sample reservoir and the Al block at the exit. During separation, flow from the sample reservoir is gated to the sample waste reservoir (SW) using the voltages as shown. During these operations, other sample reservoirs are floating. Sieving media is pumped through the sheath channels to give stable current. Channel lengths are indicated by double arrow lines and direction of flow during separation is indicated by solid, single arrows. B) Size-dependent separation of FITC-labeled protein ladder in microchips. Detection window was set at the end of separation channel, 300 µm away from the chip outlet. Electric field during separation was 240 V/cm. C) Relationship of MW to migration time. Reprinted with permission from ref. 140
5.2 Microchip isoelectric focusing
ME-based systems have also been used to verify the charge heterogeneity of mAbs with microchip isoelectric focusing (MIEF). Using a commercially available MCE-2010 system with whole-channel imaging from Shimadzu, Kinoshita et al. were able to analyze the charge variants of several mAbs [141]. The microchip consisted of two sample wells, one containing an anolyte and the second containing a catholyte, separated by a 2.7 cm channel. Following the separation, the whole channel was imaged with UV detection. To reduce the EOF, 0.2% hydroxyproplymethylcellulose was added to the BGE allowing greater focusing while preventing the non-specific adsorption of protein to the capillary wall. Using the optimized conditions, the authors were able to separate charge variants of three commercially available mAbs (bevacizumab, trastuzumab, and cetuximab) within 200–300 s. These separations were very reproducible (< 0.5% RSD) and were roughly 10 times faster than the corresponding CIEF assay.
To further improve the utility and throughput of MIEF assay a single-channel microchip device where separation, immobilization, and subsequent immunoblot rinse steps could all be performed has been reported [142]. Once the proteins were separated within the pH gradient they were exposed to UV light and covalently cross-linked to a light-activated volume-accessible gel present in the microchip. This technique gave similar capture efficiencies (≈ 0.01%) to previous reports where proteins were immobilized on the inner surface of the capillary [42, 143]. Wash steps were performed by electrophoretic transport on the immobilized protein without concern of sample loss. Using this technique it was possible to complete an isoform assay in less than 120 min, up to 15x faster than the conventional slab-gel followed by Western blot. Such rapid purity assays illustrate the significant advantage ME has over CE and other techniques.
5.3 Microchip electrophoresis-mass spectrometry
As with CE, even more specific and selective detection of analytes is possible by coupling ME to MS. The most common ionization interface for ME with MS is ESI, but MALDI is also possible [144]. The major benefit of ME-ESI-MS is that the flow rate on-chip is compatible with ESI and can therefore be seamlessly interfaced without disrupting the electrophoretic separation. When constructing an ME-MS interface, the geometry of the outlet and flow rate through the capillary must be taken into account given their monolithic construction and integration.
An advantage of ME over CE is that sample preparation and multiple separation methods can be integrated onto a single device prior to the ESI interface. Therefore, the excess dead volumes that are characteristic of conventional systems are eliminated, reducing the band broadening and sample dilution. The Ramsey group reported a fully integrated LC/CE microchip that terminated in an ESI source off the corner of the device in 2011 [145]. The potential combination of LC and ME for a more selective and specific separation is very powerful. In addition, the microchip flow rates are compatible with the ESI. However, a major disadvantage of fully integrated microchips is that the increased complexity of the device makes them difficult to fabricate.
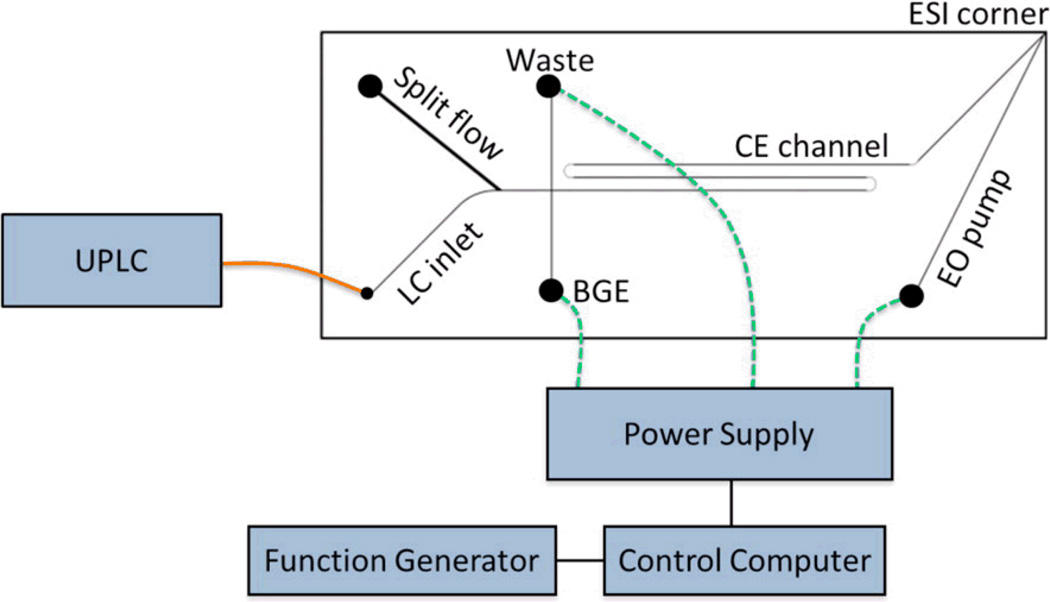
As noted in a subsequent Ramsey paper, the fully integrated chip described above could not handle pressures over 200 bar. Therefore, to improve on the earlier design, a glass microchip that could be integrated with an off-chip UPLC was designed (Fig. 9) [146]. This allowed higher pressures to be reached than were possible with the LC on-chip. The new device produced significant improvements in reproducibility and peak capacity when evaluated for the analysis of digested N-glycosylated proteins. Additionally, the authors point out the utility of the new design in its ability to integrate to existing LC equipment that is already ubiquitous in industry.
Fig. 9.
Schematic of the hybrid capillary LC microchip CE-ESI experimental setup. The orange line represents a transfer capillary connecting the LC column to the microfluidic device. The dashed green lines represent electrical connections between the high voltage power supply and the microfluidic reservoirs. Reprinted with permission from ref. 146
6. Conclusions and future perspectives
The development of protein and peptide therapeutics is a complex and high-risk venture, as products produced by recombinant expression are inherently heterogeneous. However, with advances in analytical techniques, thorough protein characterization is possible. In particular, CE-based separation techniques such as CZE, CGE, CIEF, and CEC provide versatile, efficient, and fast analyses of proteins. Additionally, CE-based techniques have the potential for high-throughput analysis using capillary arrays. The wide range of capillary-based separations can assess many aspects of protein stability, process impurities, and PTMs such as glycolysis, each of which is essential in providing a safe, effective, and quality product.
Microchip based formats have the potential for increased speed, higher throughput, and portability of CE. While the development of ME devices is still primarily an academic research area, there is considerable promise for this miniaturized technique in the future of on-site pharmaceutical analysis. Pharmaceutical applications of ME and CE to therapeutic protein analysis will be further expanded through the development and commercialization of specialty capillaries, BGEs, and detection techniques. This is especially true for CE-MS and ME-MS interfaces. With numerous reviews already existing on this topic alone, coupling of MS with these techniques show great promise in the future for therapeutic protein analysis.
Supplementary Material
Table 3.
CIEF and microchip IEF (mIEF)]
| Analyte | Mode | Capillary coating | Capillary | Detector | Ampholyte | Catholyte | Anolyte | Notes | Ref |
|---|---|---|---|---|---|---|---|---|---|
| In-house EPO, Fc-fusion protein, and IgG |
iCIEF | Fluorocarbon (Protein Simple) |
iCE280 Analyzer; 50 mm, 100 µm id |
UV 280 nm | Pharmalyte pH 3–10, 4–6.5, 5–8, and 8–10.5 |
0.1 M NaOH in 0.1% MC |
0.08 M phosphoric acid in 0.1% MC |
Wide range of Therapeutic protein applications |
[51] |
| In-house noninfectious virus-like particles |
iCIEF | Fluorocarbon (Protein Simple) |
iCE280 Analyzer; 50 mm, 100 µm id |
UV 280 nm | Pharmalyte pH 2.5–5 and 3–10 |
0.1 M NaOH in 0.1% MC |
0.08 M phosphoric acid in 0.1% MC |
Charge characterization of virus-like particles |
[52] |
| In-house vaccine carrier protein |
iCIEF | Fluorocarbon (Protein Simple) |
iCE280 Analyzer; 50 mm, 100 µm id |
UV 280 nm | Pharmalyte pH 3–10 and 4–6.5 |
0.1 M NaOH in 0.1% MC |
0.08 M phosphoric acid in 0.1% MC |
Characterization of polysaccharide vaccine carrier protein |
[53] |
| In-house IgG2κ mAb |
CIEF | PDMA | 180 mm, 50 µm id |
LIF ex. 543.5 nm / em. 590 nm |
Pharmalyte pH 3–10, 0.001% BSA |
20 mM NaOH | 20 mM phosphoric acid |
Determination of deamidation rates in mAb |
[54] |
| Trypsinogen, β-lactoglobulin, BSA, and ovalbumin |
CIEF | 4% acrylamide 0.6% cross- linker |
Various capillary lengths and id |
LIF ex. 488 nm | BioRad pH 3–10 | 20 mM NaOH | 10 mM phosphoric acid |
Microchip interface for 2D CIEF and CGE separations |
[55] |
| β-lactoglobulin A, hemoglobin A, myoglobin, α-chymotrypsinogen A, ribonulcease A, cytochrome c, lysozyme |
CIEF | Seven PEEK tubing segments, 1.55 cm, 395 µm id connected by Nafion membrane 0.2 cm, 330 µm id |
TOF and Orbitrap | Various MS-compatable carrier ampholytes |
Various electrolytes at each Nafion junction modified the local pH of the carrier ampholyte |
Segmented capillary for selective mobilization |
[60] | ||
| Insulin receptor and protein tryptic digests |
CIEF | LPA | 50 cm, 50 µm id | Sheath flow ESI- Orbitrap-MS |
Glutamate, asparagine, glycine, proline, histidine, and lysine |
0.1% Formic acid, pH 2.5 |
0.3% ammonium hydroxide, pH 11 |
Amino acids used as low MW ampholyte |
[64] |
| In-house IgG mAb |
iCIEF | Proprietary photoreactive layer |
12-channel cartidge; 50 mm, 100 µm id |
Chemiluminescence from secondary antibody |
Pharmalyte pH 5–8 (30%) and pH 8–10.5 (70%) |
0.1 M NaOH in 0.1% MC |
0.08 M phosphoric acid in 0.1% MC |
CIEF with immunoassay detection |
[65] |
| Donated mAb products | imIEF | Uncoated quartz | MCE-2010 system, 2.7 cm |
UV 280 nm | Protein Simple pH 3–10, 5–8, and 8–10.5 |
300 mM NaOH, 0.4% HPMC |
200 mM phosphoric acid, 0.4 % HPMC |
mIEF of mAb charge variants |
[141] |
| Model Proteins | iMIF | 100 µm id | Immunoblot | Polyprotic carboxylic amino acids |
20 mM lysine | 20 mM arginine | Integrated microchip for separation and immunoblot |
[142] | |
Capillary: actual length, inner diameter. Bovine serum albumin (BSA), Linear polyacrylamide (LPA), Methylcellulose (MC), Polydimethylacrylamide (PDMA), Polyvinyl alcohol (PVA),
Table 6.
CE-based analysis of protein glycosylation
| Analyte | Mode | Capillary | BGE | Detection | Label | Notes | Ref. |
|---|---|---|---|---|---|---|---|
| N-glycans of mAb1 | CGE | 48 capillary array, 50 cm each |
Applied Biosystems Pop-7™ |
LIF ex. 473 nm / em. 520 nm |
APTS | High-throughput glycan analysis | [112] |
| (1) Neu5Gc and (2) α1,3-Gal ontaining N-glycans |
Partial-filling CE | 40 (30) cm, 50 µm id DB-1 capillary |
100 mM Tris-acetic acid, 0.05% HPC, pH 7.0 |
LIF ex. 488 nm / em. 520 nm |
APTS | Reaction with (1) anti-Neu5Gc or (2) α-galactosidase inject prior to sample |
[115] |
| Non-human -glycans |
CGE | 40 (30) cm, 100 µm id DB-1 capillary |
100 mM Tris-borate, 5% PEG, pH 8.3 |
LIF ex. 325 nm / em. 405 nm |
2-AA | Analysis of commercially available mAbs |
[116] |
| α1,3-Gal containing N-glycans |
CGE | 60 (50) cm, 50 µm id eCAP NCHO coated |
Beckman Coulter Carbohydrate Separation Gel Buffer-N |
LIF ex. 488 nm / em. 520 nm |
APTS and AMAC |
Ultrasensitive detection method | [117] |
| N-glycans of mAb | CGE | 50 (40) cm, 50 µm id PVA coated |
Beckman Coulter Carbohydrate Separation Buffer Or 40 mM EACA-acetate, 0.2% HPMC |
LIF ex. 488 nm / em. 520 nm |
APTS | Glycans of mAbs from NS0 cells | [118] |
| N-glycans of fusion protein |
CE | 90, 60, or 43 cm | 0.7 M ammonia and 0.1 M EACA in 70% MeOH |
TOF-MS | APTS | Alkaline CE-MS method | [119] |
| N-glycans | (1) CGE (2) CE |
Various capillary lengths and coatings |
(1) Beckman glycan separation buffer or POP-7 polymer (2) 40 mM EACA, 131 mM acetic acid, pH 4 |
(1) LIF ex. 488 nm / em. 512 nm (2) TOF-MS |
APTS | CGE-LIF and CE-MS methods compared to the CE-MS method in ref [119] |
[120] |
| N-glycans of mAbs | CE | 50 (40) cm, 50 µm id N-CHO coated |
Beckman Coulter Carbohydrate Separation Gel Buffer |
LIF ex. 488 nm / em. 520 nm |
APTS | CE-LIF as an orthogonal technique to MS |
[121] |
Capillary: Actual length (effective length), inner diameter, coating
2-aminoacridone (AMAC), 2-aminobenzoic acid (2-AA), 9-aminopyrene-1,3,6-trisulfonic acid (APTS), ε-aminocaproic acid (EACA), Galα1–3Gal (α1,3-Gal), Hydroxypropyl cellulose (HPC), Hydroxypropylmethylcellulose (HPMC), N-Glycolylneuraminic acid (Neu5Gc), Polyvinyl alcohol (PVA), Poly(ethylene glycol) (PEG),
Table 7.
CE-based analysis of biosimilars
| Analyte | Mode | Capillary | Coating | BGE | Detection | Notes | Ref. |
|---|---|---|---|---|---|---|---|
| EPO glycoforms | CZE | 60 cm, 50 µm id | UltraTrol™ LN | 1 M acetic acid | TOF-MS | Multivariate statistical approach for glycoform analysis |
[125] |
| (1) Rituximab. trastuzumab, and ranibizumab (2) Infliximab and bevacizumab |
CZE |
40.2 (30.2) cm, 50 µm id |
Polyacrylamide |
(1) 200 mM EACA-acetic acid, 30 mM lithium acetate, 0.05% HPMC, pH 4.8 (2) 150 mM EACE-acetic acid, 20 mM lithium acetate, 0.05% HPMC, pH 5.5 |
UV 214 nm |
CZE methods tested against orthogonal techniques for mAb characterization |
[126] |
| Rituximab | CGE | NR | NR | NR | UV 214 nm | Size heterogeneity of mAbs | [127] |
| Anti-α1-antitrypsin mAb |
iCIEF | iCE280 Analyzer; 50 mm, 100 µm id |
Fluorocarbon (Protein Simple) |
Pharmalyte pH 5–8 | UV 280 nm | Interlaboratory study for robustness |
[128] |
Capillary: Actual length (effective length), inner diameter
ε-aminocaproic acid (EACA), Hydroxypropylmethyl cellulose (HPMC), Not reported (NP)
Acknowledgments
The authors would like to thank the University of Kansas for continued support and Nancy Harmony for proofreading and editorial assistance. We gratefully acknowledge funding from the National Institutes of Health, grant number NINDS R01-NS042929. Additionally, JSC and NJO were participants of the Biotech Training Grant NIGMS T32-GM008359
List of abbreviations in alphabetical order
- ViOcIm+
1-vinyl-3-octylimidazolium
- APTS
8-aminopyrene-1,3,6-trisulfonic acid
- BGE
background electrolyte
- NTf2−
bis-trifluoromethanesulfonylimide
- BSA
bovine serum albumin
- CEC
capillary electrochromatography
- CE
capillary electrophoresis
- CGE
capillary gel electrophoresis
- CIEF
capillary isoelectric focusing
- CITP
capillary isotachophoresis
- CZE
capillary zone electrophoresis
- DR
diazoresin
- EOF
electroosmotic flow
- ESI
electrospray ionization
- EPO
erythropoietin
- α1,3-Gal
galactose-α-1,3-galactose
- AuNP
gold nanoparticles
- G
graphene
- GO
graphene oxide
- HILIC
hydrophilic interaction chromatography
- HPMC
hydroxyproplymethylcellulose
- iCIEF
imaging capillary isoelectric focusing
- IL
ionic liquid
- pI
isoelectric point
- LIF
laser-induced fluorescence
- LC
liquid chromatography
- MS
mass spectrometry
- MALDI
matrix-assisted laser desorption ionization
- ME
microchip electrophoresis
- MGE
microchip gel electrophoresis
- MIEF
microchip isoelectric focusing
- MW
molecular weight
- mAbs
monoclonal antibodies
- NP
nanoparticles
- Neu5Gc
N-glycolylneuraminic acid
- [NMP]+CH3SO3™
N-methyl-2-pyrrolidonium methyl sulfonate
- OTCEC
open-tubular capillary electrochromatography
- PETA
pentaerythritol
- PLB
phospholipid bilayers
- PAMAM-SNP
polyamidoamine-grafted silica nanoparticles
- PB
polybrene
- PEG
polyethylene glycol
- PVA
polyvinyl alcohol
- PTMs
post translational modifications
- PSP
pseudostationary phase
- QC
quaternized celluoses
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SNP
silica nanoparticles
- SDS
sodium dodecyl sulfate
- SPB
stabilized phospholipid bilayer
- SBE β-CD
sulfobutyl ether β-cyclodextrins
Footnotes
The authors declare no conflicts of interest.
References
- 1.Manning MC, Chou DK, Murphy BM, Payne RW, Katayama DS. Pharm Res. 2010;27:544–575. doi: 10.1007/s11095-009-0045-6. [DOI] [PubMed] [Google Scholar]
- 2.Fekete S, Gassner A-L, Rudaz S, Schappler J, Guillarme D. TrAC, Trends Anal. Chem. 2013;42:74–83. [Google Scholar]
- 3.Little MJ, Paquette DM, Roos PK. Electrophoresis. 2006;27:2477–2485. doi: 10.1002/elps.200500951. [DOI] [PubMed] [Google Scholar]
- 4.Staub A, Guillarme D, Schappler J, Veuthey J-L, Rudaz S. J. Pharm. Biomed. Anal. 2011;55:810–822. doi: 10.1016/j.jpba.2011.01.031. [DOI] [PubMed] [Google Scholar]
- 5.Zhao SS, Chen DDY. Electrophoresis. 2014;35:96–108. doi: 10.1002/elps.201300372. [DOI] [PubMed] [Google Scholar]
- 6.Marie A-L, Przybylski C, Gonnet F, Daniel R, Urbain R, Chevreux G, Jorieux S, Taverna M. Anal. Chim. Acta. 2013;800:103–110. doi: 10.1016/j.aca.2013.09.023. [DOI] [PubMed] [Google Scholar]
- 7.Shi Y, Li Z, Qiao Y, Lin J. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2012;906:63–68. doi: 10.1016/j.jchromb.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 8.Creamer JS, Krauss ST, Lunte SM. Electrophoresis. 2013 doi: 10.1002/elps.201300394. Ahead of Print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jorgenson JW, Lukacs KD. Science. 1983;222:266–272. doi: 10.1126/science.6623076. [DOI] [PubMed] [Google Scholar]
- 10.Lukacs KD, Jorgenson JWHRCCC. J. High Resolut. Chromatogr. Chromatogr. Commun. 1985;8:407–411. [Google Scholar]
- 11.Corradini D. J. Chromatogr. B: Biomed. Sci. Appl. 1997;699:221–256. doi: 10.1016/s0378-4347(97)00301-0. [DOI] [PubMed] [Google Scholar]
- 12.Watzig H, Degenhardt M, Kunkel A. Electrophoresis. 1998;19:2695–2752. doi: 10.1002/elps.1150191603. [DOI] [PubMed] [Google Scholar]
- 13.Stutz H. Electrophoresis. 2009;30:2032–2061. doi: 10.1002/elps.200900015. [DOI] [PubMed] [Google Scholar]
- 14.Lucy CA, MacDonald AM, Gulcev MD. J. Chromatogr. A. 2008;1184:81–105. doi: 10.1016/j.chroma.2007.10.114. [DOI] [PubMed] [Google Scholar]
- 15.Corradini D, Nicoletti I, Bonn GK. Electrophoresis. 2009;30:1869–1876. doi: 10.1002/elps.200800447. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Han H, Wang Q, Liu X, Jiang S. Anal. Chim. Acta. 2010;674:243–248. doi: 10.1016/j.aca.2010.06.044. [DOI] [PubMed] [Google Scholar]
- 17.Wu X, Wei W, Su Q, Xu L, Chen G. Electrophoresis. 2008;29:2356–2362. doi: 10.1002/elps.200700746. [DOI] [PubMed] [Google Scholar]
- 18.Guo X-F, Chen H-Y, Zhou X-H, Wang H, Zhang H-S. Electrophoresis. 2013;34:3287–3292. doi: 10.1002/elps.201300369. [DOI] [PubMed] [Google Scholar]
- 19.Cao F, Luo Z, Zhou D, Zeng R, Wang Y. Electrophoresis. 2011;32:1148–1155. doi: 10.1002/elps.201000528. [DOI] [PubMed] [Google Scholar]
- 20.Fu X, Huang L, Gao F, Li W, Pang N, Zhai M, Liu H, Wu M. Electrophoresis. 2007;28:1958–1963. doi: 10.1002/elps.200600558. [DOI] [PubMed] [Google Scholar]
- 21.Kato M, Imamura E, Sakai-Kato K, Nakajima T, Toyo'oka T. Electrophoresis. 2006;27:1895–1899. doi: 10.1002/elps.200500429. [DOI] [PubMed] [Google Scholar]
- 22.Zhao L, Zhou J, Xie H, Huang D, Zhou P. Electrophoresis. 2012;33:1703–1708. doi: 10.1002/elps.201100678. [DOI] [PubMed] [Google Scholar]
- 23.Zhao L, Zhou J, Zhou H, Yang Q, Zhou P. Electrophoresis. 2013;34:1593–1599. doi: 10.1002/elps.201200528. [DOI] [PubMed] [Google Scholar]
- 24.de Jong S, Epelbaum N, Liyanage R, Krylov SN. Electrophoresis. 2012;33:2584–2590. doi: 10.1002/elps.201200153. [DOI] [PubMed] [Google Scholar]
- 25.Cooper BT, Sanzgiri RD, Maxey SB. Analyst (Cambridge, U. K.) 2012;137:5777–5784. doi: 10.1039/c2an36060h. [DOI] [PubMed] [Google Scholar]
- 26.Gassner A-L, Rudaz S, Schappler J. Electrophoresis. 2013;34:2718–2724. doi: 10.1002/elps.201300070. [DOI] [PubMed] [Google Scholar]
- 27.Mansfield E, Ross EE, Aspinwall CA. Anal. Chem. (Washington, DC, U. S.) 2007;79:3135–3141. doi: 10.1021/ac0618829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adem SM, Mansfield E, Keogh JP, Hall HK, Aspinwall CA. Anal. Chim. Acta. 2013;772:93–98. doi: 10.1016/j.aca.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu B, Liu P, Cong H, Tang J, Zhang L. Electrophoresis. 2012;33:3066–3072. doi: 10.1002/elps.201200245. [DOI] [PubMed] [Google Scholar]
- 30.Yu B, Cui W, Cong H, Jiao M, Liu P, Yang S. RSC Adv. 2013;3:20010–20015. [Google Scholar]
- 31.Castelletti L, Verzola B, Gelfi C, Stoyanov A, Righetti PG. J. Chromatogr. A. 2000;894:281–289. doi: 10.1016/s0021-9673(00)00664-6. [DOI] [PubMed] [Google Scholar]
- 32.Verzola B, Gelfi C, Righetti PG. J. Chromatogr. A. 2000;874:293–303. doi: 10.1016/s0021-9673(00)00090-x. [DOI] [PubMed] [Google Scholar]
- 33.Verzola B, Gelfi C, Righetti PG. J. Chromatogr. A. 2000;868:85–99. doi: 10.1016/s0021-9673(99)01207-8. [DOI] [PubMed] [Google Scholar]
- 34.de Jong S, Krylov SN. Anal. Chem. (Washington, DC, U. S.) 2012;84:453–458. doi: 10.1021/ac2030333. [DOI] [PubMed] [Google Scholar]
- 35.Zhu Z, Lu JJ, Liu S. Anal. Chim. Acta. 2012;709:21–31. doi: 10.1016/j.aca.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hutterer KM, Hong RW, Lull J, Zhao X, Wang T, Pei R, Le ME, Borisov O, Piper R, Liu YD, Petty K, Apostol I, Flynn GC. MAbs. 2013;5:608–613. doi: 10.4161/mabs.24725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu C, Liu D, Liu H, Motchnik P. MAbs. 2013;5:102–113. doi: 10.4161/mabs.22965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michels DA, Parker M, Salas-Solano O. Electrophoresis. 2012;33:815–826. doi: 10.1002/elps.201100528. [DOI] [PubMed] [Google Scholar]
- 39.Shi Y, Li Z, Lin J. Anal. Methods. 2012;4:1637–1642. [Google Scholar]
- 40.Cianciulli C, Hahne T, Waetzig H. Electrophoresis. 2012;33:3276–3280. doi: 10.1002/elps.201200177. [DOI] [PubMed] [Google Scholar]
- 41.Le ME, Vizel A, Hutterer KM. Electrophoresis. 2013;34:1369–1374. doi: 10.1002/elps.201200644. [DOI] [PubMed] [Google Scholar]
- 42.Rustandi RR, Loughney JW, Hamm M, Hamm C, Lancaster C, Mach A, Ha S. Electrophoresis. 2012;33:2790–2797. doi: 10.1002/elps.201200095. [DOI] [PubMed] [Google Scholar]
- 43.Lu JJ, Zhu Z, Wang W, Liu S. Anal. Chem. (Washington, DC, U. S.) 2011;83:1784–1790. doi: 10.1021/ac103148k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Na DH, Park EJ, Kim MS, Cho CK, Woo BH, Lee HS, Lee KC. Bull. Korean Chem. Soc. 2011;32:4253–4257. [Google Scholar]
- 45.Na DH, Park EJ, Kim MS, Lee HS, Lee KC. Chromatographia. 2012;75:679–683. [Google Scholar]
- 46.Kerekgyarto M, Fekete A, Szurmai Z, Kerekgyarto J, Takacs L, Kurucz I, Guttman A. Electrophoresis. 2013;34:2379–2386. doi: 10.1002/elps.201300052. [DOI] [PubMed] [Google Scholar]
- 47.Shen Y, Xiang F, Veenstra TD, Fung EN, Smith RD. Anal. Chem. 1999;71:5348–5353. doi: 10.1021/ac9909305. [DOI] [PubMed] [Google Scholar]
- 48.Koshel BM, Wirth MJ. Proteomics. 2012;12:2918–2926. doi: 10.1002/pmic.201200213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Righetti PG, Sebastiano R, Citterio A. Proteomics. 2013;13:325–340. doi: 10.1002/pmic.201200378. [DOI] [PubMed] [Google Scholar]
- 50.Wehr T, Rodriguez-Diaz R, Zhu M. Capillary Electrophoresis of Proteins. New York: Marcel Dekker; 1998. [Google Scholar]
- 51.Anderson CL, Wang Y, Rustandi RR. Electrophoresis. 2012;33:1538–1544. doi: 10.1002/elps.201100611. [DOI] [PubMed] [Google Scholar]
- 52.Rustandi RR, Peklansky B, Anderson CL. Electrophoresis. 2013 doi: 10.1002/elps.201300386. [DOI] [PubMed] [Google Scholar]
- 53.Rustandi RR, Wang F, Hamm C, Cuciniello JJ, Marley ML. Electrophoresis. 2013 doi: 10.1002/elps.201300422. Ahead of Print. [DOI] [PubMed] [Google Scholar]
- 54.Shimura K, Hoshino M, Kamiya K, Enomoto M, Hisada S, Matsumoto H, Novotny M, Kasai K-i. Anal. Chem. (Washington, DC, U. S.) 2013;85:1705–1710. doi: 10.1021/ac3033992. [DOI] [PubMed] [Google Scholar]
- 55.Lu JJ, Wang S, Li G, Wang W, Pu Q, Liu S. Anal. Chem. (Washington, DC, U. S.) 2012;84:7001–7007. doi: 10.1021/ac3017168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang Q, Harrata AK, Lee CS. Anal. Chem. 1995;67:3515–3519. [Google Scholar]
- 57.Tang Q, Harrata AK, Lee CS. J Mass Spectrom. 1996;31:1284–1290. [Google Scholar]
- 58.Foret F, Mueller O, Thorne J, Goetzinger W, Karger BL. J. Chromatogr. A. 1995;716:157–166. doi: 10.1016/0021-9673(95)00621-s. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Z, Wang J, Hui L, Li L. Electrophoresis. 2012;33:661–665. doi: 10.1002/elps.201100447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chingin K, Astorga-Wells J, Pirmoradian NM, Lavold T, Zubarev RA. Anal. Chem. (Washington, DC, U. S.) 2012;84:6856–6862. doi: 10.1021/ac3013016. [DOI] [PubMed] [Google Scholar]
- 61.Zhong X, Maxwell EJ, Ratnayake C, Mack S, Chen DDY. Anal. Chem. (Washington, DC, U. S.) 2011;83:8748–8755. doi: 10.1021/ac202130f. [DOI] [PubMed] [Google Scholar]
- 62.Wang C, Lee CS, Smith RD, Tang K. Anal. Chem. (Washington, DC, U. S.) 2013;85:7308–7315. doi: 10.1021/ac401202c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuroda Y, Yukinaga H, Kitano M, Noguchi T, Nemati M, Shibukawa A, Nakagawa T, Matsuzaki K. J. Pharm. Biomed. Anal. 2005;37:423–428. doi: 10.1016/j.jpba.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 64.Zhu G, Sun L, Yang P, Dovichi NJ. Anal. Chim. Acta. 2012;750:207–211. doi: 10.1016/j.aca.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Michels DA, Tu AW, McElroy W, Voehringer D, Salas-Solano O. Anal. Chem. (Washington, DC, U. S.) 2012;84:5380–5386. doi: 10.1021/ac3008847. [DOI] [PubMed] [Google Scholar]
- 66.Fanali C, D'Orazio G, Fanali S. Electrophoresis. 2012;33:2553–2560. doi: 10.1002/elps.201200157. [DOI] [PubMed] [Google Scholar]
- 67.Nilsson C, Birnbaum S, Nilsson S. Electrophoresis. 2011;32:1141–1147. doi: 10.1002/elps.201000645. [DOI] [PubMed] [Google Scholar]
- 68.Gao J, Latep N, Ge Y, Tian J, Wu J, Qin W. J. Sep. Sci. 2013;36:1575–1581. doi: 10.1002/jssc.201200837. [DOI] [PubMed] [Google Scholar]
- 69.Cheong WJ, Ali F, Kim YS, Lee JW. J. Chromatogr. A. 2013;1308:1–24. doi: 10.1016/j.chroma.2013.07.107. [DOI] [PubMed] [Google Scholar]
- 70.Miksik I, Lacinova K, Zmatlikova Z, Sedlakova P, Kral V, Sykora D, Rezanka P, Kasicka V. J. Sep. Sci. 2012 doi: 10.1002/jssc.201101049. Ahead of Print. [DOI] [PubMed] [Google Scholar]
- 71.Hamer M, Yone A, Rezzano I. Electrophoresis. 2012;33:334–339. doi: 10.1002/elps.201100297. [DOI] [PubMed] [Google Scholar]
- 72.Qu Q, Gu C, Hu X. Anal. Chem. (Washington, DC, U. S.) 2012;84:8880–8890. doi: 10.1021/ac3023636. [DOI] [PubMed] [Google Scholar]
- 73.Qu Q, Gu C, Gu Z, Shen Y, Wang C, Hu X. J. Chromatogr. A. 2013;1282:95–101. doi: 10.1016/j.chroma.2013.01.055. [DOI] [PubMed] [Google Scholar]
- 74.Liu H, Li X, Huang L, Zhang L, Zhang W. Anal. Biochem. 2013;442:186–188. doi: 10.1016/j.ab.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 75.Puangpila C, Nhujak T, El RZ. Electrophoresis. 2012;33:1431–1442. doi: 10.1002/elps.201200018. [DOI] [PubMed] [Google Scholar]
- 76.Wang Y, Deng Q-L, Fang G-Z, Pan M-F, Yu Y, Wang S. Anal. Chim. Acta. 2012;712:1–8. doi: 10.1016/j.aca.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 77.Liu C-C, Deng Q-L, Fang G-Z, Liu H-L, Wu J-H, Pan M-F, Wang S. Anal. Chim. Acta. 2013;804:313–320. doi: 10.1016/j.aca.2013.10.037. [DOI] [PubMed] [Google Scholar]
- 78.Hempel G. Electrophoresis. 2000;21:691–698. doi: 10.1002/(SICI)1522-2683(20000301)21:4<691::AID-ELPS691>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 79.Swinney K, Bornhop DJ. Electrophoresis. 2000;21:1239–1250. doi: 10.1002/(SICI)1522-2683(20000401)21:7<1239::AID-ELPS1239>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 80.de Kort BJ, de Jong GJ, Somsen GW. Electrophoresis. 2012;33:2996–3001. doi: 10.1002/elps.201200303. [DOI] [PubMed] [Google Scholar]
- 81.Sarazin C, Delaunay N, Costanza C, Eudes V, Mallet J-M, Gareil P. Anal. Chem. (Washington, DC, U. S.) 2011;83:7381–7387. doi: 10.1021/ac2012834. [DOI] [PubMed] [Google Scholar]
- 82.de Kort BJ, de Jong GJ, Somsen GW. Anal. Chim. Acta. 2013;766:13–33. doi: 10.1016/j.aca.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 83.Wang H, Dou P, Lue C, Liu Z. J. Chromatogr. A. 2012;1246:48–54. doi: 10.1016/j.chroma.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 84.Walt DR. Anal. Chem. (Washington, DC, U. S.) 2013;85:1258–1263. doi: 10.1021/ac3027178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ramsay LM, Dickerson JA, Dovichi NJ. Electrophoresis. 2009;30:297–302. doi: 10.1002/elps.200800498. [DOI] [PubMed] [Google Scholar]
- 86.Haselberg R, de Jong GJ, Somsen GW. LC GC Asia Pac. 2012;15:13–18. [Google Scholar]
- 87.Pioch M, Bunz S-C, Neusuess C. Electrophoresis. 2012;33:1517–1530. doi: 10.1002/elps.201200030. [DOI] [PubMed] [Google Scholar]
- 88.Haselberg R, de Jong GJ, Somsen GW. Electrophoresis. 2013;34:99–112. doi: 10.1002/elps.201200439. [DOI] [PubMed] [Google Scholar]
- 89.Hommerson P, Khan AM, de Jong GJ, Somsen GW. Mass Spectrom. Rev. 2011;30:1096–1120. doi: 10.1002/mas.20313. [DOI] [PubMed] [Google Scholar]
- 90.Olivares JA, Nguyen NT, Yonker CR, Smith RD. Anal. Chem. 1987;59:1230–1232. [Google Scholar]
- 91.Taichrib A, Pioch M, Neusuess C. Electrophoresis. 2012;33:1356–1366. doi: 10.1002/elps.201100620. [DOI] [PubMed] [Google Scholar]
- 92.Bonvin G, Schappler J, Rudaz S. J. Chromatogr. A. 2012;1267:17–31. doi: 10.1016/j.chroma.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 93.Moini M. Anal. Chem. (Washington, DC, U. S.) 2007;79:4241–4246. doi: 10.1021/ac0704560. [DOI] [PubMed] [Google Scholar]
- 94.Whitt JT, Moini M. Anal. Chem. 2003;75:2188–2191. doi: 10.1021/ac026380j. [DOI] [PubMed] [Google Scholar]
- 95.Haselberg R, Ratnayake CK, de Jong GJ, Somsen GW. J. Chromatogr. A. 2010;1217:7605–7611. doi: 10.1016/j.chroma.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 96.Haselberg R, de Jong GJ, Somsen GW. Anal. Chem. (Washington, DC, U. S.) 2013;85:2289–2296. doi: 10.1021/ac303158f. [DOI] [PubMed] [Google Scholar]
- 97.Gahoual R, Burr A, Busnel J-M, Kuhn L, Hammann P, Beck A, Francois Y-N, Leize-Wagner E. MAbs. 2013:5. doi: 10.4161/mabs.23995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Whitmore CD, Gennaro LA. Electrophoresis. 2012;33:1550–1556. doi: 10.1002/elps.201200066. [DOI] [PubMed] [Google Scholar]
- 99.Maxwell EJ, Zhong X, Zhang H, van ZN, Chen DDY. Electrophoresis. 2010;31:1130–1137. doi: 10.1002/elps.200900517. [DOI] [PubMed] [Google Scholar]
- 100.Maxwell EJ, Zhong X, Chen DDY. Anal. Chem. (Washington, DC, U. S.) 2010;82:8377–8381. doi: 10.1021/ac1017953. [DOI] [PubMed] [Google Scholar]
- 101.Zhao SS, Zhong X, Chen DDY. Electrophoresis. 2012;33:1322–1330. doi: 10.1002/elps.201100572. [DOI] [PubMed] [Google Scholar]
- 102.Zhong X, Maxwell EJ, Chen DDY. Anal. Chem. (Washington, DC, U. S.) 2011;83:4916–4923. doi: 10.1021/ac200636y. [DOI] [PubMed] [Google Scholar]
- 103.Researchmoz.us. 2013 http://www.researchmoz.us/global-and-china-monoclonal-antibody-industry-report-2013-2017-report.html.
- 104.Rustandi RR, Anderson CL, Hamm M. Glycosylation Engineering of Biopharmaceuticals. New York: Springer; 2013. [Google Scholar]
- 105.Behne A, Muth T, Borowiak M, Reichl U, Rapp E. Electrophoresis. 2013;34:2311–2315. doi: 10.1002/elps.201200696. [DOI] [PubMed] [Google Scholar]
- 106.Wuhrer M, de BAR, Deelder AM. Mass Spectrom. Rev. 2009;28:192–206. doi: 10.1002/mas.20195. [DOI] [PubMed] [Google Scholar]
- 107.Mittermayr S, Bones J, Doherty M, Guttman A, Rudd PM. J Proteome Res. 2011;10:3820–3829. doi: 10.1021/pr200371s. [DOI] [PubMed] [Google Scholar]
- 108.Guttman A. TrAC, Trends Anal. Chem. 2013;48:132–143. [Google Scholar]
- 109.Mittermayr S, Bones J, Guttman A. Anal. Chem. (Washington, DC, U. S.) 2013;85:4228–4238. doi: 10.1021/ac4006099. [DOI] [PubMed] [Google Scholar]
- 110.Kuo C-Y, Wang S-H, Lin C, Liao SK-S, Hung W-T, Fang J-M, Yang W-B. Molecules. 2012;17:7387–7400. doi: 10.3390/molecules17067387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Callewaert N, Geysens S, Molemans F, Contreras R. Glycobiology. 2001;11:275–281. doi: 10.1093/glycob/11.4.275. [DOI] [PubMed] [Google Scholar]
- 112.Reusch D, Haberger M, Kailich T, Heidenreich A-K, Kampe M, Bulau P, Wuhrer M. MAbs. 2013:6. doi: 10.4161/mabs.26712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Croset A, Delafosse L, Gaudry J-P, Arod C, Glez L, Losberger C, Begue D, Krstanovic A, Robert F, Vilbois F, Chevalet L, Antonsson B. J. Biotechnol. 2012;161:336–348. doi: 10.1016/j.jbiotec.2012.06.038. [DOI] [PubMed] [Google Scholar]
- 114.Galili U, Rachmilewitz EA, Peleg A, Flechner I. J. Exp. Med. 1984;160:1519–1531. doi: 10.1084/jem.160.5.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yagi Y, Kakehi K, Hayakawa T, Ohyama Y, Suzuki S. Anal. Biochem. 2012;431:120–126. doi: 10.1016/j.ab.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 116.Maeda E, Kita S, Kinoshita M, Urakami K, Hayakawa T, Kakehi K. Anal. Chem. (Washington, DC, U. S.) 2012;84:2373–2379. doi: 10.1021/ac300234a. [DOI] [PubMed] [Google Scholar]
- 117.Szabo Z, Guttman A, Bones J, Shand RL, Meh D, Karger BL. Mol. Pharmaceutics. 2012;9:1612–1619. doi: 10.1021/mp200612n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hamm M, Wang Y, Rustandi RR. Pharmaceuticals. 2013;6:393–406. doi: 10.3390/ph6030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bunz S-C, Cutillo F, Neusuess C. Anal. Bioanal. Chem. 2013;405:8277–8284. doi: 10.1007/s00216-013-7231-4. [DOI] [PubMed] [Google Scholar]
- 120.Bunz S-C, Rapp E, Neusuess C. Anal. Chem. (Washington, DC, U. S.) 2013;85:10218–10224. doi: 10.1021/ac401930j. [DOI] [PubMed] [Google Scholar]
- 121.Wang Y, Santos M, Guttman A. J. Sep. Sci. 2013;36:2862–2867. doi: 10.1002/jssc.201300528. [DOI] [PubMed] [Google Scholar]
- 122.Initiative, G. a. B. 2012 http://www.gabionline.net/Reports/Biosimilars-marketed-in-Europe.
- 123.Falconer RJ, Jackson-Matthews D, Mahler SM. J. Chem. Technol. Biotechnol. 2011;86:915–922. [Google Scholar]
- 124.Beck A, Diemer H, Ayoub D, Debaene F, Wagner-Rousset E, Carapito C, Van DA, Sanglier-Cianferani S. TrAC, Trends Anal. Chem. 2013;48:81–95. [Google Scholar]
- 125.Taichrib A, Pioch M, Neusuess C. Anal. Bioanal. Chem. 2012;403:797–805. doi: 10.1007/s00216-012-5924-8. [DOI] [PubMed] [Google Scholar]
- 126.Espinosa-de IGCE, Perdomo-Abundez FC, Padilla-Calderon J, Uribe-Wiechers JM, Perez NO, Flores-Ortiz LF, Medina-Rivero E. Electrophoresis. 2013;34:1133–1140. doi: 10.1002/elps.201200575. [DOI] [PubMed] [Google Scholar]
- 127.Visser J, Feuerstein I, Stangler T, Schmiederer T, Fritsch C, Schiestl M. BioDrugs. 2013;27:495–507. doi: 10.1007/s40259-013-0036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Salas-Solano O, Kennel B, Park SS, Roby K, Sosic Z, Boumajny B, Free S, Reed-Bogan A, Michels D, McElroy W, Bonasia P, Hong M, He X, Ruesch M, Moffatt F, Kiessig S, Nunnally B. J. Sep. Sci. 2012;35:3124–3129. doi: 10.1002/jssc.201200633. [DOI] [PubMed] [Google Scholar]
- 129.Salas-Solano O, Babu K, Park SS, Zhang X, Zhang L, Sosic Z, Boumajny B, Zeng M, Cheng K-C, Reed-Bogan A, Cummins-Bitz S, Michels DA, Parker M, Bonasia P, Hong M, Cook S, Ruesch M, Lamb D, Bolyan D, Kiessig S, Allender D, Nunnally B. Chromatographia. 2011;73:1137–1144. [Google Scholar]
- 130.Culbertson CT, Mickleburgh TG, Stewart-James SA, Sellens KA, Pressnall M. Anal. Chem. (Washington, DC, U. S.) 2014;86:95–118. doi: 10.1021/ac403688g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Neuzi P, Giselbrecht S, Laenge K, Huang TJ, Manz A. Nat. Rev. Drug Discovery. 2012;11:620–632. doi: 10.1038/nrd3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mitra I, Snyder CM, Alley WR, Novotny MV, Jacobson SC. American Chemical Society. 2013 pp. ANYL-279. [Google Scholar]
- 133.Mitra I, Alley WR, Goetz JA, Vasseur JA, Novotny MV, Jacobson SC. J. Proteome Res. 2013;12:4490–4496. doi: 10.1021/pr400549e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhuang Z, Starkey JA, Mechref Y, Novotny MV, Jacobson SC. Anal. Chem. (Washington, DC, U. S.) 2007;79:7170–7175. doi: 10.1021/ac071261v. [DOI] [PubMed] [Google Scholar]
- 135.Han H, Livingston E, Chen X. Anal. Chem. (Washington, DC, U. S.) 2011;83:8184–8191. doi: 10.1021/ac201741w. [DOI] [PubMed] [Google Scholar]
- 136.Primack J, Flynn GC, Pan H. Electrophoresis. 2011;32:1129–1132. doi: 10.1002/elps.201000619. [DOI] [PubMed] [Google Scholar]
- 137.Chen X, Tang K, Lee M, Flynn GC. Electrophoresis. 2008;29:4993–5002. doi: 10.1002/elps.200800324. [DOI] [PubMed] [Google Scholar]
- 138.Han H, Chen X. Electrophoresis. 2012;33:765–772. doi: 10.1002/elps.201100511. [DOI] [PubMed] [Google Scholar]
- 139.Pan W, Chen W, Jiang X. Anal. Chem. 2010;82:3974–3976. doi: 10.1021/ac1000493. [DOI] [PubMed] [Google Scholar]
- 140.Jin S, Anderson GJ, Kennedy RT. Anal. Chem. (Washington, DC, U. S.) 2013;85:6073–6079. doi: 10.1021/ac400940x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kinoshita M, Nakatsuji Y, Suzuki S, Hayakawa T, Kakehi K. J. Chromatogr. A. 2013;1309:76–83. doi: 10.1016/j.chroma.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 142.Hughes AJ, Herr AE. Proc. Natl. Acad. Sci. U. S. A. 2012;109:21450–21455. doi: 10.1073/pnas.1207754110. S21450/21451-S21450/21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.O'Neill RA, Bhamidipati A, Bi X, Deb-Basu D, Cahill L, Ferrante J, Gentalen E, Glazer M, Gossett J, Hacker K, Kirby C, Knittle J, Loder R, Mastroieni C, MacLaren M, Mills T, Nguyen U, Parker N, Rice A, Roach D, Suich D, Voehringer D, Voss K, Yang J, Yang T, Vander Horn PB. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16153–16158. doi: 10.1073/pnas.0607973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.He X, Chen Q, Zhang Y, Lin J-M. TrAC, Trends Anal. Chem. 2014;53:84–97. [Google Scholar]
- 145.Chambers AG, Mellors JS, Henley WH, Ramsey JM. Anal. Chem. (Washington, DC, U. S.) 2011;83:842–849. doi: 10.1021/ac102437z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mellors JS, Black WA, Chambers AG, Starkey JA, Lacher NA, Ramsey JM. Anal. Chem. (Washington, DC, U. S.) 2013;85:4100–4106. doi: 10.1021/ac400205a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.