Abstract
Using Caenorhabditiselegans as a model system, Norris et al. define complex combinatorial regulation of alternative splicing at single neuron resolution and illustrate functional coherence among components of a splicing regulatory network controlled by a neuronal splicing factor.
Alternative splicing (AS) is the process by which a single gene can produce multiple mRNAs that often produce protein isoforms with differential functions. With the advent of technological advances such as splicing sensitive microarrays and high throughput sequencing, it has become evident that AS is a widespread mechanism for achieving functional complexity in eukaryotes. Nearly all human multi-exon genes are alternatively spliced and most produce multiple splice variants (Pan et al., 2008; Wang et al., 2008). The regulation of alternative splicing is directed by RNA binding proteins that bind to RNA cis-elements that are typically located within alternatively spliced exons or in the flanking intronic sequences. The splicing of alternatively spliced exons reflects the combinatorial functions of numerous splicing factors bound to these elements that can promote either splicing (inclusion) or skipping of the exon. Efforts to define a “splicing code” based on the binding sites and differential expression levels of these regulators promise to define genome-wide programs of AS in numerous different tissues and cell types (Barash et al., 2010). In this issue of Molecular Cell, Norris et al. (2014) harness genetic tools available in Caenorhabditiselegans to define combinatorial regulation of AS in the nervous system at single cell resolution(Norris et al., 2014).
While many splicing regulators are widely expressed, increasing numbers of splicing factors are being discovered whose expression is limited to specific cell types, developmental stages, or cellular conditions. Transcripts regulated by specific splicing factors comprise “splicing regulatory networks” (SRNs) that are functionally and biologically coherent. For example, targets of the neural-specific splicing factor Nova encode proteins that coordinate synaptic functions (Licatalosi and Darnell, 2010).The recent explosion of genomic analyses has led to the identification of numerous genome-wide programs of AS associated with diverse organisms, tissues, cell types, and developmental stages. However, it remains a major challenge to define the functional consequences of alterations or changes in splicing at both the single gene and systems level (Kalsotra and Cooper, 2011). While the differential functions of some splice isoforms have been described, the biological relevance of the vast majority of AS events remain unknown (Kelemen et al., 2013). AS is highly prevalent in the central nervous system, but the brain is composed of many different neural cell types. While previous studies have identified differences in splicing between the nervous system and non-neural tissues, there remain limited analyses of AS in specific neural cell populations as well as within specific cell types that comprise other tissues and organs.
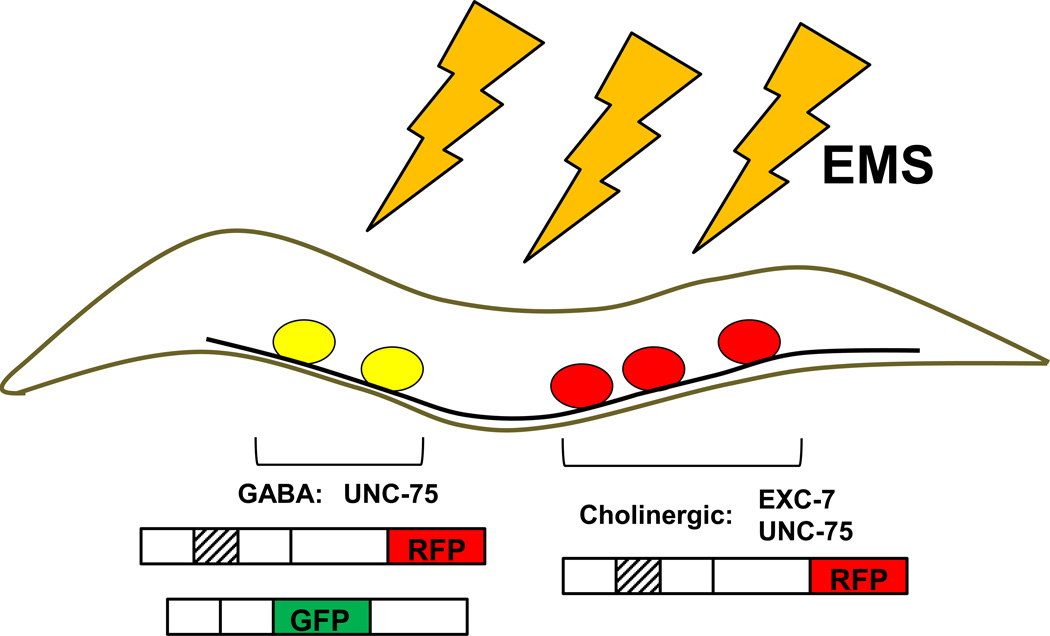
Using two-color splicing reporters similar to those first applied to nematode AS by the Kuroyanagi and Hagiwara groups (Kuroyanagi et al., 2006),Norris et al. screened a subset of conserved AS exons in genes expressed in the nervous system for differential splicing. Interestingly, 7 of the 14 AS events revealed distinct patterns of AS that differed among different classes of neurons; further evidence for the added complexity of AS within the nervous system beyond neural /non-neural AS. Among these AS exons was an alternative exon in unc-16 transcripts in neurons of the ventral nerve cord. Whereas unc-16 isoforms containing exon 16 were present in both cholinergic and GABAergic neurons, isoforms that skipped exon 16 were only identified in GABAergic neurons. To define the trans-acting factors regulating this AS event worms expressing the unc-16 reporter were subjected to EMS mutagenesis and progeny scored for changes in splicing using microscopy. These screens identified mutations in unc-75 and exc-7, orthologs of the mammalian CELF and ELAV families of splicing regulators, that led to a loss of GABAergic restricted expression of the unc-16 exon skipped isoform. In unc-75; exc-7 double mutant worms there was a complete loss of isoforms that included the exon indicating combinatorial functions of these splicing regulators to promote splicing of exon 16 in unc-16 transcripts. Whereas UNC-75 was expressed pan-neuronally, EXC-7 expression was limited to cholinergic, but not GABAergic neurons. Hence, the combined functions of both factors promote complete exon inclusion in cholinergic neurons, whereas the expression of UNC-75 alone in GABAergic neurons promotes only partial exon splicing (Figure 1).
Figure 1. Combinatorial regulation of an alternative exon in unc-16 transcripts by UNC-75 and EXC-7 in cholinergic neurons of the ventral nerve cord.
Bi-cistronic two-color splicing reporters allow single cell resolution of exon inclusion (RFP) or skipping (GFP) in the nematode nervous system. An EMS mutagenesis screen allowed visualization of increased skipping of unc-16 exon 16 in unc-75 and exc-7 mutants. In cholinergic neurons expression of both UNC-75 and EXC-7 promotes complete exon inclusion, whereas expression of UNC-75 alone leads to transcripts that include and skip the exon.
While previous studies have identified roles for both EXC-7 and UNC-75in cholinergic transmission and defined neural splicing targets for the latter, the present study explores the overlap in the SRNs regulated by both proteins (Kuroyanagi et al., 2013; Loria et al., 2003). Using RNA-Seq, the authors define SRNs directed by each splicing regulator using unc-75, exc-7, and unc-75/exc-7 double mutant animals. These studies showed combinatorial overlap in the SRNs regulated by each protein including examples of co-regulated targets indicative of broader cooperativity in splicing regulation. This overlap also correlated with functional analysis in which unc-75 mutant worms displayed defects in cholinergic transmission that were even more severe in unc-75/exc-7 double mutants. An important conceptual advance of the current work is provided by further studies exploring the relationship between phenotypic effects of unc-75 mutation and the components of the UNC-75 splicing network. The unc-75 mutants exhibit defects in cholinergic transmission that can be assayed by sensitivity to the acetyl cholinesterase inhibitor aldicarb and also display uncoordination defects. Among 118 UNC-75 regulated genes, 25% had previously been associated with loss-of-function defects in coordination or aldicarb sensitivity. To further probe the functional relevance of the UNC-75 network in neuronal processes, Norris et al tested an additional twelve UNC-75 regulated genes for which mutant strains were available. Of these, the majority exhibited locomotion and/or aldicarb sensitivity defects. While further explorations of this network using RNAi might further extend the significance of these observations, several of the genes that exhibited these functional defects had no previously known functions. These findings thereby revealed genes with roles in nervous system functions. Thus, an important take home message of the study is that experimental interrogation of biologically coherent SRNs can broadly define new genes involved in synaptic transmission as well as genes in SRNs related to other biological processes. While further studies using methods such as crosslinking immunoprecipitation coupled with high throughput sequencing (CLIP-Seq; also known as HITS-CLIP) are needed to further define direct vs indirect targets for the UNC-75 network, Norris et al. provides a blueprint to direct functional analysis of other SRNs from worms to mammals.
While studies using loss-of-function mutants provide further evidence that SRNs can define functionally related gene products, a greater challenge is to provide evidence that the different protein isoforms within the network are functionally distinct. Using transgene rescue, the authors tested two isoforms (A and B) of the UNC-75 regulated target unc-64 and showed that the A isoform, which is lost in unc-75 mutants, did not fully rescue the locomotion defect. While this differential function cannot be directly linked to the locomotion defects observed in unc-75 mutants, this example illustrates the challenge of defining isoform-specific functions on a broader scale. Further investigations to mechanistically dissect the differential functions of AS variants will lead to wider appreciation of the concept that characterizations of a given gene’s function need to account for potential isoform-specific differences. While systematic determinations of isoform specific activities for components of a given SRN remains a major task, particularly in mammalian systems, the present study helps illustrate that more focused studies of isoform-specific functions of AS genes that impact development and disease are clearly needed.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA. Nat Rev Genet. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, Stamm S. Gene. 2013;514:1–30. doi: 10.1016/j.gene.2012.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H, Kobayashi T, Mitani S, Hagiwara M. Nat Methods. 2006;3:909–915. doi: 10.1038/nmeth944. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Watanabe Y, Suzuki Y, Hagiwara M. Nucleic Acids Res. 2013;41:4015–4025. doi: 10.1093/nar/gkt097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria PM, Duke A, Rand JB, Hobert O. Curr Biol. 2003;13:1317–1323. doi: 10.1016/s0960-9822(03)00532-3. [DOI] [PubMed] [Google Scholar]
- Norris AD, Shangbang G, Norris ML, Ray D, Ramani AK, Fraser AG, Morris Q, Hughes TR, Zhen M, Calarco JA. Molecular Cell. 2014 doi: 10.1016/j.molcel.2014.05.004. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]