Abstract
The aldehyde dehydrogenase (ALDH) superfamily is composed of nicotinamide adenine dinucleotide (phosphate) (NAD(P)+)-dependent enzymes that catalyze the oxidation of aldehydes to their corresponding carboxylic acids. To date, 24 ALDH gene families have been identified in the eukaryotic genome. In addition to aldehyde metabolizing capacity, ALDHs have additional catalytic (e.g. esterase and reductase) and non-catalytic activities. The latter include functioning as structural elements in the eye (crystallins) and as binding molecules to endobiotics and xenobiotics. Mutations in human ALDH genes and subsequent inborn errors in aldehyde metabolism are the molecular basis of several diseases. Most recently ALDH polymorphisms have been associated with gout and osteoporosis. Aldehyde dehydrogenase enzymes also play important roles in embryogenesis and development, neurotransmission, oxidative stress and cancer. This article serves as a comprehensive review of the current state of knowledge regarding the ALDH superfamily and the contribution of ALDHs to various physiological and pathophysiological processes.
Keywords: Aldehyde dehydrogenase, Gene superfamily, Function, Crystallin, Metabolic disease, Stem cell
1. Aldehyde dehydrogenase superfamily
1.1. Introduction
The ALDH enzymes have usually been defined by their capacity to catalyze NAD(P)+-dependent irreversible oxidation of a wide spectrum of aliphatic and aromatic aldehydes, many of which are generated during the metabolism of endogenous and exogenous compounds (Fig. 1) [1]. The ALDH proteins are found to be present in all subcellular compartments, including cytosol, mitochondria, endoplasmic reticulum and nucleus; several isozymes have been found in more than one of these locations [2,3]. Most of the ALDHs have a wide tissue distribution. Some isozymes display distinct substrate specificity. ALDHs serve to protect cells from the cytotoxic effects of aldehydes by converting them to their respective carboxylic acids [1]. Consequently, they are regarded as being detoxification enzymes. However, some of the carboxylic acids generated by ALDHs function as important molecules in cellular physiology. Examples of these include retinoic acid (RA; essential for development), betaine (osmolyte) and γ-aminobutyric acid (GABA; neurotransmitter).
Fig. 1.
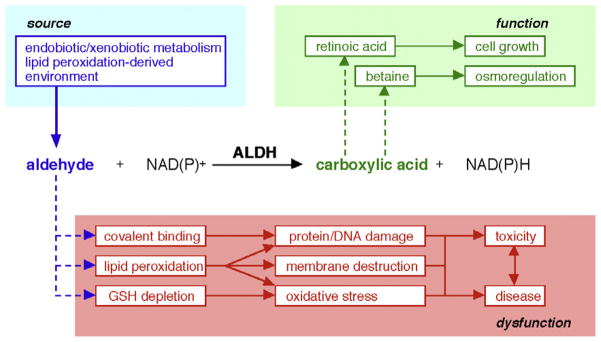
Illustration of ALDH catalytic activity.
1.2. Nomenclature
A standardized gene nomenclature system based on divergent evolution was established for the ALDH superfamily at the Ninth International Symposium on Enzymology and Molecular Biology of Carbonyl Metabolism, held June 20–24, 1998, in Varallo Sesia, Italy and it was published in 1999 [4]. Analyses of the sequence of the ALDH isozymes suggest the evolutionary divergence of the ALDH genes from a common ancestor approximately 3 billion years ago [5]. Based on the approved nomenclature system, ALDH proteins belonging to the same ALDH family (e.g. ALDH1 family) are defined as having >40% amino acid identity while two members of the same subfamily (e.g. ALDH1A subfamily) exhibit >60% amino acid identity [4]. A database and website (www.aldh.org) maintained by the laboratory of Dr. Vasilis Vasiliou at the University of Colorado Denver is dedicated to providing a detailed, up-to-date resource for the ALDH gene superfamily [6].
The eukaryotic ALDH gene superfamily currently consists of 24 families that contain putatively functional genes with distinct chromosomal locations. It is anticipated that the number of ALDH families will expand after the inclusion of the bacterial ALDHs (Vasiliou et al., unpublished). Human genes are found in ALDH1–9, ALDH16 and ALDH18 families, which also contain genes from other species including plant and fungi [7].
1.3. ALDH families
The ALDH1A subfamily comprises ALDH1A1, ALDH1A2 and ALDH1A3, all of which synthesize RA from retinaldehyde and, as such, are crucial in regulating RA signaling [4]. These isozymes have a high affinity for the oxidation of both all-trans- and 9-cis-retinal and exhibit a Km for these molecules in the low micromolar range [8]. Although ALDH2 officially qualifies as a member of the ALDH1 family, its longstanding name of “ALDH2” (associated with ethanol metabolism) has been grandfathered into the ALDH nomenclature system based on evolutionary divergence. ALDH2 is a mitochondrial enzyme that is primarily involved in the metabolism of acetaldehyde generated during alcohol metabolism [1]. ALDH1B1, a mitochondrial enzyme very similar to ALDH2 (75% identical to ALDH2), also contributes to acetaldehyde oxidation, albeit with lower affinity than ALDH2 [9]. ALDH1B1 is a potential colon cancer biomarker [10]. The ALDH1L1 gene codes for the cytosolic 10-FTHF dehydrogenase (FDH), which converts 10-formyltetrahydrofolate (10-FTHF) to tetrahydrofolate [11]. The ALDH1L2 gene is very similar to ALDH1L1 and encodes the mitochondrial FDH that has enzymatic properties similar to its cytosolic counterpart. Unlike ALDH1L1, ALDH1L2 does not metabolize short-chain aldehyde substrates [12]. The ALDH3A subfamily contains the dioxin- inducible ALDH3A1 and ALDH3A2 enzymes, both of which are involved in the oxidation of medium- and long-chain aliphatic and aromatic aldehydes [13,14]. The ALDH3B subfamily consists of two structurally-related genes, ALDH3B1 and ALDH3B2. ALDH3B1 enzyme metabolizes aldehydes generated during lipid peroxidation [15]. As yet, no functional data for ALDH3B2 gene product exists.
The mitochondrial ALDH4A1, also known as pyrroline-5-carboxylate (P5C) dehydrogenase and glutamate-γ-semialdehyde dehydrogenase, catalyzes the irreversible NAD+-dependent conversion of P5C to the neurotransmitter, glutamate [16]. The ALDH5A1 isozyme, also known as succinic semialdehyde (SSA) dehydrogenase, catalyzes the NAD+-dependent conversion of SSA to succinate in the last step GABA catabolism [17]. ALDH6A1 is a mitochondrial enzyme also known as acetyl CoA-dependent methylmalonate semialdehyde (MMS) dehydrogenase. ALDH6A1 is involved in valine and pyrimidine catabolism and catalyzes the oxidative decarboxylation of malonate semialdehyde and MMS to acetyl-CoA and propionyl-CoA, respectively [18]. Human ALDH7A1 has a primary role in the pipecolic acid pathway of lysine catabolism, catalyzing the oxidation of alpha-aminoadipic semialdehyde (AASA) to alpha-aminoadipate [3,19]. ALDH8A1 is a cytosolic enzyme that appears to metabolize retinaldehyde to retinoic acid [20,21]. ALDH9A1 codes for an enzyme that participates in the metabolism of γ-aminobutyraldehyde and aminoaldehydes derived from polyamines [22]. The ALDH16A1 gene encodes an 802-amino acid protein with as-yet unknown function that most likely does not possess catalytic activity (Vasiliou et al., in this volume). ALDH16A1 is a novel and rather unique member of the ALDH superfamily in that it contains two ALDH active site domains (as opposed to one in the other members of the superfamily), four transmembrane domains and a coiled-coil domain. Interestingly, the ALDH16 enzyme active site in frog and many invertebrates (e.g. sea squirt, sea anemone, sea urchin, lancelet, and Trichoplax adhaerens), as well as in bacteria, contains the catalyticallyimportant cysteine residue (Cys-302); in contrast, this residue is absent from the mammalian and fish orthologous protein. The Vasiliou et al. chapter in this issue describes this ALDH as a protein interacting molecule [23].
The ALDH10 family contains plant genes encoding enzymes known as aminoaldehyde dehydrogenases (AMADHs), 4-aminobutyraldehyde dehydrogenases, 4-guanidinobutyraldehyde dehydrogenases, and betaine aldehyde dehydrogenases (BADHs). The ALDH11 family of genes codes for the cytosolic non-phosphorylating glyceraldehyde 3-phosphate dehydrogenases (GAPNs). These enzymes catalyze the irreversible NADP+-dependent oxidation of glyceraldehyde 3-phosphate (GAP) to 3-phosphoglycerate and NADPH [24]. The ALDH12 family contains mostly plant genes encoding Δ-1-pyrroline-5-carboxylate dehydrogenases (P5CDH), enzymes involved in the conversion of proline and arginine to glutamate [25]. The ALDH13 family contains a gene found only in Entamoeba histolytica most likely involved in acetaldehyde metabolism [26]. The ALDH14 and ALDH15 families contain yeast and fungi ALDHs [27]. ALDH14 encodes a mitochondrial enzyme that appears to be similar to the microsomal ALDH3A2 [5]. The ALDH15 gene appears to be similar to plant ALDH21 and may play a role in protection against oxidative stress [25]. The ALDH17 gene family comprises genes in Drosophila that code for enzymes with a possible catalytic function as P5CDHs. The ALDH19 family contains plant and bacterial γ-glutamyl phosphate reductases similar to human ALDH18A1. The ALDH21, ALDH22, ALDH23, and ALDH24 families consist of plant genes [25]. No information is currently available regarding the biological role(s) of these enzymes.
2. Other functions of ALDH proteins
2.1. Catalytic functions
Many ALDH proteins possess multiple additional catalytic and non-catalytic functions (Fig. 2). Several ALDHs, including ALDH2, ALDH1A1, ALDH1L1, ALDH1L2 and ALDH9A1, function as esterases [1,28], the catalytic activity of which are physiologically significant in bioactivation of organic nitrates. ALDH2, ALDH1A1 and ALDH1B1 exhibit nitrate reductase activity. In this respect, ALDH2 plays a key role in the bioactivation of therapeutic nitrates, such as nitroglycerin [29].
Fig. 2.
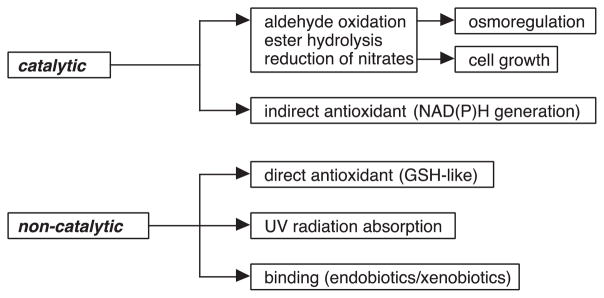
Catalytic and non-catalytic functions of ALDH proteins.
2.2. Non-catalytic functions
The non-catalytic functions of ALDH proteins range from serving a structural function in the eye to acting as binding proteins in cells. In the eye, several ALDHs have been identified as lens and corneal crystallins. ALDH1 proteins are expressed at very high levels (5–20% of water soluble fraction, which is the definition of crystallin) in the lens of various animal species including human [30–33]. In the cornea, both ALDH1A1 and ALDH3A1 have been identified as corneal crystallins in mammalian species [34,35]. The function of these ALDHs in the ocular tissue may include: (i) contribution to transparency by maximizing light transmission and refraction to the retina [36], and (ii) protection against oxidative damage by absorbing ultraviolet light, metabolizing cytotoxic aldehydes [37], producing NADPH for regeneration of reduced glutathione [38] and physicochemically scavenging hydroxyl radicals [38].
Several ALDHs have been identified as binding proteins for endogenous and exogenous compounds. ALDH1A1 is an androgen-, thyroid hormone- and cholesterol-binding protein [39]. In addition, it interacts with several drugs, specifically quinolone, daunorubicin and flavopiridol [39]. Such interactions may be due to the coiled-coil domains present in the human ALDH1A1 protein at residues 81–99, 114–144 and 172–183, all of which reside within the nucleotide-binding domain. The coiled-coil domain is a highly stable oligomerization motif found in a diversity of proteins that function in gene regulation, cell communication, membrane fusion and drug extrusion [40]. The coil-coiled domain is present in several ALDHs including ALDH3A1 and ALDH16A1. The most intriguing ALDH as a protein-binding molecule appears to be ALDH16A1 (described in detail in this issue). As noted above, ALDH16A1 is a novel member of the ALDH superfamily. The species dependence of the critical cysteine residue (Cys-302) in the active site makes it is likely that ALDH16A1 is a binding protein, at least in mammals. In support of this contention, ALDH16A1 has been shown to interact with maspardin, a protein associated with Mast syndrome [41]. In addition, experimental and predictive computational- based molecular modeling evidence from our laboratory suggests that human ALDH16A1 interacts or has the potential to interact with several other proteins associated with uric acid formation, diabetes, vesicular transport and protein degradation. The increasing availability of the crystal structures of ALDHs (Table 1) is facilitating the use of molecular modeling as a tool to explore catalytic and non-catalytic functions of these enzymes.
Table 1.
Crystal structures of ALDHs.
| ALDH enzyme | Species | PDB ID |
|---|---|---|
| ALDH1A1 | SHEEP | 1BXS |
| ALDH1A2 | RAT | 1BI9 |
| ALDH1L1 | HUMAN | 2BW0 |
| ALDH2 | HUMAN | 3N80 |
| ALDH3A1 | HUMAN | 3SZA |
| ALDH4A1 | HUMAN | 3V9G |
| ALDH5A1 | HUMAN | 2W8N |
| ALDH7A1 | HUMAN | 2J6L |
| ALDH10A1 | PLANT | 4A0M |
| ALDH18A1 | HUMAN | 2H5G |
3. Corneal and lens crystallins
3.1. ALDH1A1, ALDH1A8, ALDH1A9 and ALDH3A1
Some members of the ALDH superfamily have been identified as crystallins in the cornea and lens of both vertebrates and invertebrates [2]. The homodimeric ALDH3A1 is the first enzyme to be categorized as a corneal crystallin [42]. It accumulates in high abundance in the cornea of most mammalian species, representing 5–50% of total soluble proteins depending on the species. Rabbits exceptionally express ALDH1A1, with no detectable amounts of ALDH3A1, in the cornea [34]. Other vertebrates, including chicken, frog and fish, also express ALDH1A1 rather than ALDH3A1 in the cornea [43]. The homotetrameric ALDH1A1 is primarily considered as a lens crystallin in human, where it constitutes 2% of the soluble proteins in lens epithelium [33]. Other members of the ALDH1 family are identified as lens crystallins in the elephant shrew (ALDH1A8/η-crystallin) [44], cephalopods (ALDH1C/Ω-crystallins) [32] and scallop (ALDH1A9/Ω-crystallin) [31].
3.2. ALDH1A1 and ALDH3A1 as protective elements of the eye
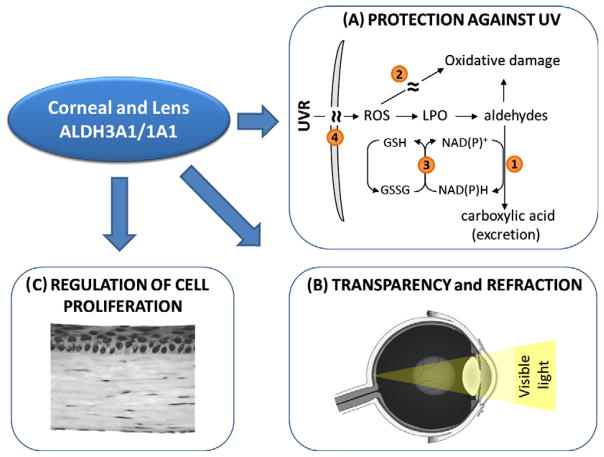
Numerous lines of evidence support the notion that ALDH isozymes, being corneal and lens crystallins, confer the necessary protective properties of these ocular tissues. An important protective function for ALDH3A1 and ALDH1A1 in the eye is supported by the observation that spontaneous cataracts develop in Aldh3a1−/− knockout and Aldh1a1−/−/Aldh3a1−/− double knockout mice by one month of age, and in Aldh1a1−/− knockout mice by 6–9 months [45]. In the same line, these Aldh-null mice are highly susceptible to UV-induced cataract formation [45]. Owning to their catalytic and non-catalytic functions, ALDH3A1 and ALDH1A1 play a key role in protecting the eye from ultraviolet radiation (UVR) induced damage (Fig. 3A). First, ALDH3A1 and ALDH1A1 metabolize toxic aldehydes produced by UV-induced lipid peroxidation. Human ALDH3A1 has high affinity for 4-hydroxynonenal (4-HNE), albeit it metabolizes malondialdehyde (MDA) poorly [13]. The presence of ALDH1A1 in the cornea and lens compensates for the absence of ALDH3A1 by oxidizing both 4-HNE and MDA [46]. Second, ALDH3A1 and ALDH1A1 act as antioxidants by serving as direct targets for UV-induced free radicals, thereby providing a passive protective effect to other proteins [47]. Third, NAD(P)H produced during ALDH-mediated metabolism contributes to the antioxidant arsenal of the cornea and lens. NAD(P)H can directly absorb UVR and it is the reducing agent used in the regeneration of antioxidant glutathione [47]. It also helps to maintain a reducing potential for various redox-active enzymes, which are involved in protecting eye tissues [48]. Fourth, ALDH3A1 is a major UVR filter. UVR-induced modifications to proteins may lead to enzyme inactivation, partial unfolding and non-native aggregation [49], all of which may be responsible for the accumulation of aggregated proteins in the lens during cataract formation [50]. Studies indicate that direct absorption of UVR energy by ALDH3A1 protects other corneal proteins at the expense of their own molecular inactivation [51].
Fig. 3.
Functions of corneal and lens crystallins. (A) Owning to their catalytic and non-catalytic functions, ALDH3A1 and ALDH1A1 proteins protect inner ocular tissues from ultraviolet radiation (UVR) through: ➊ metabolizing toxic aldehydes, ➋ acting as a direct scavengers of reactive oxygen species, ➌ reproducing regenerating antioxidant NADPH and GSH, and ➍ directly absorbing UVR. (B) These corneal crystallins contribute to cellular transparency in corneal stromal keratocytes, supporting a structural role of these ALDH proteins. (C) A putative regulatory function of ALDH3A1 on corneal cell proliferation has also been proposed.
3.3. ALDH1A1 and ALDH3A1 as structural elements of the eye
Studies using cell culture and experimental animals suggest that ALDH1A1 and ALDH3A1 may function as the structural elements of the cornea, contributing to its transparent and refractive properties (Fig. 3B). In rabbit, the development of postnatal corneal transparency is associated with stromal keratocyte quiescence and increased ALDH1A1 expression; loss of ALDH1A1 expression in the corneal stroma is associated with injury-induced corneal haze [52]. In rat and mouse, ALDH3A1 is undetectable in embryo eyes, but drastically increases in the cornea around 9–14 days after birth, at which time the eye opens [53,54]. In addition, increased light scattering and corneal haze have been observed in ALDH3A1 knockout mice (Vasiliou et al., unpublished), which provide an important piece of evidence supporting a structural role of ALDH3A1 in the cornea in vivo. A putative regulatory function of ALDH3A1 on corneal cell proliferation (Fig. 3C) has been proposed based on studies showing (i) a nuclear localization of ALDH3A1 and ALDH1A1 proteins in transfected human corneal epithelial cells [55] and in transfected and normal rabbit corneal keratocytes [2,56], and (ii) an inverse relationship between ALDH3A1 expression and cell proliferation rate in corneal cells in vitro [55] and in vivo (Koppaka et al., unpublished). Such inhibitory effect of ALDH3A1 on cellular proliferation may have important implications for a regulatory role of ALDH3A1 in the cornea. It may serve as an additional mechanism, by which this crystalline protein participates in maintaining the corneal homeostasis by controlling the mitotic phenotypes of corneal epithelial cells and fibroblasts in response to physiological or stress signals.
4. Metabolic diseases
The importance of ALDH proteins in biological processes is perhaps best exemplified by the associations between ALDH gene mutations or polymorphisms and distinct disease phenotypes in humans and rodents. In most instances, the mechanisms by which deficiencies in the ALDH isozymes contribute to the diseases remain to be defined. Aldh1a1 knockout mice are sensitive to UV light and age-related cataract formation [45]. On the other hand, these same mice are protected from obesity and diabetes [57], possibly due to the recently observed function of ALDH1A1 in regulating gluconeogenesis and lipid metabolism [58] and in thermogenic programming in white adipose tissue [59]. Aldh1a2 knockout mice are embryonic lethal [60]; mutations in ALDH1A2 gene are associated with spina bifida [61] and may play a possible causal role in rare cases of human congenital heart disease [62]. Aldh1a3 knockout mice are also lethal due to defects in nasal development [63]. Mutations in ALDH1B1 are associated with hypertension and ethanol sensitivity [64,65]; decreased ALDH1B1 expression is found in patients with ethylmalonic encephalopathy, an inherited and severe metabolic disorder [66]. ALDH1B1 has also been proposed as a susceptibility locus for mood disorder in the Finnish population [67]. NEUT2 mice, which do not express cytosolic ALDH1L1 due to a micro-deletion within chromosome 6 that results in the loss of Aldh1l1, have reduced reproductive efficiency [68,69].
The most common ALDH polymorphism, ALDH2*2, renders ALDH2 catalytically inactive. By promoting the accumulation of acetaldehyde after ethanol consumption and the consequent unpleasant physiological effects, it is associated with lower consumption of alcohol and a lower incidence of alcoholism. The ALDH2*2 allele has been associated with several pathophysiological conditions related or unrelated to ethanol, including myocardial infarction and hypertension [70,71], increased risk for cancers (reviewed in [1]), liver cirrhosis [72] and late-onset Alzheimer’s disease [73].
Aldh3a1 knockout mice are susceptible to cataract formation [45] and mutations in the ALDH3A2 gene cause Sjögren–Larsson syndrome (SLS) in humans [74]. Mutations in ALDH4A1 gene are associated with type II hyperprolinemia [34], and mutations in ALDH5A1 cause γ-hydroxybutyric aciduria [35]. A deficiency in ALDH6A1 is associated with psychomotor delay and methylmalonic aciduria [75] and a mutation in the ALDH6A1 gene causes 3-hydroxyisobutyric aciduria [76]. Patients suffering from pyridoxine-dependent seizures have a wide spectrum of mutations in the ALDH7A1 gene [36]. ALDH7A1 has also been identified as susceptibility gene for osteoporosis [77] A recent study in 6017 Icelandic subjects identified a rare missense SNP in the ALDH16A1 gene that is associated with gout and elevated serum uric acid levels [78]. Finally, polymorphisms in ALDH18A1 are associated with hyperammonemia [1].
5. Hematopoietic stem cells
Hematopoietic stem cells (HSCs), rare cells found primarily in bone marrow, generate all hematopoietic lineages, including myeloid cells, lymphocytes, red blood cells and platelets for months to decades [79]. Accordingly, HSCs play a central role in generating and sustaining hematopoiesis for life and are the key cells needed for transfer in blood and marrow transplants used in the treatment of leukemia and other blood diseases. HSCs possess high levels of ALDH catalytic activity [80,81], making them less vulnerable to the cytotoxic effects of the chemotherapeutic agent, cyclophosphamide. Aldefluor®, a fluorescent compound activated by ALDH activity, was found to identify and select for human HSCs as well as mature myeloid progenitors from bone marrow, placenta and peripheral blood and, consequently, has been used as a marker for counting the number of HSCs and myeloid progenitors in these tissues for blood and marrow transplant purposes [76,82–87]. Aldefluor® has also been used to study leukemia and leukemic stem cells (LSCs) and preliminary evidence suggests that levels of activity in these cells may correlate with response to treatment [88–90].
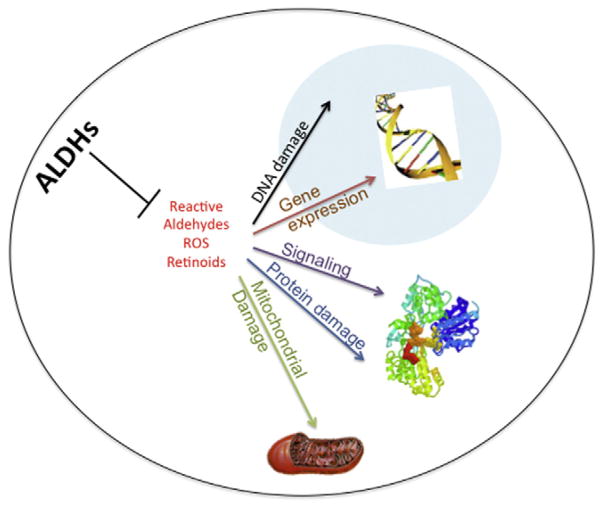
The high level of ALDH activity is suggestive of ALDH serving an important function in HSCs. It is possible that, by regulating intracellular levels of reactive oxygen species (ROS) and/or reactive aldehydes, ALDHs may modulate redox-dependent signal transduction pathways involved in fate decision processes in HSCs [91,92] (Fig. 4). In such a scheme, perturbations in the ALDH-dependent homeostatic control response may lead to the accumulation of ROS, reactive aldehydes and other compounds which, at higher (or inappropriate) levels, could foster myelodysplasia and leukemia through protein and DNA damage. Alternatively, ALDH enzymes could modulate HSCs by an effect on retinoic acid metabolism. For this reason, ALDH1A1 has been the focus of most interest in HSCs because of its involvement in retinoic acid metabolism. However, knockout of ALDH1A1 in mice fails to affect HSCs or hematopoiesis, likely due to compensation by other upregulated ALDH isozymes (or related enzymes) [92,93]. Clearly, further research needs to be conducted to elucidate why HSCs possess high ALDH activity and which ALDH isozymes contribute to the activity. The information derived from such studies will provide valuable insights into the involvement of the ALDH isozymes in normal HSC biology and leukemagenesis and potentially identify new therapeutic opportunities. For example, inhibition of ALDH activity may be useful for expanding HSCs, an intervention that could have implications for radioprotection, transplant and regenerative medicine purposes [84,94,95].
Fig. 4.
Potential mechanisms by which the ALDHs may play a role in normal HSC biology and leukemagenesis. The ALDH proteins as well as their metabolic products including retinoids, ROS, reactive aldehydes and others may directly and indirectly influence a variety of cellular processes including signal transduction, energy metabolism, gene expression and DNA integrity.
6. Cancer stem cells
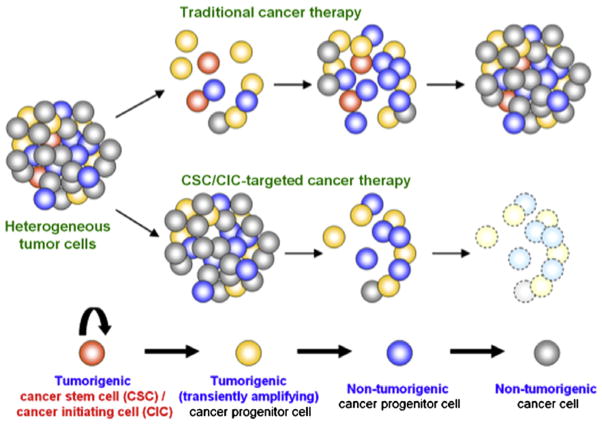
The milieu of a cancerous tumor consists of heterogenous cell populations. Several recent reports suggest that cancers harbor a small population of cells that possess increased capabilities for self-renewal, tumor initiation, propagation, treatment resistance and disease recurrence. These cells have been termed cancer stem cells (CSCs). CSCs are defined by their functional properties of (i) tumorigenesis (i.e., forming tumors in vivo), (ii) self-renewal (i.e., serially transplantable and (iii) differentiation (i.e., generating heterogeneous lineages recapitulating an original tumor) [96]. The CSC hypothesis [97,98] provides a rational basis for understanding some of the complexity of cancer biology (Fig. 5). For example, their existence may explain tumor recurrence after conventional chemotherapy and radiation therapy. In such a scheme, non-CSCs would be targeted by the treatment. The ability of CSCs to remain viable because of their intrinsic survival mechanisms would allow tumor initiation and propagation to be resurrected [99].
Fig. 5.
Cancer stem cell (CSC) hypothesis. Tumor initiation and growth result from the formation of cancer stem cells (CSCs) or cancer initiating cells (CICs). Tumor recurrence after traditional cancer therapy (such as chemotherapy or radiation) occurs from chemoresistant or radioresistant CSC/CICs. In contrast, CSC/CIC-directed therapeutics targets CSC/CICs and eradicate tumor cells.
High ALDH enzyme activities have been used as a CSC marker for many solid tumors including breast, lung, liver, colon, pancreatic, ovarian, head and neck, prostate [100–113] and melanoma [114,115]. While the Aldefluor® assay can be used to identify CSCs in a freshly dispersed population of cells from a tumor, it cannot be applied for immunohistochemical analyses of formalin-fixed cancer tissues. Given that ALDH1A1 has been considered to be the key isozyme responsible for positive Aldefluor® staining in stem cells [116], tumor specimens have been probed using ALDH1A1 antibodies. Such studies have failed to show a consistent association between ALDH1A1 expression and patient prognosis in several cancers, including breast [108,117–120], ovarian [121,122] and prostate cancers [103,123]. Since the human ALDH superfamily comprises many isozymes [7,27] each often possessing unique tissue distribution, subcellular localization and substrate specificity [1], it is not unreasonable to speculate that the isozyme expressed in each cancer type could differ based on organ- and/or tissue-specificity [1]. Supporting such a speculation are the reports of elevated expression of ALDH1B1 in colon cancer [10], ALDH3B1 in lung, breast, ovarian and colon cancer [124], ALDH3A1 in lung cancer [125], and ALDH7A1 in prostate cancer [103]. Such information will be critical for understanding the biological significance of the ALDH isozymes in the tumor cells and, potentially, for facilitating the development of therapies to treat the cancer.
Recently, ALDH1A3 was found to be expressed in Aldefluor®-positive CSCs from human breast cancer [120] and melanoma [115]. As such, it is unlikely that the ALDH enzymatic activity measured by Aldefluor® is due only to ALDH1A1. Furthermore, Marcato and colleagues found that the tumor grade and metastasis correlated with ALDH1A3 expression but not with ALDH1A1 in human breast cancer [120]. This finding underscores the importance of defining the ALDH isozyme responsible for the high ALDH activity in each cancer type. ALDH1A isozymes (including ALDH1A1 and ALDH1A3) oxidize retinaldehyde to RA. RA regulates the expression of a variety of genes through RAR and RXR, nuclear receptors that control the transcription of target genes by interacting with specific DNA sequences known as RA response elements (RAREs) [126]. Located in the promoter region of target genes, RAREs consist of two core hexameric motifs, PuG(G/T)TCA, separated by spacer nucleotides [126,127]. In ALDH-positive CSCs from human melanoma, RA-driven target genes with RAREs and genes associated with stem cell function have been elucidated [115]. In the same study, the ALDH1A isozymes were shown to be essential to the function of CSCs, making ALDH1A isozymes putative therapeutic targets [115]. For example, silencing ALDH1A genes (ALDH1A1 and ALDH1A3) decreased cell viability and increased apoptosis in ALDH-positive human melanoma CSCs in vitro. Knockdown of ALDH1A reduced tumorigenesis in vivo. In addition, ALDH-positive human melanoma CSCs were resistant to chemotherapeutic agents; resistance was attributable to the expression of ALDH1A genes. These findings provide compelling evidence in favor of the ALDH isozymes functioning as key molecules governing cell proliferation, survival and chemoresistance of CSCs, i.e., they seem more than simply CSC markers. The capacity of the ALDH isozymes to metabolize cytotoxic aldehydes (arising physiologically or as a result of chemotherapy, radiation or oxidative stress) may also contribute to the survival and drug resistance of CSCs. To prevent cancer recurrence, eradication of CSCs must be an essential part of cancer treatment in addition to eliminating non-stem cancer cells [128]. Given the accumulating and evolving evidence, selective suppression of the ALDH isozymes and genes may be a promising frontier for CSC-directed therapeutics in human cancers. It is hoped that this may spur the development of novel and highly specific inhibitors [129,130].
7. Conclusions
The ALDHs are a family of proteins with diverse biological functions and properties that can (and often do) extend beyond their catalytic activity. Research from the past decade has elucidated potential roles for the ALDH isozymes that would not have been predicted from existing knowledge. The development and availability of isozyme-specific enzyme inhibitors should greatly advance our understanding of the physiological and pathophysiological actions of the ALDHs. Further, they will allow assessment of the relative contribution of catalytic actions versus non-catalytic actions (e.g. protein–protein interactions) to any ALDH isozyme effects. If our past experience with the ALDHs is any guide, we should expect the unexpected from this fascinating protein family.
Acknowledgments
We would like to thank our colleagues for critically reviewing this manuscript. This work was supported, in part, by the following NIH grants; EY17963 and EY11490.
Abbreviations
- AASA
alpha-aminoadipic semialdehyde
- ALDH
aldehyde dehydrogenase
- AMADH
aminoaldehyde dehydrogenase
- BADH
betaine aldehyde dehydrogenase
- CIC
cancer initiating cells
- CSC
cancer stem cells
- 10-FTHF
10-formyltetrahydrofolate
- FDH
10-FTHF dehydrogenase
- GABA
γ-aminobutyric acid
- GAP
glyceraldehyde 3-phosphate
- GAPN
GAP dehydrogenase
- 4-HNE
4-Hydroxynonenal
- HSC
hematopoietic stem cell
- LSC
leukemic stem cell
- MDA
malondialdehyde
- MMS
methylmalonate semialdehyde
- NAD(P)+
nicotinamide adenine dinucleotide (phosphate)
- P5C
pyrroline-5-carboxylate
- P5CDH
P5C dehydrogenase
- RA
retinoic acid
- ROS
reactive oxygen species
- SLS
Sjögren–Larsson syndrome
- SNP
single nucleotide polymorphism
- SP
side population
- SSA
succinic semialdehyde
- TIC
tumor initiating cells
- UVR
ultraviolet radiation
References
- 1.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stagos D, Chen Y, Cantore M, Jester JV, Vasiliou V. Corneal aldehyde dehydrogenases: multiple functions and novel nuclear localization. Brain Res Bull. 2010;81:211–218. doi: 10.1016/j.brainresbull.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brocker C, Lassen N, Estey T, Pappa A, Cantore M, Orlova VV, Chavakis T, Kavanagh KL, Oppermann U, Vasiliou V. Aldehyde dehydrogenase 7A1 (ALDH7A1) is a novel enzyme involved in cellular defense against hyperosmotic stress. J Biol Chem. 2010;285:18452–18463. doi: 10.1074/jbc.M109.077925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasiliou V, Bairoch A, Tipton KF, Nebert DW. Eukaryotic aldehyde dehydrogenase (ALDH) genes: human polymorphisms, and recommended nomenclature based on divergent evolution and chromosomal mapping. Pharmacogenetics. 1999;9:421–434. [PubMed] [Google Scholar]
- 5.Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, Nebert DW, Vasiliou V. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum Genomics. 2011;5:283–303. doi: 10.1186/1479-7364-5-4-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black W, Vasiliou V. The aldehyde dehydrogenase gene superfamily resource center. Hum Genomics. 2009;4:136–142. doi: 10.1186/1479-7364-4-2-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasiliou V, Nebert DW. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum Genomics. 2005;2:138–143. doi: 10.1186/1479-7364-2-2-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duester G. Families of retinoid dehydrogenases regulating vitamin A function: production of visual pigment and retinoic acid. Eur J Biochem. 2000;267:4315–4324. doi: 10.1046/j.1432-1327.2000.01497.x. [DOI] [PubMed] [Google Scholar]
- 9.Stagos D, Chen Y, Brocker C, Donald E, Jackson BC, Orlicky DJ, Thompson DC, Vasiliou V. Aldehyde dehydrogenase 1B1: molecular cloning and characterization of a novel mitochondrial acetaldehyde-metabolizing enzyme. Drug Metab Dispos. 2010;38:1679–1687. doi: 10.1124/dmd.110.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Orlicky DJ, Matsumoto A, Singh S, Thompson DC, Vasiliou V. Aldehyde dehydrogenase 1B1 (ALDH1B1) is a potential biomarker for human colon cancer. Biochem Biophys Res Commun. 2011;405:173–179. doi: 10.1016/j.bbrc.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsybovsky Y, Krupenko SA. Conserved catalytic residues of the ALDH1L1 aldehyde dehydrogenase domain control binding and discharging of the coenzyme. J Biol Chem. 2011;286:23357–23367. doi: 10.1074/jbc.M111.221069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strickland KC, Krupenko NI, Dubard ME, Hu CJ, Tsybovsky Y, Krupenko SA. Enzymatic properties of ALDH1L2, a mitochondrial 10-formyltetrahydrofolate dehydrogenase. Chem Biol Interact. 2011;191:129–136. doi: 10.1016/j.cbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pappa A, Estey T, Manzer R, Brown D, Vasiliou V. Human aldehyde dehydrogenase 3A1 (ALDH3A1): biochemical characterization and immunohistochemical localization in the cornea. Biochem J. 2003;376:615–623. doi: 10.1042/BJ20030810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rizzo WB, Heinz E, Simon M, Craft DA. Microsomal fatty aldehyde dehydrogenase catalyzes the oxidation of aliphatic aldehyde derived from ether glycerolipid catabolism: implications for Sjogren–Larsson syndrome. Biochim Biophys Acta. 2000;1535:1–9. doi: 10.1016/s0925-4439(00)00077-6. [DOI] [PubMed] [Google Scholar]
- 15.Marchitti SA, Brocker C, Orlicky DJ, Vasiliou V. Molecular characterization, expression analysis, and role of ALDH3B1 in the cellular protection against oxidative stress. Free Radic Biol Med. 2010;49:1432–1443. doi: 10.1016/j.freeradbiomed.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forte-McRobbie CM, Pietruszko R. Purification and characterization of human liver “high Km” aldehyde dehydrogenase and its identification as glutamic gamma-semialdehyde dehydrogenase. J Biol Chem. 1986;261:2154–2163. [PubMed] [Google Scholar]
- 17.Kim YG, Lee S, Kwon OS, Park SY, Lee SJ, Park BJ, Kim KJ. Redox-switch modulation of human SSADH by dynamic catalytic loop. EMBO J. 2009;28:959–968. doi: 10.1038/emboj.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambliss KL, Gray RG, Rylance G, Pollitt RJ, Gibson KM. Molecular characterization of methylmalonate semialdehyde dehydrogenase deficiency. J Inherit Metab Dis. 2000;23:497–504. doi: 10.1023/a:1005616315087. [DOI] [PubMed] [Google Scholar]
- 19.Mills PB, Struys E, Jakobs C, Plecko B, Baxter P, Baumgartner M, Willemsen MA, Omran H, Tacke U, Uhlenberg B, Weschke B, Clayton PT. Mutations in antiquitin in individuals with pyridoxine-dependent seizures. Nat Med. 2006;12:307–309. doi: 10.1038/nm1366. [DOI] [PubMed] [Google Scholar]
- 20.Lin M, Zhang M, Abraham M, Smith SM, Napoli JL. Mouse retinal dehydrogenase 4 (RALDH4), molecular cloning, cellular expression, and activity in 9-cis-retinoic acid biosynthesis in intact cells. J Biol Chem. 2003;278:9856–9861. doi: 10.1074/jbc.M211417200. [DOI] [PubMed] [Google Scholar]
- 21.Sima A, Parisotto M, Mader S, Bhat PV. Kinetic characterization of recombinant mouse retinal dehydrogenase types 3 and 4 for retinal substrates. Biochim Biophys Acta. 2009;1790:1660–1664. doi: 10.1016/j.bbagen.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Lin SW, Chen JC, Hsu LC, Hsieh CL, Yoshida A. Human gamma-aminobutyraldehyde dehydrogenase (ALDH9): cDNA sequence, genomic organization, polymorphism, chromosomal localization, and tissue expression. Genomics. 1996;34:376–380. doi: 10.1006/geno.1996.0300. [DOI] [PubMed] [Google Scholar]
- 23.Vasiliou V, Sandoval M, Backos DS, Jackson BS, Chen Y, Reigan R, Lanaspa MA, Richard J, Johnson RJ, Koppaka V, Thompson DC. ALDH16A1 is a novel protein that may be involved in the etiology of gout via protein–protein interactions with HPRT1. doi: 10.1016/j.cbi.2012.12.018. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valverde F, Losada M, Serrano A. Engineering a central metabolic pathway: glycolysis with no net phosphorylation in an Escherichia coli gap mutant complemented with a plant GapN gene. FEBS Lett. 1999;449:153–158. doi: 10.1016/s0014-5793(99)00430-5. [DOI] [PubMed] [Google Scholar]
- 25.Brocker C, Vasiliou M, Carpenter S, Carpenter C, Zhang Y, Wang X, Kotchoni SO, Wood AJ, Kirch HH, Kopecny D, Nebert DW, Vasiliou V. Aldehyde dehydrogenase (ALDH) superfamily in plants: gene nomenclature and comparative genomics. Planta. 2012 doi: 10.1007/s00425-012-1749-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samuelson J, Zhang WW, Kumar A, Descoteaux S, Shen PS, Bailey G. Primary structures of alcohol and aldehyde dehydrogenase genes of Entamoeba histolytica. Arch Med Res. 1992;23:31–33. [PubMed] [Google Scholar]
- 27.Sophos NA, Vasiliou V. Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem Biol Interact. 2003;143–144:5–22. doi: 10.1016/s0009-2797(02)00163-1. [DOI] [PubMed] [Google Scholar]
- 28.Blatter EE, Abriola DP, Pietruszko R. Aldehyde dehydrogenase. Covalent intermediate in aldehyde dehydrogenation and ester hydrolysis. Biochem J. 1992;282(Pt 2):353–360. doi: 10.1042/bj2820353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z, Zhang J, Stamler JS. Identification of the enzymatic mechanism of nitroglycerin bioactivation. Proc Natl Acad Sci U S A. 2002;99:8306–8311. doi: 10.1073/pnas.122225199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piatigorsky J, Wistow GJ. Enzyme/crystallins: gene sharing as an evolutionary strategy. Cell. 1989;57:197–199. doi: 10.1016/0092-8674(89)90956-2. [DOI] [PubMed] [Google Scholar]
- 31.Horwitz J, Ding L, Vasiliou V, Cantore M, Piatigorsky J. Scallop lens Omega-crystallin (ALDH1A9): a novel tetrameric aldehyde dehydrogenase. Biochem Biophys Res Commun. 2006;348:1302–1309. doi: 10.1016/j.bbrc.2006.07.197. [DOI] [PubMed] [Google Scholar]
- 32.Zinovieva RD, Tomarev SI, Piatigorsky J. Aldehyde dehydrogenase-derived omega-crystallins of squid and octopus. Specialization for lens expression. J Biol Chem. 1993;268:11449–11455. [PubMed] [Google Scholar]
- 33.King G, Holmes R. Human corneal and lens aldehyde dehydrogenases. Purification and properties of human lens ALDH1 and differential expression as major soluble proteins in human lens (ALDH1) and cornea (ALDH3) Adv Exp Med Biol. 1997;414:19–27. [PubMed] [Google Scholar]
- 34.Jester JV, Moller-Pedersen T, Huang J, Sax CM, Kays WT, Cavangh HD, Petroll WM, Piatigorsky J. The cellular basis of corneal transparency: evidence for ‘corneal crystallins’. J Cell Sci. 1999;112(Pt 5):613–622. doi: 10.1242/jcs.112.5.613. [DOI] [PubMed] [Google Scholar]
- 35.Estey T, Piatigorsky J, Lassen N, Vasiliou V. ALDH3A1: a corneal crystallin with diverse functions. Exp Eye Res. 2007;84:3–12. doi: 10.1016/j.exer.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 36.Jester JV. Corneal crystallins and the development of cellular transparency. Semin Cell Dev Biol. 2008;19:82–93. doi: 10.1016/j.semcdb.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchitti SA, Chen Y, Thompson DC, Vasiliou V. Ultraviolet radiation: cellular antioxidant response and the role of ocular aldehyde dehydrogenase enzymes. Eye Contact Lens. 2011;37:206–213. doi: 10.1097/ICL.0b013e3182212642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lassen N, Pappa A, Black WJ, Jester JV, Day BJ, Min E, Vasiliou V. Antioxidant function of corneal ALDH3A1 in cultured stromal fibroblasts. Free Radic Biol Med. 2006;41:1459–1469. doi: 10.1016/j.freeradbiomed.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y, Koppaka V, Thompson DC, Vasiliou V. Focus on molecules: ALDH1A1: from lens and corneal crystallin to stem cell marker. Exp Eye Res. 2011 doi: 10.1016/j.exer.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mason JM, Arndt KM. Coiled coil domains: stability, specificity, and biological implications. Chembiochem: Eur J Chem Biol. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- 41.Hanna MC, Blackstone C. Interaction of the SPG21 protein ACP33/maspardin with the aldehyde dehydrogenase ALDH16A1. Neurogenetics. 2009;10:217–228. doi: 10.1007/s10048-009-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alexander RJ, Silverman B, Henley WL. Isolation and characterization of BCP 54, the major soluble protein of bovine cornea. Exp Eye Res. 1981;32:205–216. doi: 10.1016/0014-4835(81)90009-9. [DOI] [PubMed] [Google Scholar]
- 43.Pappa A, Sophos NA, Vasiliou V. Corneal and stomach expression of aldehyde dehydrogenases: from fish to mammals. Chem Biol Interact. 2001;130–132:181–191. doi: 10.1016/s0009-2797(00)00233-7. [DOI] [PubMed] [Google Scholar]
- 44.Wistow G, Kim H. Lens protein expression in mammals: taxon-specificity and the recruitment of crystallins. J Mol Evol. 1991;32:262–269. doi: 10.1007/BF02342749. [DOI] [PubMed] [Google Scholar]
- 45.Lassen N, Bateman JB, Estey T, Kuszak JR, Nees DW, Piatigorsky J, Duester G, Day BJ, Huang J, Hines LM, Vasiliou V. Multiple and additive functions of ALDH3A1 and ALDH1A1: cataract phenotype and ocular oxidative damage in Aldh3a1(−/−)/Aldh1a1(−/−) knock-out mice. J Biol Chem. 2007;282:25668–25676. doi: 10.1074/jbc.M702076200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manzer R, Qamar L, Estey T, Pappa A, Petersen DR, Vasiliou V. Molecular cloning and baculovirus expression of the rabbit corneal aldehyde dehydrogenase (ALDH1A1) cDNA. DNA Cell Biol. 2003;22:329–338. doi: 10.1089/104454903322216671. [DOI] [PubMed] [Google Scholar]
- 47.Lassen N, Black WJ, Estey T, Vasiliou V. The role of corneal crystallins in the cellular defense mechanisms against oxidative stress. Semin Cell Dev Biol. 2008;19:100–112. doi: 10.1016/j.semcdb.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 48.Pappa A, Chen C, Koutalos Y, Townsend AJ, Vasiliou V. Aldh3a1 protects human corneal epithelial cells from ultraviolet- and 4-hydroxy-2-nonenal-induced oxidative damage. Free Radic Biol Med. 2003;34:1178–1189. doi: 10.1016/s0891-5849(03)00070-4. [DOI] [PubMed] [Google Scholar]
- 49.Estey T, Cantore M, Weston PA, Carpenter JF, Petrash JM, Vasiliou V. Mechanisms involved in the protection of UV-induced protein inactivation by the corneal crystallin ALDH3A1. J Biol Chem. 2007;282:4382–4392. doi: 10.1074/jbc.M607546200. [DOI] [PubMed] [Google Scholar]
- 50.Bloemendal H, de Jong W, Jaenicke R, Lubsen NH, Slingsby C, Tardieu A. Ageing and vision: structure, stability and function of lens crystallins. Prog Biophys Mol Biol. 2004;86:407–485. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 51.Estey T, Chen Y, Carpenter JF, Vasiliou V. Structural and functional modifications of corneal crystallin ALDH3A1 by UVB light. PLoS One. 2010;5:e15218. doi: 10.1371/journal.pone.0015218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jester JV, Budge A, Fisher S, Huang J. Corneal keratocytes: phenotypic and species differences in abundant protein expression and in vitro light-scattering. Invest Ophthalmol Vis Sci. 2005;46:2369–2378. doi: 10.1167/iovs.04-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pappas P, Stephanou P, Vasiliou V, Karamanakos P, Marselos M. Ontogenesis and expression of ALDH activity in the skin and the eye of the rat. Adv Exp Med Biol. 1997;414:73–80. doi: 10.1007/978-1-4615-5871-2_10. [DOI] [PubMed] [Google Scholar]
- 54.Davis J, Davis D, Norman B, Piatigorsky J. Gene expression of the mouse corneal crystallin Aldh3a1: activation by Pax6, Oct1, and p300. Invest Ophthalmol Vis Sci. 2008;49:1814–1826. doi: 10.1167/iovs.07-1057. [DOI] [PubMed] [Google Scholar]
- 55.Pappa A, Brown D, Koutalos Y, DeGregori J, White C, Vasiliou V. Human aldehyde dehydrogenase 3A1 inhibits proliferation and promotes survival of human corneal epithelial cells. J Biol Chem. 2005;280:27998–28006. doi: 10.1074/jbc.M503698200. [DOI] [PubMed] [Google Scholar]
- 56.Jester JV, Brown D, Pappa A, Vasiliou V. Myofibroblast differentiation modulates keratocyte crystallin protein expression, concentration, and cellular light scattering. Invest Ophthalmol Vis Sci. 2012;53:770–778. doi: 10.1167/iovs.11-9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ziouzenkova O, Orasanu G, Sharlach M, Akiyama TE, Berger JP, Viereck J, Hamilton JA, Tang G, Dolnikowski GG, Vogel S, Duester G, Plutzky J. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat Med. 2007;13:695–702. doi: 10.1038/nm1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kiefer FW, Orasanu G, Nallamshetty S, Brown JD, Wang H, Luger P, Qi NR, Burant CF, Duester G, Plutzky J. Retinaldehyde dehydrogenase 1 coordinates hepatic gluconeogenesis and lipid metabolism. Endocrinology. 2012;153:3089–3099. doi: 10.1210/en.2011-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kiefer FW, Vernochet C, O’Brien P, Spoerl S, Brown JD, Nallamshetty S, Zeyda M, Stulnig TM, Cohen DE, Kahn CR, Plutzky J. Retinaldehyde dehydrogenase 1 regulates a thermogenic program in white adipose tissue. Nat Med. 2012 doi: 10.1038/nm.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niederreither K, Subbarayan V, Dolle P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- 61.Deak KL, Dickerson ME, Linney E, Enterline DS, George TM, Melvin EC, Graham FL, Siegel DG, Hammock P, Mehltretter L, Bassuk AG, Kessler JA, Gilbert JR, Speer MC. Analysis of ALDH1A2, CYP26A1, CYP26B1, CRABP1, and CRABP2 in human neural tube defects suggests a possible association with alleles in ALDH1A2, Birth defects research. Part A. Clin Mol Teratol. 2005;73:868–875. doi: 10.1002/bdra.20183. [DOI] [PubMed] [Google Scholar]
- 62.Pavan M, Ruiz VF, Silva FA, Sobreira TJ, Cravo RM, Vasconcelos M, Marques LP, Mesquita SM, Krieger JE, Lopes AA, Oliveira PS, Pereira AC, Xavier-Neto J. ALDH1A2 (RALDH2) genetic variation in human congenital heart disease. BMC Med Genet. 2009;10:113. doi: 10.1186/1471-2350-10-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dupe V, Matt N, Garnier JM, Chambon P, Mark M, Ghyselinck NB. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc Natl Acad Sci U S A. 2003;100:14036–14041. doi: 10.1073/pnas.2336223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Husemoen LL, Fenger M, Friedrich N, Tolstrup JS, Beenfeldt Fredriksen S, Linneberg A. The association of ADH and ALDH gene variants with alcohol drinking habits and cardiovascular disease risk factors. Alcohol Clin Exp Res. 2008;32:1984–1991. doi: 10.1111/j.1530-0277.2008.00780.x. [DOI] [PubMed] [Google Scholar]
- 65.Linneberg A, Gonzalez-Quintela A, Vidal C, Jorgensen T, Fenger M, Hansen T, Pedersen O, Husemoen LL. Genetic determinants of both ethanol and acetaldehyde metabolism influence alcohol hypersensitivity and drinking behaviour among Scandinavians. Clin Exp Allergy. 2010;40:123–130. doi: 10.1111/j.1365-2222.2009.03398.x. [DOI] [PubMed] [Google Scholar]
- 66.Palmfeldt J, Vang S, Stenbroen V, Pavlou E, Baycheva M, Buchal G, Monavari AA, Augoustides-Savvopoulou P, Mandel H, Gregersen N. Proteomics reveals that redox regulation is disrupted in patients with ethylmalonic encephalopathy. J Proteome Res. 2011;10:2389–2396. doi: 10.1021/pr101218d. [DOI] [PubMed] [Google Scholar]
- 67.Palo OM, Soronen P, Silander K, Varilo T, Tuononen K, Kieseppa T, Partonen T, Lonnqvist J, Paunio T, Peltonen L. Identification of susceptibility loci at 7q31 and 9p13 for bipolar disorder in an isolated population. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics: the Official Publication of the International Society of Psychiatric Genetics. 2010;153B:723–735. doi: 10.1002/ajmg.b.31039. [DOI] [PubMed] [Google Scholar]
- 68.Cook RJ, Champion KM, Giometti CS. Methanol toxicity and formate oxidation in NEUT2 mice. Arch Biochem Biophys. 2001;393:192–198. doi: 10.1006/abbi.2001.2485. [DOI] [PubMed] [Google Scholar]
- 69.Champion KM, Cook RJ, Tollaksen SL, Giometti CS. Identification of a heritable deficiency of the folate-dependent enzyme 10-formyltetrahydrofolate dehydrogenase in mice. Proc Natl Acad Sci U S A. 1994;91:11338–11342. doi: 10.1073/pnas.91.24.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jo SA, Kim EK, Park MH, Han C, Park HY, Jang Y, Song BJ, Jo I. A Glu487Lys polymorphism in the gene for mitochondrial aldehyde dehydrogenase 2 is associated with myocardial infarction in elderly Korean men. Clin Chim Acta. 2007;382:43–47. doi: 10.1016/j.cca.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 71.Hui P, Nakayama T, Morita A, Sato N, Hishiki M, Saito K, Yoshikawa Y, Tamura M, Sato I, Takahashi T, Soma M, Izumi Y, Ozawa Y, Cheng Z. Common single nucleotide polymorphisms in Japanese patients with essential hypertension: aldehyde dehydrogenase 2 gene as a risk factor independent of alcohol consumption. Hypertens Res. 2007;30:585–592. doi: 10.1291/hypres.30.585. [DOI] [PubMed] [Google Scholar]
- 72.Chao YC, Liou SR, Chung YY, Tang HS, Hsu CT, Li TK, Yin SJ. Polymorphism of alcohol and aldehyde dehydrogenase genes and alcoholic cirrhosis in Chinese patients. Hepatology. 1994;19:360–366. [PubMed] [Google Scholar]
- 73.Kamino K, Nagasaka K, Imagawa M, Yamamoto H, Yoneda H, Ueki A, Kitamura S, Namekata K, Miki T, Ohta S. Deficiency in mitochondrial aldehyde dehydrogenase increases the risk for late-onset Alzheimer’s disease in the Japanese population. Biochem Biophys Res Commun. 2000;273:192–196. doi: 10.1006/bbrc.2000.2923. [DOI] [PubMed] [Google Scholar]
- 74.De Laurenzi V, Rogers GR, Hamrock DJ, Marekov LN, Steinert PM, Compton JG, Markova N, Rizzo WB. Sjogren–Larsson syndrome is caused by mutations in the fatty aldehyde dehydrogenase gene. Nat Genet. 1996;12:52–57. doi: 10.1038/ng0196-52. [DOI] [PubMed] [Google Scholar]
- 75.Vasiliou V, Pappa A. Polymorphisms of human aldehyde dehydrogenases. Consequences for drug metabolism and disease. Pharmacology. 2000;61:192–198. doi: 10.1159/000028400. [DOI] [PubMed] [Google Scholar]
- 76.Sass JO, Walter M, Shield JP, Atherton AM, Garg U, Scott D, Woods CG, Smith LD. 3-Hydroxyisobutyrate aciduria and mutations in the ALDH6A1 gene coding for methylmalonate semialdehyde dehydrogenase. J Inherit Metab Dis. 2012;35:437–442. doi: 10.1007/s10545-011-9381-x. [DOI] [PubMed] [Google Scholar]
- 77.Guo Y, Tan LJ, Lei SF, Yang TL, Chen XD, Zhang F, Chen Y, Pan F, Yan H, Liu X, Tian Q, Zhang ZX, Zhou Q, Qiu C, Dong SS, Xu XH, Guo YF, Zhu XZ, Liu SL, Wang XL, Li X, Luo Y, Zhang LS, Li M, Wang JT, Wen T, Drees B, Hamilton J, Papasian CJ, Recker RR, Song XP, Cheng J, Deng HW. Genome-wide association study identifies ALDH7A1 as a novel susceptibility gene for osteoporosis. PLoS Genet. 2010;6:e1000806. doi: 10.1371/journal.pgen.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sulem P, Gudbjartsson DF, Walters GB, Helgadottir HT, Helgason A, Gudjonsson SA, Zanon C, Besenbacher S, Bjornsdottir G, Magnusson OT, Magnusson G, Hjartarson E, Saemundsdottir J, Gylfason A, Jonasdottir A, Holm H, Karason A, Rafnar T, Stefansson H, Andreassen OA, Pedersen JH, Pack AI, de Visser MC, Kiemeney LA, Geirsson AJ, Eyjolfsson GI, Olafsson I, Kong A, Masson G, Jonsson H, Thorsteinsdottir U, Jonsdottir I, Stefansson K. Identification of low-frequency variants associated with gout and serum uric acid levels. Nat Genet. 2011;43:1127–1130. doi: 10.1038/ng.972. [DOI] [PubMed] [Google Scholar]
- 79.Vasiliou V, Malamas M, Marselos M. The mechanism of alcohol intolerance produced by various therapeutic agents. Acta Pharmacol Toxicol (Copenh) 1986;58:305–310. doi: 10.1111/j.1600-0773.1986.tb00114.x. [DOI] [PubMed] [Google Scholar]
- 80.Kastan MB, Schlaffer E, Russo JE, Colvin OM, Civin CI, Hilton J. Direct demonstration of elevated aldehyde dehydrogenase in human hematopoietic progenitor cells. Blood. 1990;75:1947–1950. [PubMed] [Google Scholar]
- 81.Forsberg EC, Prohaska SS, Katzman S, Heffner GC, Stuart JM, Weissman IL. Differential expression of novel potential regulators in hematopoietic stem cells. PLoS Genet. 2005;1:e28. doi: 10.1371/journal.pgen.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fallon P, Gentry T, Balber AE, Boulware D, Janssen WE, Smilee R, Storms RW, Smith C. Mobilized peripheral blood SSCloALDHbr cells have the phenotypic and functional properties of primitive haematopoietic cells and their number correlates with engraftment following autologous transplantation. Br J Haematol. 2003;122:99–108. doi: 10.1046/j.1365-2141.2003.04357.x. [DOI] [PubMed] [Google Scholar]
- 83.Hess DA, Meyerrose TE, Wirthlin L, Craft TP, Herrbrich PE, Creer MH, Nolta JA. Functional characterization of highly purified human hematopoietic repopulating cells isolated according to aldehyde dehydrogenase activity. Blood. 2004;104:1648–1655. doi: 10.1182/blood-2004-02-0448. [DOI] [PubMed] [Google Scholar]
- 84.Muramoto GG, Russell JL, Safi R, Salter AB, Himburg HA, Daher P, Meadows SK, Doan P, Storms RW, Chao NJ, McDonnell DP, Chute JP. Inhibition of aldehyde dehydrogenase expands hematopoietic stem cells with radioprotective capacity. Stem Cells. 2010;28:523–534. doi: 10.1002/stem.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hess DA, Wirthlin L, Craft TP, Herrbrich PE, Hohm SA, Lahey R, Eades WC, Creer MH, Nolta JA. Selection based on CD133 and high aldehyde dehydrogenase activity isolates long-term reconstituting human hematopoietic stem cells. Blood. 2006;107:2162–2169. doi: 10.1182/blood-2005-06-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lioznov M, Dellbrugger C, Sputtek A, Fehse B, Kroger N, Zander AR. Transportation and cryopreservation may impair haematopoietic stem cell function and engraftment of allogeneic PBSCs, but not BM. Bone Marrow Transplant. 2008;42:121–128. doi: 10.1038/bmt.2008.93. [DOI] [PubMed] [Google Scholar]
- 87.Lioznov MV, Freiberger P, Kroger N, Zander AR, Fehse B. Aldehyde dehydrogenase activity as a marker for the quality of hematopoietic stem cell transplants. Bone Marrow Transplant. 2005;35:909–914. doi: 10.1038/sj.bmt.1704928. [DOI] [PubMed] [Google Scholar]
- 88.Cheung AM, Wan TS, Leung JC, Chan LY, Huang H, Kwong YL, Liang R, Leung AY. Aldehyde dehydrogenase activity in leukemic blasts defines a subgroup of acute myeloid leukemia with adverse prognosis and superior NOD/SCID engrafting potential. Leukemia. 2007;21:1423–1430. doi: 10.1038/sj.leu.2404721. [DOI] [PubMed] [Google Scholar]
- 89.Ran D, Schubert M, Pietsch L, Taubert I, Wuchter P, Eckstein V, Bruckner T, Zoeller M, Ho AD. Aldehyde dehydrogenase activity among primary leukemia cells is associated with stem cell features and correlates with adverse clinical outcomes. Exp Hematol. 2009;37:1423–1434. doi: 10.1016/j.exphem.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 90.Gerber JM, Smith BD, Ngwang B, Zhang H, Vala MS, Morsberger L, Galkin S, Collector MI, Perkins B, Levis MJ, Griffin CA, Sharkis SJ, Borowitz MJ, Karp JE, Jones RJ. A clinically relevant population of leukemic CD34(+)CD38(−) cells in acute myeloid leukemia. Blood. 2012;119:3571–3577. doi: 10.1182/blood-2011-06-364182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gasparetto M, Sekulovic S, Zakaryan A, Imren S, Kent DG, Humphries RK, Vasiliou V, Smith C. Varying levels of aldehyde dehydrogenase activity in adult murine marrow hematopoietic stem cells are associated with engraftment and cell cycle status. Exp Hematol. 2012 doi: 10.1016/j.exphem.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 92.Gasparetto M, Sekulovic S, Brocker C, Tang P, Zakaryan A, Xiang P, Kuchenbauer F, Wen M, Kasaian K, Witty MF, Rosten P, Chen Y, Imren S, Duester G, Thompson DC, Humphries RK, Vasiliou V, Smith C. Aldehyde dehydrogenases are regulators of hematopoietic stem cell numbers and B-cell development. Exp Hematol. 2012;40:318–329. e312. doi: 10.1016/j.exphem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 93.Levi BP, Yilmaz OH, Duester G, Morrison SJ. Aldehyde dehydrogenase 1a1 is dispensable for stem cell function in the mouse hematopoietic and nervous systems. Blood. 2009;113:1670–1680. doi: 10.1182/blood-2008-05-156752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chute JP, Muramoto GG, Whitesides J, Colvin M, Safi R, Chao NJ, McDonnell DP. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006;103:11707–11712. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ma ACH, Chung MIS, Liang R, Leung AYH. A DEAB-sensitive aldehyde dehydrogenase regulates hematopoietic stem and progenitor cells development during primitive hematopoiesis in zebrafish embryos. Leukemia. 2010 doi: 10.1038/leu.2010.206. [DOI] [PubMed] [Google Scholar]
- 96.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells–perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 97.Mackillop WJ, Ciampi A, Till JE, Buick RN. A stem cell model of human tumor growth: implications for tumor cell clonogenic assays. J Natl Cancer Inst. 1983;70:9–16. [PubMed] [Google Scholar]
- 98.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 99.Soltysova A, Altanerova V, Altaner C. Cancer stem cells. Neoplasma. 2005;52:435–440. [PubMed] [Google Scholar]
- 100.Todaro M, Iovino F, Eterno V, Cammareri P, Gambara G, Espina V, Gulotta G, Dieli F, Giordano S, De Maria R, Stassi G. Tumorigenic and metastatic activity of human thyroid cancer stem cells. Cancer Res. 2010;70:8874–8885. doi: 10.1158/0008-5472.CAN-10-1994. [DOI] [PubMed] [Google Scholar]
- 101.Ucar D, Cogle CR, Zucali JR, Ostmark B, Scott EW, Zori R, Gray BA, Moreb JS. Aldehyde dehydrogenase activity as a functional marker for lung cancer. Chem Biol Interact. 2009;178:48–55. doi: 10.1016/j.cbi.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, Wang H, Liu Z, Su Y, Stass SA, Katz RL. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7:330–338. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Lippitt JM, Guzman-Ramirez N, Hamdy FC, Eaton CL, Thalmann GN, Cecchini MG, Pelger RC, van der Pluijm G. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010;70:5163–5173. doi: 10.1158/0008-5472.CAN-09-3806. [DOI] [PubMed] [Google Scholar]
- 104.Rasheed ZA, Yang J, Wang Q, Kowalski J, Freed I, Murter C, Hong SM, Koorstra JB, Rajeshkumar NV, He X, Goggins M, Iacobuzio-Donahue C, Berman DM, Laheru D, Jimeno A, Hidalgo M, Maitra A, Matsui W. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J Natl Cancer Inst. 2010;102:340–351. doi: 10.1093/jnci/djp535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang L, Park P, Zhang H, La Marca F, Lin CY. Prospective identification of tumorigenic osteosarcoma cancer stem cells in OS99-1 cells based on high aldehyde dehydrogenase activity. Int J Cancer. 2010 doi: 10.1002/ijc.25331. [DOI] [PubMed] [Google Scholar]
- 106.Clay MR, Tabor M, Owen JH, Carey TE, Bradford CR, Wolf GT, Wicha MS, Prince ME. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck. 2010 doi: 10.1002/hed.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, Allan AL. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med. 2009;13:2236–2252. doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Carpentino JE, Hynes MJ, Appelman HD, Zheng T, Steindler DA, Scott EW, Huang EH. Aldehyde dehydrogenase-expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer. Cancer Res. 2009;69:8208–8215. doi: 10.1158/0008-5472.CAN-09-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69:3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, Houvenaeghel G, Extra JM, Bertucci F, Jacquemier J, Xerri L, Dontu G, Stassi G, Xiao Y, Barsky SH, Birnbaum D, Viens P, Wicha MS. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2009;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dylla SJ, Beviglia L, Park IK, Chartier C, Raval J, Ngan L, Pickell K, Aguilar J, Lazetic S, Smith-Berdan S, Clarke MF, Hoey T, Lewicki J, Gurney AL. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS One. 2008;3:e2428. doi: 10.1371/journal.pone.0002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Silva IA, Bai S, McLean K, Yang K, Griffith K, Thomas D, Ginestier C, Johnston C, Kueck A, Reynolds RK, Wicha MS, Buckanovich RJ. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 2011;71:3991–4001. doi: 10.1158/0008-5472.CAN-10-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Boonyaratanakornkit JB, Yue L, Strachan LR, Scalapino KJ, LeBoit PE, Lu Y, Leong SP, Smith JE, Ghadially R. Selection of tumorigenic melanoma cells using ALDH. J Invest Dermatol. 2010;130:2799–2808. doi: 10.1038/jid.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Luo Y, Dallaglio K, Chen Y, Robinson WA, Robinson SE, McCarter MD, Wng J, Gozalez R, Thompson DC, Norris DA, Roop DR, Vasiliou V, Fujita M. ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem Cells. 2012;30:2100–2113. doi: 10.1002/stem.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Burger PE, Gupta R, Xiong X, Ontiveros CS, Salm SN, Moscatelli D, Wilson EL. High aldehyde dehydrogenase activity: a novel functional marker of murine prostate stem/progenitor cells. Stem Cells. 2009;27:2220–2228. doi: 10.1002/stem.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Resetkova E, Reis-Filho JS, Jain RK, Mehta R, Thorat MA, Nakshatri H, Badve S. Prognostic impact of ALDH1 in breast cancer: a story of stem cells and tumor microenvironment. Breast Cancer Res Treat. 2009;123:97–108. doi: 10.1007/s10549-009-0619-3. [DOI] [PubMed] [Google Scholar]
- 118.Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, Houvenaeghel G, Extra JM, Bertucci F, Jacquemier J, Xerri L, Dontu G, Stassi G, Xiao Y, Barsky SH, Birnbaum D, Viens P, Wicha MS. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Neumeister V, Agarwal S, Bordeaux J, Camp RL, Rimm DL. In situ identification of putative cancer stem cells by multiplexing ALDH1, CD44, and cytokeratin identifies breast cancer patients with poor prognosis. Am J Pathol. 2010;176:2131–2138. doi: 10.2353/ajpath.2010.090712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, Helyer L, Pan L, Leidal A, Gujar S, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells. 2011;29:32–45. doi: 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- 121.Chang B, Liu G, Xue F, Rosen DG, Xiao L, Wang X, Liu J. ALDH1 expression correlates with favorable prognosis in ovarian cancers. Mod Pathol. 2009;22:817–823. doi: 10.1038/modpathol.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Deng S, Yang X, Lassus H, Liang S, Kaur S, Ye Q, Li C, Wang LP, Roby KF, Orsulic S, Connolly DC, Zhang Y, Montone K, Butzow R, Coukos G, Zhang L. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS One. 2010;5:e10277. doi: 10.1371/journal.pone.0010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li T, Su Y, Mei Y, Leng Q, Leng B, Liu Z, Stass SA, Jiang F. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients’ outcome. Lab Invest. 2010;90:234–244. doi: 10.1038/labinvest.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Marchitti SA, Orlicky DJ, Brocker C, Vasiliou V. Aldehyde dehydrogenase 3B1 (ALDH3B1): immunohistochemical tissue distribution and cellular-specific localization in normal and cancerous human tissues. J Histochem Cytochem. 2010;58:765–783. doi: 10.1369/jhc.2010.955773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Patel M, Lu L, Zander DS, Sreerama L, Coco D, Moreb JS. ALDH1A1 and ALDH3A1 expression in lung cancers: correlation with histologic type and potential precursors. Lung Cancer. 2008;59:340–349. doi: 10.1016/j.lungcan.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 126.Leid M, Kastner P, Chambon P. Multiplicity generates diversity in the retinoic acid signalling pathways. Trends Biochem Sci. 1992;17:427–433. doi: 10.1016/0968-0004(92)90014-z. [DOI] [PubMed] [Google Scholar]
- 127.Balmer JE, Blomhoff R. A robust characterization of retinoic acid response elements based on a comparison of sites in three species. J Steroid Biochem Mol Biol. 2005;96:347–354. doi: 10.1016/j.jsbmb.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 128.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 129.Khanna M, Chen CH, Kimble-Hill A, Parajuli B, Perez-Miller S, Baskaran S, Kim J, Dria K, Vasiliou V, Mochly-Rosen D, Hurley TD. Discovery of a novel class of covalent inhibitor for aldehyde dehydrogenases. J Biol Chem. 2011;286:43486–43494. doi: 10.1074/jbc.M111.293597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Koppaka V, Thompson DC, Chen Y, Ellermann M, Nicolaou KC, Juvonen RO, Petersen D, Deitrich RA, Hurley TD, Vasiliou V. Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol Rev. 2012 doi: 10.1124/pr.111.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]