Abstract
Over 100 FDA-approved medications include pharmacogenetic biomarkers in the drug label, many with cancer indications referencing germline DNA variations. With the advent of next-generation sequencing (NGS) and its rapidly increasing uptake into cancer research and clinical practice, an enormous amount of data to inform documented gene-drug associations will be collected, which must be exploited to optimize patient benefit. This state-of-the-art article focuses on the implementation of germline cancer pharmacogenetics into clinical practice. Specifically, it discusses the importance of germline variation in cancer and the role of NGS in pharmacogenetic discovery and implementation. In the context of a scenario where massive NGS-based genetic information will be increasingly available to health stakeholders, this review explores the ongoing debate over the threshold of evidence necessary for implementation, provides an overview of recommendations in cancer by professional organizations and regulatory bodies, discusses limitations of current guidelines and strategies to improve third-party coverage.
Keywords: cancer, germline, implementation, oncology, next-generation sequencing, pharmacogenetics, pharmacogenomics
Introduction
There is diffuse heterogeneity in response to cancer therapy, with only about 25% of patients responding to conventional methods of choosing chemotherapy regimens (1). Additionally, dose-limiting toxicities combined with poor target selectivity commonly result in delay or cessation of therapy owing to reduced drug efficacy in the potentially curative setting. Understanding and applying the knowledge of a patient's cancer genome to resolve these clinical problems has become increasingly utilized. In fact, prospective testing for somatic, or acquired, mutations within a tumor and appropriate selection of targeted therapies is beginning to replace standard of care administration of non-specific cytotoxic agents in many tumor types, owing to enhanced survival and reduced toxicities. For example, the v-Raf murine sarcoma viral oncogene homolog B1 (BRAF) inhibitor, vemurafenib, has replaced the standard of care, dacarbazine, for the treatment of metastatic melanoma patients harboring the BRAF V600E mutation (2). The same has been seen with crizotinib as a replacement for cytotoxic chemotherapy as standard first-line therapy in anaplastic lymphoma kinase (ALK) positive non-small-cell lung cancer (NSCLC) (3), trastuzumab in human epidermal growth factor receptor-2 (HER2) positive breast cancer (4), erlotinib and afatinib for epidermal growth factor receptor (EGFR) positive NSCLC (5, 6), and several others.
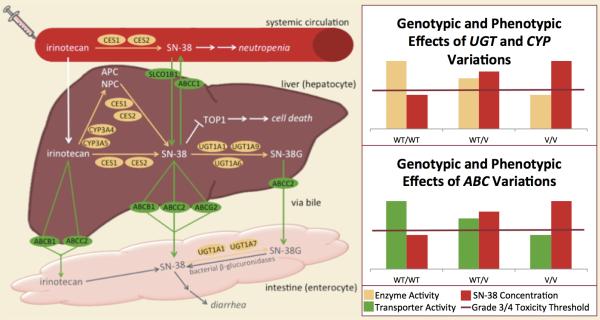
While somatic mutations in genes coding for targets of mechanisms of action of drugs are commonly used to predict pharmacodynamics and drug response, germline, or inherited, genome variation can be helpful in predicting pharmacokinetics and/or pharmacodynamics in individual patients (7) (Figure 1). Prospective identification of germline variants may aid in normalization of systemic drug exposure and minimization of drug toxicity while preserving the antitumor activity at the target site, thus enhancing clinical benefit. For example, patients carrying the uridine-diphosphate glucuronosyl transferase 1A1*28 (UGT1A1*28) allele have decreased enzymatic activity, resulting in reduced glucuronidation and impaired inactivation of 7-ethyl-10-hydroxycamptothecin (SN-38, the active component of irinotecan) (8). These patients are recommended to receive a reduced initial dose of irinotecan to minimize drug exposure and reduce severe, dose-limiting toxicities, such as grade 3/4 neutropenia (9). Likewise, patients receiving 6-mercaptopurine and harboring low activity thiopurine-S-methyltransferase (TPMT) phenotypes have increased production of the active thioguanine nucleotide metabolites, which subsequently increases the risk of myelosuppression and gastrointestinal toxicity (10-12). Six-mercaptopurine dose reduction in TPMT-deficient patients has been shown to reduce the risk of toxicity associated with high concentrations of thioguanine nucleotides without compromising efficacy (12, 13).
Figure 1. Example of the impact of germline pharmacgenetics on drug metabolism and toxicity.
Depicted is the pharmacokinetic and pharmacodynamic pathway of irinotecan. Graphs represent the effect of genotypic variation of key pharmacogenetic genes on the concentration of SN-38, the active metabolite that is also responsible for toxicities. Variations in UGT1A1, UGT1A6, UGT1A7, UGT1A9, CYP3A4 and CYP3A5 have been shown to (directly or indirectly) decrease enzymatic activity, resulting in decreased glucuronidation (inactivation) and, ultimately, an increase in SN-38 concentration. As illustrated in the graphs, UGT and CYP activity decreases in an additive manner in variant carriers (Please note that genetic associations between CYPs and irinotecan are not as strongly established as those with UGT). Along with this decrease in enzymatic activity, comes an increase in SN-38 concentration. The line on the graph represents the SN-38 concentration threshold for grade 3/4 (severe) toxicity. As depicted, variant carriers have SN-38 concentrations that more commonly cross the toxicity threshold. Likewise, variations in ABCB1, ABCC2, ABCG2, and SCLO1B1 may decrease transporter activity, also resulting prolonged exposure to SN-38 and increased side effects (Please note that genetic associations between ABC genes and irinotecan/SN-38 are not as strongly established as those with UGT)
Abbreviations – WT: wildtype; V: variant
In addition to predicting the pharmacokinetics of a drug, germline variants may also inform cancer biology (14). It is well established that immune cells can be tumor promoting through their ability to regulate angiogenesis, cell proliferation, and tumor invasiveness (14). Furthermore, neovascularization (angiogenesis) promotes tumor growth through the formation of vasculature required to provide nutrients and oxygen to cancer cells while removing wastes (e.g., carbon dioxide). In addition to the immune system and angiogenesis, other systems that do not have the typical somatic alterations of a tumor include inflammation and the stromal microenvironment. Germline variation in genes regulating these systems may affect tumor growth and survival. Initial genome-wide association studies (GWAS) of outcome of cancer patients treated with chemotherapy are corroborating the hypothesis of germline determinants of cancer outcome being related to these systems. For example, a germline GWAS of treatment response in childhood acute lymphoblastic leukemia (ALL) selected the interleukin 15 gene (IL15) a determinant of minimal residual disease, a predictor of outcome (15). Another GWAS of overall survival (OS) in pancreatic cancer patients treated with first-line gemcitabine revealed another gene in the immune system (the interleukin 17F, IL17F) as associated with OS (16). These studies are supportive of the notion that utilizing germline variation might predict not only drug behavior but also host and tumor biology, subsequently enhancing our understanding of the genetic basis of drug response in cancer.
An extensive number of reviews of the role of germline pharmacogenetics in cancer therapy are readily available in the literature (17-20). In addition, an appreciation of the clinical relevance of both germline and somatic pharmacogenetics can be gained from the number of validated, clinically significant biomarkers listed in Table 1. Despite the vast number of pharmacogenetic associations important in cancer treatment, very few pharmacogenetic tests are utilized routinely in clinical practice. This state of the art article will focus on the implementation of germline cancer pharmacogenetics into clinical practice. Specifically, we will elaborate on the importance of NGS in germline cancer pharmacogenetics implementation. We will explore the debate centered around the level of evidence required to warrant clinical implementation, provide an overview of the landscape of recommendations on pharmacogenetics implementation by professional organizations and regulatory bodies, and discuss the limitations of the current guidelines. Finally, because the uptake of pharmacogenetics into routine clinical practice is strongly influenced by third-party coverage, we will discuss limitations and strategies to improve the current reimbursement rates and, subsequently, translation of cancer pharmacogenetics into practice.
Table 1.
Examples of clinically significant cancer pharmacogenetic associations.
| Gene | Drug(s) | Genome | Association | FDA-Label?(28) |
|---|---|---|---|---|
| Abelson murine leukemia viral oncogene homolog 1 (ABL) | bosutinib, dasatinib, imatinib, nilotinib, ponatinib | Somatic | Drug activity | Yes |
| Anaplastic lymphoma receptor tyrosine kinase (ALK) | crizotinib | Somatic | Drug activity | Yes |
| Cytochrome P450 2B6 (CYP2B6) | cyclophosphamide | Germline | Nephrotoxicity risk | No (62) |
| Cytochrome P450 2D6 (CYP2D6) | tamoxifen | Germline | Disease recurrence | No (63) |
| Cytochrome P450 3A4/3A5 (CYP3A4/3A5) | cyclophosphamide | Germline | Drug activity | No (64) |
| Dihydropyrimidine dehydrogenase (DPYD) | capecitabine, fluorouracil | Germline | Stomatitis, diarrhea, and neutropenia risk | Yes |
| DNA mismatch repair genes (MLH1, MSH2, MSH6, PMS2) | fluorouracil | Germline | Drug activity | No (65) |
| Estrogen receptor 1 (ESR1) | fulvestrant, tamoxifen, toremifene | Somatic | Drug activity | Yes |
| Fc fragment of IgG receptor (FcgR) | cetuximab, rituximab, trastuzumab | Somatic | Disease progression, response | No (66) |
| Glucose-6-phosphate dehydrogenase (G6PD) | rasburicase | Germline | Hemolysis risk | Yes |
| Epidermal growth factor receptor 1 (EGFR) | afatinib, erlotinib, vandetanib | Somatic | Drug activity | Yes |
| Human epidermal growth factor receptor 2 (HER2, ERBB2) | trastuzumab, trastuzumab emtansine, lapatinib, pertuzumab | Somatic | Drug activity | Yes |
| Janus kinase 2 (JAK2) | ruxolitinib | Somatic | Drug activity | Yes |
| Kirsten rat sarcoma viral oncogene homolog (KRAS) | cetuximab, panitumumab | Somatic | Drug activity | Yes |
| Mitogen-activated protein kinase (MAP2K, MEK) | trametinib | Somatic | Drug activity | Yes |
| Promyelocytic leukemia/retinoic acid receptor, alpha fusion gene (PML/RARα) | arsenic trioxide, all trans retinoic acid (ATRA) | Somatic | Drug activity | Yes |
| Thiopurine methyltransferase (TPMT) | mercaptopurine, thioguanine, cisplatin | Germline | Myelosuppression risk, drug activity, ototoxicity risk | Yes |
| Thymidylate synthetase (TYMS) | capecitabine, fluorouracil | Germline | Drug activity | No (67, 68) |
| UDP-glucuronosyltransferase 1A1 (UGT1A1) | irinotecan, nilotinib | Germline | Neutropenia risk, hyperbilirubinemia risk, drug activity | Yes |
| V-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (KIT) | imatinib | Somatic | Drug activity | Yes |
| V-raf murine sarcoma viral oncogene homolog B (BRAF) | dabrafenib, emurafenib | Somatic | Drug activity | Yes |
Maximizing benefit from next-generation sequencing efforts
The advent of NGS has allowed sequencing of an entire human genome at a reasonable cost – the cost of sequencing one genome in 2001 was approximately $100 million, and in 2013 is less than $3000 (21). Additionally, the time required to sequence an entire human genome has also decreased dramatically; from over a decade (1990 – 2003) to complete the sequencing of the first human genome (the Human Genome Project), compared to as quickly as a one-day turnaround time offered with some 2013 technologies (e.g., Benchtop Ion Proton™ Sequencer, Life Technologies, Grand Island, NY) (22). This rapid decline in price and turnaround time has resulted in an increase in research and clinical applications of sequencing, most notably in cancer. Major academic institutions and both government-sponsored and private organizations have launched programs for NGS of the cancer genome, with the goals of describing the architecture of cancer-specific somatic alterations and of aiding clinicians in selection of targeted therapy (23).
Because tumor samples contain both acquired and inherited alterations, along with somatic DNA, cancer sequencing efforts also capture germline information. More importantly, in cancer patients, germline DNA is oftentimes also analyzed as a means to identify variants in the tumor. As discussed, this germline information plays a crucial role in optimizing the dose and selection of therapy. An additional benefit unique to NGS is the ability to discover rare variants in the genome and their impact on drug response. In a study exploring the impact of rare variants versus common variants in SLCO1B1 on methotrexate clearance, Ramsey and colleagues found that rare damaging nonsynonymous SNPs accounted for 17.8% of the gene's effects on methotrexate clearance (24). Additionally, the rare variants had larger effect sizes than the common nonsynonymous variants, with effect size being inversely proportional to minor allele frequency. Whereas this group had to perform deep resequencing of SLCO1B1 to discover these rare variants, the advent of NGS provides the opportunity to obtain comprehensive (genome-wide) catalogues of rare variants. Additionally, the larger effect sizes observed with rare variants likely contribute to the overall phenotypic variability of drug response in cancer patients treated with chemotherapy.
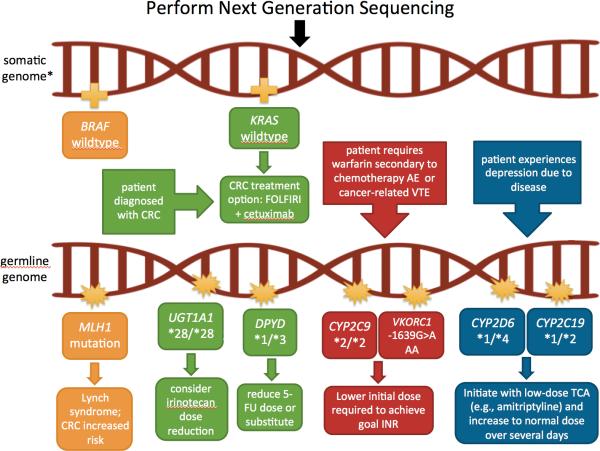
In addition to pharmacogenetic research and discovery, germline information generated through NGS has clinical applications, as it informs on drug selection and dose optimization, as well as genetic susceptibility to disease, with cascade testing for the relatives of the patient (Figure 2). In their 2010 recommendations for genetic testing for cancer susceptibility, the American Society of Clinical Oncology (ASCO) reported nine genes with well-validated germline variants predictive of cancer susceptibility (25). For example, germline variants in the adenomatous polyposis coli (APC) gene result in a hereditary condition known as familial adenomatous polyposis. Without any intervention, 100% of these patients will ultimately progress to colorectal cancer (26). Similar risks are conferred with the early onset breast cancer gene (BRCA) variants and breast and ovarian cancers (27), as well as the DNA mismatch repair gene variants, which result in Lynch syndrome and, subsequently, colorectal cancer (26).
Figure 2. Example of how next-generation sequencing can be utilized to inform therapeutic decision-making.
In this example, next-generation sequencing (NGS) is performed using both the tumor (somatic) and inherited (germline) DNA samples. NGS reveals a germline MLH1 variant, which suggests possible Lynch syndrome (see Table 1). The BRAF wildtype tumor provides further evidence for this diagnosis, as BRAF mutations are extremely uncommon with Lynch syndrome. Due to Lynch syndrome, the example patient develops colorectal cancer (CRC). NGS of a colorectal tumor reveals the patient is KRAS wildtype, thus, is predicted to respond to an epidermal growth factor receptor (EGFR) inhibitor, such as cetuximab. The clinician decides to start the patient on the first-line treatment option of FOLFIRI [folinic acid (leucovorin), fluorouracil (5-FU), and irinotecan] + cetuximab. Germline NGS information can be used to further optimize dose selection and management. Specifically, the patient is determined to carry two reduced-activity UGT1A1 alleles (*28/*28), putting him at an increased risk for severe irinotecan toxicity (neutropenia); a dose reduction is recommended. Additionally, the patient carries one inactive DPYD allele, which may warrant a 5-FU dose decrease. If the patient has encounters indications for other medications with validated pharmacogenetic associations (e.g., warfarin or amitriptyline in this case), that information will also be provided through NGS and should be readily available to inform drug decisions.
*Note: Germline variants may or may not also be present on the somatic genome.
Abbreviations – AE: adverse event; CRC: colorectal cancer; INR: international normalized ratio; TCAs: tricyclic antidepressants; TDM: therapeutic drug monitoring; VTE: venous thromboembolism
As of August 2013, there are over 100 drugs with pharmacogenetic information in the U.S. Food and Drug Administration (FDA)-approved drug labels, with 31 being cancer drugs, and 8 of the cancer drugs referencing germline variants (28). NGS is able to provide data to inform most or all of these validated gene-drug associations (i.e., some sequence information may be missed in the case of whole exome sequencing) as well as many others that are under investigation but have yet to confer a label change. Therefore, NGS of the cancer genome is likely the most effective strategy for obtaining preemptive germline assessment of actionable genotypes in cancer patients. It is important to exploit this germline information, determine which variants are validated, warranting clinical implementation, and optimize patient therapy accordingly.
With the many centers and companies now performing NGS and recommending therapy changes based on the results, collaboration to accumulate data would also help maximize the benefit of these efforts. Rather than waiting on data from prospective studies, the large sample sizes would provide the means to retrospectively analyze large patient cohorts for discovery of common and rare variants, validation, and outcomes of pharmacogenetic–based decision-making. One collaborative organization working toward maximizing benefit from NGS projects in all therapeutic areas is the Electronic Medical Records and Genomics (eMERGE) Network, which is a national consortium focused on combining DNA biorepositories with electronic medical records to facilitate large-scale, high-throughput genetic research and returning genetic testing results to patients in a clinical setting (29). Efforts such as this should be exploited from all angles, including somatic and germline variation discovery and implementation, as well as clinical and uptake outcomes. On an even broader scale, collaboration of international pharmacogenetics consortiums would provide the basis for understanding population-based genetics and the impact of race on outcomes worldwide (30). This information may be beneficial in advancing clinical uptake of pharmacogenetics throughout a wide spectrum of health care systems, including those in third-world countries.
Level of evidence to warrant implementation
Generation of clinical recommendations and guidelines is burdened by the debate surrounding the threshold of evidence required for translation of pharmacogenetics into clinical practice. The U.S. Office of Public Health Genomics’ Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group has identified the most significant challenges in developing evidence-based reviews and recommendations for genetics testing, which include: (1) uncertainty and difficulty in establishing clinical validity (i.e., how consistently and accurately the test predicts the outcome of interest), (2) lack of direct evidence of clinical utility (i.e., how likely the test is to significantly improve patient outcomes), (3) rapid development and marketing of tests, and (4) lack of robust regulatory infrastructure for genetic testing (31). These limitations contribute to the vast disparities between recommendations, and therefore must be addressed.
While prospective randomized clinical trials (RCTs) are the gold standard and often required for an intervention to be accepted into standard of care, this may not be the ideal study design to demonstrate clinical utility of pharmacogenetics. The feasibility of performing prospective, phase III, RCTs for each pharmacogenetic association discovered is unlikely due to the inherent costs, time, and large sample sizes associated with these trials, which may ultimately deprive many patients of safer and more effective treatments and dosing. To increase efficiency, the focus may be shifted toward retrospective validation and replication (32), randomized phase II studies (33), or adaptive trial designs that allow prospectively planned modifications in design after patient enrollment(34). Unfortunately, alternative approaches such as these may not completely eliminate the need for solid evidence from traditional RCTs, which we have been accustomed to for over 60 years; however, it must also be considered that many laboratory biomarkers in clinical use today have not been tested for their predictive power in randomized studies. The level of evidence considered appropriate to warrant recommendations according to the National Institute of Health's (NIH) Pharmacogenomics Research Network (PGRN) and the Clinical Pharmacogenetics Implementation Consortium (CPIC) includes a strong biological rationale for the gene-drug association, reproducible evidence linking the genetic variation to drug response, and noninferiority compared with current prescribing practice (35). The validity of this evidence threshold is supported by the pharmacogenetic association between TPMT and thiopurine toxicity, which never underwent an RCT, yet is likely the most validated and commonly utilized germline pharmacogenetic test in practice.
Another opportunity to generate sufficient evidence to warrant clinical implementation lies in the drug development process. For example, if preclinical models demonstrate that a drug is metabolized via a CYP450 enzyme or is a transporter substrate with known or suspected pharmacogenetic implications (e.g., CYP2D6 or ABCB1) then phase I, II, and III clinical trials should incorporate correlative studies to examine if the drug disposition is altered based on CYP450/transporter genotype or altered expression. This approach would allow for information to be available from the outset of the drug development process and would improve the efficiency of the current model, which has required a multitude of retrospective and prospective studies on drugs that have existed for decades, as correlative pharmacogenetic studies were traditionally never performed during the initial drug development process. The capabilities now exist to easily obtain DNA upfront and perform these studies much earlier. Additionally, it could enhance patient selection, contribute to dose optimization and therapy selection, and reduce overall healthcare costs from the beginning. For example, a UGT1A1 genotype-guided phase I study demonstrated that metastatic colorectal cancer patients lacking the ‘high-toxicity’ genotype (UGT1A1*28/*28) were able to tolerate significantly higher doses compared to the standard 180 mg/m2 administered in FOLFIRI (5-fluorouracil, leucovorin, and irinotecan) (*1/*1 and *1/*28 patients tolerated up to 370 mg/m2 and 310 mg/m2, respectively) (36). If this genetic association had been discovered from the outset, clinical studies could have been focused on tailoring the dose by genotype and testing the hypothesis of improved survival benefit with a genotype-driven dosing. Furthermore, using genotype to optimize the therapeutic dose may provide precedence for medications already on the market that have similar pharmacology and metabolism properties.
The debate of the level of evidence required for clinical utility is further complicated by the advent of NGS. Because genetic information with known and validated clinical benefits will be collected and at no additional cost and available with minimal increased effort, it can be argued that the threshold required to warrant single-gene tests greatly differs from the threshold to consider when NGS information is readily available. As mentioned previously, NGS will generate germline information along with somatic. Arguably, it is unethical to ignore this information given that phenotypes exist that predict life-threatening toxicities and/or drug efficacy. Indeed, this is the premise of the CPIC guidelines: given that genotyping information is already available, how should it be utilized by the clinician? (35). Considering the lower level of evidence that would support clinical utility in this setting, the true lack of clinical decision support (CDS) and professional organization practice guidelines is realized.
The challenge of data interpretation in facilitating translation of pharmacogenetics into clinical practice
The dissemination of pharmacogenetics into clinical practice is largely influenced by the availability of approved and validated pharmacogenetic tests, clinicians’ ability to use and understand the tests, and evidence-based recommendations to change therapeutic management over the current standards. Currently, a paucity of data exists on standardized pharmacogenetic guidelines by professional organizations, contributing to the slow clinical uptake of pharmacogenetics. A large survey of 10,303 physicians demonstrated that the vast majority (97.6%) acknowledged that genetic variations may influence drug response, but only 10.3% felt adequately informed about pharmacogenetic testing and interpretation (37). Interestingly, investigators also determined that early adopters of pharmacogenetic testing are more likely to be practicing in oncology. Thus, despite the large number of FDA-approved pharmacogenetic tests and drug label indications, readily available consensus guidelines and the lack of physician confidence present barriers to widespread pharmacogenetics implementation into clinical practice.
There are two aspects of data interpretation that affect the translation of pharmacogenetics into clinical practice: (1) interpretation of published research results and (2) clinician interpretation of reported genetic results. Firstly, the lack of standardization in conducting pharmacogenetic studies contributes to inconsistencies in results, which makes interpretation challenging or even impossible, and contributes to lack of replication in many instances. Inconsistences between studies include inaccurate or incomplete genotyping (i.e., failure to include all known functional variants or genotyping of tumor tissue rather than germline DNA), presence of concomitant medications that may affect drug disposition and/or response, thus altering observed ‘phenotype’, and the common lack of control groups, which complicates the differentiation between predictive (associated with response to treatment) and prognostic (associated with disease outcome in the absence of treatment, i.e., disease severity) genotypes (38). The discordance between positive associations reported due to these inconsistences contributes to the complexity of data interpretation by researchers, professional organizations, consortia, and clinicians alike.
Once a pharmacogenetic association has proven validity and clinical utility and is ready for clinical implementation, physicians must be willing and able to incorporate it into their practice. Despite their interest, unfortunately, many physicians lack the confidence and knowledge required to accurately interpret and implement pharmacogenetics. CDS tools, which provide physician guidance (most commonly through electronic medical records) on clinical decisions when pertinent pharmacogenetic information is available, have proven to successfully enable translation of pharmacogenetics into clinical practice (39). However, in order to develop accurate recommendations to incorporate into CDS, clear and precise algorithms based on scientifically robust results must be available to the program developers; the creation of such algorithms has been facilitated by professional organizations and consortia discussed below.
An overview of pharmacogenetic recommendations by consortia, professional organizations, and regulatory bodies
The creation of professional organizations and consortia devoted to pharmacogenetic-based clinical guidelines and recommendations has provided guidance for uptake into clinical settings, mainly at large research-intensive academic hospitals, with most applications being research focused. Examples of such organizations include CPIC and the Dutch Pharmacogenetics Working Group (DPWG), both devised to create and share evidence-based clinical pharmacogenetics guidelines with therapeutic recommendations for specific gene-drug pairs (35, 40). Table 2 summarizes the germline cancer gene-drug pairs covered by each set of currently available guidelines, as well as a summary of the specific recommendations provided. Of note, there also exists a Japanese regulatory agency and recommendations provided by the Japanese Pharmacogenomics Discussion Group (PDG), which brings together members of the Pharmaceutical and Medical Device Agency (PMDA) to exchange and share data with the goal of maintaining consistency in consultations and promoting appropriate pharmacogenetic clinical trials, but they are not readily available in English (41). While these organizations have provided helpful guidelines for the most well validated genetic associations, shortcomings remain.
Table 2.
Summary of worldwide recommendations by professional organizations and regulatory bodies for germline cancer pharmacogenetics.
| Regulatory Body | Type of Initiative | Date of Initiative | Oncology Germline Gene-Drug Pairs Addressed | Recommendation Overview |
|---|---|---|---|---|
| ASCO | Professional Organization Recommendations | GI cancer: 2006 (47) | DPYD-fluorouracil and capecitabine | Insufficient evidence to recommend testing to monitor or predict response to therapy |
| CPIC | Pharmacogenetic Guideline Consortium | Published 2011(69) Updated 2013(48) |
TPMT-thiopurines | Advocate for TPMT testing before initiation Provide dosing recommendation tables by phenotype for each of the thiopurines In general, intermediate metabolizers should be started at 30-70% of target dose; poor metabolizers warrant alternate agent consideration or drastically (10-fold) reduced doses |
| Submitted | DPYD-fluorouracil and capecitabine | Recommendation pending | ||
| Underway | G6PD-rasburicase | Recommendation pending | ||
| DPWG | Pharmacogenetic Guideline Consortium | Published 2008(40) Updated 2011(42) |
CYP2D6-tamoxifen | Intermediate metabolizers: consider aromatase inhibitor for postmenopausal women, avoid concomitant CPY2D6 inhibitors Poor metabolizers: consider aromatase inhibitor for postmenopausal women |
| DPYD- fluorouracil, capecitabine and tegafur | Intermediate metabolizers: reduce dose by 50% or select alternative agent Poor metabolizers: select alternative drug |
|||
| TPMT-thiopurines | Intermediate metabolizers: select alternative drug or reduce dose by 50%, except thioguanine – select alternative agent Poor metabolizers: select alternative drug or reduce dose by 90%, except thioguanine – select alternative agent |
|||
| UGT1A1-irinotecan | Reduce starting dose by 30% in *28 homozygotes receiving >250 mg/m2 | |||
| EGAPP(43) | Genomic Applications Working Group | 2009(49) | UGT1A1-irinotecan | Evidence insufficient to recommend for or against routine use of UGT1A1 genotyping in patients receiving irinotecan May choose lower dose or alternative drug if found to be *28/*28 |
| EMA(46) | Regulatory – Drug Label Indications | DPYD-capecitabine, tegafur | Contraindicated if known DPD deficiency due to increased toxicity | |
| G6PD-rasburicase | Contraindicated in patients with G6PD deficiencies due to risk of hemolytic anemia or methemoglobinemia | |||
| TPMT-mercaptopurine | Patients with little or no inherited TPMT activity are at increased risk for severe toxicity from conventional doses, and generally require severe dose reductions. TPMT genotyping or phenotyping can be used to identify patients with absent or reduced TPMT activity. |
|||
| UGT1A1-erlotinib | Use with caution in patients with low expression of UGT1A1 due to inhibitory effects on glucoronidation | |||
| FDA(70) | Regulatory – Drug Label Indications | 2003 | DPYD-capecitabine | Contraindicated with DPD deficiency due to potentially fatal toxicity |
| 2004 | DPYD-fluorouracil | Should not be used in patients with DPD deficiency due to potential toxicities | ||
| 2002 | G6PD-rasburicase | Contraindicated in patients with G6PD deficiency due to risk of hemolysis. Screen all higher risk patients for G6PD deficiency (e.g., African or Mediterranean ancestry) prior to initiation. | ||
| 2011 | TPMT-cisplatin | Variants in TPMT associated with increased risk of ototoxicity. All pediatric patients should undergo audiometric testing at baseline, prior to each dose, and for several years post therapy. | ||
| TPMT-mercaptopurine | Recommends, but does not require genetic testing. Homozygous-deficient patients accumulate excessive drug concentrations, which increase the risk for toxicity. They generally require substantial dose reductions. Heterozygous patients accumulate higher concentrations than people with normal TPMT activity and are also more likely to experience toxicity. Most tolerate normal doses. |
|||
| 2001 | TPMT-thioguanine | Individuals with an inherited deficiency of TPMT are at an increased risk for myelosuppression. There are laboratories that offer testing for TPMT deficiency. | ||
| 2005 | UGT1A1-irinotecan | UGT1A1*28 homozygotes are at an increased risk for neutropenia. Consider dose reduction by at least one level; precise dosing unknown. | ||
| 2007 | UGT1A1-nilotinib | UGT1A1*28 patients may be at increased risk for hyperbilirubinemia. | ||
| NCCN | Clinical Practice Guidelines | Breast: 2013(71) | CYP2D6-tamoxifen | Provides brief summary of conflicting literature. Does not recommend CYP2D6 testing to determine the optimal adjuvant endocrine strategy. |
| NHL: 12/2012(72) | G6PD-rasburicase | Patients with G6PD deficiency may have increased adverse reactions (methemoglobinemia and severe hemolysis) | ||
| ALL: 2012(73) | TPMT-thiopurines | Consider testing for TPMT polymorphisms in patients receiving 6-mercaptopurine maintenance therapy due to increased risk of hematopoietic toxicity in variant carriers | ||
| CRC: 2012(74), 2013(44) | UGT1A1-irinotecan | Genetic polymorphisms in UGT1A1 may lead to drug accumulation and possible severe toxicity. Mentions commercial tests available and FDA-label warning for UGT1A1*28 carriers. State guidelines for use in clinical practice not available. |
||
| PharmGKB(75) | Web-based Database | 2010 – present | Summary of CPIC and DPWG recommendations FDA and EMA label excerpts |
Abbreviations – ASCO: American Society of Clinical Oncology; CPIC: Clinical Pharmacogenetics Implementation Consortium; DPWG: Dutch Pharmacogenetics Working Group; EGAPP: Evaluation of Genomic Applications in Practice and Prevention; EMA: European Medicines Agency; FDA: U.S. Food and Drug Administration; NCCN: National Comprehensive Cancer Network; PharmGKB: Pharmacogenomics Knowledgebase; CYP2D6: cytochrome P450, family 2, subfamily D, polypeptide 6; DPYD: dihydropyrimidine dehydrogenase; G6PD: glucose-6-phosphate dehydrogenase; TPMT: thiopurine S-methyltransferase; UGT1A1: UDP glucuronosyltransferase 1 family, polypeptide A1
Notably, the guidelines for somatic mutations tend to be consistent across the different regulating bodies. For example, guidelines are in agreement that crizotinib is first-line for patients with the ALK positive NSCLC. The same can be said for vemurafenib in melanoma patients harboring the BRAF V600E mutation, trastuzumab in HER2-positive breast cancer patients, as well as the many other drugs with somatic pharmacogenetic implications. This consistency in guidelines can be explained by the fact that all are targeted agents, which only received approval in tumor types expressing the biomarker of interest. In contrast, the retrospective discovery of germline pharmacogenetic markers has contributed to complexity in the clinical recommendations.
While consortia, professional organizations and regulatory bodies (i.e., the FDA) are universally concordant in their recommendations for drugs associated with somatic mutations, extensive discordance exists between the recommendations based on germline pharmacogenetics (Table 2). Firstly, the gene-drug pairs covered within each set of guidelines varies. For example, while DPWG, EGAPP, FDA, and National Comprehensive Cancer Network (NCCN) guidelines exist for UGT1A1-irinotecan gene-drug pair (28, 42-45), the ASCO, CPIC, and the European Medicines Agency (EMA) do not currently provide guidelines for this specific gene-drug pair. Secondly, the extent and specifics of the therapeutic recommendations provided in the guidelines are also discordant. While the DPWG provides broad dose adjustment guidelines for capecitabine based on DPD deficient phenotype (42), EMA and FDA simply contraindicate the drug in the instance of DPD deficiency (28, 46), and ASCO states that there is insufficient evidence to recommend testing or monitoring (47). In the case of NGS, when DPYD genotype will be available, the question of how to adjust therapy accordingly arises. Likewise, although the FDA and the majority of the professional organizations and consortia address TPMT testing with thiopurine administration, the specifics of the recommendations vary. Of note, while the CPIC and DPWG recommend dose decreases (30-70% and 50%, respectively) to avoid the risk of severe myelosuppression in patients of the intermediate metabolizer phenotype (42, 48), the FDA label states that these patients usually tolerate normal doses (45). Although both sets of recommendations may be correct depending on the regimen and its recommended starting dose (i.e., when the protocol starting dosage of mercaptopurine (MP) is 75 mg/m2 per day, then dose reductions in heterozygotes is likely necessary, but when the protocol starting dose of MP is 50 mg/m2, patients are much more likely to tolerate normal doses), these discrepancies likely prove confusing to unaware clinicians who are trying to optimally dose TPMT intermediate metabolizers. Thirdly, aside from the discordance among recommendations, an additional shortcoming is the language included in some of the recommendations. For example, the FDA and EMA state that capecitabine is contraindicated in DPD deficient patients, but does not implicitly state the diagnostic criteria for DPD deficiency (i.e., enzyme expression below a certain level or harboring one or two null alleles?) (28, 46). The vague nature of this recommendation imposes challenges for clinicians attempting to apply pharmacogenetics into clinical practice. Similarly, although EGAPP, EMA, FDA, and NCCN do not specifically recommend UGT1A1 testing in all patients receiving irinotecan, they do provide general dosing recommendations for UGT1A1 *28/*28 patients (44-46, 49). In the advent of NGS, this information will be available and should be acted upon accordingly. The FDA’s recommendation of reducing the dose of irinotecan by “one level” in UGT1A1 *28/*28 patients receiving irinotecan is subject to interpretation and, without more detailed recommendations, may result in under- or over-dosing of these patients (28). Importantly, the only cancer drugs with specific germline genotype- and phenotype-guided dosing guidelines are thiopurines (provided in the CPIC and DPWG guidelines), and irinotecan (provided in the DPWG guidelines).
The disparity noted between these guidelines can partially be explained by variations in their review processes. Interestingly, EGAPP went through several iterations of their review process methods before even identifying which topics they would focus on for in-depth reviews and recommendations (31). Another explanation for discordance lies in the evaluation criteria. While clinical validity of biomarkers is easily evaluable and available, analytic validity (i.e., how accurately and reliably the test measures the genotype of interest) and clinical utility are rarely directly available (31). For example, while HER2 status has been clinically validated (consistently associated with response to trastuzumab), the analytic validity is variable depending on which assay is used (i.e., fluorescence in situ hybridization, FISH, or immunohistochemistry, IHC), and clinical utility relies on an assessment of harms versus benefits, which varies by genotype (50). The lack of objective, standardized measures of these variables, especially clinical utility, results in the need for some subjectivity when defining clinical significance, and therefore providing recommendations. A universal evidence-based approach to evaluating pharmacogenetic literature and developing recommendations and guidelines would facilitate simplified translation of pharmacogenetics into clinical practice (50). Importantly, these methods should be established in the context of NGS to align with the paradigm shift in practice (31).
While the guidelines produced by these professional organizations and consortia are a step in the right direction, with the universal goal of facilitating implementation of pharmacogenetics into clinical practice, the discrepancies between the guidelines complicate translation for clinicians. Additionally, the language used in the recommendations may be difficult for physicians without an adequate genetics background to understand. For example, while detailed dosing recommendations provide a thorough summary of the literature, the complexity introduced with specific phenotyping criteria has the potential to overwhelm and subsequently deter clinicians without extensive training from adding pharmacogenetic tests to their regular practice. Concordant, succinct guidelines, with detailed but clear recommendations would greatly simplify the transition to pharmacogenetic-guided clinical practice. Furthermore, recommendations on diagnostic assays or methods of detection would provide clinicians with knowledge on how to obtain standardized genetic results for interpretation.
Insurance coverage: More than simply cost effectiveness analyses
In the United States, clinical adoption of pharmacogenetics is heavily influenced by the presence of regulatory recommendations and third-party payment (51). Overall, reimbursement for pharmacogenetic testing has been inconsistent, and the uncertainty regarding payment represents a major barrier to utilization in clinical practice (52). Another key issue in reimbursement is determining when pharmacogenetic testing is no longer investigative, but has become clinically validated, for a specific indication (52). As previously discussed, regulatory agencies require a high threshold of evidence for clinical recommendations, as do third-party payers who impose the additional requirement of a reasonable cost, which may explain the high variable payer response in the marketplace (51). Particularly, an especially high threshold exists for the more expensive genetic-driven prescriptions for patients whom conventional therapies are predicted to be ineffective or too toxic. Furthermore, because the current procedural terminology (CPT) codes for germline pharmacogenetics are limited to single-gene tests and do not include multi-gene, exome or genome sequencing panels, the tests may be seemingly less important to third-party payers and, therefore, even more difficult to get covered (53).
Few studies have evaluated the cost effectiveness of cancer pharmacogenetics in practice. Of note, a cost implications analysis of reactive versus prospective DPYD genotyping in 134 colorectal cancer patients receiving fluorouracil-based therapy revealed the potential for a total cost saving of €131,165 (~$173,000) through avoiding 5 hospitalizations by preemptively genotyping the patients (54). Similarly, a cost effectiveness analysis of screening for KRAS and BRAF mutations in colorectal cancer patients to direct treatment with cetuximab compared to the base strategy (no anti-EGFR therapy) reported an incremental cost effectiveness ratio of approximately $650,000 per additional year of life; the addition of KRAS testing saves approximately $7500 per patient (55). A critical and systematic review of the cost effectiveness of pharmacogenetics revealed that one of the most common biomarkers evaluated was TPMT; these studies were focused on a number of indications for thiopurines, including cancer, inflammatory bowel disease, Crohn's disease, and rheumatoid arthritis (56). Nonetheless, the review indicated that TPMT genotyping demonstrated clinical validity and likely demonstrated clinical utility. All six of the studies included in the analyses reported genotype-guided dosing of thiopurines to be cost effective when compared to standard dosing. Likewise, UGT1A1*28 genotyping for irinotecan therapy was also determined to be clinically valid; however, the clinical utility of the test was classified as unclear (note: only two studies utilizing UGT1A1 genotyping were included) (56). Both of the cost effectiveness studies assessing preemptive UGT1A1 genotyping demonstrated potential cost effectiveness for the test. Factors that influenced cost effectiveness analyses included race and whether or not efficacy decreases with reduced doses in heterozygotes. Specifically, UGT1A1*28 genotyping was shown to be cost saving for Africans and Caucasians but not Asians, likely due to the low genotype frequency (observed MAF in Asians: 0.02) (56); the therapeutic efficacy, defined as survival benefit, of irinotecan in UGT1A1 *28/*28 patients after dose reduction had to be ≥98.4% of full-dose efficacy for genotype-guided dosing to remain cost-saving (56). This example also illustrates the compounded difficulty of proving cost effectiveness with low frequency pharmacogenetic variants due to the high number needed to screen. However, when the information is readily available due to NGS, the burden of proof is significantly lowered.
Perhaps more relevant in the time of NGS is a recent cost effectiveness analysis of a 21-gene assay for guiding adjuvant chemotherapy decisions in breast cancer (Oncotype DX, Genomic Health, Redwood City, CA), which demonstrated cost effectiveness for intermediate- and high-risk patients (57). Because this test interrogates more than one gene at a lower pricing threshold, these results may provide better predictions of the cost-effectiveness of NGS or other multiple marker tests. That is, compared to a single marker test which costs around $400-$500, whole genome sequencing can be completed for around $3000 and will essentially include most, if not all, germline genetic results ever needed for medication therapy management. In fact, approximately 90% of all bases within the human exome, regardless of allele frequency, can be captured using current sequencing technologies (58). The large number of clinically relevant pharmacogenetic genes interrogated through relatively low-cost NGS further decreases the cost-effectiveness burden, increasing the willingness to pay for a comprehensive genetic test (59). Of note, when NGS is not yet performed under Clinical Laboratory Improvement Amendments (CLIA) conditions (60), additional costs currently associated with NGS include the requirement for sequencing of biomarkers (or other molecular assays) used for clinical decision-making to be performed under CLIA. As NGS increasingly becomes more commonplace, limitations associated with availability of CLIA-certified laboratories will rapidly subside, and FDA-approved next generation sequencers are likely to be available in the near future.
As with the level of evidence for clinical implementation debate, a similar challenge lies in clearly demonstrating cost effectiveness of implementation through comparative effectiveness trials (61). From a reimbursement perspective, in order to truly evaluate cost savings, head-to-head studies with and without pharmacogenetic-guided therapy must be conducted (51). Aside from the low percentage of funding available for this type of research, this prospective design may not be necessary. For example, retrospective review of data collected on drugs, pharmacogenetic test usage, and inferred costs from health insurance and payers’ databases may provide an adequate source of cost information for evaluation. It must also be taken into consideration that, while the initial drug cost may be higher, especially in the cancer setting, the money saved by prescribing the optimal therapy from the beginning (i.e., decreased doctors’ visits and toxicities, improved outcomes) must not be disregarded.
Pharmacogenetic tests are most likely to be cost effective for medications with serious risks (i.e., high genotype relative risk or high rates) of toxicity or inefficacy and that are more expensive, such as chemotherapy agents. Once the price of NGS drops to well less than $1000 in the near future, the cost-effectiveness debate will likely shift from a focus on clinical outcomes to a focus on the cost associated with setting up the infrastructure required to analyze, store the resulting data, and reporting the results of the tests.
As the cost of testing decreases and effectiveness becomes well documented, reimbursement will be more widely adopted. While it may not be cost effective now to genotype single variants, as technologies continue to improve and the price for pharmacogenetic tests continue to drop, the cost effectiveness burden will also continue to decrease rapidly. Furthermore, as NGS becomes increasingly more common, the potential for derived clinical benefit from genotyping also multiplies.
CONCLUSION
While most clinical and industry efforts are focused on exploring somatic alterations as a means to target driver mutations, this is not the only key to successfully optimize therapy. Germline variation can be used to predict and reduce drug toxicity, enhance clinical efficacy, and inform the potential biology of the tumor. Similarly, researchers and clinicians must take advantage of the information gained from the numerous sequencing efforts underway (30). Not only do these projects promote research and discovery, but the information generated should not be ignored clinically. With NGS technologies, validated pharmacogenetic gene-drug pairs should be interrogated and acted upon as indicated. Concordant detailed guidelines would greatly enhance translation into clinical practice.
In order for a genetic test to be adopted into clinical practice, it must provide reliable, actionable, and predictive information that the clinician would not have otherwise known. Before clinical implementation of a pharmacogenetic gene-drug pair, robust clinical evidence is necessary; however, reliance on prospective RCTs as the only way to justify implementation is unrealistic, and the delay associated with construction, conduction, and interpretation of results could potentially deprive patients of life-saving or life-extending therapies. Rather, the DNA samples provided from patients entered into cancer clinical trials and the drug development process should be exploited to retrospectively discover and validate pharmacogenetic associations. Previously the discussion had been focused on the future of pharmacogenetics, when all patients will have pre-emptive genotyping performed in anticipation of their future medical needs. However, we need to take advantage of what is happening now – NGS is increasingly common, particularly in the realm of cancer. Utilization of these sequencing efforts to facilitate pharmacogenetics implementation will provide the basis for demonstration of uptake and feasibility of mass pre-emptive implementation.
Footnotes
CONFLICTS OF INTEREST
Dr. Innocenti receives royalties from the commercialization of the UGT1A1*28 genetic test.
REFERENCES
- 1.Spear BB, Heath-Chiozzi M, Huff J. Clinical application of pharmacogenetics. Trends Mol Med. 2001;7:201–4. doi: 10.1016/s1471-4914(01)01986-4. [DOI] [PubMed] [Google Scholar]
- 2.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaw AT, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–94. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 4.Slamon DJ, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–92. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 5.Rosell R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–46. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 6.Yang JC, et al. Symptom Control and Quality of Life in LUX-Lung 3: A Phase III Study of Afatinib or Cisplatin/Pemetrexed in Patients With Advanced Lung Adenocarcinoma With EGFR Mutations. J Clin Oncol. 2013 doi: 10.1200/JCO.2012.46.1764. [DOI] [PubMed] [Google Scholar]
- 7.Evans WE. Pharmacogenomics: Translating Functional Genomics into Rational Therapeutics. Science. 1999;286:487–91. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 8.Iyer L, et al. Phenotype-genotype correlation of in vitro SN-38 (active metabolite of irinotecan) and bilirubin glucuronidation in human liver tissue with UGT1A1 promoter polymorphism. Clin Pharmacol Ther. 1999;65:576–82. doi: 10.1016/S0009-9236(99)70078-0. [DOI] [PubMed] [Google Scholar]
- 9.Innocenti F. Genetic Variants in the UDP-glucuronosyltransferase 1A1 Gene Predict the Risk of Severe Neutropenia of Irinotecan. Journal of Clinical Oncology. 2004;22:1382–8. doi: 10.1200/JCO.2004.07.173. [DOI] [PubMed] [Google Scholar]
- 10.Lennard L, Van Loon JA, Weinshilboum RM. Pharmacogenetics of acute azathioprine toxicity: relationship to thiopurine methyltransferase genetic polymorphism. Clin Pharmacol Ther. 1989;46:149–54. doi: 10.1038/clpt.1989.119. [DOI] [PubMed] [Google Scholar]
- 11.Black AJ, et al. Thiopurine methyltransferase genotype predicts therapy-limiting severe toxicity from azathioprine. Ann Intern Med. 1998;129:716–8. doi: 10.7326/0003-4819-129-9-199811010-00007. [DOI] [PubMed] [Google Scholar]
- 12.Relling MV, et al. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst. 1999;91:2001–8. doi: 10.1093/jnci/91.23.2001. [DOI] [PubMed] [Google Scholar]
- 13.Relling MV, Pui CH, Cheng C, Evans WE. Thiopurine methyltransferase in acute lymphoblastic leukemia. Blood. 2006;107:843–4. doi: 10.1182/blood-2005-08-3379. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan D, Weinberg Robert A. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Yang JJ, et al. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA. 2009;301:393–403. doi: 10.1001/jama.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Innocenti F, et al. A Genome-Wide Association Study of Overall Survival in Pancreatic Cancer Patients Treated with Gemcitabine in CALGB 80303. Clinical Cancer Research. 2011;18:577–84. doi: 10.1158/1078-0432.CCR-11-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Relling MV, Dervieux T. Pharmacogenetics and cancer therapy. Nat Rev Cancer. 2001;1:99–108. doi: 10.1038/35101056. [DOI] [PubMed] [Google Scholar]
- 18.Coate L, Cuffe S, Horgan A, Hung RJ, Christiani D, Liu G. Germline genetic variation, cancer outcome, and pharmacogenetics. J Clin Oncol. 2010;28:4029–37. doi: 10.1200/JCO.2009.27.2336. [DOI] [PubMed] [Google Scholar]
- 19.Pinto N, Cohn SL, Dolan ME. Using germline genomics to individualize pediatric cancer treatments. Clin Cancer Res. 2012;18:2791–800. doi: 10.1158/1078-0432.CCR-11-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crona D, Innocenti F. Can knowledge of germline markers of toxicity optimize dosing and efficacy of cancer therapy? Biomark Med. 2012;6:349–62. doi: 10.2217/bmm.12.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wetterstrand K. [July 25 2013];DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) 2013 < www.genome.gov/sequencingcosts>.
- 22.Technologies L. [August 1 2013];Life Technologies Introduces the Benchtop Ion Proton™ Sequencer; Designed to Decode a Human Genome in One Day for $1,000. 2012 < http://www.lifetechnologies.com/us/en/home/about-us/news-gallery/press-releases/2012/life-techologies-itroduces-the-bechtop-io-proto.html.html>.
- 23.Simon R, Roychowdhury S. Implementing personalized cancer genomics in clinical trials. Nat Rev Drug Discov. 2013;12:358–69. doi: 10.1038/nrd3979. [DOI] [PubMed] [Google Scholar]
- 24.Ramsey LB, et al. Rare versus common variants in pharmacogenetics: SLCO1B1 variation and methotrexate disposition. Genome Res. 2012;22:1–8. doi: 10.1101/gr.129668.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robson ME, Storm CD, Weitzel J, Wollins DS, Offit K. American Society of Clinical Oncology Policy Statement Update: Genetic and Genomic Testing for Cancer Susceptibility. Journal of Clinical Oncology. 2010;28:893–901. doi: 10.1200/JCO.2009.27.0660. [DOI] [PubMed] [Google Scholar]
- 26.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–60. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 28. [July 1 2013];Table of Pharmacogenomic Biomarkers in Drug Labels. < http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm>.
- 29.McCarty CA, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kilpivaara O, Aaltonen LA. Diagnostic Cancer Genome Sequencing and the Contribution of Germline Variants. Science. 2013;339:1559–62. doi: 10.1126/science.1233899. [DOI] [PubMed] [Google Scholar]
- 31.Veenstra DL, et al. Improving the efficiency and relevance of evidence-based recommendations in the era of whole-genome sequencing: an EGAPP methods update. Genet Med. 2013;15:14–24. doi: 10.1038/gim.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLeod HL. Cancer pharmacogenomics: early promise, but concerted effort needed. Science. 2013;339:1563–6. doi: 10.1126/science.1234139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Donnell PH, Stadler WM. Pharmacogenomics in early-phase oncology clinical trials: is there a sweet spot in phase II? Clin Cancer Res. 2012;18:2809–16. doi: 10.1158/1078-0432.CCR-11-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Baan FH, Knol MJ, Klungel OH, Egberts AC, Grobbee DE, Roes KC. Potential of adaptive clinical trial designs in pharmacogenetic research. Pharmacogenomics. 2012;13:571–8. doi: 10.2217/pgs.12.10. [DOI] [PubMed] [Google Scholar]
- 35.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89:464–7. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toffoli G, et al. Genotype-Driven Phase I Study of Irinotecan Administered in Combination With Fluorouracil/Leucovorin in Patients With Metastatic Colorectal Cancer. Journal of Clinical Oncology. 2009;28:866–71. doi: 10.1200/JCO.2009.23.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanek EJ, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin Pharmacol Ther. 2012;91:450–8. doi: 10.1038/clpt.2011.306. [DOI] [PubMed] [Google Scholar]
- 38.O'Donnell PH, Ratain MJ. Germline pharmacogenomics in oncology: Decoding the patient for targeting therapy. Molecular Oncology. 2012;6:251–9. doi: 10.1016/j.molonc.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bell GC, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc. 2013 doi: 10.1136/amiajnl-2013-001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swen JJ, et al. Pharmacogenetics: from bench to byte. Clin Pharmacol Ther. 2008;83:781–7. doi: 10.1038/sj.clpt.6100507. [DOI] [PubMed] [Google Scholar]
- 41. [July 1 2013];Record of Consultations on Pharmacogenomics/Biomarkers. 2013 < http://www.pmda.go.jp/english/service/biomarker.html>.
- 42.Swen JJ, et al. Pharmacogenetics: From Bench to Byte—An Update of Guidelines. Clinical Pharmacology & Therapeutics. 2011;89:662–73. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- 43. [July 1 2013];Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. 2013 doi: 10.1097/GIM.0b013e318184137c. < http://www.egappreviews.org/>. [DOI] [PMC free article] [PubMed]
- 44.Benson AB, 3rd, et al. Metastatic colon cancer, version 3.2013: featured updates to the NCCN Guidelines. J Natl Compr Canc Netw. 2013;11:141–52. doi: 10.6004/jnccn.2013.0022. quiz 52. [DOI] [PubMed] [Google Scholar]
- 45.Administration, U.F.a.D. Camptosar (irinotecan) FDA label. Washington, D.C.: 2012. [Google Scholar]
- 46. [July 1 2013];European Public Assessment Reports. 2013 < http://www.ema.europa.eu/ema/>.
- 47.Locker GY, et al. ASCO 2006 Update of Recommendations for the Use of Tumor Markers in Gastrointestinal Cancer. Journal of Clinical Oncology. 2006;24:5313–27. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 48.Relling MV, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for Thiopurine Methyltransferase Genotype and Thiopurine Dosing: 2013 Update. Clinical Pharmacology & Therapeutics. 2013;93:324–5. doi: 10.1038/clpt.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Recommendations from the EGAPP Working Group: can UGT1A1 genotyping reduce morbidity and mortality in patients with metastatic colorectal cancer treated with irinotecan? Genetics in Medicine. 2009;11:15–20. doi: 10.1097/GIM.0b013e31818efd9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goddard KA, et al. Building the evidence base for decision making in cancer genomic medicine using comparative effectiveness research. Genet Med. 2012;14:633–42. doi: 10.1038/gim.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deverka PA, Vernon J, McLeod HL. Economic Opportunities and Challenges for Pharmacogenomics. Annual Review of Pharmacology and Toxicology. 2010;50:423–37. doi: 10.1146/annurev.pharmtox.010909.105805. [DOI] [PubMed] [Google Scholar]
- 52.Lam YW. Scientific challenges and implementation barriers to translation of pharmacogenomics in clinical practice. ISRN Pharmacol. 2013;2013:641089. doi: 10.1155/2013/641089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faruki H. Genomic Testing: The Clinical Laboratory Perspective. Clinical Pharmacology & Therapeutics. 2013;94:190–2. doi: 10.1038/clpt.2013.61. [DOI] [PubMed] [Google Scholar]
- 54.Ahmed G, et al. Cost implications of reactive versus prospective testing for dihydropyrimidine dehydrogenase (DPD) deficiency in patients with colorectal cancer. Journal of Clinical Oncology. 2013;31 doi: 10.1177/1559325818803042. abstr 3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Behl AS, et al. Cost-effectiveness analysis of screening for KRAS and BRAF mutations in metastatic colorectal cancer. J Natl Cancer Inst. 2012;104:1785–95. doi: 10.1093/jnci/djs433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong WB, Carlson JJ, Thariani R, Veenstra DL. Cost effectiveness of pharmacogenomics: a critical and systematic review. Pharmacoeconomics. 2010;28:1001–13. doi: 10.2165/11537410-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 57.Paulden M, Franek J, Pham B, Bedard PL, Trudeau M, Krahn M. Cost-Effectiveness of the 21-Gene Assay for Guiding Adjuvant Chemotherapy Decisions in Early Breast Cancer. Value in Health. 2013 doi: 10.1016/j.jval.2013.03.1625. [DOI] [PubMed] [Google Scholar]
- 58.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hurley JE. An overview of the normative economics of the health sector. In: Culyer A, Newhouse J, editors. Handbook of Health Economics. Vol. 1. Elsevier; 2000. pp. 55–118. [Google Scholar]
- 60.Meric-Bernstam F, Mills GB. Overcoming implementation challenges of personalized cancer therapy. Nature Reviews Clinical Oncology. 2012;9:542–8. doi: 10.1038/nrclinonc.2012.127. [DOI] [PubMed] [Google Scholar]
- 61.Epstein RS, Teagarden JR. Comparative effectiveness research and personalized medicine: catalyzing or colliding? Pharmacoeconomics. 2010;28:905–13. doi: 10.2165/11535830-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 62.Rocha V, et al. Association of drug metabolism gene polymorphisms with toxicities, graft-versus-host disease and survival after HLA-identical sibling hematopoietic stem cell transplantation for patients with leukemia. Leukemia. 2009;23:545–56. doi: 10.1038/leu.2008.323. [DOI] [PubMed] [Google Scholar]
- 63.Nowell SA, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005;91:249–58. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 64.Su HI, et al. Association of cyclophosphamide drug-metabolizing enzyme polymorphisms and chemotherapy-related ovarian failure in breast cancer survivors. Fertil Steril. 2010;94:645–54. doi: 10.1016/j.fertnstert.2009.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwab M, et al. Role of genetic and nongenetic factors for fluorouracil treatment-related severe toxicity: a prospective clinical trial by the German 5-FU Toxicity Study Group. J Clin Oncol. 2008;26:2131–8. doi: 10.1200/JCO.2006.10.4182. [DOI] [PubMed] [Google Scholar]
- 66.Mellor JD, Brown MP, Irving HR, Zalcberg JR, Dobrovic A. A critical review of the role of Fc gamma receptor polymorphisms in the response to monoclonal antibodies in cancer. J Hematol Oncol. 2013;6:1. doi: 10.1186/1756-8722-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pullarkat ST, et al. Thymidylate synthase gene polymorphism determines response and toxicity of 5-FU chemotherapy. Pharmacogenomics J. 2001;1:65–70. doi: 10.1038/sj.tpj.6500012. [DOI] [PubMed] [Google Scholar]
- 68.Salgado J, Zabalegui N, Gil C, Monreal I, Rodriguez J, Garcia-Foncillas J. Polymorphisms in the thymidylate synthase and dihydropyrimidine dehydrogenase genes predict response and toxicity to capecitabine-raltitrexed in colorectal cancer. Oncol Rep. 2007;17:325–8. [PubMed] [Google Scholar]
- 69.Relling MV, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for Thiopurine Methyltransferase Genotype and Thiopurine Dosing. Clinical Pharmacology & Therapeutics. 2011;89:387–91. doi: 10.1038/clpt.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. [July 1 2013];Drugs at FDA: FDA Approved Drug Products. 2013 < http://www.accessdata.fda.gov/scripts/cder/drugsatfda/>.
- 71.Theriault RL, et al. Breast Cancer, Version 3.2013. J Natl Compr Canc Netw. 2013;11:753–61. doi: 10.6004/jnccn.2013.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zelenetz AD, et al. Non-Hodgkin's Lymphomas, version 3.2012. J Natl Compr Canc Netw. 2012;10:1487–98. doi: 10.6004/jnccn.2012.0155. [DOI] [PubMed] [Google Scholar]
- 73.Alvarnas JC, et al. Acute lymphoblastic leukemia. J Natl Compr Canc Netw. 2012;10:858–914. doi: 10.6004/jnccn.2012.0089. [DOI] [PubMed] [Google Scholar]
- 74.Benson AB, 3rd, et al. Rectal cancer. J Natl Compr Canc Netw. 2012;10:1528–64. doi: 10.6004/jnccn.2012.0158. [DOI] [PubMed] [Google Scholar]
- 75. [July 1 2013];PharmGKB: The Pharmacogenomics Knowledgebase. 2013 < http://www.pharmgkb.org/>.