Abstract
A systems biology approach was applied to investigate the mechanisms of chromosomal instability in melanoma cell lines. Chromosomal instability was quantified using array comparative genomic hybridization to identify somatic copy number alterations (deletions and duplications). Primary human melanocytes displayed an average of 8.5 alterations per cell primarily representing known polymorphisms. Melanoma cell lines displayed 25 to 131 alterations per cell, with an average of 68, indicative of chromosomal instability. Copy number alterations included approximately equal numbers of deletions and duplications with greater numbers of hemizygous (−1,+1) alterations than homozygous (−2,+2). Melanoma oncogenes, such as BRAF and MITF, and tumor suppressor genes, such as CDKN2A/B and PTEN, were included in these alterations. Duplications and deletions were functional as there were significant correlations between DNA copy number and mRNA expression for these genes. Spectral karyotype analysis of three lines confirmed extensive chromosomal instability with polyploidy, aneuploidy, deletions, duplications and chromosome rearrangements. Bioinformatic analysis identified a signature of gene expression that was correlated with chromosomal instability but this signature provided no clues to the mechanisms of instability. The signature failed to generate a significant (P=0.105) prediction of melanoma progression in a separate dataset. Chromosomal instability was not correlated with elements of DNA damage response such as radiosensitivity, nucleotide excision repair, expression of the DNA damage response biomarkers γH2AX and P-CHEK2, nor G1 or G2 checkpoint function. Chromosomal instability in melanoma cell lines appears to influence gene function but it is not simply explained by alterations in the system of DNA damage response.
Keywords: melanoma, genetic instability, DNA damage response, checkpoint, repair, prognosis
Introduction
Melanoma is malignant neoplasm derived from transformed melanocytes. Melanocytes occupy many niches within the body and primary melanoma can occur in the skin, eye, bowel and brain. Because of its propensity for metastatic dissemination and the paucity of effective treatments for metastatic disease, melanoma displays greater lethality than the more common skin cancers, squamous cell and basal cell carcinoma [Jerant et al. 2000].
Cutaneous melanoma (CM) is associated with skin type and sunlight exposure [Gilchrest et al. 1999]. People with fair skin that is prone to burning have increased risk of CM. Molecular epidemiology studies have associated specific melanoma subtypes with sunlight exposure; CM with an activating mutation in the BRAF oncogene are associated with sunlight exposure in the first three decades of life while CM with mutation of NRAS are associated with sun exposure in later decades [Thomas et al. 2007]. The number of benign melanocytic nevi (moles) on skin is also a risk factor for CM [Thomas et al. 2007]. Moles that develop in childhood on sun-exposed skin may be precursors of CM. Most moles carry activating mutations in the BRAF or NRAS oncogenes [Poynter et al. 2006; Blokx et al. 2010; Yeh et al. 2013] although the codon mutations are not the C to T transitions at CC and CT dinucleotides that are signatures of UV-light induced mutation [Thomas et al. 2006]. Thus, sunburns in skin with developing nevi may cause CM.
Solar radiation in the UV region of the electromagnetic spectrum is damaging to skin. Photons in the UVB range of energies (290–320 nm) penetrate the atmosphere and stratum corneum to damage cells in the basal layer of the skin where melanocytes reside. Studies with an HGF-overexpressing mouse demonstrated that a single dose of UVB accelerated development of melanoma significantly while UVA photons (320–400 nm) did not [De Fabo et al. 2004]. A more recent study demonstrated that 350 nm UVA can initiate melanoma in mice with pigmented melanocytes [Noonan et al. 2012]. A model has emerged that UVB photons damage DNA to produce promutagenic cyclobutane pyrimidine dimers (CPD) and 6–4 pyrimidine-pyrimidone (6-4PP) photoproducts. UVA photons act on melanin to induce reactive oxygen species that produce promutagenic 8-oxo-deoxyguanosine (8oxoG) lesions in DNA. Melanomas on sunlight-exposed regions of skin contain very high frequencies of base substitution mutations with the C to T signature characteristic of UV-induced DNA damage [Hodis et al. 2012]. Xeroderma pigmentosum patients with inherited defects in repair or replication of UV-damaged DNA have a thousand-fold increased risk of development of melanoma [Kraemer et al. 1994], further establishing that sunlight-induced DNA damage causes CM.
Melanoma is much like other solid tissue malignancies displaying all of the hallmarks of cancer [Hanahan and Weinberg 2011]. Melanomas are proliferative lesions that invade dermis and metastasize through lymphatics and blood. In comparison to normal melanocytes, melanoma cells display increased expression of telomerase and immortality, reduced requirement for growth factors, attenuation or ablation of DNA damage checkpoint function, resistance to apoptosis and a severely disordered genome with numerous structural and numerical chromosomal aberrations. Global analysis of gene expression revealed hundreds to thousands of transcripts with altered expression in melanomas relative to benign moles or isolated melanocytes [Haqq et al. 2005]. As genomic instability is an enabling hallmark feature of melanoma that can account for its great variety of phenotypic alterations, the mechanisms of genomic instability are of considerable interest.
The presence of activating mutations in oncogenes in melanomas and nevi has emphasized study of the effects of oncogenes on melanocytes. As is true for skin fibroblasts, transformation of melanocytes with oncogenic BRAF and NRAS causes oncogene-induced senescence (OIS). OIS as induced in skin fibroblasts by oncogenic HRAS is a DNA damage response (DDR) with evidence for replication stress and activation of ATR- and ATM-dependent checkpoint signaling pathways. A recent study linked HRAS-induced OIS to repression of BRIP1- and BRCA1-dependent DNA repair [Tu et al. 2011]. OIS as induced by NRAS in melanocytes appears to include elements of the DDR and melanocytes arrest growth behind RB- and p53-dependent barriers [Haferkamp et al. 2009a; Haferkamp et al. 2009b]. Transformation of melanocytes with oncogenic BRAF triggers a growth arrest that is independent of p53 and does not include induction of p21Waf1 [Michaloglou et al. 2005]. A recent study demonstrated that BRAF-induced OIS in melanocytes was enforced by PTEN acting to inhibit AKT [Vredeveld et al. 2012]. However, BRAF-transformed melanocytes display increased expression of γH2AX as a marker of the DDR [Suram et al. 2012]. Expression of γH2AX appeared to be concentrated at telomeres as though oncogenic BRAF induced damage during replication of telomeric repeat DNA [Suram et al. 2012]. As OIS is a barrier to melanomagenesis, oncogene-transformed melanocytes must bypass or overcome the barrier through other genetic and phenotypic alterations. BRAF-mutant melanomas appear to inactivate PTEN by mutation or gene deletion [Daniotti et al. 2004; Tsao et al. 2004]. NRAS- and BRAF-mutant melanomas also display inactivation of the CDKN2A/B loci that encode three tumor suppressors, p16INK4A, p15INK4B and p19ARF [Daniotti et al. 2004]. The CDKN2A locus is inactivated in melanoma by mutation and deletion [Hodis et al. 2012].
This paper is concerned with the mechanisms of genetic instability in melanoma that might increase the likelihood that proliferating melanocytes in sunlight-exposed skin acquire the mutations and chromosomal aberrations that activate oncogenes and inactivate tumor suppressors. A systems biology approach was followed that attempted to quantify many of the parts in the system of response to DNA damage and the ways that they interact in primary melanocyte cultures and melanoma cell lines. Common aberrations in melanoma were identified and associated with elements of DDR. The results of this analysis indicate that genetic instability in melanoma is a complex process associated with many defects within the system of DDR but correlated with none.
Methods
For quantification of chromosomal instability, somatic copy number alterations were determined using the Agilent 180K CGH array (G4449A). Melanocytes and melanoma cell lines were grown as described [Sambade et al. 2011; Carson et al. 2012]. Cell lines were shown to be free of mycoplasma contamination using a commercial kit. Cells were harvested in log phase growth (60–80% confluent) and processed for isolation of DNA using Qiagen kits. DNA was submitted to the UNC-CH Genomics Core Facility for labeling with Cy5-dUTP. A normal diploid human lymphoblastoid line DNA sample was labeled with Cy3-dUTP. Equal amounts of the Cy5-labeled sample DNA and Cy3-labeled control DNA were mixed and hybridized to the DNA microarray. After hybridization and washing, the array was scanned and pixel intensities for each array element were returned as log2 Cy5/Cy3 ratios. Analyses were performed using BRB-ArrayTools (version 4.2.1) developed by Dr. Richard Simon and the BRB-ArrayTools Development Team (http://linus.nci.nih.gov/BRB-ArrayTools.html). BRB ArrayTools extracted the log2 ratios from the Agilent scanner file and then applied the Genomic Identification of Significant Targets in Cancer (GISTIC) bioinformatic tool for segmentation analysis [Beroukhim et al. 2007]. Segments were identified by genome map position with increased or decreased copy number in the melanocytes and melanoma cell lines relative to the diploid lymphoblast reference genome. A chromosomal instability index (CIN) was determined for NHM and melanoma cell lines by summing all segments with non-diploid DNA content.
Global gene expression data were obtained for the melanoma cell lines using an Agilent 44K printed oligonucleotide array [Carson et al. 2012; Omolo et al. 2013]. The data were analyzed by BRB Array Tools using Quantitative Trait Analysis (QTA) to identify genes whose expression was correlated with CIN based on their Spearman Correlation Coefficient. A second Bayesian analysis tool identified genes that were correlated with CIN based on their low false discovery rate (FDR) [Kaufmann et al. 2008]. The ability of these gene lists to predict CIN was determined using the Least Absolute Shrinkage and Selection Operator (LASSO) with leave-one-out cross-validation [Carson et al. 2012].
Survival risk prediction (SRP) was done using an independent melanoma gene expression database linked to patient survival. Gene expression in primary melanomas was determined using the Agilent 44K printed array with determination of transcript expression in the melanomas normalized to expression in a reference standard [Winnepenninckx et al. 2006]. Median Cy5 and Cy3 pixel intensities were recovered from a public database and imported to BRB-ArrayTools version 4.3.2 which normalized the data by median centering. QTA and Bayes analysis gene signatures, that were correlated with CIN, G1 checkpoint function and G2 checkpoint function in the melanoma cell lines, were combined and used to stratify patients into three groups, one with high (>50%) risk of development of a distant metastasis, another with low (<25%) risk, and a third with medium risk (otherwise). A permutation test was performed by randomly permuting sample identity and determining whether the separation of low-, medium- and high-risk groups equaled or exceeded the separation seen with proper identification of samples. The permutation P-value was the fraction of 1000 permutations where random labeling produced separation equal to or greater than the proper labeling. A permutation P-value <0.05 was considered to be statistically significant.
Spectral karyotypic analysis was done on one NHM culture and three melanoma cell lines by the Cytogenetics Core at the Van Andel Institute, Grand Rapids, MI. Analysis of BRAF and NRAS mutation status, global analysis of gene expression, ionizing radiation radiosensitivity, nucleotide excision repair, expression of DDR biomarkers γH2AX and P-CHEK2, and G1 and G2 checkpoint status of the melanoma cell lines has been reported [Gaddameedhi et al. 2010; Sambade et al. 2011; Carson et al. 2012; Nikolaishvilli-Feinberg et al. 2013; Omolo et al. 2013]. The sensitivity of melanoma lines to UVC-induced inactivation of colony formation was done as previously described [Boyer et al. 1990].
Results
DNA copy number variation as a measure of chromosomal instability in melanocytes and melanoma cell lines
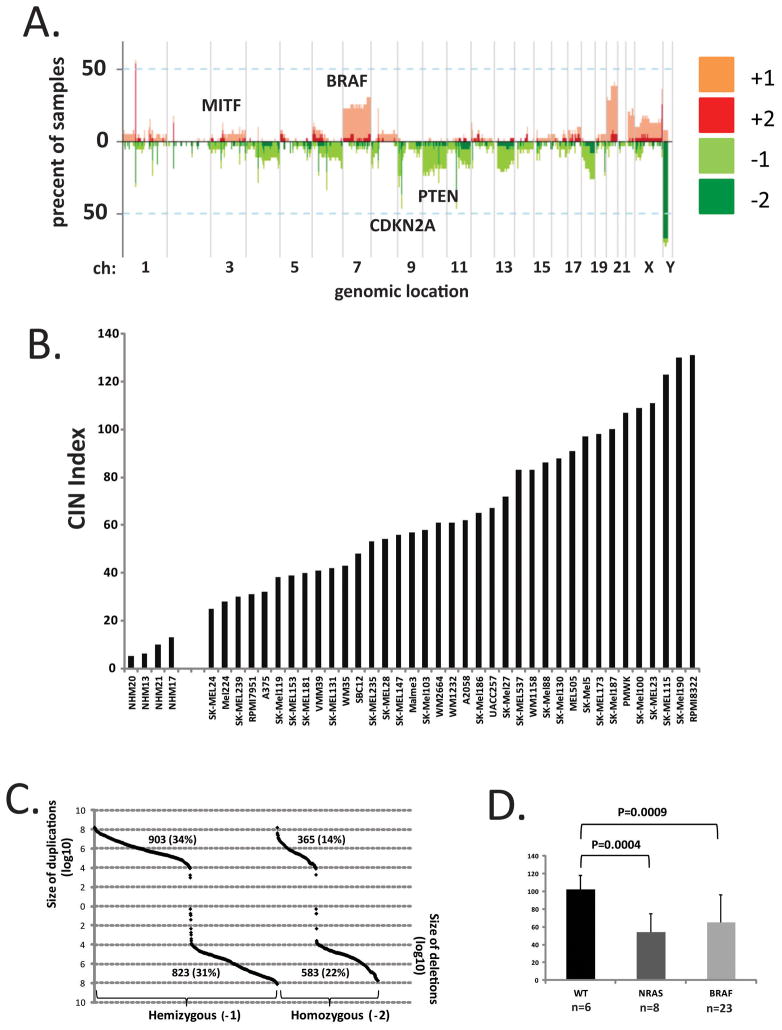
Array-based comparative genomic hybridization (arrayCGH) was applied to identify regions of the genome with somatic copy number alterations (SCNA) using GISTIC. Copy number was assessed by comparison to a reference diploid human lymphoblastoid cell line. The landscape of SCNAs seen in this sample of melanoma cell lines (Figure 1A) was quite similar to those reported previously for primary melanomas, short-term melanoma cultures and melanoma cell lines [Lin et al. 2008; Akavia et al. 2010]. Prominent features included common focal amplification of a segment of chromosome 3 that contains the master melanocyte transcriptional regulator MITF and duplication of chromosome 7 that contains the BRAF oncogene. Common deletions included segments in chromosome 9 containing CDKN2A/B and chromosome 10 containing PTEN.
Figure 1.
Landscape of SCNA in melanoma. A. GISTIC determined the frequencies of melanoma cell lines with SCNA at selected regions of the genome. B. The numbers of SCNA determined in primary melanocytes (NHM) and melanoma cell lines. C. The sizes of SCNA in melanoma cell lines. D. WT melanoma subtype displayed the greatest numbers of SCNA.
The total number of SCNA was determined as an index of chromosomal instability (CIN) for four normal human melanocyte (NHM) cultures and thirty seven melanoma cell lines (Figure 1B). NHMs displayed several SCNA with an average CIN index of 8.5 per culture. SCNA in NHMs ranged in size between 6 kbp and 2000 kbp. Among the thirty four SCNA observed in NHMs, nine were recurrent and seen in more than one culture. One SCNA that mapped to chromosome 1p31.1, approximately 27 kbp in size, was present in all four primary melanocyte cultures, three with homozygous deletion and one with homozygous duplication. Another recurrent SCNA mapped to the X chromosome and had two extra copies in two cultures and one extra copy in a third. A third duplication of a segment of chromosome 12 was seen in two cultures. The majority of individual and recurrent duplications or deletions seen in NHMs are listed in the Database of Genomic Variants (http://dgv.tcag.ca/dgv/app/home) and therefore likely correspond to polymorphic regions of the genome that have undergone amplification or deletion during human evolution.
In comparison to NHMs melanoma cell lines displayed increased SCNA across the genome (Figure 1B). The average CIN index in melanoma lines was 68 (P=2.8 X 10−14 in comparison to NHMs, Students T-test). There was substantial variability in CIN index among the melanoma cell lines with a low value of 25 seen in the SK-Mel24 line and a high of 131 seen in RPMI-8322. The sizes of SCNA that were scored by GISTIC also displayed great variation in the melanoma cell lines with a minimum size of 2 bp and a maximum size of 1.6 × 108 bp (Figure 1C). Duplications and deletions most commonly occupied segments of 104–108 bp. The total numbers of duplications and deletions were similar but hemizygous losses and gains were more frequent than homozygous losses and gains.
A common mutation seen in melanoma is the V600E codon change in BRAF. Mutations in NRAS are less common but mutually exclusive with BRAF mutations [Hodis et al. 2012]. Melanomas with wildtype BRAF and NRAS have been described and appear to represent a unique subtype, termed WT here. The CIN index was determined for melanoma lines in the three subtypes. The WT melanoma lines displayed a significantly greater CIN index compared to the BRAF- and NRAS-mutant subtypes (Figure 1D).
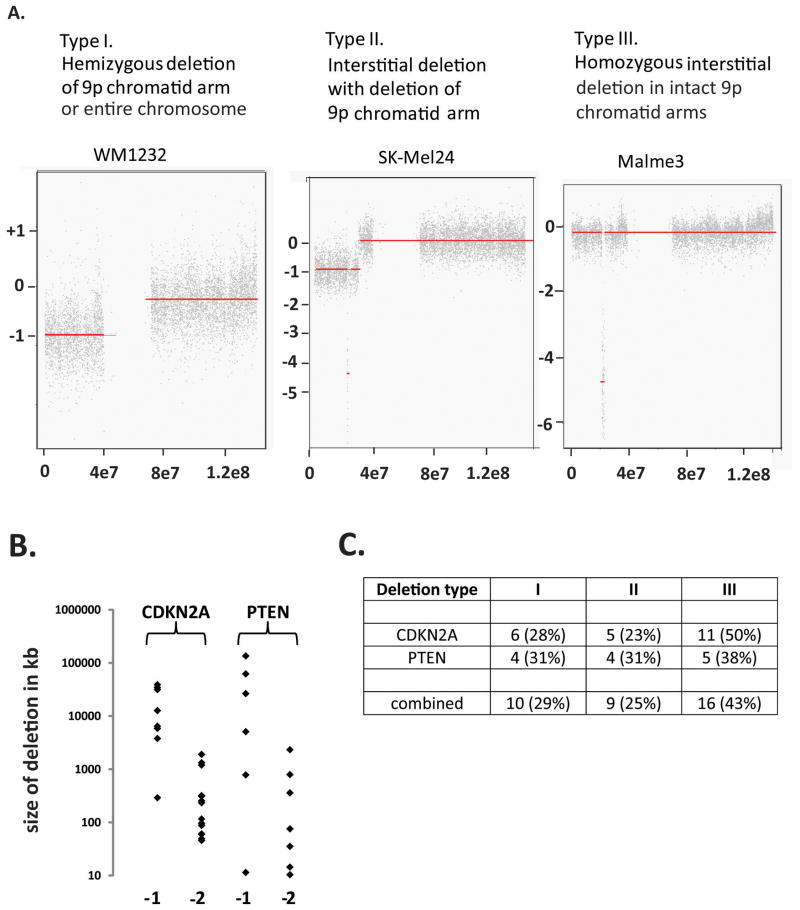
Examination of chromosome structure as determined by arrayCGH revealed three patterns of segmental deletions and duplications that are commonly observed in cancer [Bignell et al. 2010] (Figure 2). In pattern I terminal segments of chromosomes were lost or gained (terminal deletions or duplications). In pattern II smaller segments of loss or gain were present within larger segments of loss or gain (interstitial deletions or duplications within terminal deletions or duplications). Pattern III contained interstitial deletions or duplications. These three patterns were evident in chromosome 9 that contains the CDKN2A/B melanoma suppressor gene loci (Figure 2A) and in chromosome 10 that contains the PTEN tumor suppressor locus (not shown). In both the CDKN2A/B and PTEN loci, the sizes of segments associated with hemizygous (−1) and homozygous (−2) deletions were quite different, with hemizygous deletions being much larger than homozygous deletions (Figure 2B). This likely indicates that chromosomes 9 and 10 contain essential genes and homozygous deletions are only tolerated in the regions of the non-essential suppressor gene loci. Deletions in CDKN2A/B and PTEN were distributed across the three patterns with similar proportions of chromatid arm and interstitial deletions (Figure 2C).
Figure 2.
Types of allelic deletions in melanoma cell lines. A. Three types of deletions of CDKN2A were observed. B. Homozygous deletions are smaller than hemizygous deletions. C. Distribution of types of deletion in CDKN2A and PTEN (number of deletions detected and percent of total deletions).
Array-CGH technology provides information about DNA copy number and suggests alterations in chromosome structure such as terminal deletions (Figure 2A). To gain further insight to the changes in chromosome structure that are present in melanoma cell lines, spectral karyotypes were determined for one NHM culture and three melanoma cell lines. The NHM culture displayed no structural alterations in the fifteen metaphase spreads that were examined (Figure 3A). The melanoma cell lines displayed numerous structural alterations including intrachromosome deletions and duplications, interchromosomal rearrangements as well as aneusomy (chromosome losses and gains) and polyploidy. One of the melanoma lines (SK-Mel187) displayed a unimodal distribution of chromosome numbers ranging between 77 and 81. Another line (SK-Mel103) displayed a bimodal distribution of chromosomes, 82% having 59–63 chromosomes (sub-triploid) and 18% having 115–116 chromosomes. A third melanoma line, A2058, shown in Figure 3B, also displayed a biomodal distribution with 60% of metaphases having 46–48 chromosomes and 40% with 65–88 chromosomes. Inspection of A2058 showed chromosomal aneusomy in the quasi-diploid subline and the same aneusomies in the hypotetraploid subline (e.g., chromosomes 4 and 7). Interchromosome rearrangements were also evident in which portions of non-homologous chromosomes were recombined to produce marker chromosomes. Some of these rearrangements occurred before the diploid to hypotetraploid transition and so were seen in both sublines (ch1:ch6 and ch8:ch13). Other rearrangements were unique to the subline suggesting that they occurred after generation of the hypotetraploid subline (e.g. ch5:ch6). One rearrangement appeared to be reciprocal with portions of chromosomes 1 and 15 being rearranged. Three other rearrangements appeared to involve transfer of large portions of chromosomes 6, 13 and 14 to chromosomes 5, 8 and 5, respectively. A total of 8 interchromosome segments were evident in the diploid subline shown in Figure 3B and 5 interchromosome segments were evident in the hypotetraploid subline. In the two melanoma lines not shown, SK-Mel187 displayed 10 interchromosomal rearrangements and SK-Mel103 displayed as many as 33 interchromosomal rearrangements evident in the hypotriploid metaphases. The patterns of chromosomal alterations seen by SKY analysis of melanoma cell lines were congruent with those previously reported for such cells [Schulten et al. 2002]. Thus, while normal melanocytes replicated and segregated their genome with low error rates, the melanoma lines replicated with rearranged and possibly broken chromosomes as well as whole chromosome gains and losses.
Figure 3.
Spectral karyotypic analysis of normal melanocytes and the A2058 melanoma line. A. The NHM4 normal melanocytes grew without chromosomal aberrations. B. The A2058 melanoma line displayed numerical and structural chromosomal aberrations.
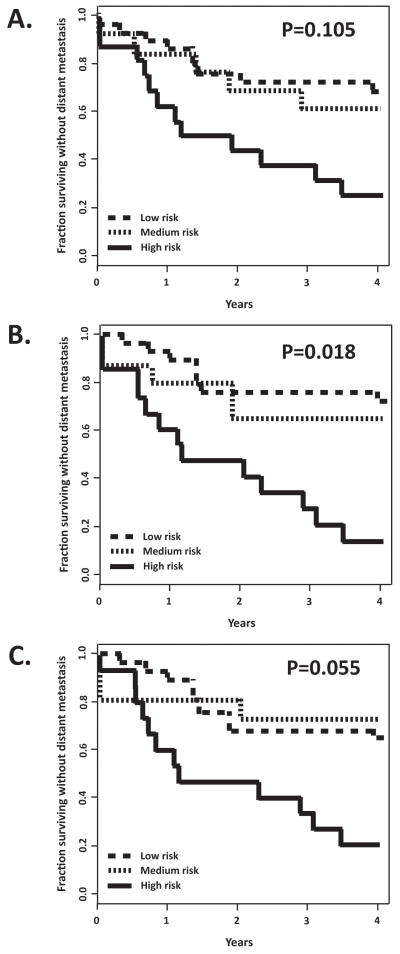
Figure 5.
CIN gene signature is not prognostic of metastatic spread in melanoma. A. The 190 gene QTA+Bayes signature composed of transcripts with high correlation with CIN (P<0.005, FDR<0.071) was applied to an independent dataset [Winnepenninckx et al. 2006] to compare outcome (4-year distant metastasis-free survival) to gene expression in the primary melanomas (using 2 principal components). Permutation analysis indicated that the observed separation of low-risk and high-risk groups was not significant (P=0.105) B. A 496 gene QTA+Bayes signature that was correlated with G1 checkpoint function in melanoma cell lines [Carson et al. 2012] was prognostic of metastasis-free survival (P=0.018). C. A 593 gene QTA+Bayes signature that was correlated with G2 checkpoint function [Omolo et al. 2013] was marginally not prognostic (P=0.055).
Allelic deletions and duplications are functional
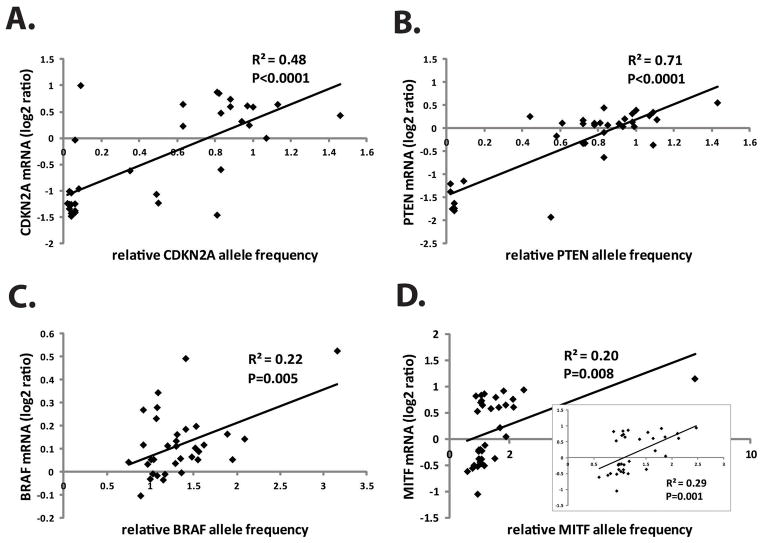
To determine the relationship between DNA SCNA and gene expression, allele copy number was compared to mRNA abundance as determined by DNA microarray analysis (34 of the melanoma lines were utilized for this analysis) [Carson et al. 2012]. For the tumor suppressor loci CKDN2A and PTEN, mRNA expression was highly correlated with allele copy number (CDKN2A DNA vs mRNA, R2= 0.48, P=0.001; PTEN DNA vs mRNA, R2=0.71, P=0.0001) (Figure 4). Generally melanoma lines with severe reduction in CDKN2A and PTEN DNA displayed severe reductions in mRNA. Two lines displayed an anomalous pattern with very low DNA content but intermediate (SK-Mel100) to high (SK-Mel147) CDKN2A mRNA expression. SK-Mel147 was scored with a type III homozygous interstitial deletion of the C-terminal half of the locus while retaining the full complement of the N-terminal half. The Agilent probe that was scored in the mRNA analysis (A_23_P43484) mapped to the N-terminus of CDKN2A. SK-Mel100 displayed a type III homozygous interstitial deletion of the entire CDKN2A locus including the region scored by the mRNA probe.
Figure 4.
SCNA’s are functional. Gene allele frequency was correlated with expression of mRNA for CDKN2A (A), PTEN (B), BRAF (C) and MITF (D). The insert in panel D shows the scatterplot and correlation coefficient when the outlier was removed.
Chromosome 9p2.1 also contains the CDKN2 gene family member CDKN2B closely adjacent to CDKN2A. CDKN2B encodes the p15INK4B cyclin-dependent kinase inhibitor that is induced by TGF-β and by oncogenic BRAF through activation of CEBP/β [Kuilman et al. 2008]. Interstitial deletions involving CDKN2A commonly include CDKN2B in primary melanomas and melanoma cell lines [Walker et al. 1998; Sauroja et al. 2000; Dutton-Regester et al. 2012]. Accordingly CDKN2B mRNA was severely reduced in 14 of the 19 melanoma cell lines with reduced expression of CDKN2A (results not shown). One melanoma line with high levels of CDKN2A mRNA (SK-Mel147) had severely reduced levels of CDKN2B mRNA associated with homozygous deletion of the entire CDKN2B gene locus as determined by arrayCGH.
For the melanoma oncogenes BRAF and MITF that were duplicated in melanoma lines, mRNA was also significantly correlated with DNA but with lower coefficients than for the tumor suppressors (Figure 4C,D). The BRAF locus is on chr7, which displayed amplification across its length (Figure 1A), and SKY analysis (Figure 3) revealed trisomy of chr7 in two of three melanoma lines (SK-Mel103, A2058). Thirty one percent (12 of 37) of the melanoma lines displayed amplification of the BRAF locus as determined by arrayCGH. Five lines displayed duplication of the entire chromosome, four lines duplication of the centromere-distal q-arm, two lines duplication of chr7 and the centromere-distal q-arm, and one line displayed complex alterations in chr7 with large segments deleted and others duplicated. The pattern of segmental losses and gains is characteristic of chromothripsis [Hirsch et al. 2013], which may develop when chromosomes are missegregated in micronuclei [Crasta et al. 2012]. The sizes of the amplified segments containing BRAF ranged from 6 mb to 88 mb. BRAF mRNA was correlated with DNA copy number albeit modestly (R2=0.22, P=0.005).
Duplication of the MITF locus did not commonly involve the entire chr3. All duplications were interstitial and ranged in size from 188 kb to 22mb. MITF allele frequency was correlated with mRNA (R2=0.20, P=0.008) (Figure 4D). One sample (SK-Mel239) was scored with severe amplification of the locus (8.15-fold) without a similar degree of enhancement of mRNA. When this outlier was removed the correlation of MITF mRNA with DNA was improved (R2=0.29, P=0.001) (Figure 4D, inset).
The frequencies of allelic deletions and duplications among the studied loci were correlated with molecular subtype in only one allele (Table 1). Deletion of PTEN was common among BRAF-mutant melanoma lines (54%) but uncommon among NRAS-mutant melanoma lines (12%, P=0.05) as previously noted by others [Daniotti et al. 2004; Tsao et al. 2004; Jonsson et al. 2007; Hodis et al. 2012].
Table 1.
Association of deletions and duplications with melanoma subtype.
| Frequency with del/dup | ||||
|---|---|---|---|---|
| WT, n=6 | NRAS mutant, n=8 | BRAF mutant, n=23 | Total, n=37 | |
| CDKN2A, del | 3 (50%) | 4 (50%) | 15 (64%) | 22 (58%) |
| PTEN, del | 1 (17%) | 1 (12%) | 12 (54%)* | 14 (38%) |
| BRAF, dup | 1 (17%) | 3 (38%) | 8 (35%) | 12 (31%) |
| MITF, dup | 0 | 2 (25%) | 9 (39%) | 11 (28%) |
Del, deletion; dup, duplication;
P=0.05, BRAF mutant vs NRAS mutant
Allelic deletions and duplications in melanoma tumor suppressors and oncogenes were functional, causing significant alterations in the levels of mRNA. Deletion of CDKN2A/B with reduced expression of mRNA was common across the melanoma subtypes while deletion of PTEN with reduced expression of mRNA was largely restricted to BRAF-mutant melanoma lines. Duplication of MITF in 28% of melanoma cell lines was associated with enhanced mRNA expression. Notably, two lines displayed hemizygous deletion of MITF (type I p-arm deletion) and 40% of the lines displayed reduced expression of MITF mRNA as well as reduced expression of its targets in melanogenesis SILV and TYR (results not shown). The presence of a melanin-negative subtype of melanoma has long been noted anatomically and histologically, and gene expression analysis has associated the MITF-negative subtype with relatively low expression of growth genes and high expression of invasion genes [Hoek et al. 2006]. The WT subtype with high CIN described here and in other studies [Shields et al. 2007; Carson et al. 2012] is represented entirely within the MITF-negative subtype.
Correlation of CIN with gene expression
CIN represents a quantitative trait that varied substantially among the melanoma cell lines (>5-fold). The BRB Array Tools QTA tool was applied to identify genes that were correlated with CIN. We first determined the effect of P-value cutoff on the numbers of CIN-correlated genes and the ability of the returned gene set to predict CIN using the Least Absolute Shrinkage and Selection Operator (LASSO) in a leave-one-out, cross-validation analysis (LOOCV) (Table 2). The best LASSO score (R2=0.42) was seen with a P-value cutoff of 0.005, generating a list of 129 transcripts. A second analytic tool used Bayesian statistics to identify transcripts that were correlated with CIN based on false-discovery rate (FDR) [Kaufmann et al. 2008]. We found 130 transcripts with a Bayes-determined FDR <0.071. These 130 transcripts predicted CIN with a LASSO score of 0.39. A total of 69 transcripts were common to the QTA and Bayes lists with P-value <0.05 and FDR<0.071. This shorter list of CIN-correlated transcripts did not predict CIN well (LASSO R2=0.13). A 190 transcript list that combined the QTA and Bayes lists (Supplemental table) performed best in the LASSO analysis with R2=0.52.
Table 2.
Quantitative trait analysis identifies probe lists that are correlated with CIN.
| Significance levela | 0.001 | 0.002 | 0.003 | 0.004 | 0.005 | 0.006 | 0.007 | 0.008 | 0.009 | 0.010 |
| Probe listb | 27 | 54 | 76 | 99 | 129 | 143 | 166 | 194 | 222 | 243 |
| LOOCV-R2,c | 0.34 | 0.39 | 0.28 | 0.37 | 0.42 | 0.37 | 0.35 | 0.09 | 0.34 | 0.31 |
| Estimated error rate | 0.66 | 0.61 | 0.72 | 0.63 | 0.58 | 0.63 | 0.65 | 0.91 | 0.66 | 0.69 |
P-value cutoff for the Spearman coefficient
Number of probes with coefficients less than the cutoff
Correlation coefficient for the relationship between the observed CIN and the predicted CIN using LASSO
To determine the biological significance of the CIN-correlated gene signatures, an independent dataset that reported gene expression in primary melanomas was examined. A Survival Risk Prediction (SRP) tool was applied to test whether the CIN-correlated gene signature was prognostic of melanoma outcome (4-yr distant metastasis-free survival). As seen in Figure 5A, the 190 probe CIN signature separated samples into low risk, medium risk and high risk groups with differences in outcome. A permutation P value was determined by randomly altering sample labels and determining the fraction of permutations with separation of risk groups equivalent to or greater than obtained with proper sample labeling. The permutation P-value for the 190 probe CIN signature list was 0.105, not achieving statistical significance. For comparison SRP was done using two other gene signatures, one that was predictive of DNA damage G1 checkpoint function [Carson et al. 2012] and one that was predictive of G2 checkpoint function [Omolo et al. 2013] in the melanoma cell lines. The G1 checkpoint signature produced a significant separation of risk groups with a permutation P-value of 0.018. The G2 checkpoint signature just failed to achieve a significant prediction of outcome (P=0.055). Thus, while the G1 checkpoint signature produced a significant stratification of risk among the melanoma patients, the CIN and G2 checkpoint signatures did not.
Gene ontology analysis was done to test whether any biological categories were over-represented in the 190 probe CIN-correlated gene signature. None passed filtration after the Benjamini correction for false-discovery. Further inspection of the gene list revealed several genes of interest because of their potential roles in carcinogenesis (e.g. APC2, HMG20B, CEBP/A, CRY1, ODC1, FBOX9, RGEF18). DNA damage response (DDR) genes were notably absent from the list.
Correlation of CIN with DDR
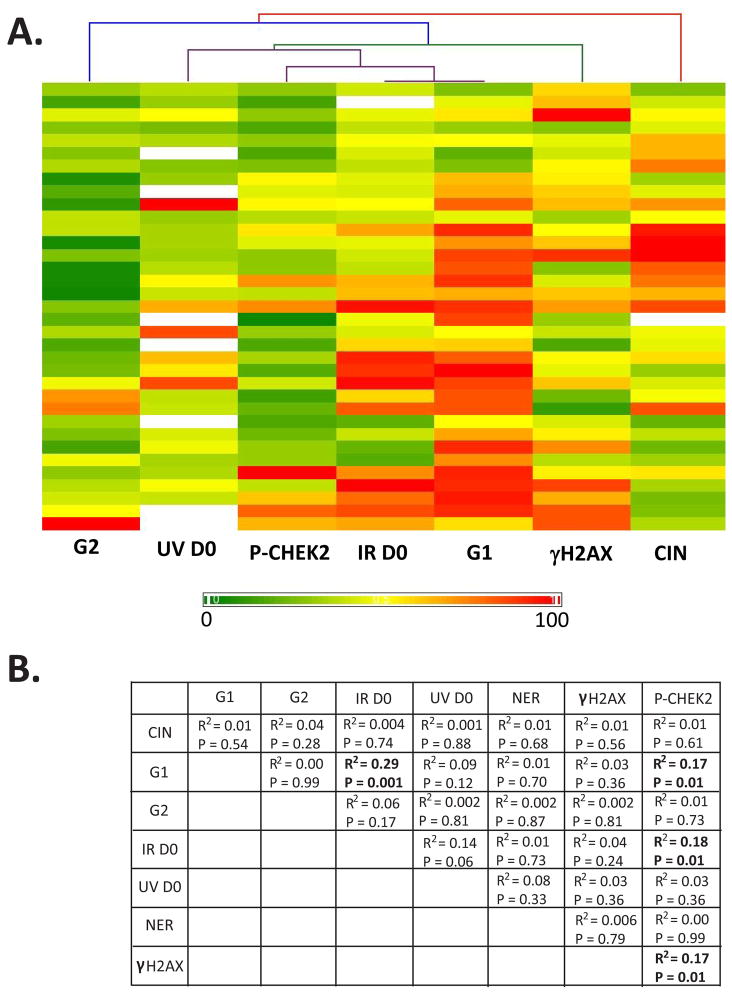
The melanoma cell lines in the CIN dataset have been examined for a variety of features of DDR including IR [Sambade et al. 2011] and UV radiosensitivity, nucleotide excision repair (NER) [Gaddameedhi et al. 2010], and DNA damage G1 [Carson et al. 2012] and G2 [Omolo et al. 2013] checkpoint functions. Immuno-fluorescence microscopy also quantified the basal levels of expression of two markers of DDR, γH2AX and P-CHEK2 [Nikolaishvilli-Feinberg et al. 2013]. We asked whether any element of DDR was correlated with CIN and displayed the results using a hierarchical cluster heat map (Figure 6A) and a table (Figure 6B). As shown in Figure 6, the highest correlation (R2 = 0.29, P=0.001 using the Pearson Correlation test) was between IR D0 and G1 checkpoint function, reflecting the resistance of p53-defective melanoma cell lines to inactivation of colony formation by IR [Sambade et al. 2011]. The correlation between IR D0 and UV D0 just failed to achieve significance (P=0.06). Expression of P-CHEK2, the activated form of a transducer checkpoint kinase that phosphorylates and activates p53 [Hirao et al. 2000], was correlated with G1 checkpoint function (R2= 0.17, P=0.01) and IR D0 (R2=0.18, P=0.01). Expression of γH2AX was correlated with P-CHEK2 (R2=0.17, P=0.01) but not with any other trait. Remarkably, CIN was placed on the extreme edge of the cluster analysis with the least correlation with the other parameters. NER was quantified for 16 melanoma cell lines as the percentage of cyclobutane pyrimidine dimers that were removed within 12 h of UV-irradiation [Gaddameedhi et al. 2010]. Dimer repair also was not correlated with CIN (Figure 6B, R2=0.01, P=0.68). Thus, no measured element of DDR was correlated with CIN in melanoma cell lines.
Figure 6.
CIN was not correlated with DDR. Melanoma cell lines were scored for expression of γH2AX and P-CHEK2 as biomarkers of DNA damage (double-strand breaks and replication stress) [Nikolaishvilli-Feinberg et al. 2013], for G1 and G2 checkpoint function [Carson et al. 2012; Omolo et al. 2013], NER [Gaddameedhi et al. 2010] and for sensitivity to inactivation of colony formation by UVC and IR [Sambade et al. 2011]. A. Maximal values for each parameter were expressed at 100% and all other values expressed as a percentage of the maximum. Thus, the color scale trends from 0 to 100%. Hierarchical clustering showed the strong correlation between p53-dependent G1 checkpoint function and sensitivity to IR, the correlation between expression of P-CHEK2 and expression of γH2AX and sensitivity to IR, and the marginal correlation between UV and IR sensitivity. B. Correlations (R2) and P values were determined for each pair of parameters.
Discussion
Melanoma cell lines displayed genomic instability as evidenced by significantly increased frequencies of SCNA and expression of markers of a persistent DDR (P-CHEK2, γH2AX). The lack of correlation between CIN and DDR markers suggests that conditions during the evolution of the melanoma cell lines leading to alterations in the genome were not preserved in the current state of the melanoma cell lines. Thus, quantitative analysis of DDR markers such as γH2AX and P-CHEK2 in melanoma in situ may not reflect the degree of genomic destabilization present in the cancer. Genomic instability may be a dynamic process that waxes and wanes during melanoma development and disease progression. A signature of gene expression that was correlated with CIN in melanoma lines was not prognostic of melanoma progression in a separate patient series. An analysis of SCNA in metastatic melanomas demonstrated that genomic instability was correlated with patient survival, with SCNA being higher in patients with low survival than in patients with high survival [Hirsch et al. 2013].
The mechanisms of genomic instability in melanoma are likely to be complex and multifactorial (Figure 7). Our current results and previous observations support a model in which chromosomal alterations (deletions, duplications and rearrangements) in melanoma derive from 1) telomere biology, 2) oncogene-induced replication stress and 3) UV-induced DNA damage. These components can explain the similar sizes and numbers of segmental deletions and duplications seen by array CGH and the rearrangements seen by spectral karyotype analysis.
Figure 7.
Mechanisms of chromosomal instability during melanomagenesis. A. Telomere crisis produces chromatid arm deletions and duplications. B. Replication stress induced by oncogenic BRAF induces breakage at common fragile sites. C. UV-induced DNA damage produces interstitial deletions and duplications.
One contributor to chromosomal instability in melanoma may be the telomere erosion that occurs in telomerase-negative cells such as melanocytes (Figure 7A) (see [Muraki et al. 2012] for a recent review). While telomerase activity is readily detected in metastatic melanoma specimens, activity is lower in primary melanomas and virtually undetectable in nevi [Miracco et al. 2000]. Thus, developing nevi and melanomas may pass through a condition of active cell proliferation with low or no expression of telomerase. Erosion of telomeric DNA to below a limit should trigger replicative senescence. Mutations and epigenetic alterations that inactivate p53 signaling allow telomerase-negative proliferative clones to bypass this senescence barrier and enter a phase of telomere crisis when chromosomes undergo cycles of bridge formation and breakage (Figure 7A). Telomere crisis will generate a transient phase of severe chromosomal instability prior to establishment of the immortal clone upon expression of telomerase [Simpson et al. 2005]. The studies of Soo et al. suggest that immortality is acquired late in melanomagenesis during metastatic spread [Soo et al. 2011]. Bypass of p53-dependent replicative senescence with ensuing telomere crisis may be a common event in development of melanoma [Carson et al. 2012; Satyamoorthy et al. 2000; Yu et al. 2009].
A second contribution to genetic instability in melanoma may derive from activation of the melanoma oncogenes, NRAS and BRAF. Activation of these oncogenes is associated with oncogene-induced senescence in primary melanocytes [Michaloglou et al. 2005; Haferkamp et al. 2009a]. Numerous studies suggest that oncogene-induced senescence is a DDR, with the oncogenes triggering replication stress that subsequently leads to chromosome breakage [Halazonetis et al. 2008]. Growth arrest in melanocytes with mutant NRAS appeared to be through pRB- and p53-dependent pathways and was associated with activation of a DDR, whereas arrest of melanocytes with mutant BRAF was independent of pRB and p53 [Haferkamp et al. 2009b]. While an initial study did not find evidence for DNA damage signaling in BRAF-induced senescence [Michaloglou et al. 2005], a more recent report demonstrated induction of γH2AX as a marker of DNA damage in melanocytes transformed with oncogenic BRAF [Suram et al. 2012]. We have shown that expression of oncogenic BRAF in melanoma cell lines induced a 5- to 8-fold increase in chromatid breaks and exchanges (WK Kaufmann and DA Simpson, unpublished) suggesting that the oncogene may be clastogenic. Melanocytic nevi with oncogenic BRAF also displayed increased expression of γH2AX associated with telomeric loci and it was concluded that replication forks in BRAF-transformed melanocytes were stalled in telomeres [Suram et al. 2012]. As stalled replication forks are prone to collapse and conversion to DNA double-strand breaks (dsb), such a phenomenon would accelerate telomere erosion and exacerbate chromosomal instability as occurs during telomere crisis as discussed above.
Oncogene-induced replication stress also may concentrate chromatid breaks at fragile sites. Fragile sites represent portions of chromosomes that are prone to breakage when cells are incubated with DNA metabolic poisons that slow DNA replication [Arlt et al. 2003; Hashash et al. 2012]. It is thought that certain DNA sequences are replicated late in S phase due to a paucity of origins of replication, thereby requiring replication by forks entering the fragile DNA from its flanks. Replicative DNA polymerases have difficulty completing replication of fragile site DNA sequences [Bergoglio et al. 2013; Walsh et al. 2013] and so replication forks may not complete replication of fragile DNA during the normal time allotted for S phase. It appears that regions of unreplicated fragile DNA escape notice by the replication checkpoint and cells enter mitosis with this unreplicated DNA [Smith-Roe et al. 2013]. Chromatid breaks at fragile sites may occur as unreplicated DNA is condensed and compacted in mitosis. A recent study of fragile DNA in yeast indicated that topoisomerase II and condensin subunits were required for DNA breakage in mitosis [Hashash et al. 2012], as though condensation at junctions of replicated and unreplicated DNA caused chromatid breaks at the junctions (as shown in Figure 7B). Oncogene-induced replication stress in telomeric DNA may contribute to telomere erosion and dysfunction while such stress in fragile DNA may contribute to interstitial chromatid breakage. It is notable that 15 sites of segmental deletions seen in the current analysis mapped to regions of the genome that contain fragile sites or sites of common homozygous deletion in cancer [Bignell et al. 2010] (see Supplement for locations of all significant losses and gains).
Analysis of melanoma gene expression and disease progression identified a set of 254 genes that were prognostic of 4 year metastasis-free survival [Winnepenninckx et al. 2006]. This gene set was enriched in many DDR genes, especially those that are associated with DNA dsb repair and stabilization of stalled DNA replication forks [Kauffmann et al. 2008]. However, metastatic spread was associated with enhanced expression of DNA repair genes, not reduced expression, suggesting that over-expression of such genes may contribute to genetic instability. It is well established that DNA repair factor stoichiometry is important for genome stability, as overexpression of the DNA repair factor AAG produces more genetic instability than deletion of the factor [Fu et al. 2012]. Over-expression of DNA dsb repair factors especially during S phase may interfere with proper regulation of DNA replication and exacerbate responses to forks stalling at natural barriers.
Remarkably, although elements of DDR such as NER influence melanoma development, no element of DDR that was measured here was correlated with CIN. Melanomas have been shown to express γH2AX as a marker of ongoing or persistent DNA damage [Gorgoulis et al. 2005; Wasco et al. 2008; Nikolaishvilli-Feinberg et al. 2013]. Melanoma cell lines display remarkably high levels of this phospho-protein [Warters et al. 2005] as our studies reproduced. While γH2AX was originally demonstrated as a sensitive marker of DNA dsb [Rogakou et al. 1998], subsequent studies have shown that it is not specific, also being produced at stalled replication forks [Ward and Chen 2001] and sites of NER [Marti et al. 2006]. The melanoma cell lines also expressed increased levels of the activated form of the transducer checkpoint kinase (P-CHEK2) and this was correlated with γH2AX. Replication stress and DNA dsb, therefore, may trigger activation of CHEK2 in melanoma cell lines. The levels of P-CHEK2 also were correlated with IR radiosensitivity and p53-dependent G1 checkpoint function. As P-CHEK2 will normally activate p53 to induce apoptosis or inhibit cell division [Hirao et al. 2002], the continued growth of melanoma cell lines with activated P-CHEK2 may require inactivation of p53 signaling. Thus, melanoma cells with high levels of P-CHEK2 corresponded to lines with inactivation of p53-dependent G1 checkpoint function. Melanoma lines with inactivation of p53 function were also radio-resistant [Sambade et al. 2011; Carson et al. 2012], consistent with inactivation of p53-dependent apoptosis or growth arrest. A p53-independent G2 checkpoint was dysfunctional in one-third of melanoma lines [Omolo et al. 2013]. Attenuation of signaling in this pathway of DDR was attributed in part to mutations in BRAF and NRAS, and in part to a complex signature of gene expression that repressed many regulators of mitosis such as CKS1B and WDHD1 (also known as AND1) [Omolo et al. 2013]. Melanoma cell lines grow with high levels of DNA damage and expression of DDR biomarkers because p53-dependent and p53-independent DNA damage checkpoint functions are attenuated. As the basal levels of γH2AX and P-CHEK2 were not correlated with CIN, it would appear that CIN in melanoma cell lines is a residue of cell biology that preceded establishment of the checkpoint-defective melanoma cell lines.
SCNA in melanoma included nearly equivalent numbers and sizes of deletions and duplications. This feature suggests a common origin. We propose that the deletions and duplications that occupy large and small segments of chromatid arms are consequences of chromatid breaks and exchanges that develop 1) at unprotected telomeres during telomere crisis and 2) at sites of stalled DNA replication forks. Repair of unprotected telomeres in sister chromatids by the alternative non-homologous endjoining (alt-NHEJ) pathway [Muraki et al. 2012] may produce isodicentric chromosomes [Kaufmann et al. 1997; Filatov et al. 1998]. Segregation of such dicentrics at anaphase can break chromosomes anywhere between the centromeres, yielding arm duplications and deletions of similar sizes. This mechanism can account for the large SCNA. The small SCNA that occupy more discreet segments may derive from slowing or stalling of DNA replication. Two aspects of melanomagenesis can produce DNA replication stalling, UV-induced DNA damage [Kaufmann and Wilson 1994; Kaufmann 2010] and oncogene-induced replication stress. When two neighboring stalled forks collapse to DNA dsb, erroneous repair by classical or alternative NHEJ may delete the interstitial segment from one chromatid and add the segment to another. The net result is inheritance of chromosomes with interstitial deletions or duplications. The development of UV-induced SCNA may be significantly enhanced in melanoma precursors with activated oncogenes and attenuated DNA damage response signaling. We have shown that oncogenic BRAF attenuates DNA damage G2 checkpoint function [Omolo et al. 2013] and sensitizes melanoma cells to UVB-induced chromosomal aberrations (DAS, WKK, in preparation). Oncogene-induced replication stress may also stall DNA replication at fragile sites leading to chromatid breakage in mitosis that duplicates and deletes gene segments. The combination of telomere crisis, oncogene-induced replication stress and UV-induced chromosomal damage can explain the common forms of chromosomal instability seen in melanoma.
Supplementary Material
Acknowledgments
Supported in part by PHS grants ES10126, ES007126, CA106209, CA16086 and ES014635.
Footnotes
Statement of authors’ contributions. The generation of the data reported here was done by Drs. Simpson, Carson, Shields, Sambade and Thomas. Statistical and bioinformatics analyses were done by Mr. Filgo and Drs. Omolo, Carson, Kaufmann and Ibrahim. The manuscript was drafted by Dr. Kaufmann and edited by Dr. Thomas.
References
- Akavia UD, Litvin O, Kim J, Sanchez-Garcia F, Kotliar D, Causton HC, Pochanard P, Mozes E, Garraway LA, Pe’er D. An integrated approach to uncover drivers of cancer. Cell. 2010;143(6):1005–1017. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Casper AM, Glover TW. Common fragile sites. Cytogenet Genome Res. 2003;100(1–4):92–100. doi: 10.1159/000072843. [DOI] [PubMed] [Google Scholar]
- Bergoglio V, Boyer AS, Walsh E, Naim V, Legube G, Lee MY, Rey L, Rosselli F, Cazaux C, Eckert KA, Hoffmann JS. DNA synthesis by Pol eta promotes fragile site stability by preventing under-replicated DNA in mitosis. J Cell Biol. 2013;201(3):395–408. doi: 10.1083/jcb.201207066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, Du J, Kau T, Thomas RK, Shah K, Soto H, Perner S, Prensner J, Debiasi RM, Demichelis F, Hatton C, Rubin MA, Garraway LA, Nelson SF, Liau L, Mischel PS, Cloughesy TF, Meyerson M, Golub TA, Lander ES, Mellinghoff IK, Sellers WR. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104(50):20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C, Widaa S, Hinton J, Fahey C, Fu B, Swamy S, Dalgliesh GL, Teh BT, Deloukas P, Yang F, Campbell PJ, Futreal PA, Stratton MR. Signatures of mutation and selection in the cancer genome. Nature. 2010;463(7283):893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokx WA, van Dijk MC, Ruiter DJ. Molecular cytogenetics of cutaneous melanocytic lesions - diagnostic, prognostic and therapeutic aspects. Histopathology. 2010;56(1):121–132. doi: 10.1111/j.1365-2559.2009.03452.x. [DOI] [PubMed] [Google Scholar]
- Boyer JC, Kaufmann WK, Brylawski BP, Cordeiro-Stone M. Defective postreplication repair in xeroderma pigmentosum variant fibroblasts. Cancer Res. 1990;50(9):2593–2598. [PubMed] [Google Scholar]
- Carson C, Omolo B, Chu H, Zhou Y, Sambade MJ, Peters EC, Tompkins P, Simpson DA, Thomas NE, Fan C, Sarasin A, Dessen P, Shields JM, Ibrahim JG, Kaufmann WK. A prognostic signature of defective p53-dependent G1 checkpoint function in melanoma cell lines. Pigment Cell Melanoma Res. 2012;25(4):514–526. doi: 10.1111/j.1755-148X.2012.01010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482(7383):53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniotti M, Oggionni M, Ranzani T, Vallacchi V, Campi V, Di Stasi D, Torre GD, Perrone F, Luoni C, Suardi S, Frattini M, Pilotti S, Anichini A, Tragni G, Parmiani G, Pierotti MA, Rodolfo M. BRAF alterations are associated with complex mutational profiles in malignant melanoma. Oncogene. 2004;23(35):5968–5977. doi: 10.1038/sj.onc.1207780. [DOI] [PubMed] [Google Scholar]
- De Fabo EC, Noonan FP, Fears T, Merlino G. Ultraviolet B but not ultraviolet A radiation initiates melanoma. Cancer Res. 2004;64(18):6372–6376. doi: 10.1158/0008-5472.CAN-04-1454. [DOI] [PubMed] [Google Scholar]
- Dutton-Regester K, Aoude LG, Nancarrow DJ, Stark MS, O’Connor L, Lanagan C, Pupo GM, Tembe V, Carter CD, O’Rourke M, Scolyer RA, Mann GJ, Schmidt CW, Herington A, Hayward NK. Identification of TFG (TRK-fused gene) as a putative metastatic melanoma tumor suppressor gene. Genes Chromosomes Cancer. 2012;51(5):452–461. doi: 10.1002/gcc.21932. [DOI] [PubMed] [Google Scholar]
- Filatov L, Golubovskaya V, Hurt JC, Byrd LL, Phillips JM, Kaufmann WK. Chromosomal instability is correlated with telomere erosion and inactivation of G2 checkpoint function in human fibroblasts expressing human papillomavirus type 16 E6 oncoprotein. Oncogene. 1998;16(14):1825–1838. doi: 10.1038/sj.onc.1201711. [DOI] [PubMed] [Google Scholar]
- Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer. 2012;12(2):104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddameedhi S, Kemp MG, Reardon JT, Shields JM, Smith-Roe SL, Kaufmann WK, Sancar A. Similar nucleotide excision repair capacity in melanocytes and melanoma cells. Cancer Res. 2010;70(12):4922–4930. doi: 10.1158/0008-5472.CAN-10-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrest BA, Eller MS, Geller AC, Yaar M. The pathogenesis of melanoma induced by ultraviolet radiation. N Engl J Med. 1999;340(17):1341–1348. doi: 10.1056/NEJM199904293401707. [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Haferkamp S, Scurr LL, Becker TM, Frausto M, Kefford RF, Rizos H. Oncogene-induced senescence does not require the p16(INK4a) or p14ARF melanoma tumor suppressors. J Invest Dermatol. 2009a;129(8):1983–1991. doi: 10.1038/jid.2009.5. [DOI] [PubMed] [Google Scholar]
- Haferkamp S, Tran SL, Becker TM, Scurr LL, Kefford RF, Rizos H. The relative contributions of the p53 and pRb pathways in oncogene-induced melanocyte senescence. Aging (Albany NY) 2009b;1(6):542–556. doi: 10.18632/aging.100051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319(5868):1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Haqq C, Nosrati M, Sudilovsky D, Crothers J, Khodabakhsh D, Pulliam BL, Federman S, Miller JR, 3rd, Allen RE, Singer MI, Leong SP, Ljung BM, Sagebiel RW, Kashani-Sabet M. The gene expression signatures of melanoma progression. Proc Natl Acad Sci U S A. 2005;102(17):6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashash N, Johnson AL, Cha RS. Topoisomerase II- and condensin-dependent breakage of MEC1ATR-sensitive fragile sites occurs independently of spindle tension, anaphase, or cytokinesis. PLoS Genet. 2012;8(10):e1002978. doi: 10.1371/journal.pgen.1002978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao A, Cheung A, Duncan G, Girard PM, Elia AJ, Wakeham A, Okada H, Sarkissian T, Wong JA, Sakai T, De Stanchina E, Bristow RG, Suda T, Lowe SW, Jeggo PA, Elledge SJ, Mak TW. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol Cell Biol. 2002;22(18):6521–6532. doi: 10.1128/MCB.22.18.6521-6532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287(5459):1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- Hirsch D, Kemmerling R, Davis S, Camps J, Meltzer PS, Ried T, Gaiser T. Chromothripsis and focal copy number alterations determine poor outcome in malignant melanoma. Cancer Res. 2013;73(5):1454–1460. doi: 10.1158/0008-5472.CAN-12-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, Weber BL, Nathanson KL, Phillips DJ, Herlyn M, Schadendorf D, Dummer R. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19(4):290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- Jerant AF, Johnson JT, Sheridan CD, Caffrey TJ. Early detection and treatment of skin cancer. Am Fam Physician. 2000;62(2):357–368. 375–356, 381–352. [PubMed] [Google Scholar]
- Jonsson G, Dahl C, Staaf J, Sandberg T, Bendahl PO, Ringner M, Guldberg P, Borg A. Genomic profiling of malignant melanoma using tiling-resolution arrayCGH. Oncogene. 2007;26(32):4738–4748. doi: 10.1038/sj.onc.1210252. [DOI] [PubMed] [Google Scholar]
- Kauffmann A, Rosselli F, Lazar V, Winnepenninckx V, Mansuet-Lupo A, Dessen P, van den Oord JJ, Spatz A, Sarasin A. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27(5):565–573. doi: 10.1038/sj.onc.1210700. [DOI] [PubMed] [Google Scholar]
- Kaufmann WK. The human intra-S checkpoint response to UVC-induced DNA damage. Carcinogenesis. 2010;31(5):751–765. doi: 10.1093/carcin/bgp230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WK, Nevis KR, Qu P, Ibrahim JG, Zhou T, Zhou Y, Simpson DA, Helms-Deaton J, Cordeiro-Stone M, Moore DT, Thomas NE, Hao H, Liu Z, Shields JM, Scott GA, Sharpless NE. Defective cell cycle checkpoint functions in melanoma are associated with altered patterns of gene expression. J Invest Dermatol. 2008;128(1):175–187. doi: 10.1038/sj.jid.5700935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WK, Schwartz JL, Hurt JC, Byrd LL, Galloway DA, Levedakou E, Paules RS. Inactivation of G2 checkpoint function and chromosomal destabilization are linked in human fibroblasts expressing human papillomavirus type 16 E6. Cell Growth Differ. 1997;8(10):1105–1114. [PubMed] [Google Scholar]
- Kaufmann WK, Wilson SJ. G1 arrest and cell-cycle-dependent clastogenesis in UV-irradiated human fibroblasts. Mutat Res. 1994;314(1):67–76. doi: 10.1016/0921-8777(94)90062-0. [DOI] [PubMed] [Google Scholar]
- Kraemer KH, Lee MM, Andrews AD, Lambert WC. The role of sunlight and DNA repair in melanoma and nonmelanoma skin cancer. The xeroderma pigmentosum paradigm. Arch Dermatol. 1994;130(8):1018–1021. [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133(6):1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, Laine E, Greulich H, Tseng H, Gates C, Hodi FS, Dranoff G, Sellers WR, Thomas RK, Meyerson M, Golub TR, Dummer R, Herlyn M, Getz G, Garraway LA. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68(3):664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc Natl Acad Sci U S A. 2006;103(26):9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Miracco C, Pacenti L, Santopietro R, Laurini L, Biagioli M, Luzi P. Evaluation of telomerase activity in cutaneous melanocytic proliferations. Hum Pathol. 2000;31(9):1018–1021. doi: 10.1053/hupa.2000.9779. [DOI] [PubMed] [Google Scholar]
- Muraki K, Nyhan K, Han L, Murnane JP. Mechanisms of telomere loss and their consequences for chromosome instability. Front Oncol. 2012;2:135. doi: 10.3389/fonc.2012.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaishvilli-Feinberg N, Cohen SM, Midkiff B, Zhou Y, Olorvida M, Ibrahim JG, Omolo B, Shields JM, Thomas NE, Groben PA, Kaufmann WK, Miller CR. Development of DNA damage response signaling biomarkers using automated, quantitative image analysis. J Histochem Cytochem. 2013 doi: 10.1369/0022155413516469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan FP, Zaidi MR, Wolnicka-Glubisz A, Anver MR, Bahn J, Wielgus A, Cadet J, Douki T, Mouret S, Tucker MA, Popratiloff A, Merlino G, De Fabo EC. Melanoma induction by ultraviolet A but not ultraviolet B radiation requires melanin pigment. Nat Commun. 2012;3:884. doi: 10.1038/ncomms1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omolo B, Carson C, Chu H, Zhou Y, Simpson DA, Hesse JE, Paules RS, Nyhan KC, Ibrahim JG, Kaufmann WK. A prognostic signature of G 2 checkpoint function in melanoma cell lines. Cell Cycle. 2013;12(7) doi: 10.4161/cc.24067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poynter JN, Elder JT, Fullen DR, Nair RP, Soengas MS, Johnson TM, Redman B, Thomas NE, Gruber SB. BRAF and NRAS mutations in melanoma and melanocytic nevi. Melanoma Res. 2006;16(4):267–273. doi: 10.1097/01.cmr.0000222600.73179.f3. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Sambade MJ, Peters EC, Thomas NE, Kaufmann WK, Kimple RJ, Shields JM. Melanoma cells show a heterogeneous range of sensitivity to ionizing radiation and are radiosensitized by inhibition of B-RAF with PLX-4032. Radiother Oncol. 2011 doi: 10.1016/j.radonc.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyamoorthy K, Chehab NH, Waterman MJ, Lien MC, El-Deiry WS, Herlyn M, Halazonetis TD. Aberrant regulation and function of wild-type p53 in radioresistant melanoma cells. Cell Growth Differ. 2000;11(9):467–474. [PubMed] [Google Scholar]
- Sauroja I, Smeds J, Vlaykova T, Kumar R, Talve L, Hahka-Kemppinen M, Punnonen K, Jansen CT, Hemminki K, Pyrhonen S. Analysis of G(1)/S checkpoint regulators in metastatic melanoma. Genes Chromosomes Cancer. 2000;28(4):404–414. doi: 10.1002/1098-2264(200008)28:4<404::aid-gcc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Schulten HJ, Gunawan B, Otto F, Hassmann R, Hallermann C, Noebel A, Fuzesi L. Cytogenetic characterization of complex karyotypes in seven established melanoma cell lines by multiplex fluorescence in situ hybridization and DAPI banding. Cancer Genet Cytogenet. 2002;133(2):134–141. doi: 10.1016/s0165-4608(01)00584-2. [DOI] [PubMed] [Google Scholar]
- Shields JM, Thomas NE, Cregger M, Berger AJ, Leslie M, Torrice C, Hao H, Penland S, Arbiser J, Scott G, Zhou T, Bar-Eli M, Bear JE, Der CJ, Kaufmann WK, Rimm DL, Sharpless NE. Lack of Extracellular Signal-Regulated Kinase Mitogen-Activated Protein Kinase Signaling Shows a New Type of Melanoma. Cancer Res. 2007;67(4):1502–1512. doi: 10.1158/0008-5472.CAN-06-3311. [DOI] [PubMed] [Google Scholar]
- Simpson DA, Livanos E, Heffernan TP, Kaufmann WK. Telomerase expression is sufficient for chromosomal integrity in cells lacking p53 dependent G1 checkpoint function. J Carcinog. 2005;4:18. doi: 10.1186/1477-3163-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Roe SL, Patel SS, Zhou Y, Simpson DA, Rao S, Ibrahim JG, Cordeiro-Stone M, Kaufmann WK. Separation of intra-S checkpoint protein contributions to DNA replication fork protection and genomic stability in normal human fibroblasts. Cell Cycle. 2013;12(2):332–345. doi: 10.4161/cc.23177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo JK, Mackenzie Ross AD, Kallenberg DM, Milagre C, Heung Chong W, Chow J, Hill L, Hoare S, Collinson RS, Hossain M, Keith WN, Marais R, Bennett DC. Malignancy without immortality? Cellular immortalization as a possible late event in melanoma progression. Pigment Cell Melanoma Res. 2011;24(3):490–503. doi: 10.1111/j.1755-148X.2011.00850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suram A, Kaplunov J, Patel PL, Ruan H, Cerutti A, Boccardi V, Fumagalli M, Di Micco R, Mirani N, Gurung RL, Hande MP, d’Adda di Fagagna F, Herbig U. Oncogene-induced telomere dysfunction enforces cellular senescence in human cancer precursor lesions. Embo J. 2012;31(13):2839–2851. doi: 10.1038/emboj.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas NE, Berwick M, Cordeiro-Stone M. Could BRAF mutations in melanocytic lesions arise from DNA damage induced by ultraviolet radiation? J Invest Dermatol. 2006;126(8):1693–1696. doi: 10.1038/sj.jid.5700458. [DOI] [PubMed] [Google Scholar]
- Thomas NE, Edmiston SN, Alexander A, Millikan RC, Groben PA, Hao H, Tolbert D, Berwick M, Busam K, Begg CB, Mattingly D, Ollila DW, Tse CK, Hummer A, Lee-Taylor J, Conway K. Number of nevi and early-life ambient UV exposure are associated with BRAF-mutant melanoma. Cancer Epidemiol Biomarkers Prev. 2007;16(5):991–997. doi: 10.1158/1055-9965.EPI-06-1038. [DOI] [PubMed] [Google Scholar]
- Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol. 2004;122(2):337–341. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z, Aird KM, Bitler BG, Nicodemus JP, Beeharry N, Xia B, Yen TJ, Zhang R. Oncogenic RAS regulates BRIP1 expression to induce dissociation of BRCA1 from chromatin, inhibit DNA repair, and promote senescence. Dev Cell. 2011;21(6):1077–1091. doi: 10.1016/j.devcel.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vredeveld LC, Possik PA, Smit MA, Meissl K, Michaloglou C, Horlings HM, Ajouaou A, Kortman PC, Dankort D, McMahon M, Mooi WJ, Peeper DS. Abrogation of BRAFV600E-induced senescence by PI3K pathway activation contributes to melanomagenesis. Genes Dev. 2012;26(10):1055–1069. doi: 10.1101/gad.187252.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GJ, Flores JF, Glendening JM, Lin AH, Markl ID, Fountain JW. Virtually 100% of melanoma cell lines harbor alterations at the DNA level within CDKN2A, CDKN2B, or one of their downstream targets. Genes Chromosomes Cancer. 1998;22(2):157–163. doi: 10.1002/(sici)1098-2264(199806)22:2<157::aid-gcc11>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Walsh E, Wang X, Lee MY, Eckert KA. Mechanism of replicative DNA polymerase delta pausing and a potential role for DNA polymerase kappa in common fragile site replication. J Mol Biol. 2013;425(2):232–243. doi: 10.1016/j.jmb.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem. 2001;276(51):47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- Warters RL, Adamson PJ, Pond CD, Leachman SA. Melanoma cells express elevated levels of phosphorylated histone H2AX foci. J Invest Dermatol. 2005;124(4):807–817. doi: 10.1111/j.0022-202X.2005.23674.x. [DOI] [PubMed] [Google Scholar]
- Wasco MJ, Pu RT, Yu L, Su L, Ma L. Expression of gamma-H2AX in melanocytic lesions. Hum Pathol. 2008;39(11):1614–1620. doi: 10.1016/j.humpath.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR, Avril MF, Ortiz Romero PL, Robert T, Balacescu O, Eggermont AM, Lenoir G, Sarasin A, Tursz T, van den Oord JJ, Spatz A. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98(7):472–482. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- Yeh I, von Deimling A, Bastian BC. Clonal BRAF Mutations in Melanocytic Nevi and Initiating Role of BRAF in Melanocytic Neoplasia. J Natl Cancer Inst. 2013;105(12):917–919. doi: 10.1093/jnci/djt119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, McDaid R, Lee J, Possik P, Li L, Kumar SM, Elder DE, Van Belle P, Gimotty P, Guerra M, Hammond R, Nathanson KL, Dalla Palma M, Herlyn M, Xu X. The role of BRAF mutation and p53 inactivation during transformation of a subpopulation of primary human melanocytes. Am J Pathol. 2009;174(6):2367–2377. doi: 10.2353/ajpath.2009.081057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.