Abstract
The explosion of genomic, transcriptomic, proteomic, metabolomic, and other omics data is challenging the research community to develop rational models for their organization and interpretation to generate novel biological knowledge. The development and use of gene regulatory networks to mechanistically interpret this data is an important development in molecular biology, usually captured under the banner of systems biology. As a result, the repertoire of methods for the reconstruction of comprehensive and cell-context-specific maps of regulatory interactions, or interactomes, has also exploded in the past few years. In this review, we focus on Network Biology and more specifically on methods for reverse engineering transcriptional, post-transcriptional, and post-translational human interaction networks and show how their interrogation is starting to impact our understanding of cellular pathophysiology and one’s ability to predict cellular phenotypes from genome-wide molecular observations.
INTRODUCTION
Systems Biology, a relatively young area in the biological sciences, is growing exponentially as demonstrated by the increase in the number of its related publications over the last 10 years (Figure 1). Despite numerous attempts, the field has successfully resisted pigeonholing and it has thus been difficult to capture its essence under a single, comprehensive, and broadly accepted definition. Rather, individual researchers, meetings, and specialized publications use the term in a wide and often orthogonal variety of acceptions, with flavors ranging from integrative genomics, to model-based biology, to various combinations of high-throughput experimental and computational biology, just to cite a few.
FIGURE 1.
The number of PubMed publications including the term ‘systems biology’ in their title or abstract, since 1999 (2011 data extrapolated from publications from January to September).
Fortunately, lack of a unifying definition has not affected the field, which is growing robustly as the sum of these heterogeneous and more narrowly defined areas. One area in particular, however, is capturing the bulk of work in the discipline with the ultimate objective of reconstructing (or reverse-engineering) accurate models of gene regulation and of interrogating them to elucidate both physiological and pathological mechanisms. As gene regulatory models are often depicted as graphical networks of molecular interactions, with nodes representing individual gene-products and arcs their interactions, this domain of investigation has become best known as Network Biology and has come, perhaps, to constitute the most eidetic and representative subfield of Systems Biology. In this article, we concentrate on Network Biology to provide a few tangible and illustrative examples of how reconstruction, modeling, and interrogation of regulatory molecular interaction networks, or interactomes, is starting to impact our understanding of cellular pathophysiology and our ability to predict cellular phenotypes from genome-wide molecular observables.
Early network biology approaches have been successfully applied to the study of a number of prokaryotic and lower eukaryotic model systems1–8 as well as a few higher eukaryotic model organisms9–11. While understanding these model organisms continues to enrich our knowledgebase, we are entering a stage in the natural progression of biology where, to paraphrase Sydney Brenner,12 ‘humans are the new model organism.’ As a result, we will try whenever possible to highlight the impact of this emergent discipline on the study of human physiology and human disease, referring to progress in model organisms mostly on an historical basis.
The genome-wide molecular profile resources from large-scale studies in humans have grown dramatically in the last few years, thanks to the systematic efforts by the research community and international funding agencies, such as the International Human Genome Sequencing Consortium,13 The Cancer Genome Atlas (TCGA) Research Network,14 dbGaP,15 and the International Network of Cancer Genome Projects.16 Adding to this is the increased availability of a variety of new high-throughput profiling technologies including Next-Gen sequencing, robotic-based perturbation and profiling of cellular systems, high-throughput tandem mass spectrometry, and high-throughput single cell imaging, just to name a few. These studies have provided us with wonderful lists of the molecular componentry that determine cellular function and behavior. Yet none of the studies has provided us with the systematic understanding of how these parts may interact together to allow behavior and function to emerge. To use a simple metaphor, if one compared the cell to an automobile, we would now know many of its individual mechanical, electrical, and structural components but we would still lack the blueprints necessary to build its most critical large-scale subassemblies, such as the carburetor or the differential, let alone the full vehicle. To a large extent, network biologists are trying to reconstruct the ‘assembly manuals’ of a number of distinct cellular contexts and to use them to elucidate the molecular mechanisms underlying cell autonomous function and behavior as well as non-cell-autonomous interactions with the environment (other cells, nutrients, exogenous stimuli, etc.).
Fortunately, a number of genome-wide technologies and methodologies have recently emerged to analyze changes in the relative abundance of molecular species in the cell (RNA, proteins, lipids, metabolites, etc.) across distinct cellular contexts, as well as to directly measure the repertoire of molecular interactions in the cell, including those involved in transcriptional, post-transcriptional, post-translational, and metabolic regulation. These are finally starting to produce quite accurate maps, both functional and physical, of how biological molecules work together to implement biological functions. These maps, or interactomes, while typically impossible to interpret visually (hence the moniker ‘fuzzyballs’ used at times to describe them), provide a machine-readable representation of the complex tangle of molecular interactions in the cell that can be increasingly interrogated by computational algorithms to reveal key mechanisms of regulation presiding over physiological functions and their dysregulated counterpart in disease. We will cover several of these approaches in the next section.
Thus, we can think of network biology as the natural, integrative model-based complement of large-scale data emerging from community-based molecular profiling efforts. By allowing the assembly and interrogation of complex human cell regulatory models, using these data, we are starting to elucidate mechanisms of differentiation/maturation,17,18 neoplastic transformation/progression,19–21 disease initiation,22,23 and response to exogenous stimuli,24 including mechanisms determining sensitivity and resistance to small-molecule therapeutic agents,25 that have been experimentally validated in vitro and in vivo. More importantly, network biology tools are starting to offer us the opportunity to carry out virtual experiments that would otherwise be unethical, prohibitively expensive, or simply unfeasible in the lab or in the clinic, but that result in a prioritization of hypothesis generating predictions that could translate to testable models in tissue culture, mouse, and clinical studies.
The implications of these methodologies for the study and treatment of human disease is profound and may allow us to predict more efficiently and accurately disease genes, biomarkers, and therapeutic targets using the combined power of in silico and experimental biology. Ultimately, network biology is hypothesis generating rather than hypothesis driven, allowing the rapid generation of hundreds to thousands of testable hypotheses that can be prioritized for experimental validation. Indeed, some of the most successful practitioners of this emerging field have developed integrated lab environments where they can progress from the production of large-scale molecular profile data by experimental assays, to computational model-based analysis resulting in testable hypotheses, then to experimental validation, and finally to reintroduction of these data into the original computational models to improve their predictive capacity.
WHAT IS A GENE REGULATORY NETWORK MODEL?
What constitutes a gene regulatory model? By ‘model’ we mean a computable representation, based on empirical data that allows the inference of measurable macroscopic ‘dependent’ variables as a function of other ‘independent’ variables. A gene regulatory model is one where the dependent variables represent the output of regulatory processes in the cell, such as the expression of the target of a transcription factor (TF) or the viability of a cellular phenotype, as a function of independent variables that represent the input of the regulatory process, such as the gene expression or protein abundance of the TF, as well as small-molecule, RNAi, or environmental perturbations. Inherent to a good regulatory model is an ability to generate hypothesis that can be experimentally tested. Indeed, computational models are largely irrelevant if their predictions are strictly theoretical and cannot be tested in the biological context of interest. This emerging paradigm is at the base of the increasingly interdisciplinary nature of the field, as the successful systems biologist must increasingly understand both the computational principles necessary to generate valuable testable hypotheses and the experimental techniques necessary to validate them.
Regulatory models can have multiple incarnations. The simplest form is that of a purely topological interaction model (i.e., noncausal), typically represented as an adirected graph of interactions (e.g., physical interactions between proteins in complexes or nonphysical synthetic–lethality interactions).26–29 While this type of model cannot be used to predict causal determinants of cellular phenotypes, it constitutes a valuable tool for the statistical integration of multiple data modalities to identify gene products whose concerted activity is involved in the implementation of a specific phenotype. For instance, such models originating from yeast-2-hybrid (Y2H), complex structure determination, nuclear pull-down experiments, and computational inference algorithms have been used to infer key components of the cellular machinery involved in the presentation of both physiological and pathological phenotypes.27,30,31
In contrast, influence maps represent direct or indirect causal regulatory relationships between gene products in the cell. They are intrinsically more difficult to assemble, because causality is not easily inferred from molecular profile data, but also obviously more relevant to the elucidation of cause-effect mechanisms. ‘Influences’ do not strictly represent physical interactions (e.g., a TF binding a target) but rather causal ones, such that modulation of the first gene product produces a statistically significant change in the other(s). Interestingly, some indirect influences may be far more predictive than direct interactions. For instance, if one considers the activity of the MYC TF in a human B cell, modulation of one of its upstream transcriptional regulators may not produce significant changes in MYC transcriptional activity either because (1) the effect may be countered by the concerted activity of other MYC activators and repressors or (2) the transcriptional effect may be compensated post-transcriptionally or post-translationally through a variety of feedback-loop mechanisms. However, signals on the B-cell receptor (BCR) pathway, an indirect regulator of MYC activity, will reliably cause MYC degradation and loss of transcriptional activity for a variety of B-cell subtypes, both normal and tumor related, likely due to careful coordination of an entire complement of regulatory events.32 Thus, the inference of highly predictive influences can be as relevant as the inference of physical regulatory relationships.
Physical regulatory maps constitute the next level of regulatory model complexity. They represent strictly physical interactions that can be experimentally validated, such as a TF binding to the promoter of a target gene and activating or repressing its expression,33 a kinase phosphorylating a protein substrate at a specific residue to induce its functional activation or degradation,34 or a microRNA affecting the mRNA stability of a target by hybridizing to its 3′ untranslated region (UTR) region.35 For instance, in Figure 2, we show predictions by the ARACNe algorithm for direct physical interactions between the TF BCL6 and its transcriptional targets in B cells,36 as validated by both silencing of the TF followed by gene expression profiling (Figure 2(a)) and arraybased chromatine immunoprecipitation (ChIP-chip) (Figure 2(b)).
FIGURE 2.
Experimental validation of ARACNe-inferred targets of BCL6 by (a) shRNA-mediated silencing, followed by gene expression profiling and (b) ChIP-chip promoter analysis. Targets are sorted left to right from the most statistically significant in terms of (a) differential expression and (b) probability of binding Bcl6 in the region surrounding the gene transcription start site.36 Red bars represent ARACNe-inferred targets. Enrichment is computed by gene set enrichment analysis.37
ODE kinetic models (ordinary differential equation) such as xi = f (x1, x2, …, xN) − βxi add yet another layer of detail, allowing not only for the definition of causal interactions, but also quantitative model changes for an endogenous variable (e.g., the mRNA concentration of a gene) as a function of a number of endogenous and exogenous variables over time.38,39
Even more complicated (and thus less tractable), PDE kinetic models (partial differential equation) represent the concentration gradient of each relevant molecular species as a function of time, space, and other endogenous/exogenous variables. These models, such as reaction-diffusion models, are used, for instance, to represent molecular species that have a gradient rather than fixed concentration in space.40,41
Finally, sometimes the availability of specific molecular species is so low that it can no longer be effectively represented as a continuous concentration and must rather be accounted for as a discrete molecular population. In these cases, a variety of stochastic models have been developed, such as the Gillespie algorithm and its derivatives.42 These are among the more computational intensive and complex models used in network biology.
Regardless of the level of complexity, a key common property of regulatory models is that they are exquisitely context specific and highly complex, defying any attempt to represent them as universal and relatively linear chains of events, as for instance in canonical cancer pathway representations.43 Different cells express distinct protein isoforms, and the molecular targets of a TF critically depend on a number of cell specific variables, such as the presence of individual cofactors and signals, as well as on the organization and chemical modifications of the chromatin. This contributes to rather remarkable changes in the molecular-interaction model and substantially different behavior of different cellular contexts, when exposed to similar stimuli or perturbations.4,24,44 As a result, the traditional notion of relatively simple and linear human pathways is useful as a conceptual tool but is both unrealistic and potentially misleading. It is purely based on our need to present regulatory control in a way that can be visually and intuitively understood and easily generalized. As a result, representations based on canonical pathway depictions (see, for instance, Figure 3(a)) rarely constitute appropriate models for the computational inference and/or modeling of cell behavior and are likely to be increasingly supplanted by more complex models that can only be interpreted computationally45 (Figure 3(b)). Over the next few years, as network analysis tool become more mature, we expect to see a gradual migration away from pathway biology into network biology.
FIGURE 3.
(a) Canonical non-Smad pathway tumor growth factor (TGF)-β signaling.46 (b) Transcriptional regulatory module controlling the mesenchymal signature of high-grade glioma, computationally inferred and validated by biochemical and functional assays.19
WHY STUDY REGULATORY NETWORK MODELS?
The rationale for the use of regulatory network models as tools to dissect human pathophysiology may not be obvious. After all, the premise of Genome wide association studies (GWAS) was that sampling the full repertoire of germline variants and somatic alterations in large populations or disease families would be sufficient to identify gene–disease relationship through relatively straightforward statistical analysis. Indeed, it has been commonly accepted that if a gene is causally related to the presentation of a trait or disease, the subpopulation with the corresponding phenotype should co-segregate with some germline variants or somatic mutation of that gene. Unfortunately, while this paradigm may hold true for simple Mendelian traits, this appears not to be the case for complex traits and diseases.47–52 Rather, only a very small component of the heritability of common complex diseases is explained by GWAS-identified allelic variants.53 Similarly, somatic alterations account for only a small fraction of all cancer subtype cases. The majority of these phenotypes are either associated with extremely rare variants/alterations that are difficult to identify and validate, or show no statistical association with any genetic event.
Recent approaches suggest that the unexplained variance may be accounted for by the ability of master regulator genes, within cell regulatory networks, to integrate an entire spectrum of genetic and epigenetic variants,19,54 where, in isolation, any one variant may not be statistically significant in a GWAS analysis. Dissecting and interrogating the underlying regulatory logic of the cell is therefore becoming a critical step in elucidating the mechanisms associated with presentation of traits and diseases that are not amenable to statistical approaches.
Let us use a simple metaphor to illustrate this concept. Imagine, for instance, that someone was set on polluting a large river by dumping poison into one or more of its tributaries. A smart polluter would choose a different tributary every day, thus eluding the efforts of law enforcement agents to catch him. A more naïve polluter would instead keep dumping poison in the same place and would eventually get caught. Given a disease gene in a particular cellular context—much like the network of tributaries of a river—there are hundreds of ways to dysregulate its activity without directly altering the gene itself. For instance, one may target one of its many transcriptional, post-translational, or microRNA regulators, each one possibly in several different ways, including epigenetic silencing, loss/gain of function mutation(s), translocation, deletion, etc. If one further considers that many gene combinations with low individual effect can produce large synergistic joint effects (i.e., synthetic lethality) and that there are a predicted >200 million two-gene and >1.2 trillion three-gene combinations, the genetic means to a phenotypic end are virtually unlimited. Since nature is neither smart nor naïve but rather opportunistic, one can safely assume that given a large enough cohort, all of these potential mechanisms of dysregulation will be represented, each with a different probability, with the exception of a few that may decrease fitness or produce lethal phenotypes. Thus, while several individuals may co-segregate on some frequent genetic alterations, including those of the actual disease genes, a large number of cases (often the majority) will be represented at very low frequency in the population. Simple reasoning indicates that a large number of individuals presenting a trait or a phenotype may do so because of a unique pattern of dysregulation never exactly replicated elsewhere, thus defying any statistical based approach to its identification.
By providing a reading chart of the ‘tributaries’ of a disease gene, i.e., its upstream regulatory mechanisms and its downstream effectors, the study of cell context-specific regulatory networks is providing new insight into genes that would have otherwise gone under the radar of traditional genomic analyses. Specifically, regulatory networks provide a natural framework for the integration of all potential events contributing to the presentation of a phenotype.23,55,56 For instance, it is relatively easy to separate events that distribute randomly over a regulatory network topology from those that cluster around specific pathways or subnetworks, although the concept of a pathway within a regulatory network may not be obvious.57,58 In particular, any gene that causally regulates a known disease gene, either directly or indirectly, and is altered in individuals with the disease is likely to be involved in its etiology. For instance, consider the PTEN gene, which has an extremely tightly regulated expression and is an established tumor suppressor. Recent results implicated its non-coding pseudo-gene PTENP1 as a candidate tumor suppressor, even in the absence of any genetic alteration evidence from cancer patients. The PTENP1 3′ UTR acts as a decoy for the same microRNA program that targets the PTEN 3′ UTR,59 titrating the machinery away from PTEN, and thus derepressing PTEN and enhancing its tumor suppressor activity in certain contexts. Conversely, genomic deletions of PTENP1 were observed in human cancers and were correlated with decreased PTEN expression.
If the regulatory networks that are relevant in the context of a specific disease are reasonably well characterized, this type of approach can be used directly to reduce genome-wide resequencing efforts to a relatively small number of high-probability candidate genes, thus dramatically reducing statistical correction for multiple hypothesis testing and allowing identification of important genes that are altered only in a handful of cases. For instance, identification of an entire set of regulators upstream of BLIMP1, a gene in the BCR pathway which is necessary for B-cell exit from the germinal center, have been implicated in diffuse large B-cell lymphoma (DLBCL) using similar reasoning.54,60,61 Many of these regulators would have eluded genome-wide association studies because of the much larger number of potential loci considered in GWAS analyses.
Furthermore, regulatory-network-based approaches may dramatically increase our ability to identify valuable therapeutic targets and disease biomarkers by extending them from a small set of alterationharboring genes to a much broader set of genes that act as master integrators of a large spectrum of aberrant signals originating from upstream genetic alterations. That is, a nonmutated target or substrate of multiple oncogenes may constitute a better therapeutic target and biomarker than any of the oncogenes whose signals it integrates. For instance, we have shown that C/EBPβ and Stat3 are synergistic master regulators of the mesenchymal subtype of glioblastoma (GBM), which accounts for about 60–70% of all GBM cases. Experimental validation assays have shown that these two genes constitute optimal biomarkers and valuable genetic targets when inhibited in combination, as they abrogate tumorigenesis in vivo and can effectively discriminate between worst and best prognosis in GBM patients.19 This result could not have been produced by genetic analysis precisely because these genes are not mutated in this tumor subtype. Similarly, activated B-cell-like (ABC)-DLBCL cells are addicted to Nf-κB, which integrates signals originating from an entire spectrum of upstream genetic alterations, even though it is not itself altered.54 Thus, targeted therapies for GBM and ABC-DLBCL may emerge, which, rather than focusing on oncogenes, target nononcogene dependencies of the tumor cell. There is an increasing wealth of similar examples that span a number of relevant disease areas, such as, in diabetes and obesity,62 in complex neurodegenerative disease,63 and in host–pathogen interactions.64 All these advances are predicated on network biology approaches to infer regulatory networks and interrogate them to identify genes causally related to the presentation of a pathophysiologic phenotype.
Biomarker identification constitutes a particularly interesting and useful area for the use of regulatory networks. Existing biomarkers fall roughly into two categories: they are either based on genetic alteration that have been shown to be causally related to the disease (e.g., EGFR in lung cancer or SOD1 in amyotrophic lateral sclerosis, ALS) or on genes/proteins/metabolites that have maximum differential expression in disease versus normal samples (e.g., prostate-specific antigen, PSA, in prostate cancer). The former, with the exception of rare Mendelian diseases, tends to have relatively small frequency in the population affected by the disease (i.e., <30% for EGFR and <2% for SOD165) and therefore their prognostic value is not easily interpreted. For instance, none of the major lesions in GBM appears to co-segregate with the most or least aggressive subtypes respectively.66 Biomarkers in the second class, on the other hand, are rarely stable because genes that display large expression changes tend to be further downstream in regulatory pathways and are thus more pleiotropically regulated. The direct result of these observations is that biomarker identification within a population rarely leads to clinical validation in follow-up studies. On the other hand, availability of candidate biomarkers that are causally related to the presentation of the disease phenotype, via predictive regulatory networks, may allow for much higher validation rates. For instance, C/EBPβ and Stat3, which were computationally inferred and experimentally validated as master regulators of the mesenchymal subtype of GBM, were validated as strong predictors of patient outcome in follow-up studies, using a new cohort of 62 GBM patients.19
Elucidation of these integrative regulatory layers may also provide the basis to understand disease and therapeutic response heterogeneity, both within an individual and across populations. This understanding is critically required to decrease failure rates of clinical trials, which often result because pharmacological effects are diluted by nonresponders that could not be identified on a quantitative basis or because of toxicity and serious adverse events that could not be predicted given the genetic and epigenetic makeup of the individual study subject.
We suggest that the limiting factors in achieving such a vision of predictive medicine are precisely related to the lack of accuracy and poor specificity of our current regulatory network models and to our still limited ability to interrogate them to elucidate key biological mechanisms. As such, this review attempts to cover our current progress in the reverse engineering and interrogation of regulatory network models.
REVERSE ENGINEERING OF REGULATORY NETWORK MODELS
For the purpose of this discussion, we concentrate specifically on relatively poorly characterized molecular-interaction networks presiding over regulation of gene-product abundance and activity. As a result, we do not cover metabolic networks, which, while extremely relevant to the study of traits and disease, have been the subject of intense and long investigation, across a number of organisms, and are relatively well covered in reviews, see for instance, Refs 67–70.
Regulatory networks presiding over gene products include at least three distinct layers: transcriptional, post-transcriptional, and post-translational regulation. Transcriptional interactions determine regulation of mRNA transcription by DNA-binding proteins called TFs, their cofactors, and their modulators, which may or may not bind DNA directly. Post-transcriptional interactions regulate mRNA stability and translation, via microRNA and other noncoding RNAs. Finally, post-translational interactions include both transient protein–protein interactions (PPIs) involved in signal transduction and stable PPIs involved in stable molecular complex formation, such as those in ribosomal subunits. Recently, another nonphysical layer has become quite valuable to the study of synergistic trait regulation. This includes synthetic–lethal and synthetic function interactions.26 Regulatory networks, across all layers but especially the transcriptional layer, are exquisitely context specific and thus a universal model cannot be effectively represented without introducing additional highly complex layers of context-specificity logic. As a result, if a regulatory model is required to elucidate a specific molecular mechanism, it should be assembled within the cellular context of interest or a closely related one.
Current State and Future Perspectives
Unfortunately, existing regulatory network models for higher eukaryotes are largely incomplete, lack context specificity, and, with few exceptions,24 address only individual molecular-interaction layers: generally either the transcriptional interaction71 or the PPI layer.28 Indeed, the vast majority of these models are assembled from the literature or from ex vivo data, such as Y2H studies, and are thus both biased and not specific to the cellular context of interest. Not surprisingly, there are only a handful of examples where unbiased computational interrogation of these models has led to the elucidation of novel biological mechanisms that were experimentally validated. Similarly, paracrine and endocrine regulatory processes spanning multiple cell types, such as those driven by stroma-tumor,72 gutbone, 73 and glia-motor neuron74 interactions, are virtually unmapped at a genome-wide level. Finally, several potentially informative data modalities are still poorly integrated into efforts to dissect molecular interactions. For instance, data on structure-based specificity of protein–DNA interactions and PPIs have not been systematically integrated with functional data to reverse-engineer regulatory networks for higher eukaryotes.
Yet, in the few examples where a systematic effort has been made to dissect context-specific networks, the models that have emerged have demonstrated significant value in the elucidation of cellular function and of its dysregulation in disease, see for instance, Refs 20, 71, 75–77. Additionally, integrative genetical-genomics models that use genetics to inform causality in regulatory models have been successfully used to elucidate determinants of mammalian traits, which have been experimentally validated.62 In these models, genetic variability is used as a perturbation that can help elucidate both the nature and the directionality of underlying regulatory interactions. For instance, if high expression of gene A co-segregates with functional inactivation of gene B by mutation or deletion, this may suggest that B is a repressor of A, assuming that expression changes are less likely to induce genetic alteration events than the opposite.
Yet, this area of investigation is just in its infancy and significant improvements are necessary before these tools and methodologies may be routinely used by biologists for the elucidation of physiological and pathological mechanisms. In the following paragraphs we explore the general principles associated with the dissection of individual layers of gene-product regulation and with their integration in a complete, hybrid regulatory model.
General Principles of Reverse Engineering
Before engaging in a reverse-engineering project, one should be aware of a few general principles that are, to a large extent, independent of the specific algorithm or regulatory layer of interest. For instance, reverse-engineering methods typically rely on indirect rather than direct evidence of physical interactions. Indeed, while direct protein–DNA or protein–protein binding data (as available from chromatin-immunoprecipitation or Y2H assays, for instance) may be taken as evidence of physical interaction, they do not necessarily imply functional regulation nor do they capture the functional properties of the underlying process. For instance, protein binding to a specific regulatory DNA region or to a cognate protein domain in these ex vivo experiments does not guarantee that the protein is involved in the regulation of those targets. Indeed, abrogation of regulation following mutation of the DNA- or protein-binding site is necessary to support claims of direct regulation. This is not generally achievable in a high-throughput fashion.
In the absence of direct regulatory evidence, a relationship such as ‘TF regulates target’ (TF → t) can only be inferred if the endogenous changes in the independent variable (e.g., the TF’s protein concentration) are relatively large, compared to experimental and measurement noise, across a relatively large number of observations, such that their functional dependency may be assessed. A key requirement of computational reverse engineering is thus the availability of multiple measurements, sampling a significant range of concentrations of the relevant moieties (e.g., mRNA abundance, protein concentration). The minimum number of observations (i.e., gene expression profiles) and the dynamic range of the independent variables (e.g., TF expression range) depend on the measurement error, on the total amount of information transmitted from each independent to each dependent variable, on the number of other independent variables affecting the state of a dependent variable, on the saturated versus linear kinetics of the interaction, and on the specific algorithm used for the analysis, among others.
Given a cellular system of interest, there are basically three distinct approaches to ensure that key gene products have sufficient dynamic range, leading to appropriate sampling of the variable space. First, one may use the natural (physiologic or pathologic) endogenous variability of samples obtained from organisms characterized by a distinct genotypic makeup32,33; second, one may perturb individual gene products using ectopic silencing or expression of individual genes; finally, one may use small-molecule or environmental perturbations, which typically affect a large number of target and off-target molecular species in the cell.78 Key issues related to each approach include nature of genotypic differences, strength of the perturbation, and time point for assay measurements following the perturbation. In general, however, the more orthogonal and nonlocal (i.e., network wide) are the effects, the more effectively this can lead to successful reverse engineering of the underlying regulatory logic. In general, it should not be surprising that methods that sample the naturally occurring phenotypic variability of the cell will be better suited at dissecting physiological interactions while those using perturbations will be more effective in dissecting interactions supporting the response to nonphysiological stimuli.
Another critical issue is that of the specific cellular population to sample. An often forgotten fact is that regulatory networks are exquisitely cell-context dependent.4,24,32 As a result, databases and resources aimed at representing interactions in multiple cellular contexts are useful to provide a space of interactions that may exist but do not necessarily exist in a specific context. In general, our results suggest that interrogation of generic (i.e., non-context-specific) molecular-interaction networks and pathways is not helpful in the elucidation of key determinants of human cellular phenotype. As a result, only regulatory networks that are specifically assembled for a given cellular context can be effectively interrogated.
A final point, which should be made for completeness, is that reverse engineering is still fairly dependent on the availability of genomewide profiles of gene expression. This has important implications in two areas and suggests current limitations and applicability of available reverse-engineering approaches. First, the relationship between mRNA xmRNA and protein concentration xp, for a given gene x, in a feedback-loop rich system, is often a nonmonotonic relationship during dynamic transients (i.e., when dxmRNA/dt ≠ 0). However, it becomes generally monotonic near equilibrium (i.e., when dxmRNA/dt ≈ 0). As a result, even though the values may be completely different, at or close to equilibrium, a gene’s mRNA constitutes a reasonably good proxy for its protein concentration for reverse-engineering purposes, unless the protein is extensively post-transcriptionally and post-translationally modulated. For instance, since mutual information is invariant under monotonic transformation of each variable, the mutual information between a TF’s protein and a target mRNA would be identical to that between the TF’s mRNA and the mRNA of the target. This is a reason for the unexpected success of information-theoretic methods in reverse-engineering systems that are relatively close to equilibrium, such as cancer cells. Unfortunately, this approximation is completely violated if the system is operating far from equilibrium, such as in diabetes, obesity, and other complex metabolic diseases. Indeed, while cancer can be thought as a cellular system operating close to equilibrium (other than for a small subset of cell-cycle-related events, which are fast compared to the kinetic constants), metabolic diseases often underlie the inability of the cell to cope with rapid changes in metabolites, requiring understanding of regulatory network models operating in a transient regime, far from equilibrium.
Transcriptional Regulation
Reverse engineering of transcriptional networks is perhaps the most well transited area in systems biology, with early contributions in the first few years of the 2000s. As for most regulatory networks, reverse engineering of transcriptional interactions was attempted first in yeast and in prokaryotes,79,80 with relatively good results, based on validation of a few predicted interactions. However, not until 2005 have reverse-engineering algorithms been developed and validated for the dissection of transcriptional networks in mammalian cells.33 Until now, while hundreds of algorithms for the reverse engineering of transcriptional networks have been proposed, only a handful have been experimentally validated. Thus, rather than discussing the value of each one, we may consider broad categories. Yet, from the efforts of the dialogue for reverse engineering assessments and methods (DREAM) community,81 aimed at creating objective experimental and synthetic benchmarks for the comparison of reverse-engineering algorithm performance, one thing is clear, i.e., different algorithms may produce optimal results dependent on a variety of conditions, including experimental noise and experimental variability of the data. There are basically five approach categories: (1) use of DNA-binding information from experimental assays of DNA-binding profile analysis,82 (2) optimization-based approaches, which search for the regulatory network that is most likely to explain the observed data,1 (3) regression-based approaches, which attempt to evaluate the kinetic or simplified-kinetics (e.g., linear models) parameters of an interaction network by fitting the model to the available data,6 (4) probabilistic and information-theoretic approaches,33,83,84 and (5) integrative approaches, which use a variety of (partially) independent clues to infer the overall probability of a specific interaction model.17 Of these, only the latter two categories have been extensively validated in mammalian cells by using an ‘opportunistic’ approach where the validation focuses on the relevant components of the network that are the most phenotypically important in a given context.19
While the first approach (1) was very popular until recently, it is clear that, with a few exceptions, such as NOTCH1 binding sites in T cells,85 binding data alone provides virtually no information about context-specific regulatory events. Additionally, through genome-wide assays (ChIP-chip and ChIP-Seq) overlap between experimental binding sites and canonical DNA-binding profiles is partial at best, with the canonical binding profile typically accounting for 20–40% of the experimentally ascertained sites.36 Indeed, use of DNA-binding profiles is being increasingly combined with other clues, from (4) in particular, to identify genes that are both bound and functionally regulated by a TF.17 Unfortunately, such an approach can only be implemented on a TF by TF basis and is thus mostly useful for validation purposes, following genome-wide analysis.
It should be noted that, depending on the problem at hand, each category may provide specific advantages and disadvantages. For instance, if the regulatory model is small, including only a handful of TFs, or if the perturbations are very local in nature, then optimization and regression approaches are very effective. Similarly, for genome-wide regulatory networks, where the variable space is reasonably well sampled, probabilistic and information-theoretic methods appear to have an edge.
Post-Translational Regulation
While recent advances in the reverse engineering of transcriptional networks in mammals33,86,87 have started to unravel their remarkable complexity, very little progress has been possible in the reverse engineering of signal transduction networks, specifically with respect to specific post-translational modification events, such as phosphorylation, acetylation, and ubiquitination.88
Interestingly, the large-scale reprogramming of the cell’s transcriptional logic, as a function of post-translational regulation, was studied in yeast.4,89 Yet, the repertoire of proteins that effect these events have never been fully dissected, especially within specific cellular contexts. Indeed, compared to tools such as ChIP-chip or reverse-engineering algorithms for the analysis of transcriptional networks,33,90 only a couple of experimentally validated algorithms exist for the dissection of signaling networks in a mammalian context. The first, NetWorkin, was used to infer substrates of 73 kinases based on phosphosite sequence information.34 The second, MINDy, was used to infer multivariate interactions where a modulator gene, M, affects the ability of a TF to regulate its targets [ti].32 Interestingly, the two algorithms were combined to provide the first genome-wide map of the regulatory interactions between signaling protein and TFs in human B cells.45 The map was shown to be highly predictive. For instance, its interrogation using a gene expression signature induced by the shRNA-mediated silencing of the kinase STK38 could infer STK38 as the 5th most likely silenced gene, out of 772 signaling genes. MINDy predictions were extensively validated,32 for instance, new modulators of MYC inferred and experimentally validated include the STK38 kinase, HDAC1 histone deacetylase, and TFs MEF2B and BHLHE40.
Post-Transcriptional Regulation
This layer represents the regulatory activity of short (16–21 bp) RNA molecules, called mature microRNAs, which originate from the highly specific, DICER-mediated cleaving of short RNA hairpin structures, resulting from the DROSHA-mediated processing of longer precursor RNA molecules. Such a regulatory activity is supported by RISC-complex-mediated partial hybridization of the mature microRNA with complementary sites in the 3′ UTR region of messenger RNA encoding specific gene transcripts. Such a process, which leads to both reduction in transcript translation efficiency and transcript degradation, ultimately reduces the concentration of the corresponding protein isoforms.
The determination of miRNA targets is still in its infancy, with different methods producing wildly different results. microRNA targets have been mostly inferred based on sequence information, either alone91,92 or in combination with phylogenetic conservation.93 Additionally, as most miRNA targets are only moderately affected at the RNA level, many reverse-engineering algorithms that rely on available gene expression data could not be directly extended to the inference of miRNA targets. Indeed, established algorithms such as TargetScan,92 miRanda,91 rna22,94 and PicTar93 produce minimally overlapping miRNA target predictions.
For instance, in Figure 4 we show the overlap of miRanda, TargetScan, and PicTar target predictions for two miRNA (mir-15a and mir-16-1) that were implicated in chronic lymphocytic leukemia (CLL) tumorigenesis.95 Not only is the overlap between algorithm predictions minimal, but the size of the miRNA regulon varies widely. Among these algorithms, PicTar appears to be the one producing the best predictions. However, its utilization is limited by the requirements that microRNA binding sites in the 3′ UTR of target genes must be highly conserved across vertebrate organisms for the algorithm to predict them effectively. We expect that evidence integration approaches5,17 will become increasingly useful in the context of microRNA target prediction as several, seemingly independent clues, both from sequence and expression data, may contribute to their identification.
FIGURE 4.
Overlap of mir-15a/16-1 target predictions using three distinct algorithms.
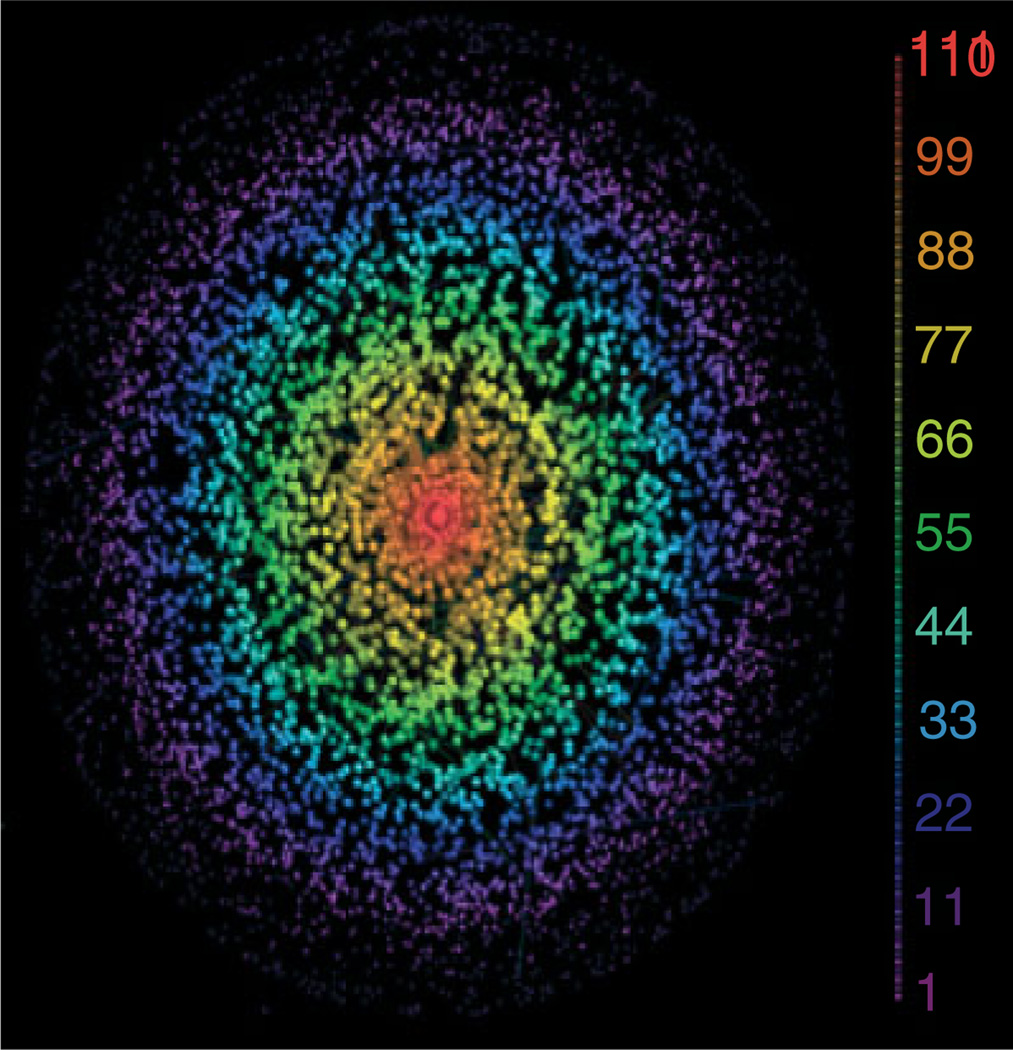
As our knowledge on non-coding RNAs evolves, we expect to discover and infer more post-transcriptionally mediated interactions, as illustrated for instance by the recent analysis of miR-mediated interactions regulating thousands of genes in GBM.96 The Hermes algorithm, derived from MINDy, was used for the systematic inference of genes that can modulate the activity of a microRNA (miR) on its targets. This led to the discovery of ~250,000 RNA–RNA interactions among ~7000 genes, the miR program-mediated regulatory (mPR) network. These interactions include both sponge-mediated ones, where two RNAs would be regulated by the same large set of miRs, such that one can titrate the miR molecules that would affect the other,59 as well as non-sponge interactions where the miR activity modulation is effected by a protein (Figure 5).
FIGURE 5.
Network of miRNA program-mediated RNA–RNA interactions in glioblastoma. Each node is an RNA with a color and a size describing its connectivity. Nodes near the center of the graph are contained within more tightly regulated, dense subgraphs, with the densest subgraph shown in red.
Protein–Protein Interactions in Stable Complexes
Virtually every process in the cell is performed by relatively stable macromolecular complexes that are themselves regulated by transient PPIs. Human disease can ensue when the interaction properties of these complexes are altered and the resulting network topology is disrupted.
The most widely used experimental means to generate large-scale identification of PPIs in human cells has been yeast 2 hybrid (Y2H) technology and tandem affinity purification coupled with mass spectrometry (TAP-MS), see reviews.97,98 These ex vivo experimental methods tend to have high false positive and false negative rates and are unlikely to generalize to all cellular contexts beyond those in which they were assessed. They are thus effective at providing an initial, albeit sparse, snapshot of PPI networks. Recent studies are starting to provide conceptual frameworks to interpret phenotypic outcomes as a function of protein–protein network dysregulation.99 Moreover, methods have been developed for the integration of protein subnetworks with other data such as genomewide linkage and association studies. For example, systematic studies have provided a draft of protein complexes associated with specific human pathologies. Proteins were ranked by the phenotype similarity score of their associated diseases and of those of their direct network neighbors.57 Another approach integrated a large-scale human PPI network and a set of genes linked to ataxia to demonstrate a potential gain in statistical power.100
REFERENCES
- 1.Friedman N. Inferring cellular networks using probabilistic graphical models. Science. 2004;303:799–805. doi: 10.1126/science.1094068. [DOI] [PubMed] [Google Scholar]
- 2.Pe’er D, Regev A, Elidan G, Friedman N. Inferring subnetworks from perturbed expression profiles. Bioinformatics. 2001;17(suppl 1):S215–S224. doi: 10.1093/bioinformatics/17.suppl_1.s215. [DOI] [PubMed] [Google Scholar]
- 3.Fiedler D, Braberg H, Mehta M, Chechik G, Cagney G, Mukherjee P, Silva AC, Shales M, Collins SR, van Wageningen S, et al. Functional organization of the S. cerevisiae phosphorylation network. Cell. 2009;136:952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- 5.Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, Chung S, Emili A, Snyder M, Greenblatt JF, Gerstein M. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 6.Yeung MK, Tegner J, Collins JJ. Reverse engineering gene networks using singular value decomposition and robust regression. Proc Natl Acad Sci U S A. 2002;99:6163–6168. doi: 10.1073/pnas.092576199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 8.Bussemaker HJ, Li H, Siggia ED. Regulatory element detection using correlation with expression. Nat Genet. 2001;27:167–171. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- 9.Yuh CH, Bolouri H, Davidson EH. Genomic cisregulatory logic: experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- 10.Tegner J, Yeung MK, Hasty J, Collins JJ. Reverse engineering gene networks: integrating genetic perturbations with dynamical modeling. Proc Natl Acad Sci U S A. 2003;100:5944–5949. doi: 10.1073/pnas.0933416100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J, Lum PY, Lamb J, GuhaThakurta D, Edwards SW, Thieringer R, Berger JP, Wu MS, Thompson J, Sachs AB, et al. An integrative genomics approach to the reconstruction of gene networks in segregating populations. Cytogenet Genome Res. 2004;105:363–374. doi: 10.1159/000078209. [DOI] [PubMed] [Google Scholar]
- 12.Friedberg EC. An interview with Sydney Brenner. Nat Rev Mol Cell Biol. 2008;9:8–9. doi: 10.1038/nrm2320. [DOI] [PubMed] [Google Scholar]
- 13.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 14.TCGA-Consortium. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International Cancer Genome Consortium. Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabe RR, Bhan MK, Calvo F, Eerola I, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lefebvre C, Rajbhandari P, Alvarez MJ, Bandaru P, Lim WK, Sato M, Wang K, Sumazin P, Kustagi M, Bisikirska BC, et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol Syst Biol. 2010;6:377. doi: 10.1038/msb.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, D’Arca D, Lim WK, Brahmachary M, Carro MS, Ludwig T, Cardo CC, Guillemot F, Aldape K, Califano A, et al. The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev Cell. 2009;17:210–221. doi: 10.1016/j.devcel.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akavia UD, Litvin O, Kim J, Sanchez-Garcia F, Kotliar D, Causton HC, Pochanard P, Mozes E, Garraway LA, Pe’er D. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–1017. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagg S, Skogsberg J, Lundstrom J, Noori P, Nilsson R, Zhong H, Maleki S, Shang MM, Brinne B, Bradshaw M, et al. Multi-organ expression profiling uncovers a gene module in coronary artery disease involving transendothelial migration of leukocytes and LIM domain binding 2: the Stockholm Atherosclerosis Gene Expression (STAGE) study. PLoS Genet. 2009;5:e1000754. doi: 10.1371/journal.pgen.1000754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, Torosyan G, Majid S, Falkard B, Kleinhanz RR, et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 2009;41:415–423. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mani KM, Lefebvre C, Wang K, Lim WK, Basso K, Dalla-Favera R, Califano A. A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Mol Syst Biol. 2008;4:169. doi: 10.1038/msb.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, Sulis ML, Barnes K, Sawai C, Homminga I, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15:50–58. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bandyopadhyay S, Chiang CY, Srivastava J, Gersten M, White S, Bell R, Kurschner C, Martin CH, Smoot M, Sahasrabudhe S, et al. A human MAP kinase interactome. Nat Methods. 2010;7:801–805. doi: 10.1038/nmeth.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteomescale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 29.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong Q, Simonis N, Li QR, Charloteaux B, Heuze F, Klitgord N, Tam S, Yu H, Venkatesan K, Mou D, et al. Edgetic perturbation models of human inherited disorders. Mol Syst Biol. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang D, Lee IY, Yoo H, Gehlenborg N, Cho JH, Petritis B, Baxter D, Pitstick R, Young R, Spicer D, et al. A systems approach to prion disease. Mol Syst Biol. 2009;5:252. doi: 10.1038/msb.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang K, Saito M, Bisikirska BC, Alvarez MJ, Lim WK, Rajbhandari P, Shen Q, Nemenman I, Basso K, Margolin AA, et al. Genome-wide identification of post-translational modulators of transcription factor activity in human B cells. Nat Biotechnol. 2009;27:829–839. doi: 10.1038/nbt.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 34.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 36.Basso K, Saito M, Sumazin P, Margolin AA, Wang K, Lim WK, Kitagawa Y, Schneider C, Alvarez MJ, Califano A, et al. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood. 2010;115:975–984. doi: 10.1182/blood-2009-06-227017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polynikis A, Hogan SJ, di Bernardo M. Comparing different ODE modelling approaches for gene regulatory networks. J Theor Biol. 2009;261:511–530. doi: 10.1016/j.jtbi.2009.07.040. [DOI] [PubMed] [Google Scholar]
- 39.Aswani A, Keranen SV, Brown J, Fowlkes CC, Knowles DW, Biggin MD, Bickel P, Tomlin CJ. Nonparametric identification of regulatory interactions from spatial and temporal gene expression data. BMC Bioinformatics. 2010;11:413. doi: 10.1186/1471-2105-11-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ballaro B, Reas PG, Riccardi R. Mathematical models for excitable systems in biology and medicine. Riv Biol. 2007;100:247–266. [PubMed] [Google Scholar]
- 41.Newman SA, Christley S, Glimm T, Hentschel HG, Kazmierczak B, Zhang YT, Zhu J, Alber M. Multiscale models for vertebrate limb development. Curr Top Dev Biol. 2008;81:311–340. doi: 10.1016/S0070-2153(07)81011-8. [DOI] [PubMed] [Google Scholar]
- 42.Gillespie DT. Stochastic simulation of chemical kinetics. Annu Rev Phys Chem. 2007;58:35–55. doi: 10.1146/annurev.physchem.58.032806.104637. [DOI] [PubMed] [Google Scholar]
- 43.Menashe I, Maeder D, Garcia-Closas M, Figueroa JD, Bhattacharjee S, Rotunno M, Kraft P, Hunter DJ, Chanock SJ, Rosenberg PS, et al. Pathway analysis of breast cancer genome-wide association study highlights three pathways and one canonical signaling cascade. Cancer Res. 2010;70:4453–4459. doi: 10.1158/0008-5472.CAN-09-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bandyopadhyay S, Mehta M, Kuo D, Sung MK, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, et al. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang K, Alvarez MJ, Bisikirska BC, Linding R, Basso K, Dalla Favera R, Califano A. Dissecting the interface between signaling and transcriptional regulation in human B cells. Pac Symp Biocomput. 2009:264–275. [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kraft P, Hunter DJ. Genetic risk prediction–are we there yet? N Engl J Med. 2009;360:1701–1703. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- 48.Hardy J, Singleton A. Genomewide association studies and human disease. N Engl JMed. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 50.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, Tuomi T, Berglund G, Altshuler D, Nilsson P, Groop L. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl JMed. 2008;359:2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 52.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 54.Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, et al. Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong H, Yang X, Kaplan LM, Molony C, Schadt EE. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am J Hum Genet. 2010;86:581–591. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong H, Beaulaurier J, Lum PY, Molony C, Yang X, Macneil DJ, Weingarth DT, Zhang B, Greenawalt D, Dobrin R, et al. Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet. 2010;6:e1000932. doi: 10.1371/journal.pgen.1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lage K, Karlberg EO, Storling ZM, Olason PI, Pedersen AG, Rigina O, Hinsby AM, Tumer Z, Pociot F, Tommerup N, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 58.Jia P, Zheng S, Long J, Zheng W, Zhao Z. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011;27:95–102. doi: 10.1093/bioinformatics/btq615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mandelbaum J, Bhagat G, Tang H, Mo T, Brahmachary M, Shen Q, Chadburn A, Rajewsky K, Tarakhovsky A, Pasqualucci L, et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18:568–579. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, Kohlhammer H, Xu W, Yang Y, Zhao H, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, Torosyan G, Majid S, Falkard B, Kleinhanz RR, et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 2009 doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iossifov I, Zheng T, Baron M, Gilliam TC, Rzhetsky A. Genetic-linkage mapping of complex hereditary disorders to a whole-genome molecular-interaction network. Genome Res. 2008;18:1150–1162. doi: 10.1101/gr.075622.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wijesekera LC, Leigh PN. Amyotrophic lateral sclerosis. Orphanet J Rare Dis. 2009;4:3. doi: 10.1186/1750-1172-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee DS, Park J, Kay KA, Christakis NA, Oltvai ZN, Barabasi AL. The implications of human metabolic network topology for disease comorbidity. Proc Natl Acad Sci U S A. 2008;105:9880–9885. doi: 10.1073/pnas.0802208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palsson B. Metabolic systems biology. FEBS Lett. 2009;583:3900–3904. doi: 10.1016/j.febslet.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goonewardena SN, Prevette LE, Desai AA. Metabolomics and atherosclerosis. Curr Atheroscler Rep. 2010;12:267–272. doi: 10.1007/s11883-010-0112-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nemenman I, Escola GS, Hlavacek WS, Unkefer PJ, Unkefer CJ, Wall ME. Reconstruction of metabolic networks from high-throughput metabolite profiling data: in silico analysis of red blood cell metabolism. Ann N Y Acad Sci. 2007;1115:102–115. doi: 10.1196/annals.1407.013. [DOI] [PubMed] [Google Scholar]
- 71.Ergun A, Lawrence CA, Kohanski MA, Brennan TA, Collins JJ. A network biology approach to prostate cancer. Mol Syst Biol. 2007;3:82. doi: 10.1038/msb4100125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reuter JA, Ortiz-Urda S, Kretz M, Garcia J, Scholl FA, Pasmooij AM, Cassarino D, Chang HY, Khavari PA. Modeling inducible human tissue neoplasia identifies an extracellular matrix interaction network involved in cancer progression. Cancer Cell. 2009;15:477–488. doi: 10.1016/j.ccr.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yadav VK, Oury F, Suda N, Liu ZW, Gao XB, Confavreux C, Klemenhagen KC, Tanaka KF, Gingrich JA, Guo XE, et al. A serotonin-dependent mechanism explains the leptin regulation of bone mass, appetite, and energy expenditure. Cell. 2009;138:976–989. doi: 10.1016/j.cell.2009.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, Wichterle H, Przedborski S. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2009 doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Networkbased classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pujana MA, Han JD, Starita LM, Stevens KN, Tewari M, Ahn JS, Rennert G, Moreno V, Kirchhoff T, Gold B, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet. 2007;39:1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- 78.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 79.Friedman N, Linial M, Nachman I, Pe’er D. Using Bayesian networks to analyze expression data. J Comput Biol. 2000;7:601–620. doi: 10.1089/106652700750050961. [DOI] [PubMed] [Google Scholar]
- 80.Lorenz DR, Cantor CR, Collins JJ. A network biology approach to aging in yeast. Proc Natl Acad Sci U S A. 2009;106:1145–1150. doi: 10.1073/pnas.0812551106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stolovitzky G, Prill RJ, Califano A. Lessons from the DREAM2 challenges. Ann N Y Acad Sci. 2009;1158:159–195. doi: 10.1111/j.1749-6632.2009.04497.x. [DOI] [PubMed] [Google Scholar]
- 82.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Barrette TR, Ghosh D, Chinnaiyan AM. Mining for regulatory programs in the cancer transcriptome. Nat Genet. 2005;37:579–583. doi: 10.1038/ng1578. [DOI] [PubMed] [Google Scholar]
- 83.Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Butte AJ, Kohane IS. Mutual information relevance networks: functional genomic clustering using pairwise entropy measurements. Pac Symp Biocomput. 2000:418–429. doi: 10.1142/9789814447331_0040. [DOI] [PubMed] [Google Scholar]
- 85.Margolin AA, Palomero T, Sumazin P, Califano A, Ferrando AA, Stolovitzky G. ChIP-on-chip significance analysis reveals large-scale binding and regulation by human transcription factor oncogenes. Proc Natl Acad Sci U S A. 2009;106:244–249. doi: 10.1073/pnas.0806445106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Elkon R, Linhart C, Sharan R, Shamir R, Shiloh Y. Genome-wide In silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 2003;13:773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stuart JM, Segal E, Koller D, Kim SK. A genecoexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 88.Zeitlinger J, Simon I, Harbison CT, Hannett NM, Volkert TL, Fink GR, Young RA. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell. 2003;113:395. doi: 10.1016/s0092-8674(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 89.Segal E, Shapira M, Regev A, Pe’er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 90.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 91.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 93.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 94.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 95.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 96.Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bruckner A, Polge C, Lentze N, Auerbach D, Schlattner U. Yeast two-hybrid, a powerful tool for systems biology. Int J Mol Sci. 2009;10:2763–2788. doi: 10.3390/ijms10062763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Volkel P, Le Faou P, Angrand PO. Interaction proteomics: characterization of protein complexes using tandem affinity purification-mass spectrometry. Biochem Soc Trans. 2010;38:883–887. doi: 10.1042/BST0380883. [DOI] [PubMed] [Google Scholar]
- 99.Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, Hao T, Zenkner M, Xin X, Goh KI, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pan W. Network-based model weighting to detect multiple loci influencing complex diseases. Hum Genet. 2008;124:225–234. doi: 10.1007/s00439-008-0545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]