Abstract
The ability to regulate gene expression in a cell-specific manner is a feature of many broadly expressed signal-dependent transcription factors, including nuclear hormone receptors and transcription factors that are activated by cell surface receptors for extracellular signals. As the most plastic cells of the hematopoietic system, macrophages are responsive to a wide spectrum of regulatory molecules and provide a robust model system for investigation of the basis for cell-specific transcriptional responses at a genome-wide level. Here, we review evidence suggesting a model in which cell-specific actions of signal-dependent transcription factors are the consequence of priming functions of lineage-determining transcription factors, focusing on recent studies in macrophages. We also discuss recent findings relating lineage-determining and signal-dependent transcription factor activity to alterations in the epigenetic landscape as well as the production and function of enhancer RNAs. These findings have implications for the understanding of how natural genetic variation impacts cell-specific programs of gene expression and suggest new approaches for altering gene expression in vivo.
Keywords: cell-specific, lineage determining transcription factors, signal dependent transcription factors, genome-wide analyses
Introduction
A central question in molecular endocrinology is how a single hormone-responsive transcription factor, reading the same DNA template, is capable of regulating different genes in different cell types. For example, activation of the glucocorticoid receptor by endogenous or synthetic glucocorticoids promotes a gluconeogenic program of gene expression in hepatocytes; mediates negative feedback control of adrenocorticotropic hormone secretion from corticotrophs of the anterior pituitary; and suppresses pro-inflammatory responses of macrophages (Lazar 2011). In each case, the same glucocorticoid receptor controls expression of a distinct set of genes that are crucial to the specific biological functions of the corresponding cell type. This ability to regulate gene expression in a cell-specific manner is shared by many broadly expressed signal-dependent transcription factors, including other nuclear hormone receptors and members of the diverse families of transcription factors that are activated by cell surface receptors for extracellular signals (e.g., STAT transcription factors, NF-κB family members, CREB, etc.). (For a complete listing of transcription factor names and abbreviations, see Table 1). The development of high-throughput sequencing methods to characterize transcription factor binding, histone modifications, and nascent RNA production at the genome-wide level has provided a powerful array of new tools for investigating the molecular basis for cell-specific transcriptional responses. These tools have been applied in a number of biologically important model systems, including models for cellular differentiation (e.g., embryonic stem cell differentiation, adipogenesis, and hematopoiesis) and signal-dependent gene expression (e.g., macrophage activation and hormone-dependent breast and/or prostate cancers). In this review, we focus on recent insights into the molecular basis of cell-specific functions of broadly expressed signal-dependent transcription factors derived from genome-wide studies in macrophages.
Table 1.
Abbreviations and full names of transcription factors cited in this review.
| Abbreviations | Transcription Factors |
|---|---|
| PU.1 or SPI1 | Spleen focus forming virus (SFFV) proviral integration oncogene SPI1 |
| AP-1 | Activator protein 1 (heterodimeric protein complex) |
| CEBPs (CEBPA and CEBPB) | CCAAT-enhancer-binding proteins (alpha and beta) |
| SOX2 | SRY (sex determining region Y)-box 2 |
| FOXD3 | Forkhead box D3 |
| JUN | Jun proto-oncogene |
| FOS | FBJ murine osteosarcoma viral oncogene homolog |
| TCF3 | Transcription factor 3 |
| EBF1 | Early B-cell factor 1 |
| FOXO1 | Forkhead box O1 |
| LXR | Liver X receptor |
| NR1D1 and NR1D2 | Nuclear receptor subfamily 1, group D, members 1 and 2 (Rev-Erb alpha and beta) |
| RELA | V-rel reticuloendotheliosis viral oncogene homolog A (avian) |
| SRF | Serum response factor |
| STATs (STAT1, STAT3, etc.) | Signal transducer and activator of transcription (1, 3, etc.) |
| SMAD3 | Mothers against decapentaplegic homolog 3 or SMAD family member 3 |
| HNF4A | Hepatocyte nuclear factor 4, alpha |
| FOXA1 | Forkhead box A1 |
| JUNB | Jun B proto-oncogene |
| IRF4 | Interferon regulatory factor 4 |
| ATF3 | Activating transcription factor 3 |
| RUNX1 | Runt-related transcription factor |
| CBFB | Core-binding factor, beta subunit |
| ETS1 | V-ets erythroblastosis virus E26 oncogene homolog 1 (avian) |
| ELF1 | E74-like factor 1 (ets domain transcription factor) |
| FOXP3 | Forkhead box P3 |
| NFAT | Nuclear factor of activated T-cells |
| BATF | Basic leucine zipper transcription factor, ATF-like |
| RORG | RAR-related orphan receptor C |
| MAF | V-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) |
| EP300 | E1A binding protein p300 |
| GATA3 and GATA4 | GATA binding protein 3 and 4 |
| TBX21 | T-box 21 |
| POU5F1 (OCT4) | POU class 5 homeobox 1 |
| MYOD1 | Myogenic differentiation 1 |
| ESR1 | Estrogen receptor 1 |
| AR | Androgen receptor |
| TFAP2C | Transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| GR | Glucocorticoid receptor |
| MLL (1/3, 2/4, 3, etc.) | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) (1/3, 2/4, 3, etc.) |
| LSD1 | Lysine (K)-specific demethylase 1A |
Macrophages were discovered in 1884 by Ilya Mechnikov, a Russian bacteriologist who later shared the 1908 Nobel Prize in Physiology or Medicine with Paul Ehrlich for their studies in innate immunity (Buryachkovskaya, et al. 2013). Classically, macrophages differentiate from bone marrow-derived monocytes; however, recent studies have found that at least some tissue resident macrophages such as microglia, Langerhans cells, and Kupffer cells are derived from hematopoietic stem cells during fetal development (Saijo and Glass 2011; Wynn, et al. 2013). As mediators of innate immunity and normal homeostatic processes, macrophages are essential to the body’s ability to control inflammation. Consequently, these immune cells have been implicated in multiple disease processes such as diabetes, rheumatoid arthritis, multiple sclerosis, and atherosclerosis (Drexler and Foxwell 2010; Glass and Saijo 2010; Olefsky and Glass 2010). Primary macrophages are easily obtainable for in vitro studies (as bone marrow-derived macrophages, thioglycollate-elicited macrophages, circulating monocytes, splenic monocytes, etc.) from wild type and genetically modified animals and can also be differentiated from human blood monocytes. The ability to obtain large numbers of these cells makes them a robust model system for contemporary ‘omics’ technologies – proteomics (Hettinger, et al. 2013; Meissner, et al. 2013), lipidomics (Maurya, et al. 2013; Spann, et al. 2012), and genomics (Heinz, et al. 2010; Kaikkonen, et al. 2013; Lam, et al. 2013; Ostuni, et al. 2013) – for investigating signal-dependent mechanisms.
As the most plastic cells of the hematopoietic system, macrophages are responsive to many regulatory molecules, including growth factors, pro- and anti-inflammatory cytokines, pathogen-associated molecular patterns (PAMPs), damage-associated molecular patterns (DAMPs) and nuclear receptor ligands (Lawrence and Natoli 2011; Murray and Wynn 2011; Shi and Pamer 2011; Sica and Mantovani 2012; Smale 2010; Van Dyken and Locksley 2013; Wynn et al. 2013). Responses to PAMPs and DAMPs are mediated by pattern recognition receptors that include the family of toll-like receptors (TLRs) (Medzhitov, et al. 2011). Toll-like receptor 4 (TLR4) recognizes lipopolysaccharide (LPS), a component of the cell wall of gram-negative bacteria. Upon binding LPS, TLR4 signal transduction activates a number of latent transcription factors, which include NF-κB and interferon regulatory factors (Medzhitov, et al. 1997; Poltorak, et al. 1998a; Poltorak, et al. 1998b). These factors subsequently enter the nucleus and regulate the expression of inflammatory response genes, which play essential roles in innate immunity and contribute to the development of adaptive immunity (Medzhitov and Horng 2009). This transcriptional response is among the most dramatic in biology with respect to both the number of genes regulated (thousands) and the magnitude of change in gene expression (>4 orders of magnitude for the most highly regulated genes) (Escoubet-Lozach, et al. 2011; Kaikkonen et al. 2013). This response is also subject to counter-regulation by anti-inflammatory signaling molecules that include ligands for nuclear hormone receptors, such as the glucocorticoid receptor, PPARs, and LXRs (Glass and Saijo 2010). These properties of the macrophage make it a particularly rich cell type for the application of genome-wide approaches.
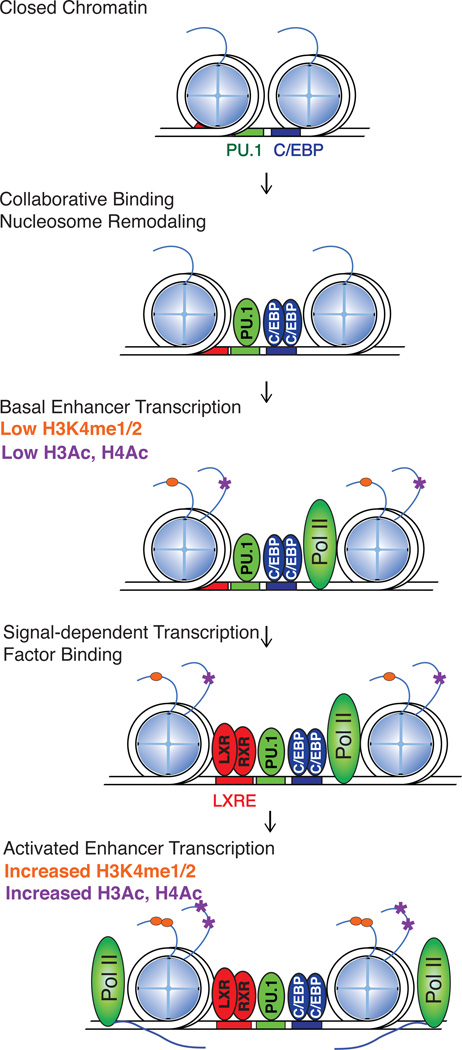
Recent studies of signal-dependent transcriptional responses in macrophages and B cells suggest a general model to explain many of the cell-specific functions of broadly expressed signal-dependent transcription factors (SDTFs) (Heinz et al. 2010) (Figure 1). In this model, relatively simple combinations of lineage determining transcription factors (LDTFs) are proposed to function collaboratively to select genomic enhancer-like regulatory regions in a cell-specific manner. The collaborative binding of these factors results in nucleosome remodeling to generate open regions of chromatin that provide access to SDTFs. Thus, signal-dependent transcription factors are directed to the vicinity of target genes in a cell-specific manner as a consequence of the priming functions of the lineage determining factors for that particular cell type.
Figure 1. Hierarchical organization of transcription factor networks.
In the poised or repressed state, closed chromatin at enhancer loci contain motifs for lineage determining transcription factors. The collaborative binding of lineage determining transcription factors such as PU.1 and CEBPs results in nucleosomal remodeling and basal enhancer transcription at sites containing low H3K4me1/2 and low H3/H4 acetylation. Next, in response to various stimuli, signal-dependent transcription factors, such as LXRs, co-bind with PU.1 and C/EBPs to activate enhancer transcription. Finally, activated eRNA transcription precedes the active deposition of H3K4me1/2 and H3/H4 acetylation. LDTF: Lineage Determining Transcription Factor; SDTF: Signal-Dependent Transcription Factor.
Setting the stage: roles of lineage determining factors
It has long been proposed that the functional output of transcription factors is determined by their competitive binding with nucleosomes. Until nucleosomes are displaced and chromatin regions are open, most transcription factors bind transiently to chromatin in a manner insufficient for efficient transcription (Lickwar, et al. 2012). For example, during hematopoietic stem cell differentiation, nucleosome remodeling by the ATP-dependent, SWI/SNF chromatin-remodeling complex allows for transcription factor binding at erythroid LDTF binding sites (Hu, et al. 2011). Furthermore, studies utilizing novel high-throughput sequencing methods have been used to define open chromatin regions, which contain important transcriptional regulatory elements that delineate cell-specific identity (Song, et al. 2011).
Recent studies of the ENCODE consortium estimate that the mammalian genome contains hundreds of thousands of enhancers (Bernstein, et al. 2012), the majority of which are selected for activity in a cell-specific manner. This process of enhancer selection is proposed to result from collaborative interactions of LDTFs, also referred to as pioneer factors or master regulators. These factors are sequence-specific DNA-binding proteins with the ability to access their binding sites even when those sites are wrapped in nucleosomes. The factors also represent placeholders that can be replaced by other transcription factors at later stages during development (Zaret and Carroll 2011). For example, in embryonic stem cells, developmental pioneer transcription factors, SOX2 and FOXD3, bind at tissue-specific elements, and are replaced by cell-specific transcription factors after differentiation (Liber, et al. 2010). Currently, two prevailing theories exist to explain how pioneer transcription factors function in defining cell-specificity during development. In the passive model, pioneer transcription factor occupancy decreases the number of binding events necessary for transcriptional activation (Zaret and Carroll 2011). In contrast, the active model suggests that pioneer transcription factors function by recruiting chromatin remodeling complexes to activate transcription (Zaret and Carroll 2011).
The roles of LDTFs in priming cell-specific regulatory sites have been well-characterized in cells of the hematopoietic lineage. The LDTF, spleen focus forming virus (SFFV) proviral integration oncogene, SPI1, more commonly referred to as PU.1, provides a particularly instructive example of a LDTF that is required for specification of more than one hematopoietic cell type; genetic deletion of PU.1 results in loss of macrophages, neutrophils, and B cells (Scott, et al. 1997; Scott, et al. 1994). In myeloid lineage precursor cells, PU.1 instructs hematopoietic progenitors to up-regulate myeloid-specific cell surface antigens and to down-regulate other cell-specific markers and transcription factors (Nerlov and Graf 1998). Recent studies have shown a dependence on cell cycle lengthening and subsequent PU.1 accumulation, which dictates myeloid differentiation from the common myeloid lymphoid progenitor (Kueh, et al. 2013). In lymphoid progenitor cells, PU.1 is required for expression of an alternative set of genes required for progression to mature B cells (Lin, et al. 2010). Thus, even a single LDTF can promote distinct programs of gene expression in different cell types.
PU.1 is a member of the E-twenty six (ETS) family of transcription factors, which bind to the canonical ETS-motif, 5’-GGAA-3’. Using genetic and genomic methods, PU.1 was recently shown to select macrophage or B cell-specific enhancers based on co-occurrence of nearby binding sites for either macrophage or B-cell LDTFs, which are selectively expressed in one or the other cell type (Heinz et al. 2010). In macrophages, PU.1 binding occurred at enhancer-like regions exhibiting nearby binding sites for other essential myeloid LDTFs, CCAAT/enhancer binding protein alpha and beta (CEBPs) and/or AP-1 transcription factors, a heterodimeric protein composted of Jun proto-oncogene (JUN) and FBJ murine osteosarcoma viral oncogene homolog (FOS) (Heinz et al. 2010). In B cells, PU.1 binds at enhancer-like regions containing binding sites for other B cell LDTFs including TCF3, EBF1, and FOXO1 (Heinz et al. 2010; Lin et al. 2010). Notably, binding of PU.1 and alternative LDTFs was mutually dependent; genetic deletion of one factor resulted in loss of binding of the other at closely spaced sites (Heinz et al. 2010). Although generally within 100 bp of each other, no strict spacing relationship was observed between PU.1 and collaborative transcription factor binding sites, suggesting a mechanism other than a ternary complex model for enhanced occupancy (Heinz et al. 2010). Gain-of-function studies indicated rapid nucleosome remodeling following collaborative binding of PU.1 to regions destined to acquire enhancer-like features in myeloid cells based on subsequent H3K4 mono-methylation (Heinz et al. 2010). Furthermore, PU.1 is required for the maintenance of the macrophage epigenome and its expression in PU.1-negative myeloid progenitors is sufficient to induce nucleosome remodeling and H3K4 mono-methylation at cell-specific sites (Heinz et al. 2010).
Signal-dependent transcription factors bind in a cell-specific manner
Genome-wide studies to date of SDTFs indicate that while binding at promoters is enriched over that expected by chance, the majority of binding sites for these factors are located at inter- and intra-genic locations that exhibit features of enhancers. Initially demonstrated for the estrogen receptor (Lupien, et al. 2008), this pattern of genome-wide binding is also observed for other members of the nuclear receptor family, including the glucocorticoid receptor (Biddie, et al. 2011), LXRs (Heinz et al. 2010), PPARG (Lefterova, et al. 2008), androgen receptor (AR) (He, et al. 2010; Lupien et al. 2008; Wang, et al. 2011), estrogen receptor related receptors (Hurtado, et al. 2011; Li, et al. 2013; Lupien et al. 2008), and nuclear receptor subfamily 1, group D, members 1 and 2 (NR1D1 and NR1D2), known colloquially as Rev-Erbs (Lam et al. 2013). Similarly, studies of transcription factors responsive to extracellular signals also primarily localize to distal enhancer-like elements, including the RELA component of NF-κB (Barish, et al. 2010; Kaikkonen et al. 2013), SRF (Sullivan, et al. 2011), FOXO1 (Fan, et al. 2010), STAT1 (Vahedi, et al. 2012), and SMAD3 (Mullen, et al. 2011). Although identification of specific enhancer-promoter interactions remain challenging at a genome-wide level, the binding of SDTFs at distal locations is correlated with changes in expression of nearby genes (Kaikkonen et al. 2013; Lam et al. 2013).
In all of the cases in which the genome-wide locations of SDTFs have been examined in macrophages, a large fraction of the DNA binding sites are observed to be in close proximity to binding sites for one or more macrophage LDTFs (PU.1, CEBPs and/or AP-1 factors) (Heinz et al. 2010). Because LDTFs select enhancers in a cell-specific manner, the co-localization of SDTFs implies a cell-specific binding pattern. Consistent with this, the genomic locations of PPARG and Rev-Erbs in macrophages are very different from their genomic locations in adipocytes (Lefterova et al. 2008) and hepatocytes (Cho, et al. 2012), respectively.
An important question is the hierarchical relationship between LDTFs and SDTFs, as well as their respective roles in the selection of cell-specific enhancers. Studies of LDTFs and SDTFs in macrophages suggest primary roles of LDTFs, with variable roles of SDTFs that depend on the specific factor analyzed. Genome-wide binding analyses of LXRs indicated that nearly all LXR binding sites were in close proximity to binding sites for macrophage LDTFs (PU.1, AP-1, C/EBPs) (Heinz et al. 2010). Genetic deletion of LXRs had virtually no effect on the overall enhancer atlas in macrophages and did not result in a loss of nearby PU.1 binding (Heinz et al. 2010). In contrast, deletion of PU.1 resulted in loss of nearby LXR binding (Heinz et al. 2010). Thus, in this case, a clear hierarchy was observed in which the binding of LXRs was dependent on the initial selection of enhancers by LDTFs.
A different picture emerged from studies of the RELA subunit of NF-κB, which is strongly activated in macrophages by signaling through TLR4. Following activation and nuclear entry, ~85% of RELA binding occurred at pre-existing enhancer-like regions of the genome that were occupied by macrophage LDTFs (Barish et al. 2010; Escoubet-Lozach et al. 2011; Kaikkonen et al. 2013). However, ~15% of the binding sites for RELA occurred at genomic regions that lacked features of enhancers, but acquired these features subsequent to RELA binding (Escoubet-Lozach et al. 2011; Kaikkonen et al. 2013). Intriguingly, these binding sites were highly enriched for nearby recognition motifs for the macrophage LDTFs AP-1 and PU.1. Subsequent experiments demonstrated that RELA binding required PU.1, while PU.1 binding required nuclear entry of RELA (Escoubet-Lozach et al. 2011; Kaikkonen et al. 2013). Thus, in this case, RELA acted as both a signal-dependent and collaborating transcription factor to open up regions of chromatin and select new enhancers. The ability of RELA but not LXR to perform this function may relate to relatively higher nuclear concentrations and/or its ability to interact with nucleosome remodeling factors.
Recent studies have utilized natural genetic variation as an in vivo mutagenesis screen to examine the collaborative binding model for LDTFs and the hierarchical relationship with NF-κB. Genome-wide transcription factor binding and gene expression analyses were used to characterize how genetic variations affect the binding of LTDFs (PU.1 and CEBPA) and the associated transcriptional output in macrophages derived from two inbred rodent strains, C57BL/6J and BALB/c (Heinz, et al. 2013). Single nucleotide variations and other non-coding genomic variants were shown to directly perturb LDTF as well as SDTF binding (Heinz et al. 2013). However, these studies also demonstrated that a variant in a binding motif for a LDTF not only affected the binding of that factor, but also the binding of nearby LDTFs, even though there were no alterations in the binding motifs for those factors. In the case of differential NF-κB binding in the two mouse strains, mutations in motifs of LDTFs were three times more likely to result in decreased RELA binding than mutations in the binding sites for RELA itself (Heinz et al. 2013). These findings provided genetic evidence that collaborative binding of LTDFs to variably spaced sequences was essential for enhancer selection and subsequent acquisition of histone modifications associated with enhancer activity (Heinz et al. 2013). These studies also provided genetic evidence for a hierarchical relationship between LDTFs and the majority of the binding sites for NF-κB.
From these studies, the authors also formulated a genetic definition of a functional binding matrix that could be used to predict whether a specific variant in a LDTF binding motif would elicit a functional consequence. These results hint at the potential power of defining functionally important non-coding regulatory variants and suggest a strategy for prioritizing natural genetic variants that may play roles in human disease (Heinz et al. 2013).
A similar study detailed the binding profile of three tissue specific transcription factors (HNF4A, CEBPA, and FOXA1) in the livers of six inbred rodents (Stefflova, et al. 2013). From these studies, the authors describe that although there are extensive binding occupancy differences in relatively similar species, single nucleotide variation plays only a modest role in these differences (Stefflova et al. 2013). Regions that are bound by combinations of LDTFs are more evolutionarily stable; however, binding by a single LDTF can become de-stabilized through the deletion of one its LDTF co-binding partners (Stefflova et al. 2013).
Lineage determining factors in other cell types
The roles of LDTFs in mediating cell-specific and signal dependent activation of transcription have been described in many cell types. Here, we provide examples derived from genome-wide studies in dendritic cells, T cells, and in model systems used to study transforming growth factor beta (TGFβ) mediated and hormone-dependent gene expression (Table 2).
Table 2.
Summary of studies demonstrating signal-dependent cell-specificity in transcription.
| Cell Type | Lineage- Determining Transcription Factors |
Stimulus | Stimulus- Activated Transcription Factors |
Cell-specific (LDTF- regulated) Gene Expression |
References |
|---|---|---|---|---|---|
| Macrophages | PU.1, CEBPs, AP-1 | LPS, GW3965 (LXR agonist) | LXRs, RELA, BCL6 | Csf1r, Cd14, Ccl3, Ccl4, Cxcl2, Abcg1 | (Barish et al. 2010; Heinz et al. 2010) |
| Dendritic cells | PU.1, CEBPB | LPS | JUNB, IRF4, ATF3 | Il1a, Ifit1, Ifit2, Ifit3, Tnf, Stat1, Nfkbiz | (Garber et al. 2012) |
| ES cells | POU5F1 | TGFB1 | SMAD3 | Sox2 | (Mullen et al. 2011) |
| Pro-B cells | PU.1 | TGFB1 | SMAD3 | Vpreb2 | (Mullen et al. 2011) |
| Myotubes | MYOD1 | TGFB1 | SMAD3 | Adora1 | (Mullen et al. 2011) |
| T cells | BATF and IRF4 | Th17 polarization | RORG, STAT3, MAF | Il17a, Il12rb1 | (Ciofani et al. 2012) |
| T cells | ? | Th1 polarization Th2 polarization | STAT4, STAT1, TBX21 STAT6, GATA3 | ? | (Vahedi et al. 2012) |
| T cells | CBFB, ETS1, ELF1 | Treg polarization | FOXP3 | Junb | (Samstein et al. 2012) |
| MCF7 cells | FOXA1, TFAP2C | Tamoxifen, 17β-estradiol | ESR1 | Ret, Xbp1, Tff1 | (Hurtado et al. 2011; Li et al. 2013; Lupien et al. 2008; Tan et al. 2011) |
| LNCaP cells | FOXA1 | 5α-dihydrotestosterone | AR | Chka | (He et al. 2010; Lupien et al. 2008; Wang et al. 2011) |
| Murine mammary epithelial cells | AP-1 | Dexamethasone | Glucocorticoid receptor | Klrb1c | (Biddie et al. 2011) |
| U2OS | GATA4 | 17β-estradiol | ESR1 | Faslg, Wnt4, Foxc1, Alpl, Runx2 | (Miranda-Carboni et al. 2011) |
Dendritic cells
Dendritic cells and macrophages derive from a common myeloid lineage. In support of a LDTF/SDTF regulatory model, H3K4me3, H3K4me1, H3K27Ac, and a comprehensive panel of myeloid transcription factors were utilized to map the promoter and enhancer landscape in dendritic cells (Garber, et al. 2012). Like in macrophages, the LDTFs in dendritic cells include PU.1 and CEBPB (Garber et al. 2012). In this model, a second tier of LDTFs (JUNB, IRF4, and ATF3) exist, termed “primers”, which co-bind with PU.1 and CEBPB at LPS-inducible genes pre-stimulation. Later, these factors become associated with specific and dynamic activation factors, mediating a signal-specific response to stimulation (Garber et al. 2012).
T cells
Three seminal papers have described the cell-specific enhancer landscape in T cells, focusing on variations amongst different subsets of T helper (Th) cells. In one study, the authors sought to define the subset-specific enhancer landscape in regulatory T (Treg) cells. LDTFs in the RUNX (Runt-related transcription factor) and ETS families, CBFB, ETS1, and ELF1, were shown to predominantly occupy quiescent Th cell enhancers (Samstein, et al. 2012). Surprisingly, the majority of FOXP3 binds to both already established and Treg polarized enhancers (Samstein et al. 2012). Only 2% of FOXP3 binding sites appear to be Treg-specific and are highly enriched for an AP-1 motif (Samstein et al. 2012). In some cases, FOXP3 was found to displace its paralog, FOXO1, or other cofactors that served as placeholders for FOXP3 binding (Samstein et al. 2012). Furthermore, during Treg polarization and TCR activation, the SDTF, FOXP3, is primarily recruited to previously established enhancers likely though AP-1 and nuclear factor of activated T-cells (NFAT) facilitated chromatin remodeling (Samstein et al. 2012). A second study describing the transcriptional network for Th17 polarization found two previously established LDTFs, BATF and IRF4, to exhibit similar binding patterns between quiescent Th cells and Th17 cells (Ciofani, et al. 2012). In the presence of Th17-polarizing cytokines, BATF and IRF4 prime pre-established enhancers for SDTF (RORG, STAT3, and MAF) recruitment (Ciofani et al. 2012). Lastly, to define the active enhancer landscape after Th1 or Th2 polarization, a comprehensive analysis of EP300 binding was conducted in the two subsets (Vahedi et al. 2012). The two closely related cell subsets display common and distinct EP300 binding (Vahedi et al. 2012). Moreover, STAT6 and GATA3 or STAT4/STAT1 and TBX21 were found to activate Th2 or Th1 subset-specific enhancers, respectively, while suppressing those of alternative fates (Vahedi et al. 2012).
Embryonic Stem Cells, Pro-B cells, Myotubes
Genome-wide binding analyses were conducted for three unique cell types, embryonic stem cells, myotubes, and pro-B cells, each in the quiescent state and after TGFβ signaling (Mullen et al. 2011). The authors found that the LDTFs for these respective cell types, POU5F1, MYOD1, and PU.1, directed the TGFβ-induced SDTF, SMAD3, to unique cell-specific enhancer sites (Mullen et al. 2011). Motif-finding at SMAD3 bound sites showed an enrichment for LDTF motifs specific to the cell type in question, highlighting the importance of LDTFs in directing the binding of SDTFs (Mullen et al. 2011).
Breast, Prostate, and Osteoblasts
FOXA1 is essential for the development and differentiation of several organs such as liver, kidney, pancreas, lung, prostate, and mammary gland (Friedman and Kaestner 2006). In breast cancer epithelial (MCF7) cells and prostate cancer (LNCaP) cells, FOXA1 creates a cell-specific enhancer network, which regulates other activating factors such as ESR1 (Hurtado et al. 2011; Lupien et al. 2008) and AR (He et al. 2010; Wang et al. 2011), respectively. In estrogen-responsive breast cancer cells, tamoxifen-mediated ESR1 activity is dependent on LDTF FOXA1. In tamoxifen-resistant cells, ESR1 binding occurs independent of ligand but is still dependent on FOXA1 (Hurtado et al. 2011). Another transcription factor implicated in breast cancer oncogenesis, TFAP2C, binds to ESR1-binding sites in a ligand-independent manner, co-localizing with FOXA1, and priming enhancer sequences for ESR1 binding (Tan, et al. 2011). Furthermore, treatment with estradiol results in a global increase of enhancer RNA (eRNA) transcription at enhancers in the vicinity of 17β-estradiol regulated genes, which promote specific E2/ESR1/eRNA-induced enhancer-promoter looping (Li et al. 2013). In addition, a recent study conducted in a murine mammary epithelial cell line found AP-1 to be an essential LDTF, mediating GR-chromatin interactions and GR-regulated transcription (Biddie et al. 2011).
In U2OS cells, an osteoblast-like cell line, ESR1 binding is preceded by the lineage determining factor, GATA4 (Miranda-Carboni, et al. 2011). GATA4 is necessary for H3K4me2 deposition at ESR1 binding sites, suggesting that it is a LDTF for ESR1 recruitment in this cell type (Miranda-Carboni et al. 2011). Furthermore, these results illustrate the cell-specific nature of priming and activating transcription factors; future studies will help clarify the differences amongst diverse cell types.
Histone modifications associated with active and poised regulatory elements
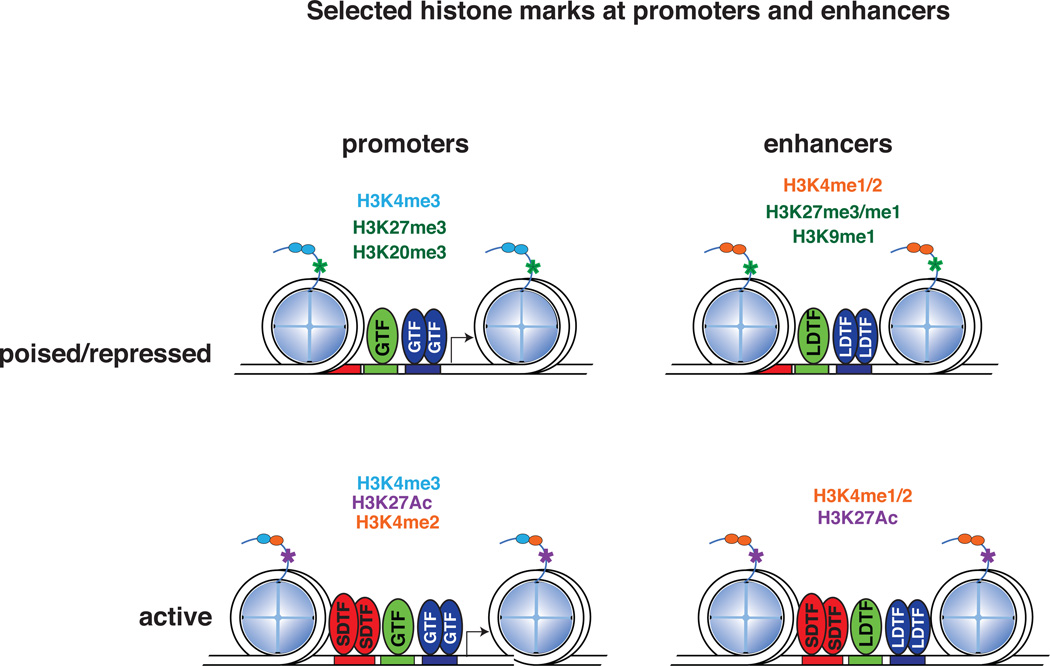
LDTFs and SDTFs bind at both enhancer and promoter elements to regulate transcription. Enhancers and promoters are associated with distinct chromatin signatures - active promoters are marked by high levels of histone H3 lysine 4 tri-methylation (H3K4me3) relative to mono-methylation (H3K4me1), whereas enhancers are marked by high levels of H3K4me1 relative to H3K4me3 (Heintzman, et al. 2007) (Figure 2). H3K4me2 occurs at both promoter and enhancer regions (Kaikkonen et al. 2013), which we define here by their distance from a transcription start site (around <2kb to indicate a promoter and >2kb to indicate an enhancer). Studies in yeast, Drosophila, and humans suggest that the SET domain-containing methyl-transferases are responsible for depositing the majority of H3K4me3 at promoters (Ardehali, et al. 2011; Hallson, et al. 2012; Nagy, et al. 2002; Wu, et al. 2008).
Figure 2. Histone marks at active and poised promoters and enhancers.
Promoters are designated by high levels of H3K4me3, while enhancers contain high levels of H3K4me1/2. In the poised or repressed state, promoters contain histone methylation marks, H3K27me3 and H3K20me3, while enhancers contain H3K27me3/me1 and H3K9me1. Active promoters and enhancers are marked by H3K4me2 and H3K27Ac. GTF: General Transcription Factor; LDTF: Lineage Determining Transcription Factor; SDTF: Signal Dependent Transcription Factor.
Mono-methylation at enhancers is dynamic and regulated in a cell-specific manner. Currently, methyl-transferases regulating H3K4me1 at mammalian enhancers include MLL1 (Jeong, et al. 2011) and MLL1/3 (Kaikkonen et al. 2013). At enhancer loci, the H3K4me1 mark functions as an active mark; its de-methylation in mouse embryonic stem cells by LSD1 results in enhancer silencing and cell differentiation. The process by which H3K4me1 is lost at enhancers is called enhancer “decommissioning” and further illustrates the fine tuning mediated by cell-specific enhancers (Whyte, et al. 2012). Not surprisingly, loss of LSD1 is associated with increased H3K4me1 and H3K4me2 at hematopoietic stem cell promoter and enhancer elements as well as subsequent gene de-repression, resulting in altered transcription and compromised differentiation (Kerenyi, et al. 2013).
While the presence of H3K4me2 dictates active transcriptional regions, active enhancers are also marked by EP300 (or CBP) histone acetyl-transferase (Ghisletti, et al. 2010; Heintzman et al. 2007) (Figure 2). EP300/CBP acetylates H3K27 (Jin, et al. 2011; Pasini, et al. 2010; Tie, et al. 2009), a mark for active transcription, which distinguishes active enhancers from poised or disengaged enhancers (Creyghton, et al. 2010; Heintzman, et al. 2009; Rada-Iglesias, et al. 2011; Zentner, et al. 2011) in human and mouse embryonic stem cells (Creyghton et al. 2010; Rada-Iglesias et al. 2011; Zentner et al. 2011), as well as adult tissues (Creyghton et al. 2010; Heintzman et al. 2009; Zentner et al. 2011). EP300
In contrast to active acetylation marks, the combination of H3K4me3 and H3K27me3 marks promoters that are “poised” for transcription (Mikkelsen, et al. 2007) (Figure 2). In mouse and human embryonic stem cells, poised enhancers are marked by H3K4me1, EP300, SMARCA4, and H3K27me3, and are bound by the Polycomb complex (Rada-Iglesias et al. 2011). Unable to drive gene expression, poised enhancers transition into active enhancers during differentiation through the acquisition of H3K27ac and the loss of H3K27me3, as well as the recruitment of tissue-specific transcription factors and RNA polymerase II (Rada-Iglesias et al. 2011). Furthermore, poised enhancers in hematopoietic stem cells contain H3K27me1 and H3K9me1 prior to their activation (Cui, et al. 2009), whereas promoters in embryonic stem cells (Rada-Iglesias et al. 2011), hematopoietic stem cells (Cui et al. 2009), and adult tissues (De Santa, et al. 2007), all contain H3K27me3 and may contain other repressive marks such as H3K20me3 (Stender, et al. 2012) (Figure 2).
Signal-dependent control of the epigenetic landscape
Investigation of macrophage differentiation and activation has provided a powerful experimental system for linking the actions of LDTFs and SDTFs to specific histone modifications and subsequent transcriptional output. Macrophage gene expression is stimulus-specific, gene-specific, cell-specific, and macrophage-subset specific. In early genome-wide studies to study the stimulus-specific nature of gene expression, microarrays were used to systematically examine the response of macrophages to various bacterial pathogens that act through TLRs, comparing the shared and differential transcriptional output (Nau, et al. 2002). These studies were the first to document the very dramatic transcriptional responses of macrophages to these signals, and revealed that these responses were finely tuned to specific pathogens.
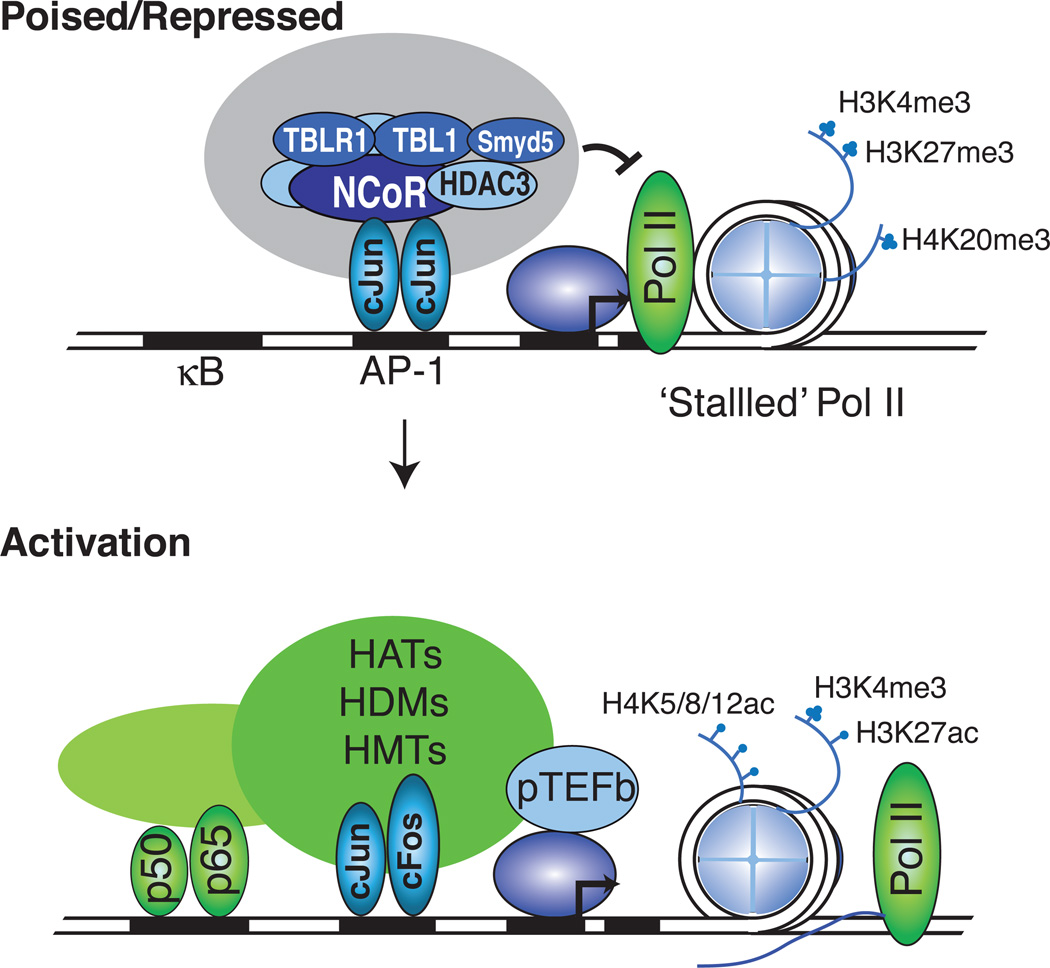
Several lines of evidence indicate that the program of macrophage activation is subject to many types of negative regulation that act prior and/or subsequent to stimulus responses. BCL6 has been shown to co-repress almost a third of the TLR4 cistrome, such that its loss results in hypersensitivity to pro-inflammatory de-repression (Barish et al. 2010). The NCOR1/HDAC3 co-repressor complex has been proposed to function as a transcriptional checkpoint for some TLR-responsive genes under basal conditions through recruitment to non-phosphorylated forms of AP-1. The HDAC3 component of this complex contributes to the repressive functions of the NCOR1 complex by removing histone acetylation marks required for transcriptional activity (Ogawa, et al. 2004) (Figure 3). Activation of inflammatory genes necessitates signal-dependent phosphorylation of c-Jun and removal of NCOR1 from AP-1 target genes (Ogawa et al. 2004). Surprisingly, loss of HDAC3 in macrophages results in decreased activation of almost half of the inflammatory program (Chen, et al. 2012). This phenomenon has been suggested to be the result of decreased Ifnb1 expression and the secondary STAT1-mediated transcriptional response, a pre-requisite for inflammatory induction (Chen et al. 2012). In addition to HDAC3, NCOR1 complexes in macrophages also contain the histone methyl-transferase SMYD5, which contributes to repression by catalyzing H4K20 tri-methylation, inhibiting the expression of TLR4 target genes (Stender et al. 2012). Signal-dependent de-methylation of H4K20me3 at promoters occurs through the recruitment of the histone de-methylase PHF2 by the RELA component of NF-κB (Stender et al. 2012) (Figure 3).
Figure 3. Mechanisms for regulating gene expression in macrophages: co-regulator exchange from co-repressor to co-activator.
In the poised or repressed state, the NCOR1/HDAC3/SMYD5 complex inhibits active transcription through de-acetylation of histone H3 and H4, as well as tri-methylation of H4K20. After signal dependent recruitment of activating transcription factors such as NF-κB, the NCOR1 complex is exchanged for co-activator complexes that remove repressive marks and acetylate histones H3 and H4, recruiting the P-TEFb complex to activate transcriptional elongation.
After co-repressor dismissal from inflammatory genes, transcriptional activation in response to TLR4 signaling occurs in two phases – primary response genes are immediately up-regulated in response to stimuli whereas secondary response genes require expression of gene products from the initial wave of transcription, such as the type I interferons. CpG islands are usually found at the promoters of primary response genes and associate with nucleosome-destabilizing elements, allowing these genes to be transcribed at low levels in the basal state. This low level of transcription produces nonfunctional transcripts that are rapidly degraded until the recruitment of the positive transcription elongation factor (P-TEFb) complex, which phosphorylates RNA polymerase II and couples transcriptional initiation with elongation (Hargreaves, et al. 2009; Ramirez-Carrozzi, et al. 2009). In contrast, promoters with low CpG content are indicative of secondary response genes, which undergo stimulus-dependent H3K4me3 and H3 acetylation, requiring selective nucleosome remodeling (Hargreaves et al. 2009; Ramirez-Carrozzi et al. 2009). Inflammatory genes in macrophages can be further classified into two categories on the basis of function and regulation – genes capable of being induced after stimulation of tolerant macrophages and those that will not be induced during re-stimulation. Genes capable of being re-stimulated exhibit more H4 acetylation and maintain H3K4me3 after re-stimulation (Foster, et al. 2007). Like the genes in naÔve macrophages, these genes are capable of recruiting the SMARCA4 and CHD4 chromatin remodeling complexes to their promoters; however, they exhibit different transcriptional requirements than those in naÔve macrophages (Foster et al. 2007).
Macrophage activation results in the sequential binding of lineage-determining and stimulus-activated transcription factors to cell-specific regions, enabling the novel deposition of cis-regulatory enhancer marks such as H3K4me1 and H3K27ac (Escoubet-Lozach et al. 2011; Kaikkonen et al. 2013; Ostuni et al. 2013). In one study, the authors removed the stimuli post-treatment in a “washout” experiment. Surprisingly, H3K4me1 did not revert to its latent state; instead, it persisted and conditioned a faster and stronger response upon re-stimulation, suggesting the existence of epigenetic memory in response to stimuli exposure (Ostuni et al. 2013).
Recent studies have reported the productive transcription of RNA polymerase II-associated non-coding RNA (De Santa, et al. 2010) (ncRNA) from cis-regulatory enhancers located both intragenically (Kowalczyk, et al. 2012) and intergenically in response to LPS stimulation (Kaikkonen et al. 2013). TLR4 signaling regulates macrophage gene expression through both a pre-existing enhancer landscape as well as the induction of ~3000 enhancer RNAs (eRNAs) from de novo enhancer regulatory regions (de novo eRNAs) (Kaikkonen et al. 2013) (Figure 1). In all, ~2200 de novo eRNAs are induced >2 fold in response to TLR4. Regions that show eRNA induction also gain H3K4me2 and are enriched for AP-1, NF-κB, CEBP, IRF/STAT, and PU.1 motifs (Kaikkonen et al. 2013). Surprisingly, the inhibition of RNA polymerase II elongation and eRNA synthesis at enhancers resulted in decreased H3K4me1 and H3K4me2 deposition without affecting H4K8ac (Kaikkonen et al. 2013). This result suggests that enhancer transcription precedes and may be essential for H3K4me1/2 deposition, at least for the set of enhancers that are established de novo following TLR4 activation. Systemic depletion of all known H3K4 methyl-transferases indicated that TLR4-induced H3K4 methylation was dependent upon the histone methyl-transferases, MLL1, MLL2/4, and MLL3 (Kaikkonen et al. 2013).
Functional roles of enhancer RNAs
It has been proposed that transcription at enhancers maybe due to non-specific interactions of RNA polymerase II with the genome, thus representing noise rather than biological function (Struhl 2007). Others have speculated that the functional effects of enhancer transcription are due to the transcription process and machinery or RNA polymerase II moving unobstructed in either direction upon binding DNA. In contrast, studies have shown that in vivo developmental enhancers have highly conserved non-coding elements (Pennacchio, et al. 2006; Visel, et al. 2008), positing that developmental enhancers may have retained their function throughout evolution.
Enhancer RNAs are believed to exert their function through two main mechanisms: in cis, the eRNAs may act on the same chromatin fiber or in trans, the eRNAs may function at distant sites in the genome. Recently, in macrophages, Rev-Erbs were described to repress key inflammatory genes through direct promoter-mediated mechanisms as well as in cis through eRNAs (Lam et al. 2013). Primarily, Rev-Erbs bind at distal elements, which are marked by PU.1 co-binding and H3K4me1 (Lam et al. 2013). The binding of Rev-Erbs at enhancer elements inhibits enhancer mediated transcription at two distal cis regulatory elements, subsequently decreasing transcription at the nearby protein coding genes, Mmp9 and Cx3cr1 (Lam et al. 2013). Genetic studies depleting either Rev-Erbs from these loci or the eRNAs themselves resulted in the subsequent de-repression of Mmp9 and Cx3cr1 mRNA (Lam et al. 2013). Similar studies in multiple cell types have found the transcription of ncRNAs, especially eRNAs, to be essential in promoting (Feng, et al. 2006; Kim, et al. 2010; Li et al. 2013; Melo, et al. 2013; Orom, et al. 2010) and inhibiting (Wang, et al. 2008) gene expression. Further studies will help elucidate the cell-specific mechanisms of enhancers in regulating diverse facets of transcription.
Conclusions
In concert, these studies suggest a general model to explain how genes with a broad distribution of expression can be regulated in a cell-specific manner and how broadly expressed SDTFs are capable of regulating discriminative responses in different cell types. Traditionally, the regulation of transcription is thought to occur primarily at the promoter. The above described studies show that LDTFs collaboratively select and prime distal enhancer regulatory elements in a cell-specific manner, genome-wide. In a signal-dependent manner or during development, LDTFs may remodel and open chromatin at promoter and enhancer regions, broadly defining the regulatory potential of genes in a cell-specific manner. For example, PU.1 binds to more than two-thirds of enhancer-like regions in macrophages, priming target genes for the subsequent recruitment of SDTFs to their vicinity, and their subsequent activation (Heinz et al. 2010). Thus, although transcription can occur at both promoters and enhancers – transcription at promoters is often ubiquitous amongst different cell-types while that at enhancers tends to be cell-specific.
Furthermore, genetic analyses have shown that mutations of SDTF motifs can only account for a small percentage of the variable gene expression that is the result of genetic variation (Heinz et al. 2013; Kasowski, et al. 2010). Understanding how mutations in LDTF and SDTF motifs result in direct and indirect effects on enhancer selection and function is essential to defining relevant non-coding variants in the genome and their impact on human disease (Heinz et al. 2013). Comprehension of how key lineage determining transcription factors modulate signal dependent mechanisms in different cell types may also have cell-specific therapeutic applications. Many human disease states could benefit from cell-specific alterations in gene expression in a manner that would decrease toxicity and increase efficacy. New technologies such as antisense oligonucleotides and small interfering RNAs can be potentially used to modulate cell-specific eRNA transcripts that are associated with malfunctioning genes (Lam et al. 2013). In principle, this methodology would enable the cell-specific regulation of aberrant gene expression implicated in disease states such as cancer and chronic inflammatory diseases without adversely modulating normal physiological expression in unaffected cell types.
Acknowledgements
We would like to thank Karmel Allison for the critical reading of this manuscript and Lynn Bautista for her expertise in figure preparation.
Funding
DXZ is supported by an American Heart Association Predoctoral Fellowship. These studies were also supported by NIH grants DK091183, DK074868 and CA17390 to CKG.
References
- Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 2011;30:2817–2828. doi: 10.1038/emboj.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, Dent AL, Tangirala RK, Evans RM. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 2010;24:2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung MH, Trump S, Lightman SL, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buryachkovskaya L, Sumarokov A, Lomakin N. Historical overview of studies on inflammation in Russia. Inflamm Res. 2013;62:441–450. doi: 10.1007/s00011-013-0597-0. [DOI] [PubMed] [Google Scholar]
- Chen X, Barozzi I, Termanini A, Prosperini E, Recchiuti A, Dalli J, Mietton F, Matteoli G, Hiebert S, Natoli G. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc Natl Acad Sci U S A. 2012;109:E2865–E2874. doi: 10.1073/pnas.1121131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Drexler SK, Foxwell BM. The role of toll-like receptors in chronic inflammation. Int J Biochem Cell Biol. 2010;42:506–518. doi: 10.1016/j.biocel.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Escoubet-Lozach L, Benner C, Kaikkonen MU, Lozach J, Heinz S, Spann NJ, Crotti A, Stender J, Ghisletti S, Reichart D, et al. Mechanisms establishing TLR4-responsive activation states of inflammatory response genes. PLoS Genet. 2011;7:e1002401. doi: 10.1371/journal.pgen.1002401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Morinaga H, Kim JJ, Bae E, Spann NJ, Heinz S, Glass CK, Olefsky JM. FoxO1 regulates Tlr4 inflammatory pathway signalling in macrophages. EMBO J. 2010;29:4223–4236. doi: 10.1038/emboj.2010.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, Robinson J, Minie B, Chevrier N, Itzhaki Z, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell. 2012;47:810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol. 2010;10:365–376. doi: 10.1038/nri2748. [DOI] [PubMed] [Google Scholar]
- Hallson G, Hollebakken RE, Li T, Syrzycka M, Kim I, Cotsworth S, Fitzpatrick KA, Sinclair DA, Honda BM. dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development. Genetics. 2012;190:91–100. doi: 10.1534/genetics.111.135863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Allison KA, Kaikkonen MU, Orozco LD, Glass CK. Impact of natural genetic variation on enhancer selection and function. Nature. 2013 doi: 10.1038/nature12615. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettinger J, Richards DM, Hansson J, Barra MM, Joschko AC, Krijgsveld J, Feuerer M. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol. 2013 doi: 10.1038/ni.2638. [DOI] [PubMed] [Google Scholar]
- Hu G, Schones DE, Cui K, Ybarra R, Northrup D, Tang Q, Gattinoni L, Restifo NP, Huang S, Zhao K. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 2011;21:1650–1658. doi: 10.1101/gr.121145.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong KW, Kim K, Situ AJ, Ulmer TS, An W, Stallcup MR. Recognition of enhancer element-specific histone methylation by TIP60 in transcriptional activation. Nat Struct Mol Biol. 2011;18:1358–1365. doi: 10.1038/nsmb.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, et al. Remodeling of the Enhancer Landscape during Macrophage Activation Is Coupled to Enhancer Transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasowski M, Grubert F, Heffelfinger C, Hariharan M, Asabere A, Waszak SM, Habegger L, Rozowsky J, Shi M, Urban AE, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerenyi MA, Shao Z, Hsu YJ, Guo G, Luc S, O'Brien K, Fujiwara Y, Peng C, Nguyen M, Orkin SH. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. Elife. 2013;2:e00633. doi: 10.7554/eLife.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk MS, Hughes JR, Garrick D, Lynch MD, Sharpe JA, Sloane-Stanley JA, McGowan SJ, De Gobbi M, Hosseini M, Vernimmen D, et al. Intragenic enhancers act as alternative promoters. Mol Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- Kueh HY, Champhekhar A, Nutt SL, Elowitz MB, Rothenberg EV. Positive Feedback Between PU.1 and the Cell Cycle Controls Myeloid Differentiation. Science. 2013 doi: 10.1126/science.1240831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- Lazar MA. Mechanism of action of hormones that act on nuclear receptors. In: Melmed S, Williams RH, editors. Williams textbook of endocrinology. edn 12. Philadelphia: Elsevier/Saunders; 2011. pp. 52–61. [Google Scholar]
- Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Jr, Liu XS, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liber D, Domaschenz R, Holmqvist PH, Mazzarella L, Georgiou A, Leleu M, Fisher AG, Labosky PA, Dillon N. Epigenetic priming of a pre-B cell-specific enhancer through binding of Sox2 and Foxd3 at the ESC stage. Cell Stem Cell. 2010;7:114–126. doi: 10.1016/j.stem.2010.05.020. [DOI] [PubMed] [Google Scholar]
- Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature. 2012;484:251–255. doi: 10.1038/nature10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurya MR, Gupta S, Li X, Fahy E, Dinasarapu AR, Sud M, Brown HA, Glass CK, Murphy RC, Russell DW, et al. Analysis of Inflammatory and Lipid Metabolic Networks across RAW264.7 and Thioglycolate-elicited Macrophages. J Lipid Res. 2013 doi: 10.1194/jlr.M040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Shevach EM, Trinchieri G, Mellor AL, Munn DH, Gordon S, Libby P, Hansson GK, Shortman K, Dong C, et al. Highlights of 10 years of immunology in Nature Reviews Immunology. Nat Rev Immunol. 2011;11:693–702. doi: 10.1038/nri3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner F, Scheltema RA, Mollenkopf HJ, Mann M. Direct proteomic quantification of the secretome of activated immune cells. Science. 2013;340:475–478. doi: 10.1126/science.1232578. [DOI] [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Carboni GA, Guemes M, Bailey S, Anaya E, Corselli M, Peault B, Krum SA. GATA4 regulates estrogen receptor-alpha-mediated osteoblast transcription. Mol Endocrinol. 2011;25:1126–1136. doi: 10.1210/me.2010-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci U S A. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nau GJ, Richmond JF, Schlesinger A, Jennings EG, Lander ES, Young RA. Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci U S A. 2002;99:1503–1508. doi: 10.1073/pnas.022649799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 1998;12:2403–2412. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Jepsen K, Sawka-Verhelle D, Perissi V, Sasik R, Rose DW, Johnson RS, Rosenfeld MG, Glass CK. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc Natl Acad Sci U S A. 2004;101:14461–14466. doi: 10.1073/pnas.0405786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, Skotte J, Wutz A, Porse B, Jensen ON, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010;38:4958–4969. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998a;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Poltorak A, Smirnova I, He X, Liu MY, Van Huffel C, McNally O, Birdwell D, Alejos E, Silva M, Du X, et al. Genetic and physical mapping of the Lps locus: identification of the toll-4 receptor as a candidate gene in the critical region. Blood Cells Mol Dis. 1998b;24:340–355. doi: 10.1006/bcmd.1998.0201. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo K, Glass CK. Microglial cell origin and phenotypes in health and disease. Nat Rev Immunol. 2011;11:775–787. doi: 10.1038/nri3086. [DOI] [PubMed] [Google Scholar]
- Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, Neph S, Sabo P, Kim JM, Liao W, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EW, Fisher RC, Olson MC, Kehrli EW, Simon MC, Singh H. PU.1 functions in a cell-autonomous manner to control the differentiation of multipotential lymphoid-myeloid progenitors. Immunity. 1997;6:437–447. doi: 10.1016/s1074-7613(00)80287-3. [DOI] [PubMed] [Google Scholar]
- Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol. 2011;11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST. Selective transcription in response to an inflammatory stimulus. Cell. 2010;140:833–844. doi: 10.1016/j.cell.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann NJ, Garmire LX, McDonald JG, Myers DS, Milne SB, Shibata N, Reichart D, Fox JN, Shaked I, Heudobler D, et al. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 2012;151:138–152. doi: 10.1016/j.cell.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefflova K, Thybert D, Wilson MD, Streeter I, Aleksic J, Karagianni P, Brazma A, Adams DJ, Talianidis I, Marioni JC, et al. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell. 2013;154:530–540. doi: 10.1016/j.cell.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender JD, Pascual G, Liu W, Kaikkonen MU, Do K, Spann NJ, Boutros M, Perrimon N, Rosenfeld MG, Glass CK. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol Cell. 2012;48:28–38. doi: 10.1016/j.molcel.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- Sullivan AL, Benner C, Heinz S, Huang W, Xie L, Miano JM, Glass CK. Serum response factor utilizes distinct promoter- and enhancer-based mechanisms to regulate cytoskeletal gene expression in macrophages. Mol Cell Biol. 2011;31:861–875. doi: 10.1128/MCB.00836-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SK, Lin ZH, Chang CW, Varang V, Chng KR, Pan YF, Yong EL, Sung WK, Cheung E. AP-2gamma regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription. EMBO J. 2011;30:2569–2581. doi: 10.1038/emboj.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, O'Shea JJ. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken SJ, Locksley RM. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu Rev Immunol. 2013;31:317–343. doi: 10.1146/annurev-immunol-032712-095906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]