Summary
Background
Trichoplax adhaerens is the best-known member of the Phylum Placozoa, one of the earliest-diverging metazoan phyla. It is a small disk-shaped animal that glides on surfaces in warm oceans to feed on algae. Prior anatomical studies of Trichoplax revealed that it has a simple three-layered organization with four somatic cell types.
Results
We reinvestigate the cellular organization of Trichoplax using advanced freezing and microscopy techniques to identify localize and count cells. Six somatic cell types are deployed in stereotyped positions. A thick ventral plate, comprising the majority of the cells, includes ciliated epithelial cells, newly identified lipophil cells packed with large lipid granules, and gland cells. Lipophils project deep into the interior where they alternate with regularly spaced fiber cells whose branches contact all other cell types, including cells of the dorsal and ventral epithelium. Crystal cells, each containing a birefringent crystal, are arrayed around the rim. Gland cells express several proteins typical of neurosecretory cells, and a subset of them, around the rim, also expresses an FMRFamide-like neuropeptide.
Conclusions
Structural analysis of Trichoplax with significantly improved techniques provides an advance in understanding its cell types and their distributions. We find two previously undetected cell types, lipohil and crystal cells, and an organized body plan in which different cell types are arranged in distinct patterns. The composition of gland cells suggests that they are neurosecretory cells and could control locomotor and feeding behavior.
Introduction
Results of recent genome projects have renewed interest in the long-standing question of how the earliest-diverging animals are related [1–7]. Genomic sequences reveal that the basal lineages Porifera, Placozoa, Ctenophora and Cnidaria share a large fraction of the genetic toolkits involved in cell fate, patterning, differentiation, and cell-cell communication in bilaterians. Whether the homologous toolkits serve similar functions and how the gene products are parsed into cell types and organ systems in early diverging taxa remain largely unknown. Moreover, adequately detailed anatomical understanding to properly exploit the genome data is lacking in some early-branching groups.
Placozoans are a case in point. Placozoans are small, flat marine animals that adhere to substrates, locomote by ciliary gliding, feed by external digestion and propagate by fission [8–11]. The sole named member of the Phylum is Trichoplax adhaerens (Schulze). Following its discovery in the late 19th century [8, 9], the group was largely unstudied until the early 1970’s, when the first electron microscopic observations were made [10–13]. The early work revealed an organized body plan with four somatic cell types: dorsal and ventral epithelial cells, both ciliated but differing in shape; gland cells, so named due to their content of secretory granules; and branching fiber cells that occupy the interior. Profiles thought to represent degenerating cells and lipid granules not associated with an identified cell type also were reported.
Early investigations left several key questions unaddressed, in part because chemical fixation obscured some types of cells. Here freezing-based preparative methods for both light and electron microscopy are applied to reinvestigate the cell types and body plan of Trichoplax. We provide an extensive description of the cell types, including two new types of cells, lipophil and crystal cells, and demonstrate that gland cells express neuronal secretory proteins. Classification, mapping, and counting the cells in Trichoplax reveal aspects of the body plan and cell types that differ materially from those previously described.
Results
Freeze-substitution protocols adapted for transmission electron microscopy (TEM) and immunofluorescence light microscopy (LM) provide improvement in both structure and sensitivity over conventional fixation. Cells exhibit smooth, unbroken membranes, well-preserved cytoplasm, cytoskeletal fibers and organelles. The improved TEM procedure reveals six distinct cell types while the improved protocol for immunofluorescence allows further distinction between cell types and a quantitative picture of their distribution and relationships (Figure S1 and Table S1).
Ventral Epithelial Cells
The pseudo-stratified ventral epithelium facing the substrate is composed of several cell types (Figure 1). Most prevalent are small, dorso-ventrally elongated ventral epithelial cells [10] with multiple microvilli and a single cilium arising from a ciliary cup (Figure S2). Ventral epithelial cells contain one large (~1 µm) flocculent inclusion usually near the nucleus, dense granules (diameter ~200–400 nm) primarily near the basal surface, and smaller clear vesicles throughout the cell.
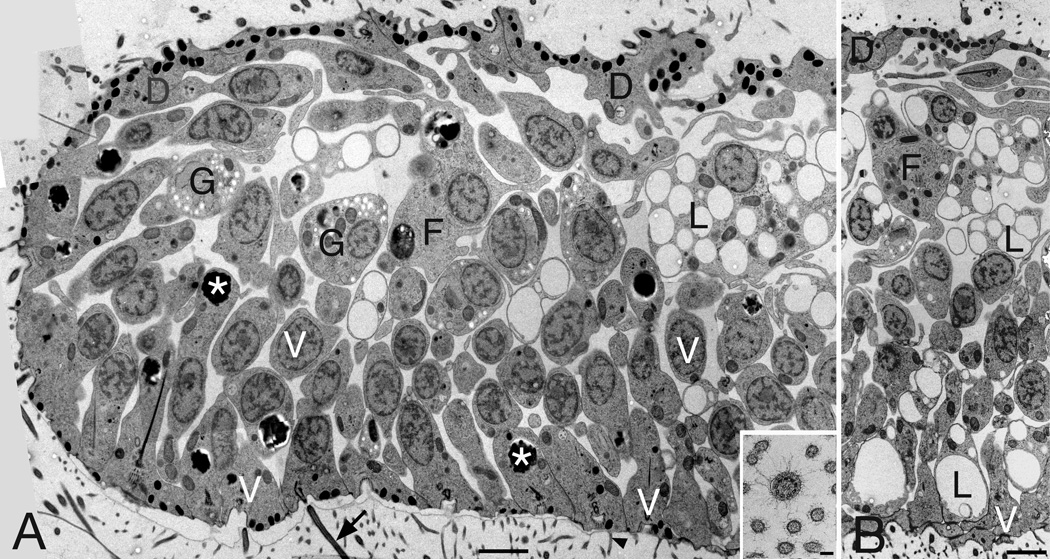
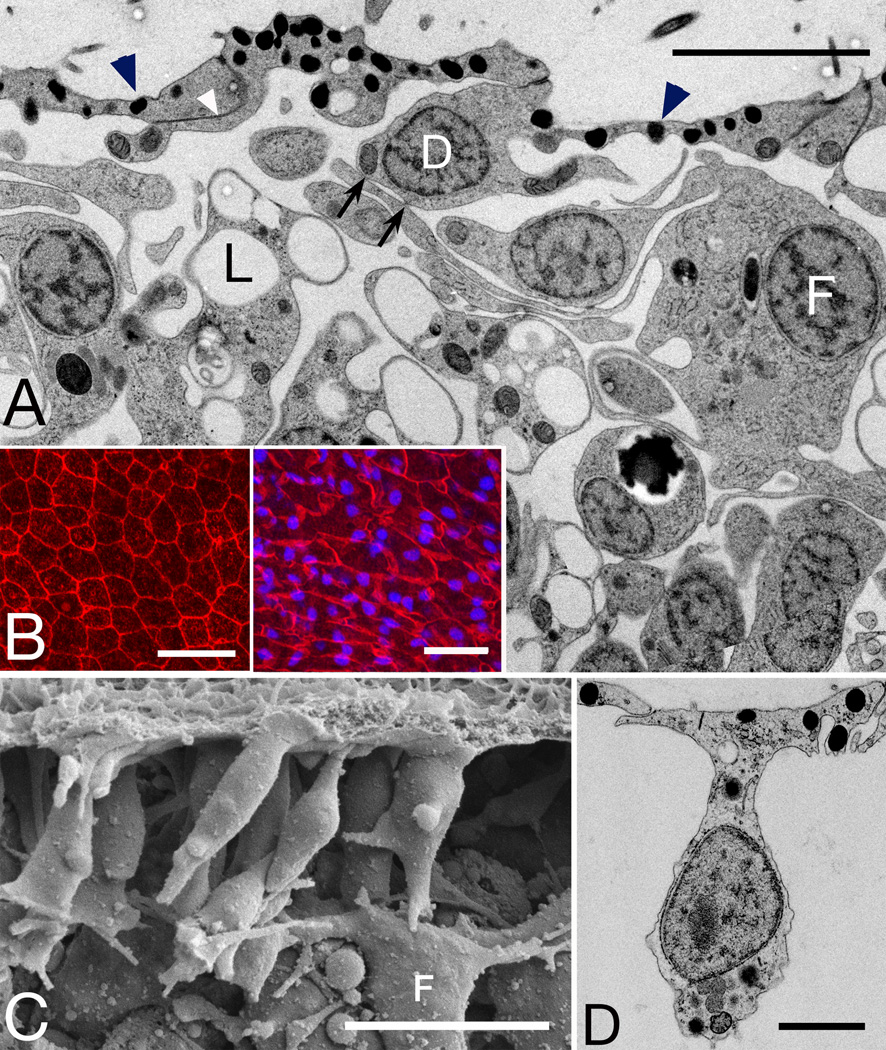
Figure 1. Trichoplax overview.
Montages of TEM micrographs show full thickness of a Trichoplax at its edge (A) and a central region 15-µm distant (B). Ventral side faces the substrate (bottom). Five of the six principle cell types are visible. A thin layer of dorsal epithelial cells (D) transition at the edge to the pseudo-columnar ventral epithelium consisting of ventral epithelial cells (V), lipophil cells (L), and gland cells (G), some cut along their long axes, others obliquely. In the interior, fiber cells with large cell bodies (F) mingle with the cell bodies of lipophil cells. Ventral epithelial cells bear typical cilium (arrow) and microvilli (arrowhead), and contain small dense granules near the ventral membrane and a large inclusion deep inside (asterisks). Inset: Cross section through a ventral cilium and surrounding microvilli. Scale bars represent 2 µm (A,B), 200 nm (inset).
Apical junctions join adjacent epithelial cells, which lack tight junctions and basal lamina characteristic of epithelia in other animals [10, 11], excepting some sponges [14]. Epithelial cells are outlined by fluorescent antibody to membrane-associated guanylate kinases (MAGUKs), proteins often associated with intercellular junctions (Figure 2D). The dense packing of the ciliated cells is evident with immunofluorescent stains for ciliary bases and cilia (Figure S1A). Ciliated ventral epithelial cells constitute 72% of the total cell population (Table S1).
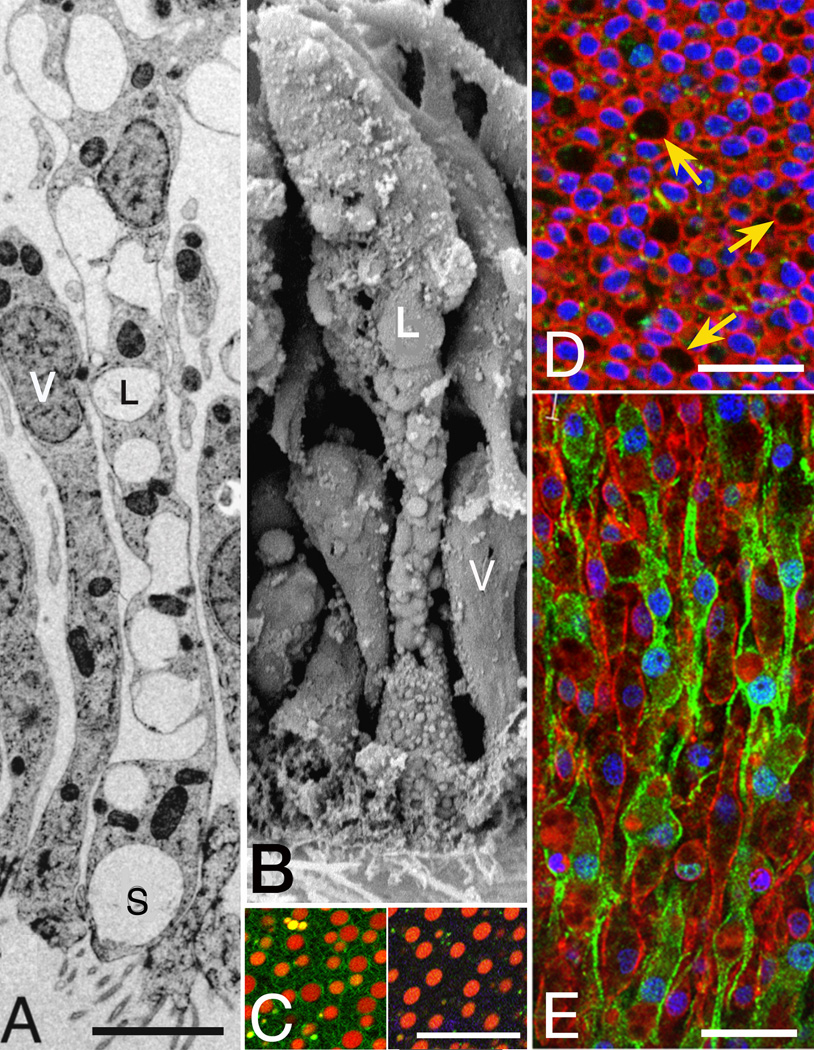
Figure 2. Lipophil Cells.
(A, B) TEM and SEM images show lipophil cells (L) interspersed with epithelial cells (V) in the ventral epithelium. (A) Lipophils are filled with numerous spherical granules (S) and consistently bear a large clear spherical granule at the ventral surface. (B) Knobby contours of lipophils (L) reflect their granular content, in contrast to the smooth surfaces of ventral epithelial cells (V). (C) Ventral surface of living Trichoplax stained with fluorescent dyes. The spherical inclusions in lipophil cells take up acidophilic Lysotracker Red (left, red; FM143 membrane dye, green) and lipophilic Nile Red (right) (D, E) Single optical sections showing immunofluorescence for MAGUKs (red) and TaCDH (green). (D) Anti-MAGUK staining near the ventral surface outlines ventral epithelial cells (small profiles, nuclei in blue) and lipophils (large profiles indicated by arrows). (E) Deep in the interior, lipophil cell bodies (nuclei outlined by anti-MAGUK staining) mingle with fiber cells outlined by anti-TaCDH. Scale bars represent 2 µm (A, B), 20 µm (C), 10 µm (D–E).
Lipophil Cells
Interspersed among the ciliated ventral epithelial cells are non-ciliated cells that each contain a large (~3 µm) membrane-enclosed spherical inclusion close to the ventral surface (Figures 1 and 2A). These inclusions correspond in size and location to large refractile inclusions at the ventral surface visible by bright field or DIC microscopy in living Trichoplax (Figure S1B), inclusions that Schulze referred to as matte-finished spheres [9]. In fortuitous sections, it is evident that the inclusions are in the ventral extensions of previously unidentified cells packed with spherical inclusions and having cell bodies lying deep in the interior (Figure 2A). In SEM, these cells are readily distinguished from other ventral epithelial cells by their larger sizes, location deeper in the interior and knobby contours (Figure 2B). The large ventral inclusions are not apparent in SEM, probably because they are extruded by chemical fixation. Indeed, granules attached to the ventral surface occasionally are observed by SEM, but the large inclusions in samples prepared by high pressure freezing and freeze substitution are exclusively intracellular. Upon OsO4 treatment, the inclusions show varying degrees of osmophilia, presumably reflecting their lipid content (not illustrated). In LM, the large inclusions stain intensely with Nile Red, a dye for neutral lipids (Figure 2C, right inset) and take up the fluorescent fatty acid analog C1-Bodipy-C12 (Figure S3B). Hence we refer to these previously uncharacterized cells as lipophil cells. Inclusions also are acidic, as they stain with the acidophilic Lysotracker Red (Figure 2C, left inset, S1B, S3B). Upon double staining, most large inclusions take up both dyes, while some smaller granules stain only with Lysotracker Red (Figure S3B). Lipophils outlined by MAGUK staining (Figures 2D–E) extend deep in the interior (continuity evident in serial optical sections) where their cell bodies are interspersed with fiber cells (see below). Large lipid inclusions are roughly evenly distributed throughout the ventral epithelium, but absent within ~20 µm of the rim (Figure S3B), and none occur in the dorsal epithelium (Figure S3A). Based on counts of large spheres stained with Lysotracker Red or Nile red, lipophils comprise 11% of the cells in Trichoplax, making them the second most prevalent cell type (Figure S1B and Table S1).
Gland cells
Gland cells, a subset of cells in the ventral epithelium thought to have a secretory function [10, 11], contain numerous 200–500 nm pale granules, pale in some cells (Figure 3C), darker in others (not illustrated), distributed throughout their cell bodies. Gland cells at the ventral surface typically manifest cilia and microvilli. Profiles deeper in the interior containing secretory granules (Figure 1) represent obliquely sectioned cells. Gland cells are more prevalent near the periphery of the ventral epithelium (within 20 µm of the edge) than further toward the center (Figure 1).
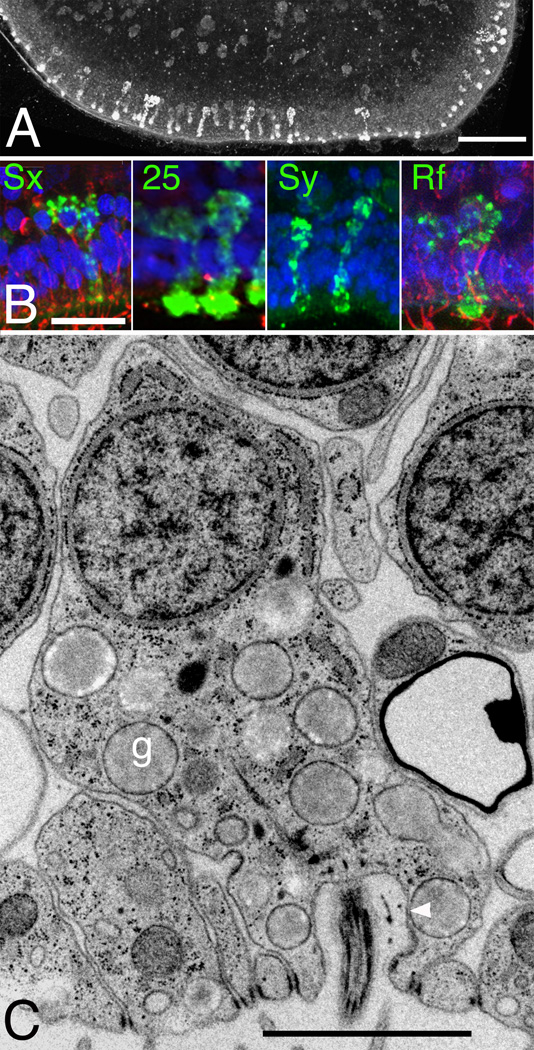
Figure 3. Gland cells.
(A, B) Projection of optical sections of the ventral epithelium. An antibody raised against syntaxin (A, B; Sx) strongly labels cells deployed near the edge (bottom) and, less intensely, scattered cells further from the edge. Their typical hourglass shape is revealed at the margin where they are tilted, while they appear circular when they are perpendicular to the imaging plane. (B) Cells of similar shape and appearance also immunolabel for SNAP-25 (25), synapsin (Sy), and FMRF-amide (Rf) (green; nuclei, blue; tubulin, red). (C) Electron micrograph of a ciliated gland cell reveals a population of membrane-enclosed granules (g) comparable in size with the stained granules in (B). Dense profile at right belongs to an adjacent lipophil cell. Arrowhead indicates contact between granule and surface. Scale bars represent 20 µm (A), 10 µm (B), 1 µm (C).
The genome of Trichoplax encodes for proteins highly homologous to proteins involved in regulated secretion in higher metazoans, including SNARE proteins for vesicular release [9]. Antibodies against the secretory SNARE proteins syntaxin1, synaptobrevin, SNAP-25 (Figures 3A–B, S1C, S3F–G) and synapsin (a peripheral membrane protein of synaptic vesicles; Figure 3B) consistently label a row of hourglass-shaped cells arrayed within ~ 20 µm from the edge and scattered cells in more central regions of the ventral epithelium. The cells further from the edge typically are less bright and appear circular in cross-section, but their hourglass shapes and position within the ventral epithelium are evident when viewed in serial optical sections (not illustrated). The labeling for syntaxin, synapsin and synaptobrevin (Figure 3B, Figure S1C and Figure S3F) is granular (~300–500 nm) and the sizes of the granules are consistent with the sizes of granules in gland cells observed by TEM. These granules are much smaller than those in lipophil cells. The distributions of cells immunostained for syntaxin, synaptobrevin and SNAP25 were counted and mapped and the results for all three were similar (see map for synaptobrevin in Figure S1C, Table S1). The labeled cells comprise about 3% of the total cell population, with roughly half concentrated in a ring around the rim. The close correspondence of the shapes, numbers and distributions of the labeled cells suggest that the antibodies all label the same cell type and that they correspond to gland cells seen by TEM. FMRFamide immunoreactive cells are arrayed around the periphery [15], and a polyclonal antibody against FMRFamide stains cells whose shapes and granular contents closely resemble those that express neurosecretory proteins (Figure 3B, SF3H). Thus, an FMRFamide-like peptide may be one of the secretory products of gland cells.
Fiber Cells
SEM micrographs reveal a population of cells, called fiber cells [10], evenly spaced throughout the interior of Trichoplax in a roughly single layer, each bearing six or more main branches and secondary ramifications that extend in all directions (Figure 4A). Surfaces are remarkably smooth, indicating a paucity of extracellular matrix material. The spacing between adjacent fiber cell bodies is uniformly smaller than the span of their processes. These long tapering processes extend around the dorsal surfaces of the ventral epithelial cells to penetrate between their underlying cell bodies. The processes also contact the underside of the dorsal epithelium (not seen in Figure 4A, but see Figure 5A,C) as well as other fiber cells and lipophils (Figure 4A). This arrangement brings essentially all cells in the interior within the reach of a fiber cell.
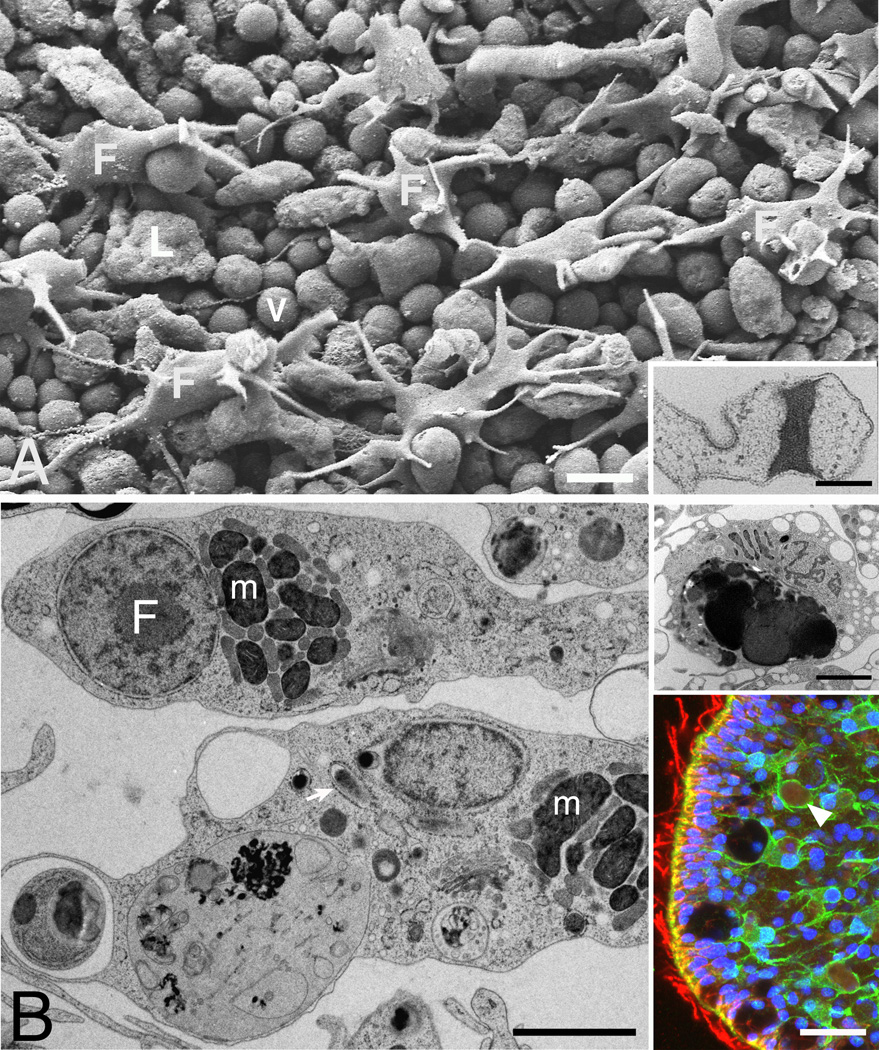
Figure 4. Fiber cells.
(A) SEM image of interior surface exposed by removing the dorsal epithelium. Regularly spaced fiber cells (F) extend multiple tapering processes around the cell bodies of underlying ventral epithelial cells (v). Lipophil (L) is identified by its content of granules. Inset. TEM shows dense septum bisecting the cytoplasm of a process resembling that of a fiber cell. The membrane is continuous across the septum. (B) TEM micrographs show characteristic inclusions of fiber cell bodies: clusters of mitochondria (m) interspersed with smaller inclusions; a rod-shaped structure within RER that may be a bacterium (arrow); and large electron dense inclusion (upper inset). Lower inset: Immunofluorescence staining for TaCDH (green) and tubulin (red) in an optical section near edge (left). TaCDH stain outlines fibers cells, identified by their branching patterns and large inclusions (dim red, arrowhead). Red channel was digitally enhanced (gamma 0.6) to accentuate the autofluorescence of the inclusions. Tubulin antibody stains microtubules in cells and cilia around the edge. Yellow represents overlapping signals of microtubules and exterior surfaces of epithelial cells. Nuclei blue. Scale bars represent 5 µm (A), 0.5 µm (A, inset), 2 µm (B), 10 µm (B, inset).
FIGURE 5. Dorsal epithelial cells.
(A) The dorsal surface is made of a single layer of thin cells containing dark granules (black arrowheads) and joined by apical junctions (white arrowhead). Small cell bodies of dorsal epithelial cells (D) are readily distinguished from a larger fiber cell (F) containing a rod-shaped inclusion and a lipophil (L) with numerous clear inclusions. Fiber cell processes contact dorsal cells (arrows). (B) En face views of dorsal epithelium after staining with rhodamine-phalloidin (left) or antibody against MAGUKs (right, red) illustrate the variable shapes of the cells. Nuclei blue. (C) SEM of dorsal epithelium broken open transversely to expose the columnar cell bodies of dorsal epithelial cells. A fiber cell (F) lies below the dorsal cells. (D) TEM micrograph shows a dorsal cell body giving rise to a flat expansion that connects to similar expansions of the adjoining cells, thus forming the thin dorsal surface of the Trichoplax. Scale bars represent 5 µm (A, C), 20 µm (B), 1 µm (D).
Additional identifying characteristics of fiber cells apparent by TEM [10, 11] (Figure 4B) are inclusions consisting of a cluster of mitochondria interspersed with paler bodies, rod-shaped inclusions (possibly symbiotic bacteria) [16] surrounded by rough endoplasmic reticulum (RER), and a large, complex electron-dense inclusion (Figure 4B, upper inset), or concrement vacuole [10], which may contain remnants of the algae that the organism feeds [17] and likely corresponds to the large inclusions seen in LM (Figure 4B, lower inset).
Fiber cells processes contact other cells, including cells of the ventral and dorsal epithelium (Figures 4A, 5A), yet do not form junctional specializations other than a faint dense material connecting the adjacent plasma membranes (Figure 5A). Dense septa previously interpreted as syncytial connections between fiber cells [11, 12] were rare (Figure 4A, inset).
Fiber cell contours are outlined by immunofluorescence with antibodies against peptides from predicted Trichoplax cadherin (TaCDH; Figure 4B, lower inset, 2E, S3E) and P2X receptor (Figure S1E). The varying outlines of the fiber cell arbors (symmetrical in Figure 4B; elongated in Figures 2E and S1E), likely reflect the momentary shape of the moving organism. Fiber cells are evenly distributed in the horizontal plane and constitute 4.4% of the population (Figure S1E and Table S1).
Dorsal Epithelial cells
The dorsal surface of Trichoplax is bounded by a thin layer of dorsal epithelial cells that are joined by junctions similar to those in the ventral epithelium and have a cilium and microvilli, although fewer microvilli than ventral cells. These cells also contain numerous dense, elliptical granules ~0.5 µm in diameter (Figures 1, 5). Fluorescent labeling for filamentous actin or MAGUKs (Figure 5B) outlines the shapes of the cells at the dorsal surface, demonstrating that the dorsal cells make a continuous pavement. The shapes vary between circular and elliptical, reflecting the overall shape of the animal. The cell bodies hang, attached by necks, from the flat disks making up the elements of the pavement (Figures 5C–D). The volume of the extracellular space below the dorsal epithelium and around their cell bodies (Figure 5C) varies between animals. Dorsal cells constitute 9% of the population (Figure S1D and Table S1).
Prior studies on Trichoplax and several recently-discovered strains of Placozoa reported lipid granules, called shiny spheres, on the dorsal surface [9–11, 13, 18]. Shiny spheres are not present at the dorsal surfaces of the Trichoplax used for our study (Figures 1, 5, and S3A). However, some cells on the dorsal surface of field-collected placozoans contain a large clear inclusion similar to the spherical inclusions at the ventral surface (Figure S2B). Cells containing these large inclusions resemble dorsal epithelium cells in that their cell bodies lie close to the dorsal surface and contain small dark granules. Like the large ventral inclusions of lipophils, those at the dorsal surface appear refractile by DIC microscopy and take up Lysotracker Red and the fluorescent fatty acid analog C1-Bodipy-C12 (Figure S3C–D). Aside from the presence of dorsal cells with large lipid inclusions, the cellular organization of this strain of Placozoa (Figure S2A–B) resembles that of Trichoplax.
Crystal Cells
Birefringent structures scattered around the margins of Trichoplax [19] are apparent under DIC or polarized LM (Figure S1F). Viewed living they modulate in intensity as they change orientation relative to the polarizer (Figure S1F, inset). At higher magnification, the crystals appear as rhomboids ~2 µm in diameter (Figure 6, inset) and are contained in cells that have a cup-shaped nucleus applied to one side of the cell body. These cells that we name crystal cells are rare but obvious by TEM because of the cup shaped nucleus and the centrally located pair of mitochondria flanking a rhomboid-shaped hole marking the location of the crystal, which typically falls out of the section (Figure 6). The cytoplasm of crystal cells is otherwise remarkably clear and free of inclusions. Crystal cells lie underneath the dorsal epithelium and adjacent to fiber cells and lipophil cells and are contacted by fiber cell processes. They are located more dorsally than gland cells and do not extend to the surface. Crystal cells do not label with antibodies against TaCDH, P2X receptor, or MAGUKs. Crystal cell numbers were estimated by counting birefringent crystals in living specimens (Figure S1F), only including animals in which crystals were visible around the entire rim (Figure S1F). Crystal cells constitute less that 0.2% of the cell population (Table S1).
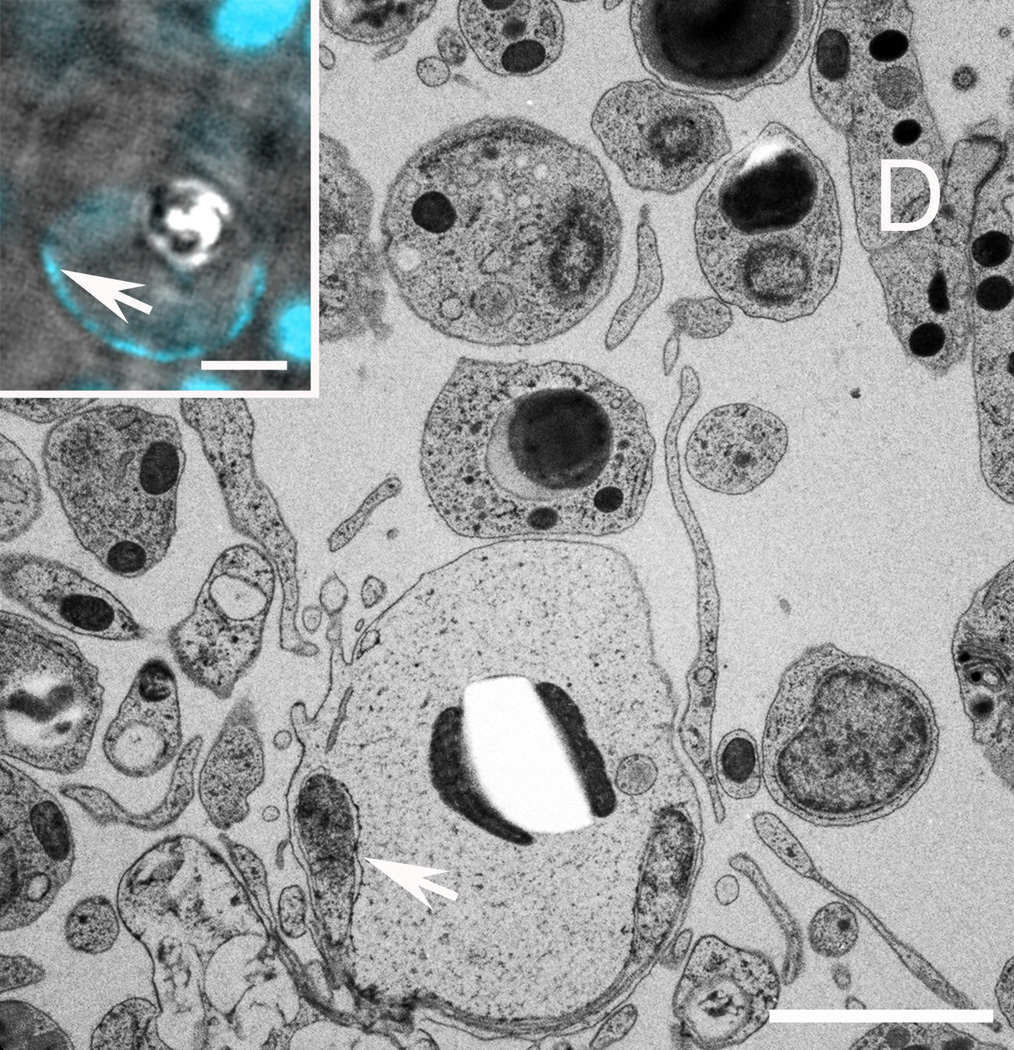
Figure 6. Crystal cells.
TEM micrograph shows crystal cell located near the dorsal surface (D, right) with flattened nucleus (arrow) and a central inclusion flanked by two mitochondria. Crystal cell inclusions do not survive sectioning for EM, leaving a clear, rhomboid hole in the section. Inset. DIC image of a crystal (bright rhomboid inclusion) in a cell in a living Trichoplax; nuclear stain (cyan, arrow) shows flattened nucleus. Scale bars represent 2 µm.
Discussion
Knowledge about the cell types and body plan of the early-diverging animal Trichoplax has not kept pace with the increasing amount of genomic information. Our improved structural methods yield a more detailed and quantitative picture of its cellular organization. The body plan that emerges (Figure 7) with six somatic cell types, rather than four, differs in significant ways from that described previously [10, 11, 18, 20].
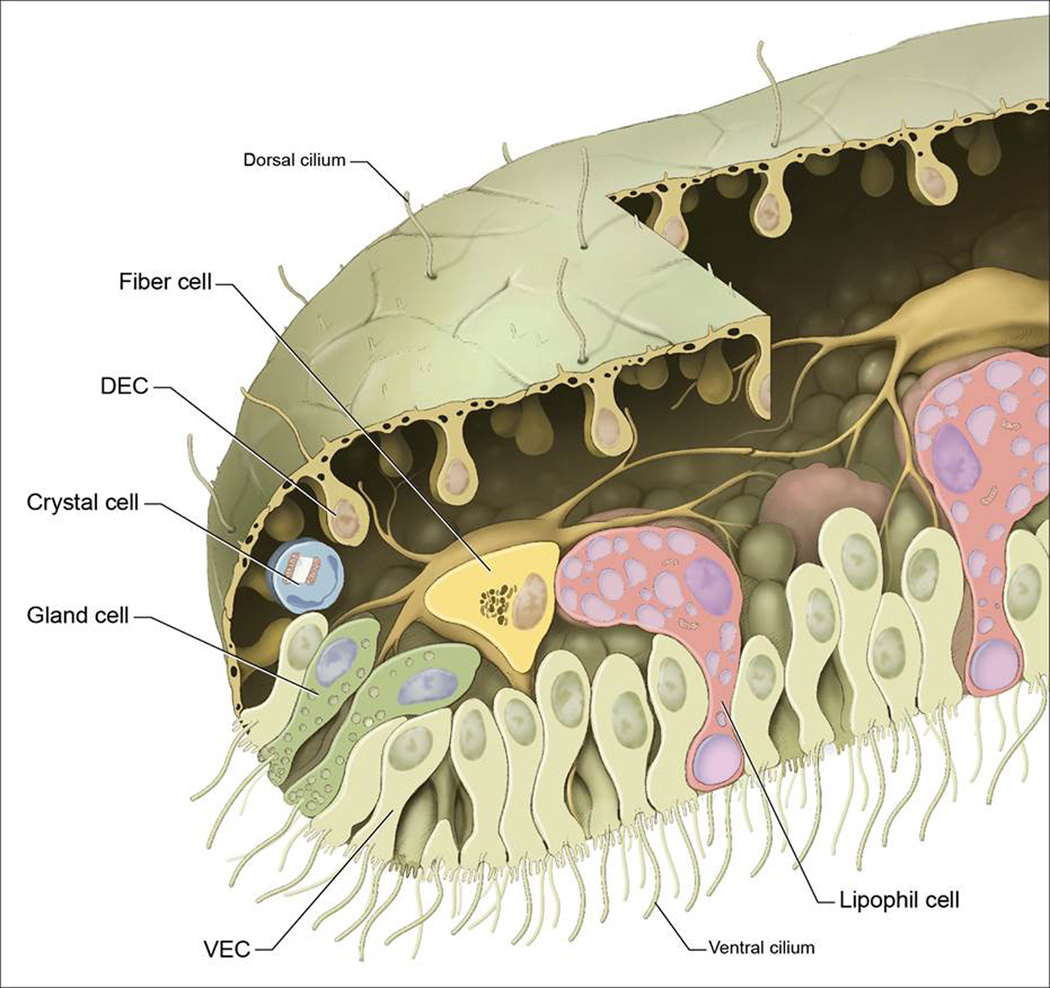
Figure 7.
Drawing summarizing Trichoplax cell types and body plan. Facing the substrate (below) is a thick ventral plate composed of: ventral epithelial cells (VEC; light yellow) each bearing a cilium and multiple microvilli; lipophil cells (brick) that contain large lipophilic inclusions, including a very large spherical inclusion near the ventral surface (lavender); and gland cells (pale green), distinguished by their contents of secretory granules and prevalence near the margin. Dorsal epithelial cells (DEC; tan) form a roof across the top from which are suspended their cell bodies surrounded by a fluid-filled space. In between the dorsal epithelium and ventral plate are fiber cells with branching processes that contact each of the other cell types. A crystal cell (pale blue) containing a birefringent crystal lies under the dorsal epithelium near the rim.
The ventral epithelium has been depicted as having only two cell types, ciliated ventral epithelial cells and gland cells, with interspersed vacuolated profiles assumed to be remains of decayed cells [10, 11]. The locations of these vacuolated profiles correspond with those of newly identified lipophil cells, the second most prevalent type of cell in Trichoplax. Lipophils are evenly distributed throughout the ventral epithelium but absent near the rim. In contrast, gland cells are prevalent around the rim but sparse in central regions, an arrangement not noted previously. The distinctive structures and distributions of the cells suggest they are deployed for different functions.
Trichoplax feeds by external digestion at its ventral surface, leaving partially decomposed algae behind [9, 11]. It pauses periodically and the duration and frequency of the pauses depend on the concentration of food, with pauses longer and more frequent when food is present [21]. Ciliated ventral epithelial cells have numerous microvilli (a point previously debated) [10, 11, 20], and in this respect, resemble cells in the digestion tracks of other animals [22]. The cells are thought to engage in vesicular uptake and uptake of ferritin has been demonstrated [20]. Their resemblance to cells of the digestive tracks of other animals suggests that they also may absorb small molecules via transporters [23]. Gland cells have been thought to secret the digestive enzymes [11] but are sparse and poorly positioned for this role. Lipophils, in view of their abundance, regular distribution in the ventral epithelium, and numerous membrane-bound inclusions seem more likely to secrete digestive enzymes.
Gland cells observed by TEM correspond in shape, granular contents and position to cells that immunolabel for neuronal secretory proteins, consistent with the suggestion that these are secretory cells. The cells around the rim also correspond in appearance and position to cells that immunolabel for the neuropeptide FMRFamide [15], consequently we suggest that gland cells are neurosecretory cells and that those arrayed around the rim likely secrete an FMRFamide-like peptide. The presence of neurosecretory cells in Trichoplax supports the idea that this cell type made an early appearance during metazoan evolution [24].
FMRFamide-like neuropeptides are found widely in the animal kingdom where they function in diverse ways including to regulate ciliary beat and feeding [25– 29]. Although gland cells do not make synapses, they might control the behavior of surrounding cells in a paracrine fashion. The Trichoplax genome contains sequences for an FMRFamide-like neuropeptide, two additional neuropeptides [30] and several classical neurotransmitters (Srivastava et al., 2008), and their expression has been confirmed [31]. Thus, gland cells may comprise a diverse set of neurosecretory cells. If they are chemosensory, as the presence of a cilium might indicate [28], then secretion could be regulated by the presence of food so as to modulate appropriately the activity of the ciliated and digestive cells.
In addition to gliding on its cilia, Trichoplax frequently changes shape, for example assuming an elliptical rather than a circular outline. It has been proposed that such shape changes are powered by contractions of fiber cell processes [9–11, 32]. Fiber cells do have long processes that contact cells in both dorsal and ventral epithelia, but these processes are thin, lack myofibrils and their contacts with other cells are not typical of adhesive contacts. In these three respects, fiber cells lack features expected of contractile cells, such as muscle cells.
Electron dense septa in fiber cell processes have been interpreted as syncytial junctions between fiber cells because the plasma membrane extends continuously across the septal region [12]. The septal junctions resemble the plug junctions that interconnect electrically-excitable cells in the syncytia of Hexactinellid sponges [33]. Membrane continuity across the septal junctions in sponges supports conduction of electrical signals, while the septa are thought to block exchange of large molecules and organelles [34, 35]. Whether the septal structures in Trichoplax support electrical conduction or are present in sufficient numbers and arrangements to be physiologically significant is uncertain.
Although we find that the dorsal epithelium of Trichoplax is composed of a single cell type and lacks lipid inclusions, previous studies reported lipid granules on the dorsal surface [10, 11, 13] and that these contain toxins for defense against predators [36]. This discrepancy could indicate that our clone has altered in culture. We confirmed that lipid inclusions are present at the dorsal surface in placozoans collected from the field and show that they reside in cells that otherwise resemble dorsal epithelial rather than lipophil cells.
Birefringent crystals are common in metazoans [37, 38], but the newly-identified crystal cells in Trichoplax bear little resemblance to other cells containing crystals. The distinctive shapes, contents, scarcity and distributions crystal cells raise the question of whether they might serve a sensory function, such as responding to gravity or light. There are anecdotal reports that Trichoplax responds to light and its genome includes most components of the genetic toolkit needed for photosensitivity [1, 39].
Small cells around the margin that express regulatory genes have been interpreted as possible stem cells [18, 32, 40–42]. Crystal cells and gland cells lie in roughly the same location but appear too differentiated to be stem cells. It is possible that stem cells, if present, lack distinctive markers that would have allowed us to distinguish them from other cells.
Analysis of Trichoplax with significantly improved structural techniques represents a significant advance in understanding its cell types and body plan. Trichoplax contains at least six somatic cell types: dorsal and ventral epithelial cells, lipophils, fiber cells, gland cells and crystal cells. Different cell types are arranged in distinct patterns–lipophils are roughly evenly interspersed within the ventral epithelium while crystal cells and most gland cells are arrayed in a ring near the edge of the animal. New details of cell structure suggest that functions previously attributed to specific cell types may need to be reassigned. Lipophils are likely candidates for digestive cells. Gland cells appear to be neurosecretory cells and could have a role in controlling the feeding and locomotor behavior. The improved structural methods and markers for specific cell types introduced in this study provide tools that will prove useful in future studies of placozoans.
Experimental Procedures
Animals
Trichoplax adhaerens of the Grell (1971) strain from Dr. Leo Buss (Yale University) and Dr. Bernd Schierwater (TiHo Hannover, Germany) were maintained in artificial seawater (ASW; Instant Ocean) with 1% Micro Algae Grow (Florida Aqua Farms Inc. Dade City, FL) as described [43, 44]. Illustrations are derived from the Trichoplax provided by Dr. Buss but observations apply to Trichoplax from both sources. They were fed red algae Rhodamonas salina (Pyrenomonas salina, Provasoli-Guillard National Center for Culture of Marine Plankton, West Boothbay, ME), Pyrenomonas Helgolandii (SAG, Göttingen) or green algae Nannochloropsis (Florida Aqua Farms, Inc.). Placozoans we discovered in a shipment of marine sponges collected from the wild (Florida Aqua Farms) were used for Figures S2 and S3C–D. These placozoans were maintained in a saltwater aquarium containing self-sustaining phytoplankton without added food.
Trichoplax were transferred to Labtech II coverslip chambers (Thermo Fisher Scientific) for observation by LM. Vital dyes were: Nile Red, C1-Bodipy 500/510-C12, Lysotracker Red, FM1-43, and Hoechst dye (Life Technologies).
Antibodies
Both custom-made and commercially available antibodies were used. Rabbit polyclonal antibodies were custom-made by New England Peptide (Fitchburg, MA) based on Trichoplax protein sequences [1, 31] highly conserved in other metazoans. These antibodies were to peptides from: predicted P2X-like purinergic receptor (TRIADDRAFT_57279), Ac-SETHKNHDAHC-amide; predicted classical cadherin TaCDH [45], Ac-LDNPLRESRTSRYC-amide; uncharacterized protein (TRIADDRAFT_54798), Ac-QPSDGDEHQASVEC-amide. The TaCDH and P2X antibodies were useful because they outlined fiber cells, and the third antibody (CilB) because it labeled the bases of cilia. Rabbit polyclonal antibodies generated against syntaxins (BoNT/C fragment of rat syntaxin1; Aa 183–240 of rat syntaxin 1A; Synaptic Systems #110402, abbreviation “Sx” in illustrations, and #110022), synaptobrevin (Synaptic Systems #104002, “Sb”, and #104102), SNAP-25 (full-length leech SNAP-25, “25”), synapsin (Synaptic Systems #106002, “Sy”; gift of Reinhard Jahn, Max Planck Institute, “Sy4”; Millipore AB1543). Antibodies against α-tubulin (T9026, Sigma-Aldrich), MAGUKs (Pan-MAGUK K28/86, NeuroMab), FMRF-amide (H-047-29, Phoenix Pharmaceuticals, Inc.) also were used. Staining with each antibody was performed in at least three animals in two or more separate experiments. Different antibodies against the same protein (three for syntaxin, two for synaptobrevin and three for synapsin) produced similar staining patterns, as expected if they recognize the same antigen. Western blots from homogenates of 250–300 Trichoplax with anti-synaptobrevin (Sb, Figure SF3H; #104102 not shown) labeled a band at 18 kDa, the expected molecular weight for Trichoplax synaptobrevin. Blots with two synapsin 1 antibodies (Sy, Sy4, Figure SF3H) labeled a band at 72 kDa. A synapsin homolog has not been identified in Trichoplax but mammalian synapsin is 80 kDa.
Immunofluorescence
Our protocol for preparing Trichoplax for immunofluorescence microscopy by freeze-substitution was adapted from a previous protocol [46]. Glass coverslips (#1.5) cleaned in nitric acid were coated with 0.1% protamine sulfate (1 hour), rinsed, coated with 1% PureCol (Advanced Biomatrix) and dried. Trichoplax in ASW were transferred to coverslips and left to adhere. Coverslips were transferred to dishes containing ASW with 0.3 M sucrose or 50% ASW and 50% 0.97 M mannitol for 1–2 minutes, until animals flattened. Coverslips were then plunged into tetrahydrafuran (Figures 2–4; S1A, C, E; S3F–G), acetone (Figure 5B, right), or MEOH (Figure S3E) at −80° on dry ice in a Styrofoam box and left for 16 hours. Coverslips were transferred to methanol with 1.6% paraformaldehyde on dry ice for 1 hour, held at −20°C for 2–3 hours, and room temperature (RT) for 2 hours. The specimens were rinsed in 100% ETOH and rehydrated in 50% ETOH and PBS each for 1 minute. Specimens were incubated in blocking buffer (BB: 3% normal goat serum, 2 % horse serum, 1% BSA in PBS) and then BB with primary antibody overnight at 4°C. Secondary antibodies were Atto 488 goat anti-mouse IgG (62197, Sigma-Aldrich) and Alexa 555 goat anti-rabbit (A-21428, Life Technologies) diluted 1:500 in BB and incubated for 2 hours at RT. Nuclei were stained with Hoechst. To visualize filamentous actin, specimens were fixed in 4% paraformaldehyde with 0.1% Triton-X in HASW (ASW with 20 mM HEPES; pH 7.6) for 30 minutes, rinsed in PBS stained with rhodamine- or Alexa 488-phalloidin (Life Technologies, 1:40) for 30 minutes. Samples on coverslips were mounted in Vectashield (Vector Laboratories) on glass slides with EM grids sandwiched between as spacers. Controls for nonspecific binding of the secondary antibodies showed no staining.
Immunofluorescence was readily distinguishable from autofluorescence, which was less bright and had broader excitation and emission spectra. Incubation of anti-FMRFamide antibody with FMRFamide peptide (Phoenix Peptide, #047-29) prior to use eliminated staining by the antibody (Figure S3H; representative of six animals in two separate experiments).
Light Microscopy
Fluorescence and DIC microscopy were performed on a laser scanning confocal microscope (LSM 510 or LSM 780, Carl Zeiss Microscopy LLC; SP5, Leica Microsystems) with 405, 488 and 561 nm illumination and a 40X NA 1.3, 63X NA 1.4, or 100X NA 1.4 objective. Single horizontal optical sections or maximum intensity projections of stacks of optical sections are illustrated. Cell maps and counts were made from image stacks reconstructed with Volocity 3D Image Analysis Software (PerkinElmer). Confocal DIC (Figure S1B; Figure 40X C-APO, NA 1.2) and polarization microscopy (Figure S1F; 10X NA 0.45) were with 633 nm illumination on a LSM510.
Scanning electron microscopy (SEM)
Trichoplax were transferred to glass coverslips, cleaned with nitric acid, before fixation in 2% OsO4 and 2% glutaraldehyde in 0.1 M Na cacodylate in 0.45 M NaCl, pH 7.4 on ice for 20 minutes [13]. The specimens were rinsed in HASW (pH 7.4) and stored in HASW with 0.3% glutaraldehyde. Secondary fixation was in 1% OsO4 for 1 hr at RT followed by dehydration in ethanols, with final transfer to three changes of hexamethyldisilazane (SPI). Coverslips were drained and residual hexamethyldisilazane dried in a 37°C oven. After mounting on SEM stubs, the tops of individual Trichoplax were removed with adhesive tape affixed to toothpicks. Specimens were immediately sputter-coated with platinum/carbon and examined at 10 KV in a Zeiss FE SEM.
Transmission electron microscopy (TEM)
Trichoplax were frozen in a high pressure freezing machine (Baltec 010, TechnoTrade) in gold specimen carriers (Leica). Trichoplax transferred to the 3.0 × 0.8 mm carrier were restrained with 1.5% low melting point agar at 30°C in ASW. The specimen was left at RT for ~ 1 minute, then hexadecene (Sigma-Aldrich) was layered over it and the chamber was covered and immediately frozen in the high pressure freezing machine.
Freeze-substitution was performed in a Leica AFS 1 by traversing a series of temperature ramps (5°C /hour) and plateaus [47]. All reagents were dissolved in HPLC grade acetone (Chemsolv, Sigma-Aldrich). Rinses were three times in dry acetone except as noted. Samples were placed under liquid nitrogen in a vial in which 4% acrolein (Sigma) was layered over previously frozen, saturated uranyl acetate (Polysciences). The temperature was ramped from −160° to −90° C, held 8 hours, ramped to −60°C and held 12 hours. Samples were rinsed and transferred to 1% OsO4 (Electron Microscopy Sciences) for 1 hour at −60 °C. Temperature was ramped to −30°C at 2°C /hour, held 8 hours, and then ramped to −10° C. Some samples were block-stained with 0.1% hafnium chloride [48] in acetone for 1 hour. Samples were rinsed, block stained with saturated uranyl acetate in acetone overnight, brought to room temperature, rinsed twice with acetone, once with methanol, once with acetone, and embedded in Epon. Embedded samples were removed from hemisected specimen carriers by cooling in liquid nitrogen and warming. Sections were examined, with or without a short application of uranyl acetate and lead citrate stains, at 120–200 KV in a JEOL 200-CX electron microscope, and photographed with an AMT camera mounted below the column.
Supplementary Material
Highlights.
Trichoplax has six cell types each arrayed in a distinct pattern
Structural cryotechniques reveal two new cell types, lipophils and crystal cells
Lipophils contain large lipid granules, crystal cells contain one birefringent crystal
Gland cells express proteins and a peptide characteristic of neurosecretory cells
Acknowledgements
We thank Leo Buss and Bernd Schierwater for providing us with Trichoplax adhaerens and cultivation advice, and Joan Barrick, Soe Thein, Vincent Schram, Carolin Wichman, Jan Hegermann, Stefan Eimer, Cordelia Imig, Klaus Hellmann and Nils Brose for support. Alan Hoofring, NIH Medical Arts, produced the graphics for the highlight and Figure 7. This work was supported by the National Institute of Neurological Disorders and Stroke intramural program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
References
- 1.Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- 2.Suga H, Chen Z, de Mendoza A, Sebé-Pedrós A, Brown MW, Kramer E, Carr M, Kerner P, Vervoort M, Sánchez-Pons N, et al. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat. Commun. 2013;4:2325. doi: 10.1038/ncomms3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 4.Ryan JF, Pang K, Schnitzler CE, Nguyen A-D, Moreland RT, Simmons DK, Koch BJ, Francis WR, Havlak P, Smith SA, et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342:1242592. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conaco C, Neveu P, Zhou H, Arcila ML, Degnan SM, Degnan BM, Kosik KS. Transcriptome profiling of the demosponge Amphimedon queenslandica reveals genome-wide events that accompany major life cycle transitions. BMC Genomics. 2012;13:209. doi: 10.1186/1471-2164-13-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, et al. The dynamic genome of Hydra. Nature. 2010;464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulze FE. Trichoplax adhaerens. nov. gen., nov. spec. Zool. Anzeiger - A J. Comp. Zool. 1883;6:92–97. [Google Scholar]
- 9.Schulze FE. Uber Trichoplax adhaerens. Phys. Abh. Kgl. Acad. Wiss. Berl. 1892:1–23. [Google Scholar]
- 10.Grell KG, Benwitz G. Die Ultrastruktur von Trichoplax adhaerens F.E. Schulze. Cytobiol 4:216–240. Cytobiologie. 1971;4:216–240. [Google Scholar]
- 11.Grell KG, Ruthmann A. Placozoa. In: Harrison FW, Westfall JA, editors. Microscopic Anatomy of Invertebrates. New York: Wiley-Liss; 1991. pp. 13–27. [Google Scholar]
- 12.Grell KG, Benwitz G. Spezifische Verbindungsstrukuren der Faserzellen von Trichoplax ad- haerens F.E. Schulze. Z. Naturforsch. 1974;29c:790. [PubMed] [Google Scholar]
- 13.Rassat J, Ruthmann A. Trichoplax adhaerens F.E. Schulze (Placozoa) in the Scanning Electron Microscope. Zoomorphologie. 1979;72:59–72. [Google Scholar]
- 14.Leys SP, Riesgo A. Epithelia, an evolutionary novelty of metazoans. J. Exp. Zool. Part B Mol. Dev. Evol. 2011;318 doi: 10.1002/jez.b.21442. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 15.Schuchert P. Trichoplax adhaerens (Phylum Placozoa) has cells that react with antibodies against the neuropeptide RFamide. Acta Zool. 1993;74:115–117. [Google Scholar]
- 16.Driscoll T, Gillespie JJ, Nordberg EK, Azad AF, Sobral BW. Bacterial DNA sifted from the Trichoplax adhaerens (Animalia:Placozoa) genome project reveals a putative rickettsial endosymbiont. Genome Biol. Evol. 2013 doi: 10.1093/gbe/evt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wenderoth H. Transepithelial cytophagy by Trichoplax adhaerens F.E. Schulze (Placozoa) feeding on yeast. Z. Naturforsch. 1986;41c:343–347. [Google Scholar]
- 18.Guidi L, Eitel M, Cesarini E, Schierwater B, Balsamo M. Ultrastructural analyses support different morphological lineages in the phylum placozoa Grell, 1971. J. Morphol. 2011;272:371–378. doi: 10.1002/jmor.10922. [DOI] [PubMed] [Google Scholar]
- 19.Pearse VB, Uehara T, Miller RL. Birefringent Granules in Placozoans. Transcations Am. Microsc. Soc. 1994;113:385–389. [Google Scholar]
- 20.Ruthmann A, Behrendt G, Wahl R. The ventral epithelium of Trichoplax adhaerens (Placozoa): Cytoskeletal structures, cell contacts and endocytosis. Zoomorphology. 1986;106:115–122. [Google Scholar]
- 21.Ueda T, Koya S, Maruyama YK. Dynamic patterns in the locomotion and feeding behaviors by the placozoan Trichoplax adhaerens. Biosystems. 1999;54:65–70. doi: 10.1016/s0303-2647(99)00066-0. [DOI] [PubMed] [Google Scholar]
- 22.Rupert EE, Fox RS, Barnes RB. Invertebrate Zoology, A functional evolutionary approach. 7th ed. Belmont, CA: Brooks Cole Thomson; 2004. [Google Scholar]
- 23.Field M, Frizzell RA, editors. Handbook of Physiology: Gastrointestinal System. Vol. 4. Bethesda, MD: American Physiological Society; 1991. [Google Scholar]
- 24.Hartenstein V. The neuroendocrine system of invertebrates: a developmental and evolutionary perspective. J. Endocrinol. 2006;190:555–570. doi: 10.1677/joe.1.06964. [DOI] [PubMed] [Google Scholar]
- 25.Kass-Simon G, Pierobon P. Cnidarian chemical neurotransmission, an updated overview. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007;146:9–25. doi: 10.1016/j.cbpa.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Katsukura Y. Control of planula migration by LWamide and RFamide neuropeptides in Hydractinia echinata. J. Exp. Biol. 2004;207:1803–1810. doi: 10.1242/jeb.00974. [DOI] [PubMed] [Google Scholar]
- 27.Conzelmann M, Offenburger S-L, Asadulina A, Keller T, Münch TA, Jékely G. Neuropeptides regulate swimming depth of Platynereis larvae. Proc. Natl. Acad. Sci. U. S. A. 2011;108:E1174–E1183. doi: 10.1073/pnas.1109085108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jékely G. Origin and early evolution of neural circuits for the control of ciliary locomotion. Proc. R. Soc. B Biol. Sci. 2011;278:914–922. doi: 10.1098/rspb.2010.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krajniak KG. Invertebrate FMRFamide Related Peptides. Protein Pept. Lett. 2013;20:647–670. doi: 10.2174/0929866511320060005. [DOI] [PubMed] [Google Scholar]
- 30.Jékely G. Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8702–8707. doi: 10.1073/pnas.1221833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ringrose JH, van den Toorn HWP, Eitel M, Post H, Neerincx P, Schierwater B, Maarten Altelaar AF, Heck AJR. Deep proteome profiling of Trichoplax adhaerens reveals remarkable features at the origin of metazoan multicellularity. Nat. Commun. 2013;4:1408. doi: 10.1038/ncomms2424. [DOI] [PubMed] [Google Scholar]
- 32.Schierwater B. My favorite animal, Trichoplax adhaerens. Bioessays. 2005;27:1294–1302. doi: 10.1002/bies.20320. [DOI] [PubMed] [Google Scholar]
- 33.Mackie GO, Singla CL. Studies on Hexactinellid Sponges. I. Histology of Rhabdocalyptus dawsoni (Lambe, 1873) Philos. Trans. R. Soc. B Biol. Sci. 1983;301:365–400. [Google Scholar]
- 34.Mackie GO, Lawn ID, Ceccatty MPD. Studies on Hexactinellid Sponges. II. Excitability, Conduction and Coordination of Responses in Rhabdocalyptus dawsoni (Lambe, 1873) Philos. Trans. R. Soc. B Biol. Sci. 1983;301:401–418. [Google Scholar]
- 35.Leys S, Mackie G, Meech R. Impulse conduction in a sponge. J. Exp. Biol. 1999;202:1139–1150. doi: 10.1242/jeb.202.9.1139. [DOI] [PubMed] [Google Scholar]
- 36.Jackson AM, Buss LW. Shiny spheres of placozoans (Trichoplax) function in anti-predator defense. Invertebr. Biol. 2009;128:205–212. [Google Scholar]
- 37.Denton EJ. Review Lecture: On the Organization of Reflecting Surfaces in Some Marine Animals. Philos. Trans. R. Soc. B Biol. Sci. 1970;258:285–313. doi: 10.1098/rstb.1970.0037. [DOI] [PubMed] [Google Scholar]
- 38.Gur D, Politi Y, Sivan B, Fratzl P, Weiner S, Addadi L. Guanine-based photonic crystals in fish scales form from an amorphous precursor. Angew. Chem. Int. Ed. Engl. 2013;52:388–391. doi: 10.1002/anie.201205336. [DOI] [PubMed] [Google Scholar]
- 39.Feuda R, Hamilton SC, Mcinerney JO, Pisani D. Metazoan opsin evolution reveals a simple route to animal vision. 2012 doi: 10.1073/pnas.1204609109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jakob W, Sagasser S, Dellaporta S, Holland P, Kuhn K, Schierwater B. The Trox-2 Hox/ParaHox gene of Trichoplax (Placozoa) marks an epithelial boundary. Dev. Genes Evol. 2004;214:170–175. doi: 10.1007/s00427-004-0390-8. [DOI] [PubMed] [Google Scholar]
- 41.Martinelli C, Spring J. Distinct expression patterns of the two T-box homologues Brachyury and Tbx2/3 in the placozoan Trichoplax adhaerens. Dev. Genes Evol. 2003;213:492–499. doi: 10.1007/s00427-003-0353-5. [DOI] [PubMed] [Google Scholar]
- 42.Martinelli C, Spring J. Expression pattern of the homeobox gene Not in the basal metazoan Trichoplax adhaerens. 2004 doi: 10.1016/j.modgep.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 43.Adamska M, Degnan BM, Green K, Zwafink C. What sponges can tell us about the evolution of developmental processes. Zoology (Jena) 2011;114:1–10. doi: 10.1016/j.zool.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Wörheide G, Dohrmann M, Erpenbeck D, Larroux C, Maldonado M, Voigt O, Borchiellini C, Lavrov DV. Deep phylogeny and evolution of sponges (phylum Porifera) Adv. Mar. Biol. 2012;61:1–78. doi: 10.1016/B978-0-12-387787-1.00007-6. [DOI] [PubMed] [Google Scholar]
- 45.Hulpiau P, Van Roy F. New Insights into the Evolution of Metazoan Cadherins. Mol. Biol. Evol. 2010;28:647–657. doi: 10.1093/molbev/msq233. [DOI] [PubMed] [Google Scholar]
- 46.Terasaki M, Reese TS. Characterization of endoplasmic reticulum by co-localization of BiP and dicarbocyanine dyes. J. Cell Sci. 1992;101(Pt 2):315–322. doi: 10.1242/jcs.101.2.315. [DOI] [PubMed] [Google Scholar]
- 47.Chen X, Winters C, Azzam R, Sourse AA, Leapman RD, Reese TS. Nanoscale Imaging of Protein Molecules at the Postsynaptic Density. In: Nägerl UV, Triller A, editors. Nanoscale Imaging of Synapses : New Concepts and Opportunities. New York: Heidelberg; 2013. pp. 1–21. [Google Scholar]
- 48.Ornberg RL, Reese TS. Beginning of exocytosis captured by rapid-freezing of Limulus amebocytes. J. Cell Biol. 1981;90:40–54. doi: 10.1083/jcb.90.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.