ABSTRACT
The mammalian gut contains a complex assembly of commensal microbes termed microbiota. Although much has been learned about the role of these microbes in health, the mechanisms underlying these functions are ill defined. We have recently shown that the mammalian gut contains thousands of small molecules, most of which are currently unidentified. Therefore, we hypothesized that these molecules function as chemical cues used by hosts and microbes during their interactions in health and disease. Thus, a search was initiated to identify molecules produced by the microbiota that are sensed by pathogens. We found that a secreted molecule produced by clostridia acts as a strong repressor of Salmonella virulence, obliterating expression of the Salmonella pathogenicity island 1 as well as host cell invasion. It has been known for decades that the microbiota protects its hosts from invading pathogens, and these data suggest that chemical sensing may be involved in this phenomenon. Further investigations should reveal the exact biological role of this molecule as well as its therapeutic potential.
IMPORTANCE
Microbes can communicate through the production and sensing of small molecules. Within the complex ecosystem formed by commensal microbes living in and on the human body, it is likely that these molecular messages are used extensively during the interactions between different microbial species as well as with host cells. Deciphering such a molecular dialect will be fundamental to our understanding of host-microbe interactions in health and disease and may prove useful for the design of new therapeutic strategies that target these mechanisms of communication.
INTRODUCTION
The human body is colonized by a complex community of commensal microbes, collectively termed microbiota (1–5). In the past few decades, a wealth of knowledge on the importance of the human microbiota has emerged. This is particularly true for the microbiota residing in the gastrointestinal tract, which is critical for the development of the immune system, production of vitamins, and protection against pathogens, together with other important roles (1, 6–8). Although many general functions of the intestinal microbiota have been identified, due to the complex nature of this microbial assembly and its interactions with the host, in most cases the mechanisms involved are still ill defined.
We have used a high-throughput metabolomics approach to study the chemical complexity of the mammalian gastrointestinal tract and to investigate the impact of the intestinal microbiota on the small-molecule composition of feces (9). The results showed that the chemical composition of the mammalian intestinal tract is highly complex, and thousands of small molecules could be detected. In nature, small molecules are often involved as chemical cues; it has been known for over a century that mammals use small molecules as tools to convey messages throughout the body (10). These small molecules, termed hormones, are used as autocrine, paracrine, and endocrine signals that allow the organism to maintain homeostasis as well as respond to external insults, such as infections (11–14). More recently, it was shown that microbes also communicate using chemical signals (15–17). Dozens of microbial species are now recognized to produce and respond to small-molecule signals. One such form of communication is termed quorum sensing, and new signals continue to be discovered (17–19). Therefore, we hypothesized that within the chemical diversity found in the gastrointestinal tract, many of the molecules could constitute chemical cues important for the communication between the gut microbiota, host cells, and invading pathogens and that the sensing events involved could be a critical factor in controlling the balance between health and disease. To address this, we studied the effect of molecules extracted from human feces on microbial gene expression using the invasive enteric pathogen Salmonella enterica serovar Typhimurium as a model. Our results showed that this pathogen responds readily to the presence of molecules from the human gut and that the expression of more than 100 genes is affected by the gut metabolome. Of note, Salmonella invasion gene expression is highly repressed by molecules from the mammalian gut, supporting the notion that chemical sensing may be critical to the control of virulence. Our studies have also determined that this biological activity is widespread in humans and can be recapitulated in the laboratory by employing isolated Clostridium species. Further studies should reveal the regulatory networks involved in sensing active molecules as well as the potential of the gut metabolome as a source of new antivirulence therapeutics.
RESULTS
The mammalian gut metabolome is rich in molecular diversity.
We have shown elsewhere that the mammalian gut metabolome contains thousands of small molecules and that many of these molecules have critical biological functions (9, 20, 21). Using direct infusion Fourier transform ion cyclotron resonance mass spectrometry (DI-FT-ICR-MS) in both negative and positive ionization modes, we detected a combined total of 2,429 metabolites in the murine gut metabolome (9). In order to assess the degree of chemical complexity and novelty of this environment and its potential as a source of new biologically active molecules, we analyzed this data set to determine the proportion of unidentified molecules. We used the metabolites detected previously to search the MassTRIX: Mass TRanslator into Pathways database (http://masstrix3.helmholtz-muenchen.de/masstrix3/) (22). Our results showed that of the 1,564 small molecules annotated as part of metabolic pathways on MassTRIX, only 200 (12.8%) were detected in our data set. When we considered the entire gut metabolome data set (over 2,000 molecules), and calculated the percentage of detected molecules represented in the metabolic pathways of MassTRIX, the result was even lower; only 8.2% of the molecules detected in the gut metabolome are predicted to be part of the annotated metabolic pathways of MassTRIX. Therefore, our results suggest that the intestinal metabolome is a poorly explored source of significant chemical diversity, and we hypothesized that many of these molecules are likely to possess important biological functions and properties.
The mammalian gut metabolome contains molecules with biological activity on Salmonella.
In order to probe the unknown functions and properties of the mammalian gut metabolome, we extracted molecules from fresh feces of a healthy donor, allowed the solvent to evaporate, and tested the effect of the dried extract on Salmonella. As a first measure of the effect of the extract on Salmonella, we compared bacterial growth in the absence and presence of the fecal extract. As can be seen in Fig. 1A, Salmonella growth showed a modest, although statistically significant, impairment in the presence of the fecal extract. Although growth levels were similar in the presence and absence of the fecal extract during the logarithmic growth phase, the bacterial culture reached significantly lower levels of growth (as measured by the optical density of the solution) in the presence of the fecal extract during the transition to stationary phase (Fig. 1B). This is not due to pH, as the pH of the solution containing the fecal extract was adjusted to match that of the culture medium alone. These data suggested that molecules that are biologically active against Salmonella are present in the fecal metabolome, although the exact reason for this effect on growth is unknown.
FIG 1 .
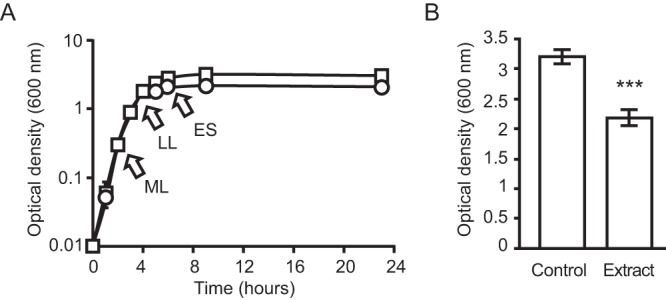
An extract from human feces is active against Salmonella. Salmonella was inoculated in LB broth with or without the addition of a dried ethyl acetate extract of human feces, and growth was monitored through measurements of optical density at 600 nm. Dried extracts were resuspended at a concentration that approximates the concentration present in feces (1×), given the weight of sample and volume of solvent used. (A) Squares represent cultures without the extract, whereas circles represent cultures containing the fecal extract. Results represent the averages of four independent measurements (n = 4), and bars (too small to be seen in most cases) show the standard errors of means. ML, mid-logarithmic growth phase; LL, late logarithmic growth phase; ES, early stationary growth phase. (B) To allow better visualization, the maximum optical density (9-h time point) achieved by each culture condition is shown. Results represent the averages of four independent measurements (n = 4), and bars show the standard errors of the means. ***, P < 0.0008.
The mammalian gut metabolome contains molecules that modulate Salmonella gene expression.
The data presented above suggested that the intestinal metabolome contains molecules active against Salmonella. Due to its chemical complexity, the intestinal metabolome is clearly an environment where microbes must sense numerous chemical cues, and many microbe-microbe and host-microbe interactions may have evolved based on specific chemical sensing events. Therefore, we hypothesized that an enteric pathogen would likely have evolved systems to sense molecules present in the intestinal tract. Conversely, it is likely that the intestinal microbiota evolved protection mechanisms against pathogens by producing molecules that could modulate their virulence mechanisms. To address this, we compared the transcriptomes of Salmonella during late logarithmic growth in the presence and absence of fecal extracts, as described above. As expected, our results revealed that many genes are differentially expressed in the presence of fecal extracts (Table 1). Specifically, 62 genes were upregulated during growth in the fecal extract, whereas 76 genes were downregulated. Among the genes activated by the fecal extract were those involved in metabolism, motility and chemotaxis, production of surface appendages (fimbriae), and phage production and transport, as well as many hypothetical proteins. Relevant to the interactions of Salmonella with its host, it is worth noting that a significant number of motility and chemotaxis genes were included within this data set. Out of the 62 genes activated by the fecal extract, 10 genes (15.6%) were involved in motility or chemotaxis. Among the genes repressed by the fecal extract, we found genes involved in the invasion of host cells, metabolism, and many hypothetical proteins, with a dramatic overrepresentation of genes involved in host cell invasion. Of the 76 genes repressed by the fecal extract, 29 (38.2%) are involved in Salmonella host cell invasion, clearly demonstrating that a major effect of the gut metabolome on Salmonella is the repression of invasion. If hypothetical proteins are disregarded, some 43.3% of the genes repressed are involved in host cell invasion. Due to the critical functions of host cell invasion genes for Salmonella pathogenesis, we focused our further studies on this virulence trait.
TABLE 1 .
Regulation of Salmonella gene expression in response to molecules from human feces
| ORFa | Gene | Annotation | Fold changeb | P valuec |
|---|---|---|---|---|
| 0113 | leuA | 2-Isopropylmalate synthase | 7.60 | ≤0.01 |
| 3753 | uhpT | Hexose phosphate transport protein | 7.06 | ≤0.05 |
| 0948 | Bacteriophage protein | 6.55 | ≤0.05 | |
| 3962 | Hypothetical protein | 6.07 | ≤0.05 | |
| 0111 | leuC | 3-Isopropylmalate dehydratase large subunit | 5.70 | ≤0.01 |
| 0112 | leuB | 3-Isopropylmalate dehydrogenase | 5.69 | ≤0.01 |
| 0015 | Bacteriophage protein | 5.35 | ≤0.05 | |
| 2692 | Putative capsid protein | 5.11 | ≤0.05 | |
| 0742 | Putative cation transporter | 4.63 | ≤0.05 | |
| 4397 | Arginine deiminase | 4.53 | ≤0.05 | |
| 3782A | ccmD1 | Protoheme transport protein D1 | 4.41 | ≤0.01 |
| 0110 | leuD | 3-Isopropylmalate dehydratase small subunit | 4.28 | ≤0.01 |
| 2129 | stcA | Putative fimbrial subunit protein | 4.22 | ≤0.05 |
| 2690 | Putative bacteriophage terminase | 4.19 | ≤0.05 | |
| 0057 | oadG | Oxaloacetate decarboxylase subunit gamma | 4.18 | ≤0.05 |
| 1797 | pagM | Virulence factor | 4.04 | ≤0.05 |
| 1469 | Putative secreted hydrolase | 3.82 | ≤0.05 | |
| 0155 | Secreted protein | 3.73 | ≤0.05 | |
| 1799 | pagK | Bacteriophage-encoded phoPQ-activated protein | 3.72 | ≤0.05 |
| 3107 | Hydrolase | 3.64 | ≤0.05 | |
| 2592 | cII | Regulatory protein CII | 3.44 | ≤0.01 |
| 3441 | nirB | Nitrite reductase large subunit | 3.30 | ≤0.05 |
| 1057 | Hypothetical protein | 3.24 | ≤0.05 | |
| 4152 | Bacteriophage protein | 3.22d | ≤0.05 | |
| 0947 | Bacteriophage protein | 3.16 | ≤0.05 | |
| 1058 | wrbA | Trp repressor binding protein | 3.12 | ≤0.05 |
| 3542 | Methyl-accepting chemotaxis citrate transducer | 3.10 | ≤0.05 | |
| 0199 | stfE | Minor fimbrial subunit | 3.09 | ≤0.05 |
| 1856 | cheA | Chemotaxis protein | 3.00 | ≤0.05 |
| 1763 | Hypothetical protein | 2.95 | ≤0.01 | |
| 0210 | htrA | Protease DO precursor; heat shock protein | 2.95 | ≤0.01 |
| 3762 | ilvB | Acetohydroxy acid synthase I, small subunit | 2.92 | ≤0.01 |
| 2596 | Pseudogene | 2.90 | ≤0.05 | |
| 1889 | fliD | Flagellar hook-associated protein | 2.89 | ≤0.01 |
| 1121 | flgL | Flagellar hook-associated protein 3 | 2.85 | ≤0.05 |
| 1795 | Hypothetical protein | 2.84 | ≤0.05 | |
| 4255 | phoN | Nonspecific acid phosphatase | 2.77 | ≤0.01 |
| 1851 | cheY | Chemotaxis protein | 2.76 | ≤0.01 |
| 2685 | Bacteriophage protein | 2.76 | ≤0.05 | |
| 1854 | tar | Methyl-accepting chemotaxis protein II | 2.75 | ≤0.05 |
| 0794 | HlyD family secretion protein | 2.75 | ≤0.01 | |
| 1966 | Bacteriophage protein | 2.75 | ≤0.05 | |
| 2547 | Putative transposase | 2.73 | ≤0.05 | |
| 1789 | Pseudogene | 2.71 | ≤0.05 | |
| 0459 | Hypothetical protein | 2.66 | ≤0.01 | |
| 2610 | Hypothetical protein | 2.65 | ≤0.05 | |
| 0347 | Putative cation efflux pump | 2.63 | ≤0.05 | |
| 1858 | motA | Motility protein A | 2.62 | ≤0.01 |
| 2021 | pduJ | Propanediol utilization protein | 2.62 | ≤0.05 |
| 2624 | yfiA | Putative sigma-54 modulation protein | 2.62 | ≤0.05 |
| 3761 | ilvN | Acetohydroxy acid synthase I, small subunit | 2.62 | ≤0.01 |
| 1235 | Hypothetical protein | 2.62 | ≤0.01 | |
| 4431 | Hypothetical protein | 2.61 | ≤0.01 | |
| 3614 | Putative HTHe-type transcriptional regulator | 2.60 | ≤0.01 | |
| 0829 | Hypothetical protein | 2.60 | ≤0.01 | |
| 1794 | Putative inner membrane protein | 2.59 | ≤0.05 | |
| 1120 | flgK | Flagellar hook-associated protein 1 | 2.56 | ≤0.05 |
| 1850 | cheZ | Chemotaxis protein | 2.56 | ≤0.01 |
| 1857 | motB | Motility protein B | 2.54 | ≤0.01 |
| 2828 | hycF | Formate hydrogen lyase subunit 6 | 2.54 | ≤0.01 |
| 2763 | Hypothetical protein | 2.52 | ≤0.05 | |
| 1243 | Hypothetical protein | 2.52 | ≤0.01 | |
| 1028 | Inner membrane protein | −6.80 | ≤0.01 | |
| 2853 | prgI | Type III secretion system apparatus | −5.72 | ≤0.01 |
| 2862 | sipD | Pathogenicity island 1 effector protein | −5.58 | ≤0.05 |
| 2863 | sipC | Pathogenicity island 1 effector protein | −5.53 | ≤0.01 |
| 2876 | invE | Cell invasion protein | −5.53 | ≤0.01 |
| 2869 | spaP | Type III secretion system secretory apparatus | −5.41 | ≤0.05 |
| 2878 | invF | AraC family regulatory protein | −5.40 | ≤0.01 |
| 2864 | sipB | Pathogenicity island 1 effector protein | −5.35 | ≤0.01 |
| 1030 | sigD | Cell invasion protein | −5.34 | ≤0.05 |
| 2865 | spaT | Type III secretion-associated chaperone | −5.08 | ≤0.01 |
| tRNA0069 | tRNA Pro anticodon TGG | −4.98 | ≤0.01 | |
| 2861 | sipA | Pathogenicity island 1 effector protein | −4.92 | ≤0.05 |
| 2860 | sipF | Acyl carrier protein | −4.91 | ≤0.05 |
| 2852 | prgJ | Type III secretion system apparatus | −4.86 | ≤0.01 |
| 4250 | Putative GerE family regulatory protein | −4.80 | ≤0.05 | |
| 3401 | rpsC | 30S ribosomal protein S3 | −4.68 | ≤0.05 |
| tRNA0029 | tRNA Cys anticodon GCA | −4.56 | ≤0.05 | |
| 2877 | invG | Type III secretion system secretory apparatus | −4.49 | ≤0.01 |
| 4251 | AraC family regulatory protein | −4.40 | ≤0.01 | |
| 0885 | tnp | Transposase for insertion element IS1541 | −4.30 | ≤0.01 |
| 1029 | pipC | Cell invasion protein | −4.30 | ≤0.01 |
| 2854 | prgH | Type III secretion apparatus component | −4.27 | ≤0.01 |
| 0291 | Hypothetical protein | −4.24 | ≤0.05 | |
| 3398 | rpsQ | 30S ribosomal protein S17 | −4.10 | ≤0.01 |
| 2856 | hilA | Invasion protein regulator | −4.10 | ≤0.01 |
| 2873 | invC | Secretory apparatus ATP synthase (associated with virulence) | −4.09 | ≤0.05 |
| 2875 | invA | Secretory apparatus of type III secretion system | −3.98 | ≤0.01 |
| 3400 | rplP | 50S ribosomal protein L16 | −3.97 | ≤0.05 |
| 1784 | sopE2 | Invasion-associated secreted effector protein | −3.87 | ≤0.01 |
| 1343 | ssaJ | Putative pathogenicity island lipoprotein | −3.86 | ≤0.05 |
| 2851 | prgK | Type III secretion system apparatus | −3.78 | ≤0.01 |
| 3399 | rpmC | 50S ribosomal protein L29 | −3.62 | ≤0.05 |
| tRNA0034 | tRNA Asn anticodon GTT | −3.59 | ≤0.01 | |
| 2674 | sopE | Invasion-associated secreted protein | −3.57 | ≤0.01 |
| 2879 | invH | Outer membrane lipoprotein | −3.51 | ≤0.01 |
| 0267 | sciH | Hypothetical protein | −3.49 | ≤0.05 |
| 2867 | spaR | Type III secretion system secretory apparatus | −3.42 | ≤0.05 |
| 1341 | ssaH | Type III secretion system apparatus | −3.42 | ≤0.01 |
| 4249 | Hypothetical protein | −3.36 | ≤0.01 | |
| 1347 | ssaM | Putative pathogenicity island protein | −3.27 | ≤0.05 |
| tRNA0040 | tRNA Val anticodon TAC | −3.22 | ≤0.05 | |
| tRNA0033 | tRNA Asn anticodon GTT | −3.22 | ≤0.05 | |
| 2840 | Hypothetical protein | −3.21 | ≤0.01 | |
| tRNA0080 | tRNA Gly anticodon GCC | −3.19 | ≤0.05 | |
| 1561 | sseJ | Translocated effector protein | −3.17 | ≤0.05 |
| 3063 | Hypothetical protein | −3.13 | ≤0.05 | |
| tRNA0050 | tRNA Ser anticodon GCT | −3.10 | ≤0.01 | |
| 2874 | invB | Chaperone protein for type III secretion system effectors | −3.10 | ≤0.01 |
| 2732 | Putative transcriptional regulator | −3.05 | ≤0.01 | |
| tRNA0081 | tRNA Gly anticodon GCC | −3.01 | ≤0.01 | |
| tRNA0066 | tRNA Arg anticodon CCG | −3.00 | ≤0.01 | |
| tRNA0041 | tRNA Val anticodon TAC | −2.98 | ≤0.05 | |
| tRNA0046 | tRNA Arg anticodon ACG | −2.95 | ≤0.01 | |
| tRNA0084 | tRNA Leu anticodon CAG | −2.94 | ≤0.01 | |
| 2428 | eutM | Ethanolamine utilization protein | −2.94 | ≤0.05 |
| 2846 | sprB | AraC family transcriptional regulator | −2.93 | ≤0.01 |
| 2850 | orgAa | Oxygen-regulated invasion protein | −2.89 | ≤0.05 |
| 2216 | Bacteriophage protein | −2.89 | ≤0.01 | |
| 4197 | siiE | Hypothetical protein | −2.85 | ≤0.05 |
| 2857 | iagB | Cell invasion protein | −2.83 | ≤0.01 |
| 0336 | Transmembrane regulator | −2.80 | ≤0.01 | |
| tRNA0011 | tRNA Gln anticodon TTG | −2.80 | ≤0.01 | |
| 1568 | Methyltransferase | −2.71 | ≤0.05 | |
| tRNA0003 | tRNA Asp anticodon GTC | −2.71 | ≤0.05 | |
| tRNA0067 | tRNA His anticodon GTG | −2.69 | ≤0.05 | |
| 1630 | Hypothetical protein | −2.67 | ≤0.05 | |
| tRNA0013 | tRNA Met anticodon CAT | −2.66 | ≤0.05 | |
| 2859 | sicP | Chaperone (associated with virulence) | −2.64 | ≤0.01 |
| 2110 | Hypothetical protein | −2.63 | ≤0.05 | |
| tRNA0007 | tRNA Gln anticodon CTG | −2.61 | ≤0.05 | |
| 1802 | Putative bacteriophage membrane protein | −2.61 | ≤0.01 | |
| 1007 | fabA | d-3-Hydroxydecanoyl-(acyl carrier protein) dehydratase | −2.59 | ≤0.01 |
| tRNA0015 | tRNA Val anticodon TAC | −2.58 | ≤0.05 | |
| tRNA0074 | tRNA Thr anticodon TGT | −2.56 | ≤0.01 | |
| 3381 | rplQ | 50S ribosomal protein L17 | −2.54 | ≤0.05 |
| 4208 | Hypothetical protein | −2.51 | ≤0.05 |
Open reading frame (ORF) designation based on the genome sequence of Salmonella enterica serovar Typhimurium strain SL1344.
Fold change values indicate regulation by the fecal extract; positive values indicate activation, whereas negative values indicate repression.
Determined by two-tailed equal-variance Student t tests.
Transcripts for this gene could not be detected in the absence of the extract. Therefore, the fold change value was calculated using the lowest expression value detected in the experiment (across all genes and samples) as the denominator.
HTH, helix-turn-helix.
The mammalian gut metabolome contains a strong inhibitor of Salmonella invasion gene expression.
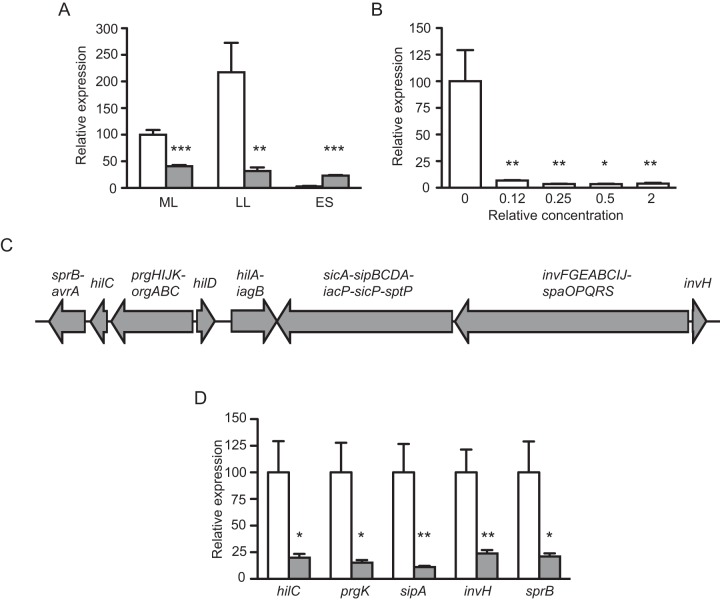
The genetic apparatus required for Salmonella host cell invasion is contained within a genomic region termed Salmonella pathogenicity island 1 (SPI-1) (23, 24). SPI-1 expression is controlled by many environmental factors, and it is generally accepted that the OmpR-ToxR family regulator HilA represents a global regulatory hub through which most environmental signals controlling SPI-1 expression are routed (25, 26). In order to confirm the observation that invasion gene expression is regulated by molecules present in the gut metabolome, we determined relative mRNA levels for HilA during Salmonella growth in the absence and presence of the fecal extract. Figure 2A shows that in culture medium without the fecal extract hilA expression is moderate in the mid-logarithmic growth phase, increases during late logarithmic growth, and is turned off in the stationary phase. However, when an extract from human feces is added to the culture medium, hilA expression is consistently low, confirming our original observation that invasion gene expression is repressed by molecules present in the gut metabolome. Although the presence of the fecal extract resulted in higher levels of hilA transcript in early stationary phase, we believe that this is likely not biologically relevant, given the low transcript levels shown at this stage of growth, both in the absence and in the presence of the extract (Fig. 2A). Besides testing the effect of a fecal extract on hilA expression over the course of bacterial growth, we also assessed the degree of activity of the extract by testing the effect of different concentrations on hilA expression. As can be seen from Fig. 2B, the fecal extract is highly active against hilA and shows full activity at levels that ranged from 2 times higher to 8 times lower than the concentration in feces.
FIG 2 .
The human gut metabolome contains a strong inhibitor of Salmonella invasion gene expression. (A) Salmonella was grown in LB broth with or without the addition of an extract from human feces at a concentration that approximates the concentration present in feces (1×), RNA was extracted, and hilA expression was assessed by real-time PCR. White bars, cultures without the fecal extract; gray bars, cultures with the fecal extract. ML, mid-logarithmic growth phase; LL, late logarithmic growth phase; ES, early stationary growth phase. Results represent the averages of four independent measurements (n = 4), except for measurements at ES in the presence of the extract, where 3 cultures were used (n = 3). Bars show the standard errors of the means. (B) Salmonella was grown in LB broth with various concentrations of the extract from human feces, RNA was extracted, and hilA expression was assessed by real-time PCR. Relative concentrations shown are in comparison with the concentration present in human feces, given the weight of sample and volume of solvent used. Results represent the averages of four independent measurements (n = 4), except for measurements at 0.5×, where 3 cultures were used (n = 3). Bars show the standard errors of the means. *, P < 0.02; **, P < 0.01; ***, P < 0.0004. (C) Schematic of the genetic locus responsible for host cell invasion by Salmonella, the Salmonella pathogenicity island 1 (SPI-1). Each arrow corresponds to a transcript, and the genes comprised within the transcript are indicated above the arrows. (D) Salmonella was grown in LB broth with or without the addition of an extract from human feces at a concentration that approximates the concentration present in feces (1×), RNA was extracted, and the expression of the indicated genes was assessed by real-time PCR. White bars, cultures without the fecal extract; gray bars, cultures with the fecal extract. Results represent the averages of four independent measurements (n = 4), and bars show the standard errors of the means. *, P < 0.02; **, P < 0.008.
Because hilA is the major regulator of invasion gene expression in Salmonella (23, 27), we predicted that the effect of the fecal extract on hilA would result in repression of the entire invasion genetic locus. To test this prediction, we designed primers toward most of the transcripts present in SPI-1 (Fig. 2C) and determined relative levels of these transcripts during Salmonella late logarithmic growth in medium with or without the fecal extract described above. As expected, every transcript tested showed drastic repression when Salmonella was grown in the presence of the fecal extract (Fig. 2D), confirming that the human gut metabolome contains a strong inhibitor of Salmonella invasion gene expression.
Bioactive molecules from human feces strongly inhibit Salmonella host cell invasion.
The results presented above suggested that the human gut metabolome contains compounds that would inhibit host cell invasion by Salmonella. To test this prediction, we grew Salmonella to the late logarithmic growth phase in culture medium with or without the fecal extract, washed the bacterial cells to remove the fecal extract, and used these cells to infect cultured mammalian cells (HeLa) using a standard gentamicin protection assay to measure invasion (28). As expected, growth in the fecal extract caused a reduction in invasion of over 96%, or 26-fold (Fig. 3). This established that the regulation of invasion gene expression elicited by molecules from the mammalian gut metabolome is translated into a drastic reduction of the invasion potential of Salmonella.
FIG 3 .
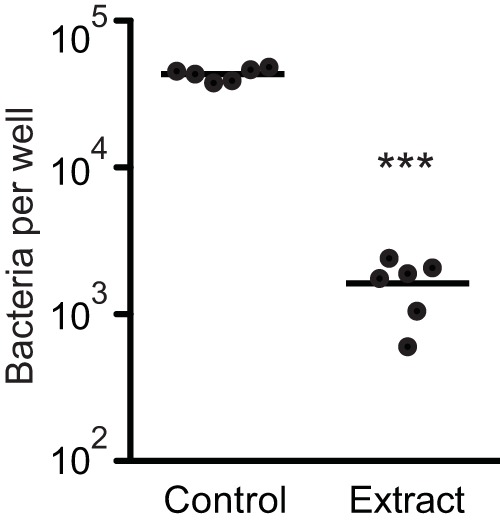
Molecules from human feces strongly repress the invasion of cultured host cells by Salmonella. HeLa cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 1% nonessential amino acids, and 1% GlutaMAX. Salmonella was grown in LB broth with or without the addition of an extract from human feces at a concentration that approximates the concentration present in feces (1×). Salmonella cultures were centrifuged, and cells were resuspended in phosphate-buffered saline and diluted in tissue culture medium. HeLa cells were infected at a multiplicity of infection of 10 for a total of 2 h at 37°C and 5% CO2. Cells were washed with buffer and lysed, and serial dilutions of the lysates were plated on LB plates for bacterial enumeration. Each dot on the graph represents the average of the results of two wells using an individual bacterial culture, for a total of 6 independent measurements (n = 6). Bars show the averages of the results obtained. ***, P < 0.0001.
The biological activity against Salmonella invasion gene expression is widespread.
Upon determining that the fecal extract used in the studies above contains an antivirulence molecule, we asked whether this activity is peculiar to the donor recruited for the initial phase of this study or is a widespread feature of the human gut metabolome. Accordingly, we recruited 9 additional volunteers, who donated fecal samples that were used for the extraction of molecules, as described above. The activity of these extracts against Salmonella hilA expression was then determined during late logarithmic growth. Donors varied with regard to gender and age (Table 2) but were all healthy and with no recent history of antibiotic use (30 days preceding sample collection). As can be seen in Fig. 4, of a total of 10 samples tested, 9 significantly inhibited hilA expression, as determined by real-time PCR (RT-PCR). Therefore, the biological activity of the gut metabolome against Salmonella invasion gene expression is a general phenomenon.
TABLE 2 .
Fecal samples used in this study
| Purpose | Donor | Gendera | Age (yr)b |
|---|---|---|---|
| Direct extraction | 1 | M | 1 |
| 2 | F | 7 | |
| 3 | M | 7 | |
| 4 | M | 11 | |
| 5 | M | 18 | |
| 6 | M | 18 | |
| 7 | M | 18 | |
| 8 | F | 1 | |
| 9 | F | 6 | |
| 10 | M | 4 | |
| Chemostat inoculation | A | F | 38 |
| B | M | 43 | |
| C | F | 42 |
F, female; M, male.
At the time of sample collection.
FIG 4 .
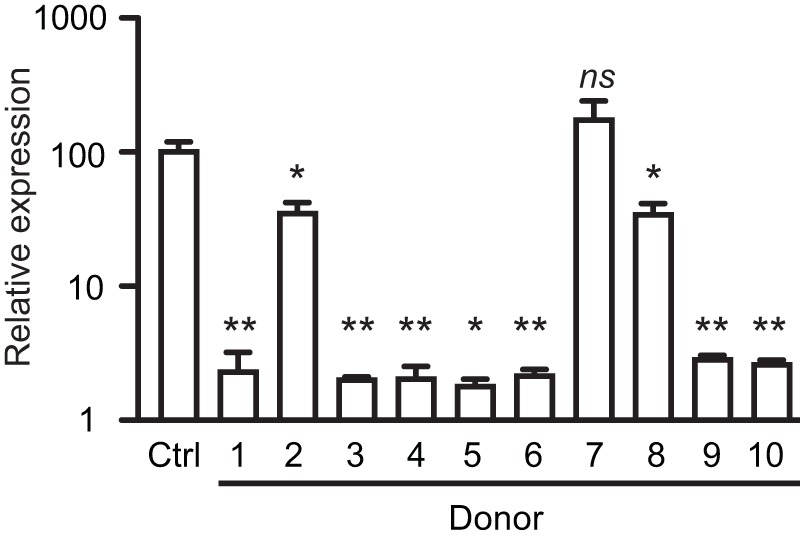
The inhibitory activity of the human gut metabolome is universal. Feces from 10 healthy subjects were extracted with ethyl acetate, and dried extracts were added to LB broth. Salmonella was grown in LB broth with or without the addition of the extracts at a concentration of approximately 0.25×, RNA was extracted, and hilA expression was assessed by real-time PCR. Results shown are the averages of three independent bacterial cultures (n = 3), except for donor 5, where 2 cultures were used (n = 2). *, P < 0.02; **, P < 0.004; ns, not significant (P > 0.05).
The murine gut metabolome contains an inhibitor of Salmonella invasion gene expression.
Collectively, our results showed that the human gut metabolome contains a molecule (or molecules) that acts as a strong inhibitor of Salmonella host cell invasion. In order to determine if this is specific to the human gut metabolome or a conserved feature among mammals, we tested the effect of molecules from the murine gut metabolome on Salmonella invasion gene expression during late logarithmic growth. Fresh feces of 129S1/SvImJ Nramp1−/− and Swiss Webster mice were extracted essentially as described for human samples and tested for effects on hilA expression. The results showed that the phenotype of hilA repression elicited by the gut metabolome is not exclusive to humans; fecal extracts from murine feces were also strong repressors of hilA (Fig. 5).
FIG 5 .
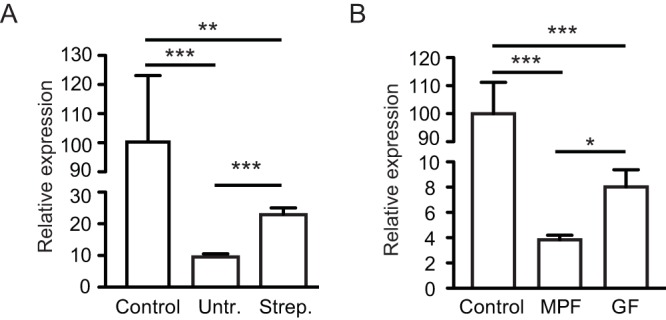
The mammalian gut microbiota is required for full inhibition of Salmonella invasion gene expression. (A) Molecules were extracted from feces of 129S1/SvImJ Nramp1−/− mice using ethyl acetate, as described in the text. Animals were then treated with 20 mg of streptomycin through oral gavage, and feces were collected and extracted again, 24 h after treatment. Salmonella was grown in LB broth with or without the addition of the dried extracts, and hilA expression was tested through RT-PCR. Results shown are the averages of 5 to 6 measurements, and bars represent the standard errors of the means. (B) Feces from conventionally raised as well as germfree Swiss Webster mice were collected and extracted with ethyl acetate, as described in the text. Salmonella was grown in LB broth with or without the addition of the dried extracts, and hilA expression was tested through RT-PCR. Results shown are the averages of 3 to 5 measurements, and bars represent the standard errors of the means. Untr., samples collected before antibiotic treatment; Strep., samples collected after streptomycin treatment; MPF, murine-pathogen-free animals (conventionally raised); GF, germfree animals. *, P < 0.04; **, P < 0.003; ***, P < 0.002.
The mammalian gut microbiota is required for full inhibition of Salmonella invasion gene expression.
After determining that the mammalian gut metabolome contains molecules that repress Salmonella invasion gene expression, we next sought to determine the source of this activity. Is this activity produced by mammalian cells or by the intestinal microbiota? To test if the microbiota was implicated in the production of the bioactive molecule, we collected feces from 129S1/SvImJ Nramp1−/− mice, extracted molecules as described above, and then treated the animals with a solution containing 20 mg of streptomycin by oral gavage. This treatment causes a reduction of approximately 95% in the microbial loads in the gastrointestinal tract (9). Fresh feces were collected 24 h after treatment, and molecules were extracted. The extracts were tested for inhibition of hilA expression during late logarithmic growth, as described above, and the results showed that antibiotic treatment caused a significant reduction in the inhibitory activity (Fig. 5A). However, one caveat of this experiment is that we cannot rule out the possibility that streptomycin itself is interfering with the inhibitory activity. Therefore, we also tested the inhibitory activity in feces of Swiss Webster germfree mice and compared their activity to that found in conventionally raised animals. Again, we found that the absence of the intestinal microbiota causes a significant reduction in the inhibitory activity (Fig. 5B). Collectively, these data show that the intestinal microbiota is at least partially responsible for the inhibitory activity. Although feces from antibiotic-treated and germfree animals still repressed hilA expression, the inhibitory activity of animals with an intact microbiota was significantly higher in both experiments.
The mammalian gut microbiota is sufficient for production of the inhibitory activity.
Although our results strongly suggested that the intestinal microbiota is involved in the production of the inhibitor of Salmonella invasion gene expression, it was still possible that the bioactive compounds were produced by host cells in response to the intestinal microbiota. In order to determine if this was the case or if the activity was independent of host factors, we set out to try to reproduce the inhibitory activity of the fecal extracts using gut microbes grown in the laboratory. To do so, we used an anaerobic, continuous-culture chemostat system to grow microbial communities from the intestinal tracts of three healthy human donors. Culture medium was inoculated with fresh fecal samples, and the communities were allowed to develop and stabilize for several weeks. The effluents from each of these laboratory-grown microbial communities were collected and extracted with ethyl acetate, as described previously. The extracts were then tested against Salmonella to determine their effect on hilA expression during late logarithmic growth. Figure 6 shows that extracts from the three microbial communities used caused significant repression of hilA expression, establishing that the intestinal microbiota is indeed responsible for production of the biological activity, independently of host factors.
FIG 6 .
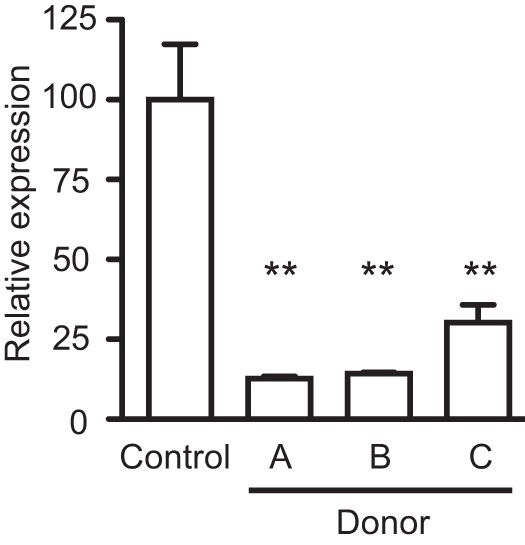
The mammalian gut microbiota is sufficient for production of the inhibitory activity. Feces from three healthy human donors were used to inoculate a bioreactor system run as a chemostat to culture microbial communities from the human gut. After appropriate incubation, effluents were collected and extracted with ethyl acetate. Dried extracts were then added to LB broth, and the medium was used to culture Salmonella. The expression of hilA in medium with or without the extracts was then monitored through RT-PCR. Results shown are the averages of 3 measurements, and bars represent the standard errors of the means. **, P < 0.01.
Closely related Clostridium species produce the inhibitory molecules.
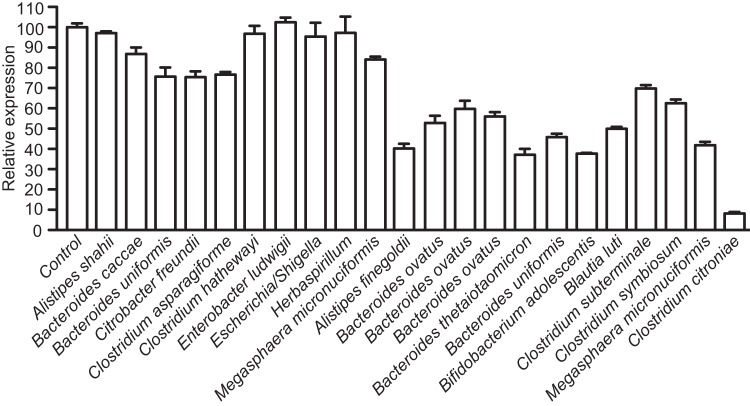
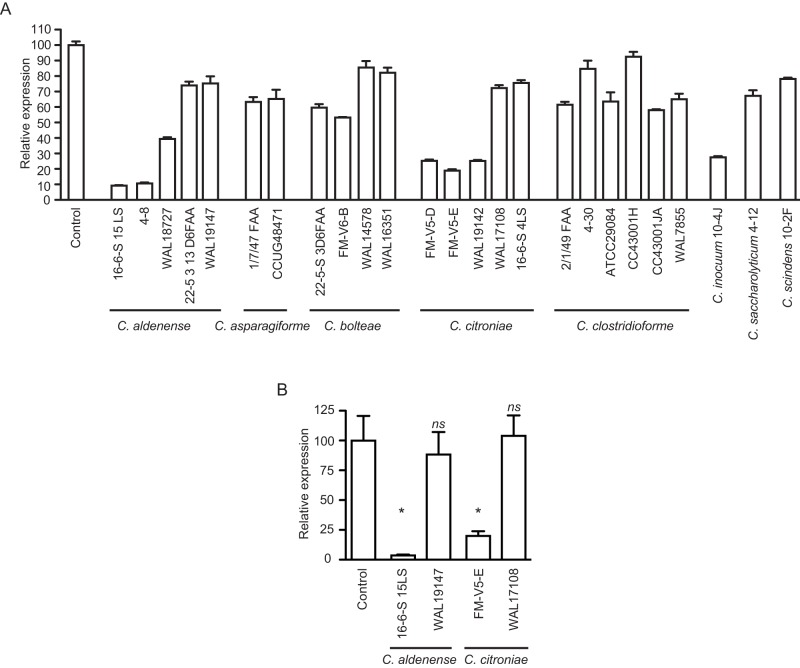
The results described above established that the inhibitory activity is produced by the intestinal microbiota in the absence of any host factors. In order to determine if individual microbial isolates can produce the bioactive molecule or if a community of microbes is required for such activity, we screened individual isolates from one of the chemostat microbial communities used (donor A, Table 2) for specific strains with inhibitory activity against hilA. This was done using a reporter strain containing a fusion between the promoter of hilA and gfp in pFPV25 (28, 29). The bacterial strains were grown in culture medium, as described in Materials and Methods, and extracted with ethyl acetate. Extracts were dried and resuspended in Luria-Bertani (LB) broth. Salmonella was then grown in LB medium supplemented with these extracts or with an ethyl acetate extract of culture medium alone to the late logarithmic growth phase, and green fluorescent protein (GFP) production was tested using flow cytometry. As can be seen in Fig. 7, most microbial isolates showed little to no inhibitory activity against hilA. However, a specific strain of Clostridium citroniae caused strong inhibition of invasion gene expression. Therefore, this determined not only that the microbiota is involved in the production of the active molecule but also that a single microbial species can produce the biological activity in the laboratory. We tested several other C. citroniae strains as well as strains of closely related species for inhibitory activity using this reporter system and found that multiple strains of C. citroniae were active. In addition, multiple isolates of Clostridium aldenense also produced active molecules, suggesting that a closely related clade within the Clostridiales cluster XIVa (otherwise known as the Lachnospiraceae family) is involved in this phenomenon (Fig. 8A). We also determined hilA mRNA levels using RT-PCR for selected C. citroniae and C. aldenense strains and confirmed that extracts from cultures of these microbes showed strong inhibitory effects on the expression of Salmonella invasion genes (Fig. 8B).
FIG 7 .
A human isolate of Clostridium citroniae produces strong activity against Salmonella invasion gene expression. Microbial isolates from a chemostat culture showing activity against hilA were tested individually for biological activity. Isolates were cultured under anaerobic conditions in Trypticase soy broth supplemented with menadione and hemin for at least 2 days, and the cultures were extracted with ethyl acetate. Dried extracts were added to LB broth, which was used to culture a Salmonella hilA::gfp reporter strain. GFP production was then monitored through flow cytometry. Results shown are the averages of three individual measurements (n = 3), except for the control culture, where six cultures were used (n = 6). Bars indicate the standard errors of the means.
FIG 8 .
Closely related Clostridium species are involved in the production of the inhibitory molecules. (A) Strains from diverse Clostridium species were tested for biological activity against hilA. Strains were cultured under anaerobic conditions in Trypticase soy broth supplemented with menadione and hemin for at least 2 days, and the cultures were extracted with ethyl acetate. Dried extracts were added to LB broth, which was used to culture a Salmonella hilA::gfp reporter strain. GFP production was then monitored through flow cytometry. Results shown are the averages of three individual measurements (n = 3), except for the control culture, where six cultures were used (n = 6). Bars indicate the standard errors of the means. (B) Production of the inhibitory molecule by select Clostridium strains was confirmed through RT-PCR targeting hilA. Results shown are the averages of 2 to 6 individual measurements, and bars show the standard errors of the means. *, P < 0.03; ns, not significant (P > 0.05).
The bioactive molecules are secreted.
Our results show that bioactive molecules produced by select species of Clostridium are sensed by Salmonella and affect the expression of virulence genes, especially those involved in host cell invasion. However, to determine if the bioactive molecules produced by Clostridium spp. are indeed secreted chemical cues, two strains of C. citroniae and C. aldenense were grown and the supernatants were separated from the bacterial cells by centrifugation and filtration. Extracts of the cells as well as the culture supernatants were tested for the ability to repress hilA expression during Salmonella late logarithmic growth using the hilA::gfp reporter strain. The results show that the biological activity is present exclusively in the extracellular fraction. That is, the inhibitory activity produced by the Clostridium strains is conferred by secreted molecules (Fig. 9).
FIG 9 .
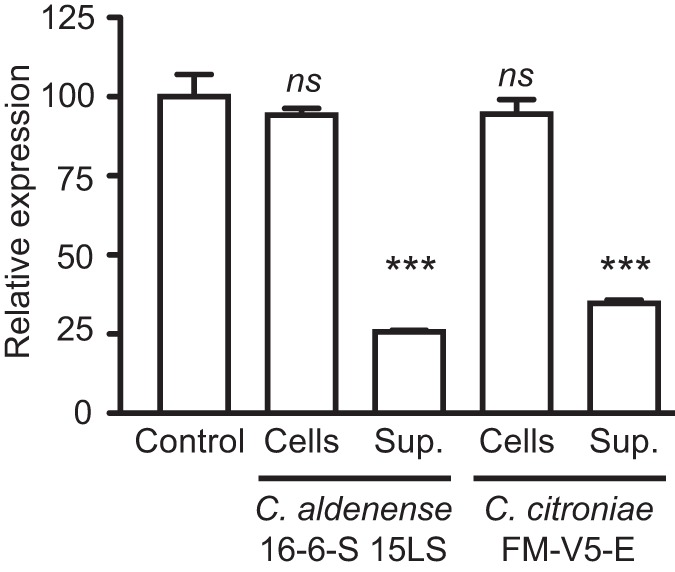
The bioactive molecules are secreted. Strains of C. citroniae and C. aldenense that showed strong activity against hilA expression were cultured under anaerobic conditions in Trypticase soy broth supplemented with menadione and hemin for at least 2 days, and the cells were separated from the supernatants through centrifugation. Both the cells and the supernatants were then extracted with ethyl acetate, and dried extracts were added to LB broth, which was used to culture the Salmonella hilA::gfp reporter strain for measurements of hilA expression. Results shown are the averages of 3 individual measurements, and bars show the standard errors of the means. ***, P < 0.0003; ns, not significant (P > 0.05).
DISCUSSION
Microbes can communicate through the production and sensing of small chemical signals. This allows them to sense their surroundings and adapt accordingly, increasing their fitness and chances of survival (15, 16, 30). Although this phenomenon has been extensively studied in recent years, most work has focused on microbes grown in isolation, without taking into account the complexity of their environments. In the context of the gastrointestinal tract, extensive microbial chemical sensing is predicted to take place; the complexity of the resident microbiota as well as its interactions with the host brings forth innumerable opportunities for the rise of such events over the course of evolution (1, 5, 30). However, only a few specific signaling events in the gastrointestinal tract have been studied to date. Perhaps the best-studied case of chemical signaling in the mammalian gut involves enterohemorrhagic Escherichia coli (EHEC). EHEC senses the mammalian hormones epinephrine and norepinephrine to activate the expression of a type 3 secretion system (T3SS), its major virulence factor (31, 32). Additionally, EHEC can also sense a microbiota-produced molecule, termed autoinducer 3 and whose chemical nature is still unknown, to activate its T3SS (31). These studies led to the hypothesis that EHEC uses these, and other, chemical cues to sense the host environment and activate the genetic loci required for successful host colonization (31). More recently, a different mechanism of microbiota-EHEC cross talk has been reported (33). Pacheco et al. demonstrated that EHEC senses fucose availability in the gastrointestinal tract and responds by regulating the expression of the T3SS (33). In this case, however, fucose acts as a repressor of virulence. It has been proposed that the release of fucose from host mucus by members of the microbiota, such as Bacteroides thetaiotaomicron, is used by EHEC as a chemical cue that it is not in close proximity to the intestinal epithelium. Under this circumstance, EHEC avoids the metabolic burden of virulence gene expression. Once reaching the surface of the intestinal epithelium, EHEC sensing of host-produced epinephrine and norepinephrine leads to the activation of the T3SS, culminating with successful colonization of the host tissue. This elegant body of work convincingly demonstrates that complex chemical sensing events are used in interactions between host, microbiota, and pathogen in the mammalian gut and begs the question of whether such phenomena are widespread among gut microbes.
It has been known for several decades that the intestinal microbiota has critical functions for human health. Early studies by Miller, Bohnhoff, and others showed that disruption of the intestinal microbiota through the use of antibiotics significantly decreases host resistance to enteric infections (34–36), a phenomenon that became known as colonization resistance (37). Many potential mechanisms for colonization resistance have been suggested, including competition for nutrients or binding sites and production of antibacterial molecules such as bacteriocins (37). Although all are plausible explanations, the exact mechanisms through which colonization resistance is conferred remain for the most part unidentified. Recently, Ng et al. shed some light on this issue (38). By studying the postantibiotic expansion of two human pathogens, Salmonella enterica and Clostridium difficile, the authors showed that access to food is an important aspect of a pathogen’s ability to colonize the host gut; the liberation of host sugars (fucose and sialic acid) by members of the gut microbiota is critical to allow pathogen proliferation (38). Although this recent study supports the original notion that colonization resistance is conferred by competition for nutrients, other mechanisms are still likely in place (39). Given the involvement of chemical signaling in host-microbe and microbe-microbe interactions in the gut, it is likely that a chemical warfare is elicited during the competition between commensals and pathogens for colonization of the mammalian gut. It is interesting that the same molecule, fucose, is used as a chemical signal and an energy source in each model of colonization resistance, suggesting that the mechanisms are interactive and that colonization resistance is multifactorial.
To shed some light into the role of chemical sensing in host-microbe and microbe-microbe interactions in the mammalian gut, we studied the effect of molecules from human feces on the transcriptome of Salmonella and found that bioactive molecules produced by C. citroniae and C. aldenense act as strong inhibitors of virulence gene expression by this pathogen. These Clostridium species are members of the Clostridiales cluster XIVa, which, together with Clostridiales cluster IV and Bacteroides, makes up the majority of the human intestinal microbiota (40). These clostridia have critical functions for health during the entire human life span; they are involved in modulating immune functions, providing energy sources to host cells and other members of the gut microbiota, and even maintaining endocrine homeostasis (41). Our studies suggest that this microbial group may also play a role in colonization resistance by producing molecules that inhibit colonization by invading pathogens.
Other studies have previously shown that metabolic products of gut microbes can impinge on Salmonella virulence gene expression. Lawhon et al. have found that acetate can act as an inducer of invasion gene expression through a BarA/SirA-independent pathway (42). BarA and SirA form a two-component regulatory system responsible for activating the expression of SPI-1 invasion genes in Salmonella. The authors showed that acetate could restore the expression of invasion genes in a BarA mutant and postulated that acetate could be used by Salmonella as a signal that it is in the distal small intestine, its preferred invasion site. Interestingly, a mixture of the short-chain fatty acids (SCFAs) acetate, propionate, and butyrate at concentrations mimicking those in the distal small intestine also induced invasion gene expression, whereas the same SCFAs at concentrations mimicking those in the large intestine repressed it, corroborating the notion that SCFAs found in the gut can be used as chemical cues by Salmonella to control its virulence (42). In a related study, while investigating the genetic requirements for the induction of SPI-1 by acetate, Huang et al. found yet another molecule with a strong effect on Salmonella invasion gene expression (43). By studying the effect of spent culture medium on the expression of an SPI-1 effector protein, the authors found that formate also acts as a strong inducer of Salmonella invasion gene expression. Although these studies established the presence of inducers of invasion gene expression in the mammalian gut, the presence of inhibitors has also been described. For instance, Gantois et al. have shown that, in contrast to acetate and formate, butyrate can act as a strong repressor of Salmonella invasion gene expression (44). More recently, Hung et al. studied the effect of another short-chain fatty acid, propionate, on Salmonella gene expression using microarrays and found that this fatty acid repressed the expression of 22 out of the 35 SPI-1 genes investigated (45). Among the repressed genes were the major regulators of SPI-1, hilA and hilD, suggesting that propionate acts as yet another intestinal chemical cue with a significant effect on the Salmonella virulence genetic program.
In the present study, we describe the presence of a strong inhibitory activity against Salmonella invasion gene expression in feces as well as culture supernatants of select members of the gut microbiota. It is possible that the inhibitory activity described in this work is related to the molecules described above. For instance, Garner et al. have previously shown that streptomycin treatment of mice significantly reduces the concentration of butyrate in the large intestine (46), and this could be related to the partial loss of the inhibitory activity observed with antibiotic-treated as well as germfree animals in our study. However, the body of literature described above makes it clear that the regulation of invasion gene expression by molecules present in the gut is complex and multifactorial. Therefore, it is possible that the activity described here represents yet another chemical cue sensed by Salmonella during colonization of the intestinal tract.
The precise biological significance of the findings described here is currently unknown. It is possible that the inhibition of virulence gene expression elicited by molecules present in the gastrointestinal tract represents a defense strategy mounted by the resident microbiota against pathogenic invaders. However, it is also possible that these phenomena are chemical sensing events devised by Salmonella, during the course of its evolution, to sense its environment. By regulating the expression of invasion genes in response to multiple intestinal molecules, Salmonella may be able to fine tune gene expression to avoid the expenditure of unnecessary virulence factor production and maximize its success during host colonization. Nevertheless, our work suggests that the gut metabolome is an underexplored source of biologically active molecules that should be mined for their antivirulence properties and perhaps other biological activities.
MATERIALS AND METHODS
Ethics statement.
Written informed consent was obtained for all experiments involving humans; all work described here was reviewed and approved by either the University of British Columbia or the University of Guelph Research Ethics Board. All experiments involving animals were reviewed and approved by the University of British Columbia Animal Care Committee and followed the NIH Guide for the Care and Use of Laboratory Animals.
Human samples.
Fecal samples were collected from healthy donors between 1 and 43 years of age using a sterile container. Samples were refrigerated and brought to the laboratory within 24 h, where they were immediately used or frozen at −20°C until used. Table 2 shows the information on the donors recruited.
Animals.
129S1/SvImJ Nramp1−/− mice were maintained at the Centre for Disease Modeling, the University of British Columbia (Vancouver, Canada). Swiss Webster mice, both conventionally raised and germfree, were kept at Taconic (Germantown, NY).
Bacterial strains and growth conditions.
Strains used in this study are shown in Table S1 in the supplemental material. All studies were performed using Salmonella enterica serovar Typhimurium SL1344 (here referred to as Salmonella) (47, 48). When indicated, this strain also harbored a reporter vector bearing a fusion between the promoter region of the hilA gene (−675 through −70, relative to the translational start codon) and the promoterless gfp gene in pFPV25 (28, 29). Details of this fusion are described elsewhere (28). Salmonella was grown in Luria-Bertani (LB) broth containing 1% (wt/vol) sodium chloride and 100 µg/ml of either streptomycin (in the case of the wild-type strain; Sigma-Aldrich, Oakville, Canada) or carbenicillin (in the case of the wild-type strain bearing the reporter plasmid; EMD Chemicals, San Diego, CA), at 37°C and with shaking (225 rpm). In all experiments, Salmonella was grown in glass culture tubes incubated at an angle and containing a limited amount of broth to allow extensive aeration of the growth medium. Where indicated, intestinal commensals were cultured under anaerobic conditions in Trypticase soy broth (Oxoid, Cambridge, England) supplemented with menadione (1 µg/ml; Sigma-Aldrich) and hemin (5 µg/ml; BDH, Radnor, PA) for at least 2 days.
Extraction of molecules.
Molecules were extracted from fecal samples or bacterial cultures using 1 volume of ethyl acetate (Sigma-Aldrich; ≥99.7% pure). Fecal samples were extracted for 10 min using a tissue homogenizer (Mixer Mill MM 301 apparatus; Retsch, Haan, Germany). Samples were then centrifuged, and the supernatant was collected. Bacterial cultures were extracted by mixing them with ethyl acetate, allowing the phases to separate, and collecting the organic solvent phase. In both cases, the solvent was evaporated on a centrifuge equipped with a vacuum pump and the dried extracts were saved at −20°C until used. For experiments, dried extracts were resuspended directly into culture medium, i.e., without the use of ethyl acetate. The medium was then filtered, and the pH was adjusted to match that of culture medium alone. This was done to avoid interference of the solvent in the experiments performed. Additionally, dried residues of ethyl acetate were used in all controls without fecal or bacterial extracts to ensure that any of the effects seen were not caused by residues of the evaporated solvent.
RNA sequencing and data analysis.
Salmonella was grown in LB broth with or without the addition of an extract from human feces. After approximately 4 h of growth, the cultures had reached the late logarithmic growth phase (Fig. 1), and bacterial RNA was stabilized by the addition of 2 volumes of RNAprotect bacterial reagent (Qiagen, Hilden, Germany) to the bacterial cultures and incubation at room temperature for approximately 5 min. Cells were pelleted by centrifugation, and RNA was isolated using the RNeasy minikit (Qiagen) with on-column DNase digestion, according to the manufacturer’s recommendations. RNA samples were quantified using a Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA), and RNA integrity was checked with the RNA6000 Nano assay using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). cDNA library preparation and sequencing reactions were conducted at Genewiz, Inc. (South Plainfield, NJ). Illumina TruSeq RNA library preparation, clustering, and sequencing reagents were used throughout the process according to the manufacturer’s recommendations (Illumina, San Diego, CA). Briefly, mRNAs were purified and fragmented, and cDNAs (first and second strands) were synthesized and end repaired. Adaptors were ligated after adenylation at the 3′ ends, and cDNA templates were enriched by PCR. cDNA libraries were validated using a high-sensitivity chip on the Agilent 2100 Bioanalyzer (Agilent Technologies) and quantified using a Qubit 2.0 fluorometer (Life Technologies) and by quantitative PCR (qPCR). Samples were clustered on a flow cell using cBOT and were then loaded on the Illumina HiSeq 2000 instrument. Raw sequence data were converted into fastq files and demultiplexed using the Illumina CASSAVA 1.8.2 program. fastq files from each sample were imported into CLC Genomics Workbench 5.5.1, and sequence reads were trimmed to remove low-quality bases at the ends. Sequence reads were mapped to the genome of Salmonella enterica serovar Typhimurium strain SL1344 downloaded from NCBI (http://www.ncbi.nlm.nih.gov/nuccore/NC_016810.1). Sequence hit account and RPKM (reads per kilobase per million) values were calculated for genes, and quantile normalization was performed for RPKM values. Two-tailed equal-variance Student t tests were conducted, and genes showing a differential expression of 2.5-fold or higher at a P value of ≤0.05 between sample groups were considered significantly regulated by the fecal extract.
Real-time PCR.
RNA preparation was performed as described above. cDNA synthesis was performed using the QuantiTect reverse transcription kit (Qiagen). Reverse transcription-PCRs (RT-PCRs) were performed using the QuantiTect SYBR green PCR kit (Qiagen) and the Applied Biosystems 7500 system (Carlsbad, CA). Reaction mixtures contained forward and reverse primers at 0.4 µM each. All results were normalized using the expression levels of the housekeeping gene gapA, encoding the glyceraldehyde-3-phosphate dehydrogenase enzyme (49) as the baseline. Averages of the data obtained with control cultures were normalized to 100, and the data from each sample were normalized accordingly.
GFP reporter assays.
For the screening of microbial producers of bioactive molecules, we used the bacterial reporter strain described above to track biological activity. GFP production was analyzed through flow cytometry of bacterial cultures using a FACSCalibur cytometer (BD Biosciences, Franklin Lakes, NJ), as indicated. In each experiment, 50,000 events were collected per sample. Because hilA expression is bimodal (50), instead of measuring the average fluorescence intensity of the sample as an indication of hilA expression, we gated the GFP-positive population of cells and calculated the percentage of the total population that it represented. This value was normalized to 100% in the control samples and used as a reference to calculate all other values.
Invasion assays.
HeLa cells were obtained from the American Type Culture Collection (Manassas, VA) and were grown in Dulbecco’s modified Eagle’s medium with a high glucose concentration, 4 mM l-glutamine, and sodium pyruvate (HyClone, Waltham, MA), supplemented with 10% fetal bovine serum (HyClone), 1% nonessential amino acids (Gibco, Carlsbad, CA) and 1% GlutaMAX (Gibco). Invasion assays were performed essentially as previously described (28). Salmonella cultures were centrifuged, and cells were resuspended in phosphate-buffered saline (PBS; HyClone) and diluted in tissue culture medium. HeLa cells were infected at a multiplicity of infection of 10 for 30 min at 37°C and 5% CO2. Cells were then washed twice with PBS and incubated at 37°C and 5% CO2 in growth medium containing 100 µg/ml gentamicin (Sigma-Aldrich) for 1.5 h. After a total of 2 h of infection, cells were washed twice with PBS and lysed in 250 µl of 1% Triton X-100 (BDH), 0.1% sodium dodecyl sulfate (Sigma-Aldrich). Serial dilutions were plated on LB plates containing 100 µg/ml of streptomycin. After overnight incubation, colonies were counted for bacterial enumeration.
Single-stage chemostat simulation of the human distal gut environment.
An Infors Multifors bioreactor system run as a chemostat was used for this work (Infors HT, Bottmingen, Switzerland) and run using a retention time of 24 h, pH 7, with growth medium supplied as previously detailed (51). Prior to inoculation, the vessel was aseptically sampled to check for absence of contaminant growth on fastidious anaerobe agar (Acumedia, Lansing, MI) supplemented with 5% defibrinated sheep blood (Hemostat Laboratories, Dixon, CA; sFAA). Fresh fecal samples derived from healthy donors were obtained and separately homogenized in prereduced growth medium (to 10% [wt/vol]) using a Tekmar stomacher lab blender (Seward, Worthing, England). Homogenates were gently centrifuged at 175 × g to sediment large particles, and 100 ml of supernatant was used to inoculate prepared chemostat vessels containing 300 ml of prereduced medium prepared as described above. Cultures were allowed to adjust to the chemostat vessel environment for 24 h in batch culture before the medium pumps were switched on. Monitoring of the stability of the microbial community to steady state was done using the method of McDonald et al. (51). Briefly, genomic DNA was extracted from daily samples drawn from the vessel and used as the template for amplification of the V3 region of the 16S rRNA genes, and subsequent separation of amplicons by percent G+C content using denaturing gradient gel electrophoresis. Similarity indices of gel profiles were determined using GeneDirectory software (Syngene, Frederick, MD), and moving window analysis was performed to ascertain the development and maintenance of steady state. At steady state, chemostat communities were removed from the vessel for extraction.
Isolation and identification of strains from chemostat communities.
Bacterial strains were isolated from chemostat cultures by performing standard dilution series in prereduced PBS and plating on prereduced sFAA with the addition of filter-sterilized chemostat effluent to 5% (vol/vol). Isolates were identified by performing 16S rRNA gene-directed PCR with crude extracts (approximately 10 µl of freshly grown cells boiled in 500 µl of PBS for 5 min at 100°C) as the template, using a universal primer set designed to amplify the V3 region of the 16S rRNA gene (52). Generated sequences were parsed against the Greengenes database (http://greengenes.lbl.gov) in order to determine their closest relatives. Identity to a species of >97% was used as a means of identification to species level.
Antibiotic treatment of mice.
Mice were treated through oral gavage with 100 µl of a 200-mg/ml sterile solution of streptomycin prepared in water.
Statistical analyses.
Data were analyzed by one-tailed, unpaired t tests with 95% confidence intervals, unless otherwise noted.
SUPPLEMENTAL MATERIAL
Bacterial strains used in this study.
ACKNOWLEDGMENTS
We are greatly thankful to Sydney Finegold, who kindly donated the WAL strains used in this study.
This work was funded by the Canadian Institutes of Health Research. L.C.M.A. was supported by postdoctoral fellowships from the Department of Foreign Affairs and International Trade Canada and the Canadian Institutes of Health Research as well as a fellowship from the Science without Borders program of the National Council of Technological and Scientific Development (CNPq-Brazil). R.B.R.F. was funded by a postdoctoral fellowship from the Canadian Institutes of Health Research and a fellowship from the Science without Borders program of the National Council of Technological and Scientific Development (CNPq-Brazil). B.B.F. is the University of British Columbia Peter Wall Distinguished Professor.
No competing financial interests exist.
Footnotes
Citation Antunes LCM, McDonald JAK, Schroeter K, Carlucci C, Ferreira RBR, Wang M, Yurist-Doutsch S, Hira G, Jacobson K, Davies J, Allen-Vercoe E, Finlay BB. 2014. Antivirulence activity of the human gut metabolome. mBio 5(4):e01183-14. doi:10.1128/mBio.01183-14.
REFERENCES
- 1. Sekirov I, Russell SL, Antunes LC, Finlay BB. 2010. Gut microbiota in health and disease. Physiol. Rev. 90:859–904. 10.1152/physrev.00045.2009 [DOI] [PubMed] [Google Scholar]
- 2. Maynard CL, Elson CO, Hatton RD, Weaver CT. 2012. Reciprocal interactions of the intestinal microbiota and immune system. Nature 489:231–241. 10.1038/nature11551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tremaroli V, Bäckhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249. 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- 4. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U. S. A. 110:3229–3236. 10.1073/pnas.1218525110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Macpherson AJ, Harris NL. 2004. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 4:478–485. 10.1038/nri1373 [DOI] [PubMed] [Google Scholar]
- 7. Blumberg R, Powrie F. 2012. Microbiota, disease, and back to health: a metastable journey. Sci. Transl. Med. 4:137rv7. 10.1126/scitranslmed.3004184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O’Hara AM, Shanahan F. 2006. The gut flora as a forgotten organ. EMBO Rep. 7:688–693. 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Antunes LC, Han J, Ferreira RB, Lolić P, Borchers CH, Finlay BB. 2011. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob. Agents Chemother. 55:1494–1503. 10.1128/AAC.01664-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Starling EH. 1905. The chemical correlation of the functions of the body. Lecture I. Lancet ii:339–341 [Google Scholar]
- 11. Boyce JA. 2008. Eicosanoids in asthma, allergic inflammation, and host defense. Curr. Mol. Med. 8:335–349. 10.2174/156652408785160989 [DOI] [PubMed] [Google Scholar]
- 12. Gilliver SC. 2010. Sex steroids as inflammatory regulators. J. Steroid Biochem. Mol. Biol. 120:105–115. 10.1016/j.jsbmb.2009.12.015 [DOI] [PubMed] [Google Scholar]
- 13. Kyrou I, Tsigos C. 2009. Stress hormones: physiological stress and regulation of metabolism. Curr. Opin. Pharmacol. 9:787–793. 10.1016/j.coph.2009.08.007 [DOI] [PubMed] [Google Scholar]
- 14. Levine MA. 2003. Normal mineral homeostasis. Interplay of parathyroid hormone and vitamin D. Endocr. Dev. 6:14–33. 10.1159/000072764 [DOI] [PubMed] [Google Scholar]
- 15. Antunes LC, Ferreira RB. 2009. Intercellular communication in bacteria. Crit. Rev. Microbiol. 35:69–80. 10.1080/10408410902733946 [DOI] [PubMed] [Google Scholar]
- 16. Antunes LC, Ferreira RB, Buckner MM, Finlay BB. 2010. Quorum sensing in bacterial virulence. Microbiology 156:2271–2282. 10.1099/mic.0.038794-0 [DOI] [PubMed] [Google Scholar]
- 17. Fuqua WC, Winans SC, Greenberg EP. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schaefer AL, Greenberg EP, Oliver CM, Oda Y, Huang JJ, Bittan-Banin G, Peres CM, Schmidt S, Juhaszova K, Sufrin JR, Harwood CS. 2008. A new class of homoserine lactone quorum-sensing signals. Nature 454:595–599. 10.1038/nature07088 [DOI] [PubMed] [Google Scholar]
- 19. Higgins DA, Pomianek ME, Kraml CM, Taylor RK, Semmelhack MF, Bassler BL. 2007. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450:883–886. 10.1038/nature06284 [DOI] [PubMed] [Google Scholar]
- 20. Antunes LC, Arena ET, Menendez A, Han J, Ferreira RB, Buckner MM, Lolic P, Madilao LL, Bohlmann J, Borchers CH, Finlay BB. 2011. Impact of Salmonella infection on host hormone metabolism revealed by metabolomics. Infect. Immun. 79:1759–1769. 10.1128/IAI.01373-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buckner MM, Antunes LC, Gill N, Russell SL, Shames SR, Finlay BB. 2013. 15-Deoxy-delta(12,14)-prostaglandin J2 inhibits macrophage colonization by Salmonella enterica serovar Typhimurium. PLoS One 8:e69759. 10.1371/journal.pone.0069759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suhre K, Schmitt-Kopplin P. 2008. MassTRIX: mass translator into pathways. Nucleic Acids Res. 36:W481–W484. 10.1093/nar/gkn194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lostroh CP, Lee CA. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3:1281–1291. 10.1016/S1286-4579(01)01488-5 [DOI] [PubMed] [Google Scholar]
- 24. Srikanth CV, Mercado-Lubo R, Hallstrom K, McCormick BA. 2011. Salmonella effector proteins and host-cell responses. Cell. Mol. Life Sci. 68:3687–3697. 10.1007/s00018-011-0841-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bajaj V, Lucas RL, Hwang C, Lee CA. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703–714. 10.1046/j.1365-2958.1996.d01-1718.x [DOI] [PubMed] [Google Scholar]
- 26. Lucas RL, Lostroh CP, DiRusso CC, Spector MP, Wanner BL, Lee CA. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:1872–1882. 10.1128/JB.182.7.1872-1882.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones BD. 2005. Salmonella invasion gene regulation: a story of environmental awareness. J. Microbiol. 43:110–117 [PubMed] [Google Scholar]
- 28. Antunes LC, Buckner MM, Auweter SD, Ferreira RB, Lolić P, Finlay BB. 2010. Inhibition of Salmonella host cell invasion by dimethyl sulfide. Appl. Environ. Microbiol. 76:5300–5304. 10.1128/AEM.00851-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Valdivia RH, Falkow S. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22:367–378. 10.1046/j.1365-2958.1996.00120.x [DOI] [PubMed] [Google Scholar]
- 30. Antunes LC, Davies JE, Finlay BB. 2011. Chemical signaling in the gastrointestinal tract. F1000 Biol. Rep. 3:4. 10.3410/B3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. U. S. A. 100:8951–8956. 10.1073/pnas.1537100100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. 2006. The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U. S. A. 103:10420–10425. 10.1073/pnas.0604343103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. 2012. Fucose sensing regulates bacterial intestinal colonization. Nature 492:113–117. 10.1038/nature11623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miller P, Bohnhoff M, Rifkind D. 1956. The effect of an antibiotic on the susceptibility of the mouse’s intestinal tract to Salmonella infection. Trans. Am. Clin. Climatol. Assoc. 68:51–55 [PMC free article] [PubMed] [Google Scholar]
- 35. Bohnhoff M, Miller CP. 1962. Enhanced susceptibility to Salmonella infection in streptomycin-treated mice. J. Infect. Dis. 111:117–127. 10.1093/infdis/111.2.117 [DOI] [PubMed] [Google Scholar]
- 36. Miller CP, Bohnhoff M. 1963. Changes in the mouse’s enteric microflora associated with enhanced susceptibility to Salmonella infection following streptomycin treatment. J. Infect. Dis. 113:59–66. 10.1093/infdis/113.1.59 [DOI] [PubMed] [Google Scholar]
- 37. Buffie CG, Pamer EG. 2013. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13:790–801. 10.1038/nri3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502:96–99. 10.1038/502S96a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Willing BP, Vacharaksa A, Croxen M, Thanachayanont T, Finlay BB. 2011. Altering host resistance to infections through microbial transplantation. PLoS One 6:e26988. 10.1371/journal.pone.0026988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hold GL, Pryde SE, Russell VJ, Furrie E, Flint HJ. 2002. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 39:33–39. 10.1111/j.1574-6941.2002.tb00904.x [DOI] [PubMed] [Google Scholar]
- 41. Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. 2013. Commensal clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 5:23. 10.1186/1757-4749-5-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lawhon SD, Maurer R, Suyemoto M, Altier C. 2002. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46:1451–1464. 10.1046/j.1365-2958.2002.03268.x [DOI] [PubMed] [Google Scholar]
- 43. Huang Y, Suyemoto M, Garner CD, Cicconi KM, Altier C. 2008. Formate acts as a diffusible signal to induce Salmonella invasion. J. Bacteriol. 190:4233–4241. 10.1128/JB.00205-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Hautefort I, Thompson A, Hinton JC, Van Immerseel F. 2006. Butyrate specifically down-regulates salmonella pathogenicity island 1 gene expression. Appl. Environ. Microbiol. 72:946–949. 10.1128/AEM.72.1.946-949.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hung CC, Garner CD, Slauch JM, Dwyer ZW, Lawhon SD, Frye JG, McClelland M, Ahmer BM, Altier C. 2013. The intestinal fatty acid propionate inhibits Salmonella invasion through the post-translational control of HilD. Mol. Microbiol. 87:1045–1060. 10.1111/mmi.12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garner CD, Antonopoulos DA, Wagner B, Duhamel GE, Keresztes I, Ross DA, Young VB, Altier C. 2009. Perturbation of the small intestine microbial ecology by streptomycin alters pathology in a Salmonella enterica serovar Typhimurium murine model of infection. Infect. Immun. 77:2691–2702. 10.1128/IAI.01570-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wray C, Sojka WJ. 1978. Experimental Salmonella typhimurium infection in calves. Res. Vet. Sci. 25:139–143 [PubMed] [Google Scholar]
- 48. Rankin JD, Taylor RJ. 1966. The estimation of doses of Salmonella typhimurium suitable for the experimental production of disease in calves. Vet. Rec. 78:706–707. 10.1136/vr.78.21.706 [DOI] [PubMed] [Google Scholar]
- 49. Nelson K, Whittam TS, Selander RK. 1991. Nucleotide polymorphism and evolution in the glyceraldehyde-3-phosphate dehydrogenase gene (gapA) in natural populations of Salmonella and Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 88:6667–6671. 10.1073/pnas.88.15.6667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Antunes LC, Wang M, Andersen SK, Ferreira RB, Kappelhoff R, Han J, Borchers CH, Finlay BB. 2012. Repression of Salmonella enterica phoP expression by small molecules from physiological bile. J. Bacteriol. 194:2286–2296. 10.1128/JB.00104-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McDonald JA, Schroeter K, Fuentes S, Heikamp-Dejong I, Khursigara CM, de Vos WM, Allen-Vercoe E. 2013. Evaluation of microbial community reproducibility, stability and composition in a human distal gut chemostat model. J. Microbiol. Methods 95:167–174. 10.1016/j.mimet.2013.08.008 [DOI] [PubMed] [Google Scholar]
- 52. Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bacterial strains used in this study.