Abstract
Anopheles gambiae s.s. Giles, An. stephensi Liston, An. freeborni Aitken, and An. quadrimaculatus Say are cultured and studied in molecular genetic and transgenic laboratories with increasing frequency. With limited research space, these mosquitoes are often maintained in the same insectary. Under these conditions, cross-contamination of colonies can occur and have devastating consequences to affected research programs. We have developed a polymerase chain reaction−based assay targeting the 28S large subunit ribosomal RNA gene to easily differentiate between these four taxa and An. funestus Giles, which occurs in sympatry with An. gambiae. The resulting assay identifies individual mosquito preparations as well as all taxa within a mixed or pooled DNA template preparation.
INTRODUCTION
Anopheles gambiae s.s. Giles, An. stephensi Liston, An. freeborni Aitken, and An. quadrimaculatus Say are cultured and studied in molecular genetic and transgenic laboratories with increasing frequency. These mosquitoes are often maintained in the same insectary due to limited research space. If not carefully maintained and monitored, cross-contamination of colonies can occur and lead to mixed colonies or effectively replaced colonies. Mixed colonies can lead to inconsistent or irreproducible experimental results. Although culture conditions for most anophelines are similar, some species, strains, or colonies develop more efficiently than others. The differences in development may allow one species to out-compete another after cross-contamination occurs, leading to the extinction and replacement of the less efficient colony. Either cross-contamination scenario can be devastating to affected research programs.
The obvious prevention to colony contamination is colony monitoring. There are reliable morphologic characteristics that can be used to differentiate between these four species; however, few laboratories have personnel trained in mosquito taxonomy. Key morphologic characteristics may also be unclear in early instar larvae or adult specimens that have not been carefully prepared. Unfortunately, questions about colony contamination usually do not arise in molecular-based or transgenic laboratories until assays fail or experimental results are significantly different from those expected. At this point, the specimens in question have most likely been destroyed during sample preparation and the use of morphologic characteristics is moot. Personnel trained in molecular techniques could easily use a molecular diagnostic for routine monitoring of mosquito colonies or to confirm species identity of extracted DNAs. To address this need, we have developed a polymerase chain reaction (PCR)−based assay to quickly and clearly differentiate between these anopheline taxa.
Many PCR-based diagnostic assays for mosquito taxa currently exist.1–3 The most useful assays are those that rely on a single PCR to produce DNA fragments that can be easily size-fractionated by electrophoresis on an agarose gel and that can clearly differentiate between the taxa involved. Using existing examples and sequence information available in the public domain, we designed a PCR-based assay that will clearly and reliably identify An. gambiae, An. stephensi, and An. freeborni or An. quadrimaculatus in both single-taxa and mixed (contaminated) DNA template preparations. Furthermore, this diagnostic procedure was expanded to include An. funestus Giles, a newly colonized species that occurs in sympatry in the field with An. gambiae. The ability to molecularly differentiate between these two taxa extends the utility of this diagnostic tool to field-collected specimens from Africa.
MATERIALS AND METHODS
Sample source and isolation of DNA
The molecular assay was developed using colony mosquitoes sourced from the National Institutes of Health (HIH) (Bethesda, MD) in May 2003. Anopheles gambiae were of the G-3 strain originating from The Gambia via the London School of Hygiene and Tropical Medicine. Anopheles freeborni were of the Marysville strain from California. The origin of the An. stephensi colony obtained from the NIH is unknown. Additional validation specimens were sourced from established colonies at Johns Hopkins Bloomberg School of Public Health (4ARR and Keele strains of An. gambiae) and Virginia Tech Polytechnic Institute and State University (An. stephensi) in January 2004. Field-collected An. gambiae were captured in the village of Banambani, Mali, in June 2002 and near Accra, Ghana in 2003. Anopheles quadrimaculatus specimens were collected in July 2003 from two locations along the Patuxent River in Maryland. Anopheles funestus specimens were obtained from a colony maintained at the National Institute of Communicable Diseases of the South African National Health Laboratory Service in Johannesburg.
Voucher specimens were mounted and taxa were validated by morphology. Following morphologic validation, genomic DNA was isolated from individual adult mosquitoes by a modified salt extraction as previously described.4 Genomic DNA was isolated from individual larval specimens using the same protocol. Individual field specimens were identified to species, An. gambiae or An. arabiensis, using the PCR-based assay developed by Scott and others.1 Anopheles gambiae specimens were identified to molecular M or S form using the PCR-based assay developed by Favia and others.2,3
Primer design and PCR
Sequences of the 28S large subunit ribosomal RNA gene were retrieved from GenBank for each taxon (An. gambiae AF417813, An. stephensi AF417820, and An. freeborni AF417824). Sequences were aligned using MultAlin (Institut National de la Recherche Agronomique, Paris, France)5 and PCR primers for the assay were manually selected using Primer36 to assist with assessment of primer Tm compatibility and dimer and hairpin formation (Table 1). Before optimization of the multiplexed PCR, species-specific amplifications were attempted and optimized. The PCR was also attempted with species-specific primers and mismatched template DNA to ensure that the derived PCR products were taxon-specific. Individual primer pairs and the multiplexed primer set were also evaluated for non-specific amplification of genomic DNA derived from Culex p. quinquefasciatus Say, Cx. restuans Theobald, Cx. p. pipiens L., Cx. salinarius Coquillett, Aedes aegypti (L.), Ae. albopictus (Skuse), Ochlerotatus triseriatus (Say), and Oc. taeniorhynchus (Wiedemann). Finally, the multiplexed PCR was optimized for single and mixed templates. All single taxa and multiplexed PCRs were conducted under the following conditions. An initial denaturation of 5 minutes at 95°C was followed by 25 cycles at 94°C for 1 minute, 55°C for 1.5 minutes, and 72°C for 2 minutes. The final 72°C extension step was 5 minutes. The multiplexed PCR was run in 20-μL reaction volumes using ≈190 ng (mean for 0.5 μL of genomic DNA extraction) of template DNA. Each 20-μL PCR consisted of 10 mM Tris, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin, 1.0 mM of dNTPs; 0.5 units of Taq polymerase, 25 pmol of the UN400F and UNREV primers, and 50 pmol of the GAMB230F and STEP148F primers. Amplification reactions without the UN400F forward primer used 50 pmol of each of the remaining three primers. DNA amplifications were completed on a PTC-200 thermal cycler (MJ Research, Watertown, MA) and evaluated after electrophoresis on 2% agarose gels stained with ethidium bromide. All gels were run with GeneRuler 100-basepair (bp) molecular mass marker (Fermentas Life Science, Hanover, MD).
Table 1.
Primers used for the multiplexed polymerase chain reaction
| Primer | Primer sequence (5′ → 3′) |
|---|---|
| UN400F | TCTGAATAGAGAGTCAAATAGTACG |
| UNREV | ATTGTGCTACATCGCCGA |
| GAMB230F | CTAACGCTCCGGCATACACT |
| STEP148F | GCGCCTTTCACACCCGAG |
Additional experiments were conducted to evaluate the sensitivity of the diagnostic assay on disproportionate ratios of contaminating templates. Paired templates (An. gambiae/ An. stephensi, An. gambiae/An. freeborni, and An. stephensi/ An. freeborni) were run together with one template concentration fixed (≈190 ng) and the other serially diluted through a series of reactions to a relative concentration of 1:256 (≈0.75 ng). Each series of template mixtures was PCR amplified as described with and without the UN400F forward primer, and the sensitivity was evaluated by visualization of corresponding species-specific fragments by agarose gel electrophoresis.
The universal primers produced amplicons of similar size for An. freeborni and An. quadrimaculatus. Therefore, PCR products for these two species were compared by single-strand conformation polymorphism (SSCP)7 and sequenced to identify nucleotide substitutions, insertions, and deletions. To ensure that any interspecies and intraspecies variation in the resulting amplicons were accounted for, the PCR products were purified with the QIAquick Gel Extraction kit (Qiagen, Inc., Valencia, CA) and cloned (TOPO TA Cloning kit; Invitrogen, Carlsbad, CA) prior to SSCP. Transformed bacteria were grown at 37°C overnight on Luria-Bertani agar plates containing carbenicillin (50 μg/μL) and transformed colonies were identified by blue-white selection. The 28S insert was recovered from individual colonies by the previously described 28S PCR, and amplicons from multiple clones from a series of individual mosquitoes representing each taxon were evaluated for interspecific and intraspecific variability by SSCP. Briefly, 4 μL of PCR product mixed with 8 μL of denaturing loading mixture was subjected to electrophoresis at 4°C on a 5% polyacrylamide gel and stained with SYBR® Green (Cambrex BioScience Rockland, Inc., Rockland, ME). Sequence information for these 28S products were derived from two clones selected from each species using the universal M13 forward and reverse primers.
The 400-bp 28S fragment recovered from An. funestus was evaluated for intraspecific polymorphism by SSCP as described earlier. Sequence information for these 28S products were similarly derived from two clones using the universal M13 forward and reverse primers.
RESULTS AND DISCUSSION
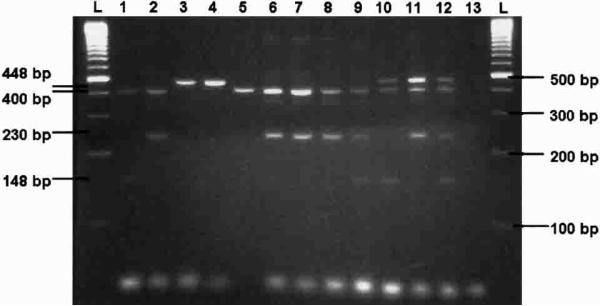
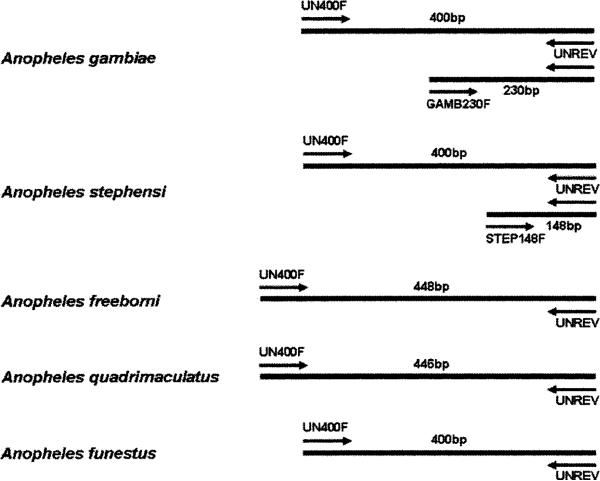
All single-taxa and multiplexed PCR resulted in products as predicted by primer design (Figures 1 and 2). Species-specific PCR products were identical whether derived from different laboratory strains or field collected material. The PCR with species-specific primers did not yield any product when conducted with template DNA from non-specific culicine taxa. These findings indicate that the PCR targets are conserved within each species and specific for the selected set of taxa. Amplification with primers UN400F and UNREV produce a 400-bp fragment for An. gambiae, An. stephensi, and An. funestus specimens. The length of the An. funestus PCR product was confirmed through sequencing (AY569553, AY570970). Conservation of this sequence was verified by SSCP. The same primer pair produces a single and differential 448-bp fragment for An. freeborni and 446-bp fragment for An. quadrimaculatus due to ≈50 bp of non-contiguous insertion sequence in the homologous fragment. In addition to the 400-bp fragment, the UNREV and GAMB230F primer pair amplify a 230-bp fragment for An. gambiae specimens and the UNREV and STEP148F primer pair amplify a 148-bp fragment for An. stephensi. The UN400F, UNREV, and GAMB230F primer set also produce 400-bp and 230-bp fragments for both the molecular M and S forms of An. gambiae s.s. and An. arabiensis. These results were predicted by the original sequence alignments and suggest that this assay will positively identify An. gambiae s.l., but will not differentiate between taxa within this complex. A reliable PCR-based assay already exists to differentiate between members of the An. gambiae species complex.1 In addition, because the GAMB230F primer does not cross-react with An. funestus, this diagnostic has field application to differentiate between An. gambiae complex and An. funestus s.s. where these species are sympatric.
Figure 1.
Ethidium bromide−stained agarose gel showing diagnostic polymerase chain reaction products (single adult specimens). Anopheles stephensi (lane 1), An. gambiae (lane 2), An. freeborni (lane 3), An. quadrimaculatus (lane 4), An. funestus (lane 5), An. arabiensis (lane 6), An. gambiae S form (lane 7), An. gambiae M form (lane 8), An. gambiae plus An. stephensi (lane 9), An. stephensi plus An. freeborni (lane 10), An. freeborni plus An. gambiae (lane 11), An. gambiae plus An. stephensi plus An. freeborni (lane 12), negative control (lane 13), and 100-basepair (bp) molecular markers (lanes L).
Figure 2.
Diagram of polymerase chain reaction (PCR) products and relative primer positions on homologous template DNA for each of the five species used in the multiplexed PCR assay. bp = base-pairs.
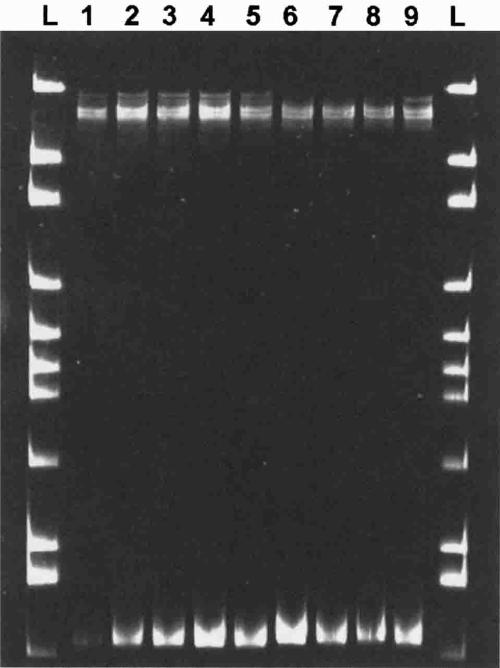
Anopheles quadrimaculatus and An. freeborni can be distinguished morphologically by the shape of the scales on the cubital wing vein;8 however, this can be an especially difficult characteristic to observe. The SSCP analysis of PCR products produced by amplification using UN400F and UNREV suggested substantial sequence variation in the 28S ribosomal DNA exists between these two species, but that the sequence is conserved within each taxa (Figure 3). The SSCP also confirmed that a slight difference in fragment size exists between these species. Differences in banding pattern for the rena-tured single strands (top set of bands) are indicative of differences in nucleotide sequence, whereas shifts in migration of the denatured single strands (lower band) indicate insertions or deletions. Sequencing of these fragments confirmed a fragment size of 446 bp for An. quadrimaculatus (AY569555, AY570972) and 448 bp for An. freeborni (AY569554, AY570971), in addition to 26 conserved base pair differences between these taxa. Although differences between these two species were not apparent by agarose gel electrophoresis, SSCP analysis was sensitive enough to detect the 2-bp difference in fragment size, and holds promise as a reliable molecular diagnostic for differentiating between these two species.
Figure 3.
Differences in banding patterns between Anopheles freeborni and An. quadrimaculatus single-strand conformation polymorphism analysis of 28S ribosomal DNA amplicons subjected to electrophoresis on a polyacrylamide gel and visualized with SYBR® Green. Colony An. freeborni sourced from the National Institutes of Health (lanes 1−5); An. quadrimaculatus collected from two different field sites in Maryland (lanes 6−7 and 8−9. respectively); 100-basepair molecular marker (lanes L).
It is envisioned that the multiplexed primer PCR reported here will be primarily used to monitor colony status as a regular control measure when colonies of more than one taxon are maintained in the same insectary. The PCR can be run on individual specimens or pooled specimens to reduce cost and labor. Both individual and pooled adult or larval DNA preparations have been successfully used for the diagnostic assay. Figures 1 and 4 illustrate that sources either combined or accidentally contaminated, as may happen in the laboratory, will produce all species-specific fragments represented in the template preparations when run with the multiplexed primer mixture. A secondary use envisioned for this PCR diagnostic will be to verify the identity of mosquito DNA extractions that may not perform as expected in molecular assays because this is most often the stage of investigation where colony contamination becomes apparent.
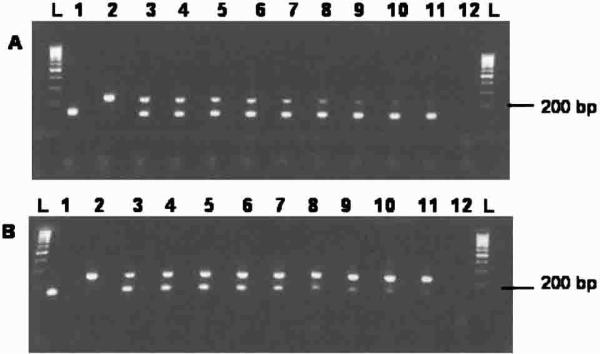
Figure 4.
Ethidium bromide−stained agarose gels comparing disproportionate ratios of Anopheles gambiae and An. stephensi mixed templates in the absence of the universal forward primer UN400F. A, Constant concentration of An. stephensi template mixed with serial dilutions of An. gambiae template. B, Constant concentration of An. gambiae template mixed with serial dilutions of An. stephensi template. An. stephensi (lane 1), An. gambiae (lane 2), mixed 1:1 (equal concentrations of both templates, lane 3), 1:2 (lane 4), 1:4 (lane 5), 1:8 (lane 6), 1:16 (lane 7), 1:32 (lane 8), 1:64 (lane 9), 1:128 (lane 10), 1:256 (lane 11), negative control (lane 12), 100-basepair (bp)-molecular marker (lanes L).
Sensitivity is an important component of the diagnostic procedure, especially when screening for colony or sample contamination. The existing assay is capable of detecting both DNA templates in a contamination scenario where the contaminant comprises only 1/256 (≈0.4%) of the total template DNA in the PCR (Figure 4). If only screening between An. gambiae and An. stephensi, the UN400F primer can simply be removed from the primer mixture. Even without the 400-bp control product, the An. gambiae (230 bp) and An. stephensi (148 bp) products are still easy to differentiate. In addition, removal of the UN400F primer appears to increase the availability of reagents within the PCR and enhance the sensitivity for these shorter products.
The PCR-based assay described here will not prevent cross-contamination of colonies from occurring; the only prevention is careful colony maintenance and monitoring. An advantage to this PCR-based assay is that it will allow the monitoring of colonies as early instar larvae, long before late instar larvae or adults are available for morphologic identification. It is our hope that this assay will be used as a tool to prevent costly research losses.
Acknowledgments
We thank M. Coetzee and R. Hunt for providing insight into the morphology of these anopheline taxa and providing An. funestus colony specimens. We also thank the laboratories of N. Kumar, M. Jacobs-Lorena, and S. Luckhart for providing colony mosquitoes for this study, and S. Shone for donating field-collected An. quadrimaculatus.
Financial support: This research was supported by financial assistance to Douglas E. Norris from the UNDP/World Bank/World Health Organization Special Program for Research and Training in Tropical Diseases (TDR) (A10360), partial support from the Johns Hopkins Malaria Research Institute, and a NIEHS training award (T32ES07141) to Rebekah J. Kent.
REFERENCES
- 1.Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- 2.Favia G, della Torre A, Bagayoko M, Lanfrancotti A, Sagnon N, Touré YT, Coluzzi M. Molecular identification of sympatric chromosomal forms of Anopheles gambiae and further evidence of their reproductive isolation. Insect Mol Biol. 1997;6:377–383. doi: 10.1046/j.1365-2583.1997.00189.x. [DOI] [PubMed] [Google Scholar]
- 3.Favia G, Lanfrancotti A, Spanos L, Siden-Kiamos I, Louis C. Molecular characterization of ribosomal DNA polymorphisms discriminating among chromosomal forms of Anopheles gambiae s.s. Insect Mol Biol. 2001;10:19–23. doi: 10.1046/j.1365-2583.2001.00236.x. [DOI] [PubMed] [Google Scholar]
- 4.Norris DE, Shurtleff AC, Touré YT, Lanzaro GC. Micro-satellite DNA polymorphism and heterozygosity among field and laboratory populations of Anopheles gambiae ss (Diptera: Culicidae). J Med Entomol. 2001;38:336–340. doi: 10.1603/0022-2585-38.2.336. [DOI] [PubMed] [Google Scholar]
- 5.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. http://prodes.toulouse.inra.fr/multalin/multalin.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press; Totowa, NJ: 2000. pp. 365–386. http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi. [DOI] [PubMed] [Google Scholar]
- 7.Hiss RH, Norris DE, Dietrich CH, Whitcomb RF, West DF, Bosio CF, Kambhampati S, Piesman J, Antolin MF, Black WC., IV Molecular taxonomy using single strand conformation polymorphism (SSCP) analysis of mitochondrial ribosomal DNA genes. Insect Mol Biol. 1994;3:171–182. doi: 10.1111/j.1365-2583.1994.tb00164.x. [DOI] [PubMed] [Google Scholar]
- 8.Darsie RF, Ward RA. Identification and geographical distribution of the mosquitoes of North America north of Mexico. Mosq Syst. 1981;1(suppl):1–313. [Google Scholar]